ABSTRACT
Several plant viruses encode elements at the 5′ end of their RNAs, which, unlike most cellular mRNAs, can initiate translation in the absence of a 5′ m7GpppG cap. Here, we describe an exceptionally long (739-nucleotide [nt]) leader sequence in triticum mosaic virus (TriMV), a recently emerged wheat pathogen that belongs to the Potyviridae family of positive-strand RNA viruses. We demonstrate that the TriMV 5′ leader drives strong cap-independent translation in both wheat germ extract and oat protoplasts through a novel, noncanonical translation mechanism. Translation preferentially initiates at the 13th start codon within the leader sequence independently of eIF4E but involves eIF4G. We truncated the 5′ leader to a 300-nucleotide sequence that drives cap-independent translation from the 5′ end. We show that within this sequence, translation activity relies on a stem-loop structure identified at nucleotide positions 469 to 490. The disruption of the stem significantly impairs the function of the 5′ untranslated region (UTR) in driving translation and competing against a capped RNA. Additionally, the TriMV 5′ UTR can direct translation from an internal position of a bicistronic mRNA, and unlike cap-driven translation, it is unimpaired when the 5′ end is blocked by a strong hairpin in a monocistronic reporter. However, the disruption of the identified stem structure eliminates such a translational advantage. Our results reveal a potent and uniquely controlled translation enhancer that may provide new insights into mechanisms of plant virus translational regulation.
IMPORTANCE Many members of the Potyviridae family rely on their 5′ end for translation. Here, we show that the 739-nucleotide-long triticum mosaic virus 5′ leader bears a powerful translation element with features distinct from those described for other plant viruses. Despite the presence of 12 AUG start codons within the TriMV 5′ UTR, translation initiates primarily at the 13th AUG codon. The TriMV 5′ UTR is capable of driving cap-independent translation in vitro and in vivo, is independent of eIF4E, and can drive internal translation initiation. A hairpin structure at nucleotide positions 469 to 490 is required for the cap-independent translation and internal translation initiation abilities of the element and plays a role in the ability of the TriMV UTR to compete against a capped RNA in vitro. Our results reveal a novel translation enhancer that may provide new insights into the large diversity of plant virus translation mechanisms.
INTRODUCTION
Translation initiation of most eukaryotic mRNAs occurs by a scanning mechanism, where the 43S ribosomal subunit enters the mRNA from an accessible 5′ end, is dependent on the 7-methyl guanosine cap structure (m7GpppG), and scans in a 5′-to-3′ direction in search of the initiation codon (1). The ribosomal subunit is recruited to the 5′ end by the cap-binding protein factor eIF4E, which is bound to the 5′ cap. eIF4E is the small subunit and cap-binding protein in the eIF4F complex. eIF4F is also comprised of the RNA helicase (eIF4A) and the large subunit and scaffold factor (eIF4G), which coordinates the attachment of the 43S ribosomal subunit to the mRNA via eIF3. In plants, eIF4E and eIF4G are also present in isoforms (eIFiso4E and eIFiso4G) (2, 3).
Many viruses deviate from canonical cap-dependent translation initiation to compete against cellular mRNAs for limited translation factors (4–7). Some plant RNA viruses have evolved diverse RNA elements in their 5′ and/or 3′ untranslated regions (UTRs) to recruit the ribosomal complex and initiate translation at the 5′-proximal AUG start codon without needing a 5′ cap. Interestingly, many plant virus translation mechanisms are unique compared to those of animal viruses. In several noncapped and nonadenylated plant viruses, including members of the Luteovirus genus of the Luteoviridae family, members of the Tombusviridae family, the umbraviruses, and satellite tobacco necrosis virus (8–10), translation is mediated by a 3′ cap-independent translation enhancer (CITE). These 3′ CITEs are ∼100 nucleotides (nt) long, are located at the 3′ end of the viral RNA, and direct translation of the 5′-end-proximal open reading frame (9, 10). Despite their divergent structures and sequences, all 3′ CITEs initiate translation by recruiting one of the components of the initiation complex (eIF4F and/or eIFiso4F) by directly binding and delivering the complex to the 5′ end via a 3′-to-5′ long-distance interaction. The ribosomal complex can then begin scanning for the 5′-proximal AUG start codon (10). This mechanism, although mediated by the 3′ UTR, requires sequences in the 5′ UTR to initiate translation.
Only a few plant viruses have been shown to rely solely on their 5′ UTRs to initiate protein expression, including some members of the largest family of plant viruses (11). Members of the Potyviridae family, which is comprised of ∼190 plant viruses distributed in at least 6 genera (6, 11–14), have a genome structure similar to that of the animal-infecting picornaviruses. Like the picornaviruses, potyviruses possess a viral protein covalently linked to their 5′ end (VPg) instead of a 5′ cap and are polyadenylated at their 3′ end, and their genomic RNA encodes a single large polyprotein that is cleaved into several functional proteins (15).
Despite their similar genome organizations, the potyviruses diverge from the animal picornaviruses in 5′-driven translation mechanisms. Potyviruses typically regulate translation by using their relatively short (<200-nt), adenosine-rich, and structurally weak 5′ UTRs (11). Although some show internal initiation abilities when tested in bicistronic constructs, potyviral leader sequences drive optimal translation when the 5′ end of the mRNA remains accessible and open to ribosomal entry, suggesting a preference for 5′-terminus-dependent initiation (11). For example, the 5′ leader of tobacco etch virus (TEV) (family Potyviridae), which is one of the better-studied potyvirus translational enhancers (5, 12, 16–18), contains two cap-independent regulatory elements (CIREs) ∼75 nt long that fold into pseudoknots, which can independently mediate translation of the downstream cistron at twice the level of the control sequence when placed into the intergenic region of a bicistronic mRNA (5, 18). This suggests that the TEV leader acts as an internal ribosome entry site. However, the addition of a strong stem-loop structure upstream of the 5′ UTR to block ribosomal scanning and 5′ entry in a monocistronic reporter RNA significantly reduced its translational ability (18). Similarly, the 5′ UTRs of other noncapped potyviruses, including turnip mosaic virus (TuMV), potato virus Y (PVY), and plum pox virus (PPV), stimulate cap-independent translation (6). The TuMV 5′ UTR drives translation at the level of a control m7GpppG-capped monocistronic RNA in vitro and in vivo, although the exact mechanism remains unclear (13). A stable stem-loop placed at the 5′ end of the mRNA sustained TuMV 5′-UTR-driven translation at 30% of the level observed without the stem-loop, but translation was abolished for the control m7GpppG-capped RNA (13). Additionally, the TuMV 5′ UTR remains translationally functional in the reverse orientation and such a reverse sequence can inhibit cap-dependent translation in trans (13). In PVY, the 184-nt-long 5′ UTR tolerates large deletions within the leader sequence, down to 55 total nucleotides, without major effects on the translation of noncapped monocistronic reporter mRNAs (14, 19). The PPV 5′ UTR is different yet in that it contains an upstream AUG (uAUG) codon that regulates the translation of the downstream correct AUG start codon via a leaky scanning mechanism (19).
Triticum mosaic virus (TriMV) is a newly emerged wheat-infecting member of the Potyviridae family (20, 21) with a 10,266-nt-long genomic RNA. Similar to other members of the family, it naturally lacks the canonical 5′ cap structure, has a VPg protein that is likely bound to its 5′ end, and is polyadenylated at its 3′ end. It encodes a single, large, 3,112-amino-acid polyprotein, which is posttranslationally cleaved into 10 characteristic potyviral mature putative proteins (20, 21). The virus varies significantly from other members of the Potyviridae in protein sequence and is therefore classified into the new genus Poacevirus (20). Interestingly, its encoded proteins share only 29 to 40% sequence homology with the proteins of the other potyviruses, and unlike the typically short and unstructured 5′ UTRs described for most potyviruses, the TriMV 5′ UTR is 739 nucleotides long with 48% GC content and a predicted structural free energy value of −205 kcal/mol (predicted by using mFOLD). Sequence analysis also revealed 12 AUG codons located in the 5′ UTR upstream of the presumed initiation site of the viral polyprotein at nucleotide positions 740 to 742 (20). Although found in several picornaviruses, this is uncommon in 5′ leader sequences of most plant RNA viruses (20).
Here, we demonstrate that the TriMV 5′ leader confers strong cap-independent translation in both oat protoplast cells and wheat germ cell extracts through a noncanonical mechanism. Translation primarily initiates at the 13th AUG codon independently of eIF4E and is dependent on at least eIF4G. We constructed a truncated sequence of ∼300 nt derived from the 5′ UTR that drives cap-independent translation from the 5′ end. In this region, we identified a stem-loop structure at nt 469 to 490 that is necessary for its activity. The disruption of the stem structure was sufficient to impair both translation facilitation and trans-inhibition against a capped RNA. Additionally, we found that the TriMV 5′ UTR drives translation internally in both a bicistronic construct and a monocistronic construct with a strong hairpin structure placed at the 5′ end. This internal initiation activity is completely dependent upon the identified stem structure. In the context of the monocistronic mRNA with the 5′ obstruction, the disruption of the stem structure abolished internal initiation, while the double compensatory mutation that restores the structure fully reestablished the activity of the TriMV 5′ UTR. In summary, our results reveal a potent and uniquely regulated translation enhancer that may provide new insights into mechanisms of plant virus translation.
MATERIALS AND METHODS
Luciferase reporter constructs.
A cDNA clone containing the 5′ UTR of TriMV (22) was used as a PCR template to generate the appropriate clones. The monocistronic TriMV firefly luciferase constructs were assembled in the T3 polymerase-driven plasmid c-myc-T3LUC(pA) (23). The 5′ UTRs of TriMV (nt 1 to 739) and the derived mutants were amplified by PCR. The PCR fragments were digested with the HindIII and NcoI restriction enzymes and ligated to the 5′ end of the luciferase reporter gene in the HindIII- and NcoI-cut cmyc-T3LUC(pA) plasmid. The cmyc-T3Luc(pA) plasmid contains a T3 RNA polymerase promoter followed by the c-myc untranslated region, which was removed by HindIII-NcoI digestion, and contains a firefly luciferase reporter gene at its 3′ end with 62 adenines downstream. Our control construct, designed to mimic a cellular mRNA, was derived from the pLGMS2 plasmid (24). This plasmid contains a firefly luciferase reporter gene flanked by 18 nt of vector sequence at its 5′ end and a 39-nt poly(A) tail at its 3′ end. It was linearized with either BamHI to include the poly(A) tail or BglII to exclude the poly(A) tail.
The TriMV and empty control bicistronic constructs were synthesized from the pDluc plasmid (W. Allen Miller, Iowa State University, Ames, IA). The plasmid contains a T7 RNA polymerase promoter, a renilla luciferase gene as the first open reading frame, an intergenic region with multiple cloning sites, a firefly luciferase gene as the second open reading frame, and a 60-nt-long poly(A) tail. The PCR-generated TriMV 5′ UTR was cloned into the intergenic region by using XhoI and BglII restriction enzymes. The control construct (empty) was digested with XhoI and BglII and then religated with the DNA polymerase I large fragment (Klenow) to make an empty cassette.
To insert a stable stem-loop at the 5′ end of pDluc (SLpDluc), we synthesized and annealed two reverse-complementary primers corresponding to the sequence of the stem-loop, CGC GCG CAC GGC CCA AGC TGG GCC GTG CGC GCC, with NcoI sticky ends. The fragment was then inserted into the plasmid by using the restriction site NcoI, located immediately after the promoter. The reverse-complementary sequences form a stable stem-loop with a ΔG of >−34 kcal (25).
To insert the same stable stem-loop at the 5′ end of the firefly luciferase monocistronic construct, we designed primers with an overlapping 15-nt sequence and 15 bases complementary to the vector sequence (forward primer GGG CCC CCC CTC GAG CGC GCG CAC G and reverse primer ACC GTC GAC CTC GAG GGC GCG CAC G). The vector was cut with XhoI to linearize the vector, and dimerized primers generated by PCR were used as the template for cloning by using the In-Fusion cloning kit from Clontech.
The TriMV and green fluorescent protein (GFP) free RNAs used in the trans-inhibition assays were in vitro transcribed by using a PCR-based template. GFP free RNA was amplified from the TuMV infectious clone p35S::TuMV-GFP (26) and corresponds to bases 1699 to 2410 in the vector sequence (bases 1 to 711 in the GFP open reading frame). Primer sequences used for amplification were forward primer TAATACGACTCACTATAGGGAGTAAAGGAGAAGAACTTTTC and reverse primer TTT GTA TAG TTC ATC CAT GCC ATG TGT AAT C. Engineered primers were used to insert a T7 RNA polymerase promoter at the 5′ end before the sequence of interest. The resulting PCR products were used as the templates for in vitro transcription. The 105-nt barley yellow dwarf virus translation element (BTE) RNA was provided by the laboratory of W. Allen Miller (Iowa State University, Ames, IA) (27).
Transcription.
All RNAs were transcribed in vitro from linearized plasmids or PCR-amplified products by using either the T7 MegaScript kit from Ambion or T3 or T7 RNA polymerase from Life Technologies. Monocistronic TriMV luciferase constructs were linearized with either SfcI to include the poly(A) tail or SpeI to exclude the poly(A) tail. The pDluc-derived constructs and the empty vector control construct were linearized with BamHI to include the poly(A) tail. The renilla luciferase control construct (pRL-null; Promega) was linearized with BamHI to include the poly(A) tail.
Reaction mixtures were assembled according to protocols of the appropriate transcription kit. All mRNAs, unless otherwise specified, were in vitro transcribed with either the 3′-0-Me-m7G(5′)ppp(5′)G or G(5′)ppp(5′)A cap analog (New England BioLabs) at 4 mM concentrations. The ApppG cap analog increases the stability of the RNA without interfering with translation initiation and has no ability to recruit translation factors. The in vitro transcription reaction was executed for 2 h at 37°C. Turbo DNase (Life Technologies) was added to the mixture after the reaction to degrade the template DNA before RNA precipitation. RNA was precipitated with ethanol by the addition of a 10% volume of 3 M ammonium acetate and a 200% volume of absolute ethanol. RNAs were washed with 70% ethanol and resuspended in RNase-free water. The RNA concentration was measured by using a Nanodrop ND-1000 spectrophotometer, and RNA quality and equal loading were verified on an agarose gel.
Translation assays and data collection.
The in vitro translation reactions were performed by using the wheat germ extract system kit (Promega). A 50-μl translation master mix was prepared by using 0.1 pmol of RNA transcript, 25 μl of wheat germ extract, 4 μl of potassium acetate, and 4 μl of amino acid mix, all of which, except for the RNA, were provided in the wheat germ extract kit (28). RNase-free water was used to bring the volume up to 50 μl. The master mix was then aliquoted for each replicate to a final volume of 10 μl, and the reaction mixtures were assembled on ice. Each assay was performed in triplicate and repeated in at least three independent experiments. Translation reactions were executed at room temperature for 45 min and stopped by placing the reaction mixtures on ice and adding 30 μl of 1× passive lysis buffer (Dual Luciferase kit; Promega) to each 10-μl reaction mixture. Luciferase activity was read for 10 s on a Centro XS3 LB 960 luminometer following the injection of 10 μl of Luciferase Assay reagent (Promega). For dual-luciferase assays, 10 μl of Stop & Glo reagent (Dual Luciferase kit; Promega) was injected after the firefly luciferase reading, and renilla luciferase activity was then read for 6 s.
In the competition assays, luciferase activity was measured for the control m7GpppG-capped and polyadenylated vector mRNA construct. Exogenously added RNA or cap analog was added to the translation reaction mixtures at the appropriate concentrations after the reporter mRNA. The in vitro translation reactions were performed as described above.
Depleted wheat germ assay extracts were prepared as described previously by Gallie and Browning (2). Wheat germ extract was self-prepared, loaded onto an m7GTP-Sepharose affinity column (GE Life Sciences), and equilibrated in a solution containing 25 mM HEPES-KOH at pH 7.6, 100 mM KCl, 1 mM MgCl2, and 1 mM dithiothreitol (DTT). The unbound fraction was then collected and aliquoted for storage at −80°C prior to use. In vitro translation was conducted in 50-μl reaction mixtures with 5 pmol of RNA and 34 μM 14C-radiolabeled leucine (PerkinElmer Life Science). The reaction mixture was incubated at room temperature for 30 min, and the amount of 14C-radiolabeled leucine incorporated into the newly synthesized proteins was measured following trichloroacetic acid (TCA) protein precipitation using a scintillation counter as previously described (29).
Translation in oat protoplasts.
Oat protoplasts were prepared from an oat cell suspension culture, as described previously (30). A total of 1 pmol of each RNA reporter construct was electroporated into ∼106 cells. To normalize RNA incorporation into cells, 0.1 pmol of capped polyadenylated renilla luciferase RNA was included in each electroporation mixture. At 4 h postelectroporation, the cells were harvested, lysed in 500 μl passive lysis buffer (Dual Luciferase kit; Promega), and centrifuged for 10 min at 15,000 × g. Luciferase activity was measured by using 100 μl of the supernatant. Fifty microliters of the luciferase assay reagent was injected into each sample, and activity was read for 10 s. Fifty microliters of Stop & Glo reagent was then injected, and renilla luciferase activity was measured for 6 s. All experiments were performed in triplicate and repeated in at least three independent experiments.
Protein expression and translation assays.
Wheat eIF4F, eIF4G, and eIF4E recombinant proteins were prepared as described previously by Mayberry et al. (31). In vitro translation assays with initiation factor proteins added in trans were performed with 10-μl wheat germ extract reaction mixtures. For the recovery assay of trans-inhibition and for the translation assay in the depleted wheat germ, increasing concentrations of eIF4F, eIF4G, or eIF4E were added to the reaction mixture, as indicated.
SHAPE assay.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) analysis of RNA was carried out with fluorescently labeled primers and resolved by two-capillary electrophoresis as described previously by Karabiber et al. (32). Briefly, 15 pmol of the TriMV firefly luciferase RNA construct was heated to 95°C for 3 min and then placed on ice for 2 min. The RNA was resuspended in 5× folding buffer [80 mM Tris-HCl (pH 8.0), 160 mM NH4Cl, 11 mM Mg(OAc)2] and then chemically modified at 37°C for 40 min with N-methylisatoic anhydride (NMIA) dissolved in dimethyl sulfoxide (DMSO) (Sigma) to a final concentration of 15 mM. For the control RNA sample, only DMSO was added. Unpaired or unconstrained nucleotides will likely react with the reagent through its 2′-hydroxyl group to form a bulky 2′-0 adduct, which prevents primer extension by reverse transcriptase. The reverse primer used for reverse transcription (AGGGCGTATCTCTTCATAGCCTT) was complementary to a sequence within the firefly luciferase coding region, beginning 70 nt downstream of the TriMV 5′ UTR. The primer was 5′-end labeled with either a JOE (6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein) NHS ester or a 6-carboxyfluorescein (FAM) fluorophore (Integrated DNA Technologies). For the primer extension, 3 pmol of modified or nonmodified RNAs was mixed with 6 pmol of JOE-labeled primers. For the RNA sequencing ladder, 3 pmol of nonmodified RNA was mixed with 6 pmol of FAM-labeled primers, and a primer extension step was performed with 10 mM dideoxynucleoside triphosphates (ddNTPs). The SHAPE reactions were resolved by capillary electrophoresis through Integrated DNA Technologies services. The capillary data were analyzed with QuSHAPE software (32) with the normalized annotation of the SHAPE reactivity for each position. The SHAPE data were then further analyzed by using VARNA software, where the normalized reactivity was taken and used for structure modeling (33).
Data analysis.
Each experiment was performed in triplicate and independently replicated at least three times. Translation measurement units were plotted with the means and the standard errors of the means (SEM) by using GraphPad Prism 5. The translation values are expressed as luciferase light units, relative luciferase activities (percent), fold increases, or firefly/renilla luciferase activity ratios. To compare the monocistronic mRNA translation measurements, luciferase units were standardized to the mean for the control in each graph (set at 100%). For comparisons of the translation levels of RNA transcripts with or without a 5′-m7GpppG cap, or with or without a 3′-polyadenylated tail, the fold increase is derived by relativizing the data to the non-m7GpppG cap or non-poly(A) tail constructs (set as 1-fold). The ability to conduct internal initiation was defined by calculating the ratio of firefly luciferase units to renilla luciferase units expressed by the bicistronic RNA. In competition assays with added free RNA, data were standardized by using the mean for the exogenously added RNA at the zero time point (100% relative translation).
A t test (see Fig. 1) or analysis of variance (ANOVA) (see Fig. 2 to 5) was performed by using GraphPad Prism 5 to compare significances of the means of the data. Significant P values are reported where appropriate.
FIG 1.
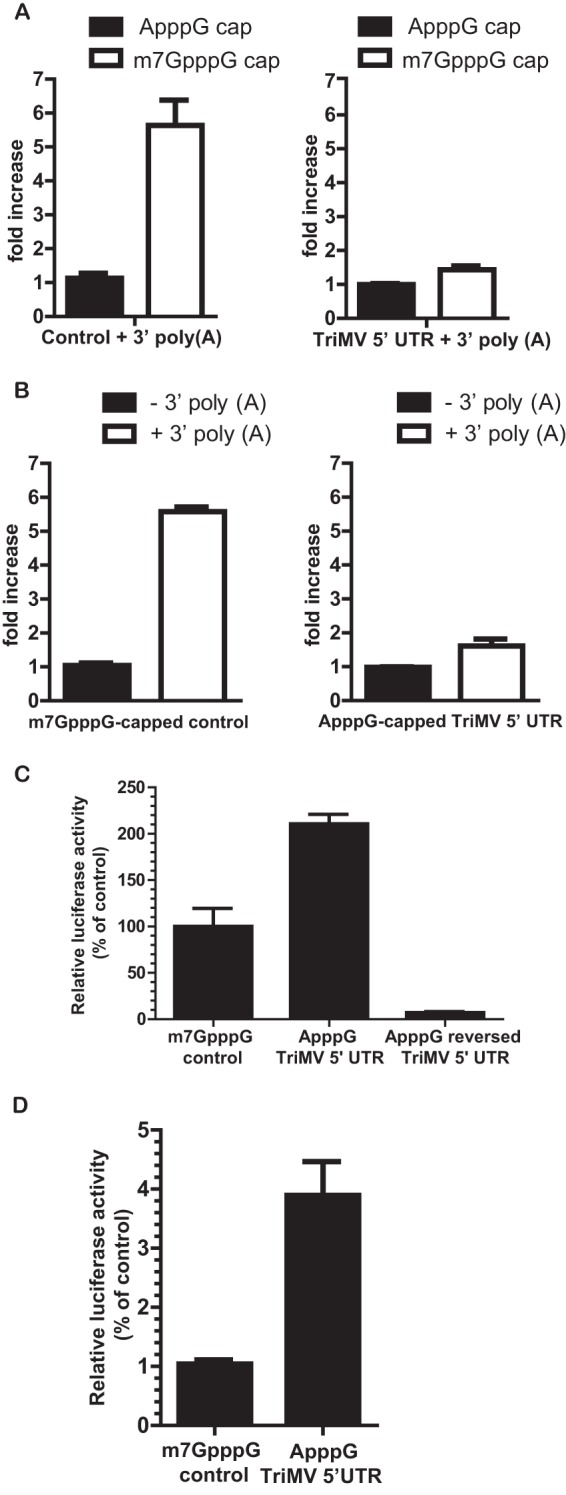
The TriMV 5′ leader supports cap-independent translation. (A) Fold increase of luciferase activity in wheat germ extract of the polyadenylated TriMV 5′-UTR reporter mRNA (right) and of the polyadenylated control vector sequence mRNA (left) with a 5′ m7GpppG cap analog or a 5′ ApppG cap at their 5′ ends. The ApppG cap analog has no function in translation. The fold increase of luciferase activity was standardized to the measured luciferase activity of the ApppG-capped and polyadenylated RNA. (B) Fold increase of luciferase activity in wheat germ extract of the ApppG-capped TriMV 5′-UTR reporter mRNA (right) and m7GpppG-capped control RNA (left) with or without a 3′ poly(A) tail. The fold increase of luciferase activity was standardized to the measured luciferase activities of the nonadenylated RNAs. (C) Luciferase activity in wheat germ extract of the mRNA transcripts containing the TriMV leader (ApppG TriMV 5′ UTR) or its reversed complementary sequence (ApppG reversed TriMV 5′ UTR) with an ApppG cap analog, relative to the control m7GpppG-capped vector sequence. All constructs are polyadenylated. Luciferase activities were standardized to the measured luciferase activities of the m7GpppG-capped control mRNA. (D) Relative luciferase activity in oat protoplasts of the reporter mRNAs containing the ApppG-capped TriMV leader or an m7GpppG-capped vector sequence as the 5′ UTR, normalized to values for an m7GpppG-capped polyadenylated renilla reporter RNA used as an internal control, which was coelectroporated at a 1:10 ratio. All transcripts are polyadenylated.
FIG 2.
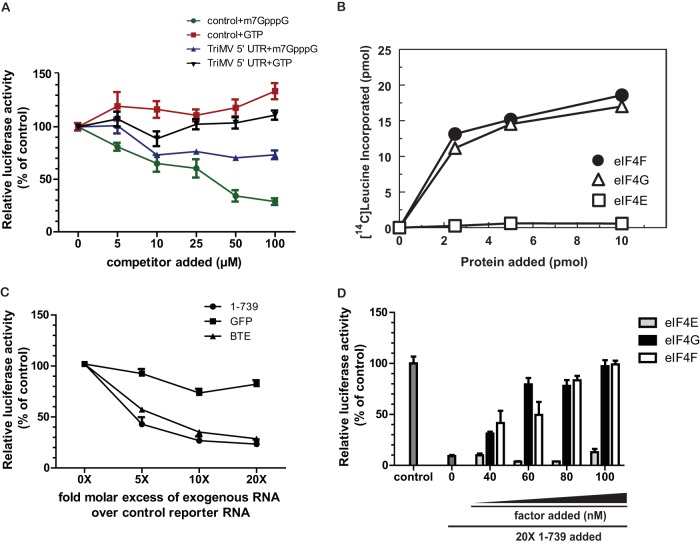
TriMV 5′-UTR translation is eIF4E independent. (A) Relative luciferase activity in wheat germ extract of the TriMV 5′ UTR and m7GpppG-capped control polyadenylated mRNAs with increasing concentrations (0 to 100 μM) of GTP or the m7GpppG cap analog. One hundred percent relative luciferase activity corresponds to the measured activity of the transcripts alone (with no competitor added). (B) Translation ability of ApppG-capped polyadenylated TriMV RNA in wheat germ extract depleted of cap-binding factors with increasing concentrations (0 to 10 pmol) of the wheat eIF4F complex or its individual subunits (eIF4G or eIF4E). The assay was performed with 14C-radiolabeled leucine, and the results display the incorporation of radiolabeled leucine (picomoles) in newly synthesized proteins following TCA protein precipitation. (C) trans-Inhibition assay with increasing molar excesses of free RNAs competing against the m7GpppG-capped and polyadenylated vector reporter in wheat germ extract. A 0- to 20-fold molar excess of the competing free RNAs corresponding to the TriMV leader (nt 1 to 739), the BTE (36), or a 700-nt RNA sequence from the GFP coding region was added to the translation reaction mixture. (D) Restoration of translation of the capped polyadenylated control reporter mRNA by the addition of increasing concentrations of purified wheat eIF4F, eIF4G, or eIF4E (0 to 100 μM) in wheat germ extract inhibited by a 20-fold molar excess of the TriMV 5′-UTR (nt 1 to 739) free RNA. The luciferase activities were standardized to the measured luciferase activities of the capped polyadenylated control mRNA alone (control) with no competitor or translation factors added.
FIG 5.
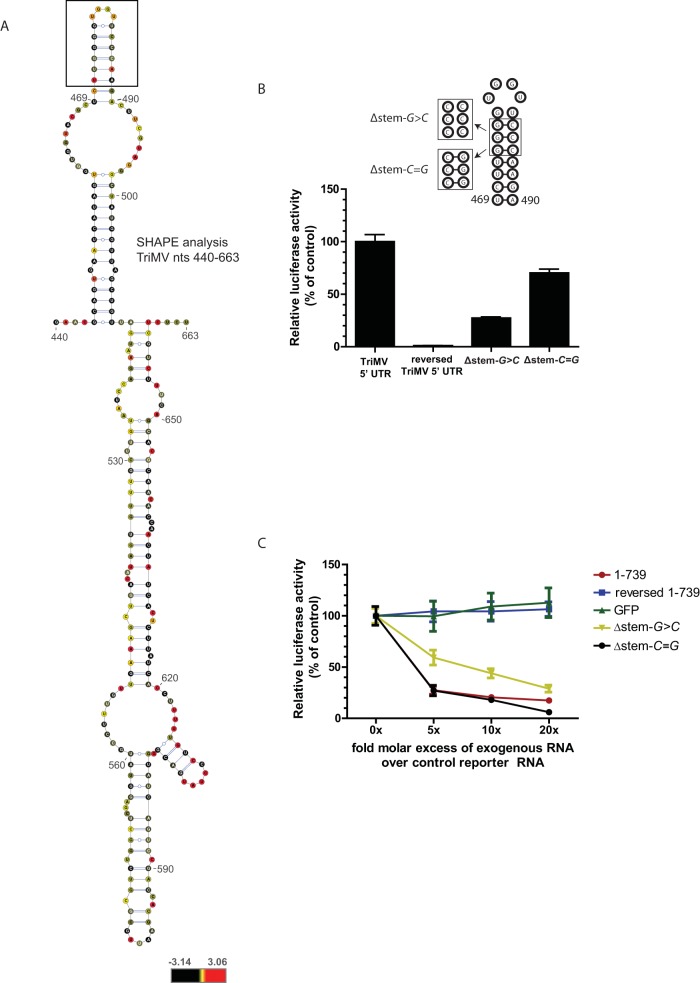
Disruption of the stem structure at nucleotide positions 469 to 490 impairs TriMV-mediated translation. (A) Structural probing results for nucleotides 440 to 610 within the TriMV 5′ UTR superimposed onto the best-fitting secondary structure by using VARNA software (33). The reactivity of each nucleotide to the methylisatoic anhydride added was analyzed with QuSHAPE software (32). Normalized SHAPE reactivity values of −3.14 to +0.4 (in black), +0.5 to +0.8 (in yellow), and +0.9 to +3.06 (in red) correspond to unreactive, moderately reactive, and highly reactive nucleotides, respectively. A stem-loop structure at nucleotide positions 469 to 490 that was predicted in silico (data not shown) is boxed. (B) Relative luciferase activity in wheat germ extract of the TriMV stem mutant RNAs compared to the wild-type and TriMV reverse sequences. The stem mutant RNAs contain either three consecutive base mutations (three guanines into three cytosines) of the stem structure at nucleotide positions 474 to 476 (construct Δstem-G>C) or a double-compensatory mutation (construct Δstem-G=C) that restores the stem structure. All transcripts are ApppG capped and polyadenylated. Mutated bases are boxed in the schematic diagram of the stem-loop structure at positions 469 to 490. (C) In vitro trans-inhibition assay of cap-dependent translation with up to a 20-fold molar excess of the competing free RNAs corresponding to the TriMV stem-loop mutants (Δstem-G>C and Δstem-G=C), the wild-type 5′ UTR (nt 1 to 739), the negative control (GFP), and the reversed complementary sequence of the TriMV 5′ UTR (nt 1 to 739).
RESULTS
The TriMV 5′ UTR drives cap-independent translation.
The genomic RNA of triticum mosaic virus (TriMV) is naturally noncapped at its 5′ end and polyadenylated at its 3′ end (21). To examine the function of the TriMV 5′ leader in translation in vitro, we first tested the dependency of the RNA sequence on the cap and the poly(A) tail in translation in wheat germ extract (22, 34, 35) (Fig. 1). We engineered a firefly luciferase reporter construct that contains the presumed TriMV 5′ UTR (nt 1 to 739) at its 5′ end and a 46-nt vector sequence at its 3′ end followed by a 62-nt poly(A) tail. We compared the level of translation of the TriMV RNA transcript with an m7GpppG cap to that of a transcript with an ApppG cap at its 5′ end. The ApppG cap analog was added for RNA stability, but unlike the m7GpppG cap, it has no function in translation. As a control, a luciferase RNA construct that contains vector sequences at its 5′ and 3′ ends followed by a 3′ poly(A) tail was used to mimic the structure of a eukaryotic mRNA. We compared the level of translation of our control RNA with an m7GpppG cap to that of the control RNA with an ApppG cap at its 5′ end. As expected, translation of the m7GpppG-capped polyadenylated control RNA was nearly 6-fold higher than that of the ApppG-capped control RNA (Fig. 1A, left). In contrast, the m7GpppG cap added to the TriMV 5′ UTR boosted translation by <0.5-fold compared to the ApppG-capped TriMV mRNA (P value of 0.005) (Fig. 1A, right). We next compared translational stimulation of the poly(A) tail by adding or removing the 3′ poly(A) tail on the m7GpppG-capped control RNA and the ApppG-capped TriMV RNA (Fig. 1B). In combination with the m7GpppG cap, the poly(A) tail increases translation of the control mRNA by 5-fold compared to that of the nonadenylated RNA (Fig. 1B, left). However, for the TriMV RNA transcript, the 3′ poly(A) tail provided an ∼0.5-fold increase in translation (P value of 0.001) (Fig. 1B, right). These results show that under our translation conditions, in which the wheat germ in vitro system can recapitulate cap and poly(A) tail dependency, the TriMV 5′ UTR alone is sufficient to initiate translation.
Next, we measured the translational activity of the TriMV reporter RNA containing an ApppG cap and a poly(A) tail and directly compared its activity with that of the m7GpppG-capped polyadenylated control RNA (Fig. 1C). The results showed that the TriMV 5′ UTR supports translation in wheat germ, with a translational output 2-fold higher than that of the control RNA. In contrast, the reversed complementary sequence of the TriMV 5′ UTR (reversed TriMV) failed to drive translation (Fig. 1C). We also tested the constructs in oat protoplast cells, which are a natural host of the virus (20) (Fig. 1D). ApppG-capped TriMV RNA and the capped polyadenylated control RNA transcripts were electroporated into ∼106 cells. For an internal control, we coelectroporated the mRNAs at a 1:10 ratio with an m7GpppG-capped polyadenylated renilla reporter RNA. Next, we measured the ratio of firefly luciferase activity/renilla luciferase activity. The results showed that the TriMV 5′-UTR RNA had 2-fold greater translation activity than the capped polyadenylated control RNA.
Taken together, the TriMV 5′ UTR is able to drive cap-independent translation both in vitro and in vivo.
TriMV 5′-UTR-mediated translation is largely eIF4E independent.
Because the TriMV 5′ UTR can drive translation without a 5′ cap, we tested whether the 5′ UTR requires the cap-binding factor eIF4E for translation initiation (Fig. 2). We first measured the translation of the ApppG-capped TriMV 5′-UTR reporter RNA in the presence of increasing concentrations of an m7GpppG cap analog (0 to 100 μM) added to the translation reaction mixture (Fig. 2A). The exogenously added m7GpppG cap analog normally inhibits cap-dependent translation by competitively sequestering eIF4E. We compared the translation of the TriMV 5′ UTR reporter RNA to that of the control m7GpppG-capped polyadenylated RNA, which is dependent upon eIF4E for translation. As a negative control, we added increasing concentrations of noncompetitive GTP in place of the m7GpppG cap analog. At a concentration of 10 μM, m7GpppG inhibited the translation of the capped control mRNA by 35%. At concentrations of 50 μM and higher, translation was reduced by 65%. However, the addition of the m7GpppG cap analog for TriMV 5′ UTR-driven translation had a less radical effect. The TriMV mRNA retained at least 75% of its translation at most concentrations of the inhibitor. We conclude that, unlike cap-driven translation, TriMV-mediated translation is largely unaffected by cap analog inhibition.
To directly test the requirement for the TriMV 5′ UTR on the cap-binding complex (eIF4F/eIFiso4F), we passed wheat germ extract through an m7GpppG-Sepharose column to make wheat germ extract translation dependent on the addition of the complex. This well-established process largely depletes the cap-binding complex from the extract, including the factors eIF4E and eIF4G and their isoforms (eIFiso4E and eIFiso4G), and limited amounts of eIF4A, eIF4B, and the poly(A) binding protein (PABP) (2, 3). This approach has been extensively used to determine the translation factor dependency of plant virus translation enhancers (36–38). We measured translation by determining the percentage of 14C-labeled leucine incorporated into the newly synthesized protein (29). We added increasing concentrations (0 to 10 pmol) of eIF4F or its individual subunits (eIF4G and eIF4E) to the depleted wheat germ extract translation reaction mixture. TriMV 5′-UTR-driven translation was fully dependent on exogenously added translation factors (Fig. 2B), and maximum translation was reached by adding the full eIF4F complex. eIF4E itself did not stimulate translation, but the addition of the large subunit eIF4G alone supported translation.
Next, we investigated the competitive ability of the TriMV 5′-UTR sequence to interfere in trans with the translation of the capped polyadenylated control RNA, presumably through sequestration of translation factors (Fig. 2C). We added free RNA consisting of the TriMV 5′-UTR sequence (nt 1 to 739), at an increasing molar excess (up to 20-fold) over the m7GpppG-capped polyadenylated control mRNA, to the in vitro translation reaction mixture. For a positive control, we used the well-characterized 105-nt barley yellow dwarf virus cap-independent translation element (BTE) RNA, which supports the translation of noncapped mRNAs both in vivo and in vitro and inhibits cap-dependent translation when added in trans (36, 39–41). For a negative control, we used a 700-nt RNA sequence from the coding region of green fluorescent protein (GFP). We determined the ability for trans-inhibition as the amount of exogenously added free RNA required to attain 50% inhibition of cap-driven translation. We relativized the translation level to the value for the control RNA treatment (no exogenous RNA), which was defined as 100% translation (0-fold molar excess). Our results revealed that the free TriMV RNA sequence spanning nt 1 to 739 added in trans interfered with cap-driven translation as strongly as BTE RNA (Fig. 2C). Similarly to BTE RNA, a 5-fold molar excess of TriMV RNA decreased translation by 50%. Even at high concentrations, the control GFP RNA showed little ability to inhibit translation, which confirms the specificity of our assay. The TriMV reverse complement RNA was unable to inhibit translation in trans (data not shown).
The BTE is known to specifically interact with the eIF4F complex in wheat germ extract. It was demonstrated previously that the addition of eIF4F reverses trans-inhibition of cap-dependent translation by the BTE (36, 41), which we replicated in our assay (data not shown). To determine whether eIF4F and/or its individual subunits (eIF4G and eIF4E) affect TriMV 5′-UTR-mediated trans-inhibition, we added increasing concentrations of eIF4F, eIF4G, or eIF4E (0 to 100 nM) to the translation reaction mixture inhibited by a 20-fold molar excess of free RNA of the TriMV region spanning nt 1 to 739 and measured the recovery of translation (Fig. 2D). We relativized recovery to 100% using the translation reaction mixture with no inhibitor added (control). Increasing concentrations of eIF4F reversed TriMV-mediated inhibition, with 100 nM being sufficient to fully recover translation. The addition of eIF4G alone restored the translation of the capped polyadenylated mRNA. At every concentration, there was no statistically significant difference between eIF4F and eIF4G (P value of 0.6881). However, the addition of eIF4E alone had no effect on translation, and inhibition by the TriMV sequence spanning nt 1 to 739 was maintained. These observations demonstrate that the TriMV 5′-UTR sequence competes for some components of the eIF4F complex but possibly not eIF4E. Together, the data are in line with a lack of full dependence upon eIF4E for TriMV translational function.
Translation initiates at the start codon beginning at nucleotide 740.
The TriMV 5′ UTR uniquely possesses 12 AUG start codons upstream of the presumed authentic start codon at nt 740 (Fig. 3A) (20). Our sequence analysis reveals that four of these AUGs (nucleotide positions 116 to 118, 146 to 148, 281 to 283, and 333 to 335) are in frame with the presumed initiation site (Fig. 3A, underlined bases). The AUGs at positions 116 to 118, 333 to 335, 501 to 503, 561 to 563, and 598 to 600 are in a good Kozak sequence context (A/C at position −2 and G at position +4) (42, 43). The AUGs at positions 281 to 283, 525 to 527, and 598 to 600 could potentially be start sites for upstream open reading frames (uORFs) of ∼33, 64, and 77 amino acids, respectively (20). If TriMV-mediated translation relies on ribosomal scanning that obeys conventional Kozak rules, ribosomes should encounter one of these upstream AUGs (uAUGs) and begin translation before reaching the start codon of the luciferase gene. To verify that the 13th AUG at nt 740, which corresponds to the start codon of the firefly luciferase gene in our reporter construct, is the authentic translation initiation site, we introduced different mutations of the 13th AUG (Fig. 3B). In one construct, we mutated the 13th AUG to an AUC non-start codon (construct AUG>AUC), and in another construct, we inserted two stop codons immediately after this AUG codon to block translation elongation (construct AUG-2XSTOP). If the 13th AUG is the correct start site, these mutations should greatly reduce the translation of the luciferase gene. As shown in Fig. 3B, the AUG-to-AUC mutation reduced translation to a level similar to that for the nonfunctional TriMV reverse RNA. The double stops placed after the AUG codon abolished the translation of luciferase. However, when we placed a stop codon immediately upstream of the 13th AUG (construct STOP-AUG) to arrest potential translation elongation from any of the in-frame AUGs, translation of the luciferase gene was not significantly affected (P value of 0.2161). Our data support that the 13th AUG at nucleotide positions 740 to 742 is the correct initiation site.
FIG 3.
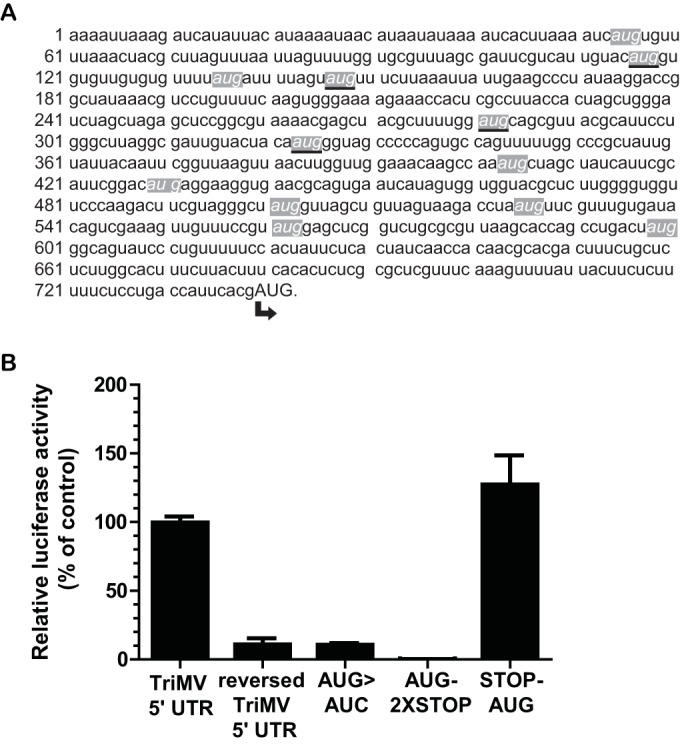
TriMV-driven translation initiates primarily at the 13th AUG codon at nucleotide position 740. (A) The TriMV 5′-UTR sequence (GenBank accession number FJ669487) contains 12 upstream AUG start codons, highlighted in italicized font in gray-shaded boxes. The upstream AUG codons that are in frame with the presumed correct initiation site at position 740 (in capital letters) are underlined. (B) Relative luciferase activity in wheat germ extract of the ApppG-capped polyadenylated TriMV-derived mutant mRNAs with the 13th AUG codon mutated to AUC (construct AUG>AUC), with two stop codons inserted just downstream of the 13th AUG (construct AUG-2XSTOP), or with a stop codon inserted just upstream of the 13th AUG (construct STOP-AUG), relative to the wild-type sequence and the reverse complement TriMV sequence.
A 300-nt region can support cap-independent translation.
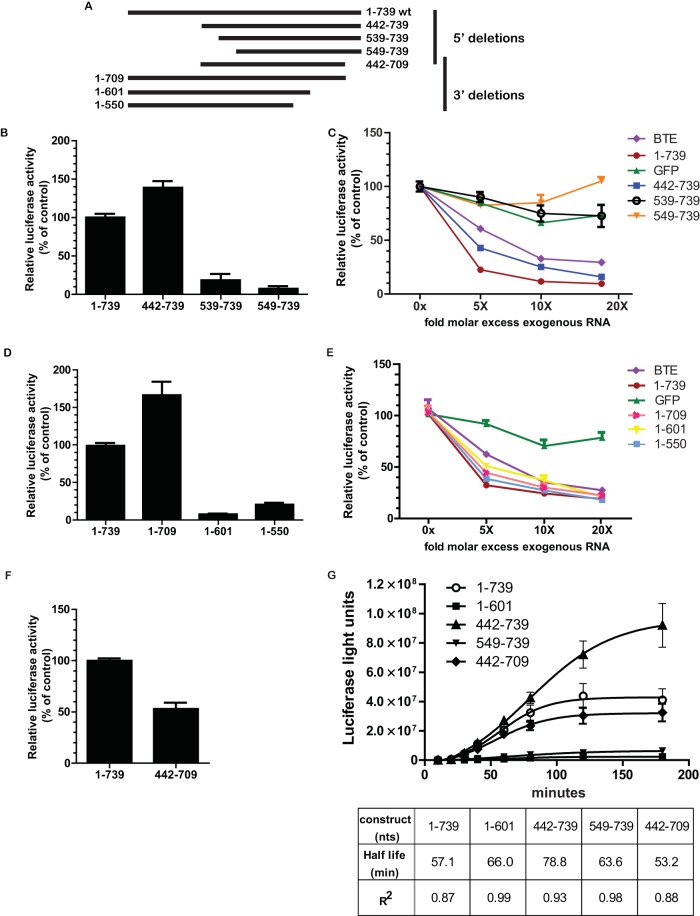
To functionally map the TriMV translation element, we roughly truncated the 739-nt TriMV 5′ UTR from its 5′ and 3′ ends and tested the mutants in the monocistronic reporter construct using an ApppG cap at the 5′ ends (Fig. 4). We compared translation of the mutants to that of the full-length TriMV 5′ UTR. As shown in Fig. 4B, deletion of ∼400 nucleotides from the 5′ end did not impair the ability of the TriMV 5′ UTR to drive cap-independent translation. The mutant TriMV 5′ UTR spanning positions 442 to 739, which has a deletion of nucleotides 1 to 441, showed a significant increase in the translation of the luciferase reporter compared to the full-length TriMV 5′ UTR (P value of 0.01). However, further 5′ deletions (nt 539 to 739 and nt 549 to 739) caused a significant reduction in translation (P values of <0.0001) (Fig. 4B).
FIG 4.
The minimal TriMV 5′ UTR necessary for cap-independent translation is within nucleotides 442 to 709. (A) Schematic representation of the 5′ and 3′ deletion mutants of the TriMV 5′ leader. The full-length TriMV 5′ UTR corresponds to nucleotides 1 to 739. (B) Relative luciferase activity in wheat germ extract of the 5′ deletion mutants derived from the TriMV 5′ leader (nt 442 to 739, 539 to 739, and 549 to 739) compared to the full-length 5′ UTR (nt 1 to 739). All transcripts are ApppG capped and 3′ polyadenylated. Luciferase light units were standardized by using the full-length TriMV 5′-UTR (nt 1 to 739) luciferase light units. (C) In vitro trans-inhibition assay of cap-dependent translation with up to a 20-fold molar excess of the competing free RNAs corresponding to the TriMV 5′ mutant sequences (nt 442 to 739, 539 to 739, and 549 to 739), the full-length sequence (nt 1 to 739), the BTE, or a 700-nt RNA sequence from the GFP coding region. (D) Relative luciferase activity in wheat germ extracts of the 3′ deletion mutants derived from the TriMV 5′ leader (nt 1 to 709, 1 to 601, and 1 to 550) compared to the full-length 5′ UTR (nt 1 to 739). All mRNAs are ApppG capped and 3′ polyadenylated. Luciferase activities are standardized by using full-length TriMV 5′-UTR luciferase light units. (E) In vitro trans-inhibition assay of cap-dependent translation with up to a 20-fold molar excess of the competing free RNAs corresponding to the TriMV 3′ mutant sequences (nt 1 to 709, 1 to 601, and 1 to 550), the full-length sequence (nt 1 to 739), the BTE, or a 700-nt RNA sequence from the GFP coding region. (F) Relative luciferase activity in wheat germ extract of deletion mutant RNA (nt 442 to 709) compared to full-length TriMV 5′-UTR RNA (nt 1 to 739). Both transcripts are ApppG capped and 3′ polyadenylated. Luciferase activities are standardized by using the measured luciferase light units for the full-length TriMV 5′ UTR. (G) Functional half-life of RNA mutants (nt 442 to 739, 549 to 739, 1 to 601, and 442 to 709) relative to the wild type (nt 1 to 739). The functional half-life of the mRNA was determined as the time needed to reach half of the maximum accumulation of luciferase light units over a 3-h course of time in wheat germ extract minus the lag time. The curves with the best fit to the experimental data points were generated by using GraphPad software.
To further test the functionality of the 5′ deletion mutants, we performed a trans-inhibition assay in wheat germ extract using the mutant free RNAs. We compared the levels of inhibition of cap-dependent translation with exogenously added deletion mutant RNAs (nt 442 to 739, 539 to 739, and 549 to 739), full-length TriMV 5′ UTR RNA (nt 1 to 739), the control BTE RNA, and the nonfunctional RNA control (GFP). The results from the trans-inhibition assays corroborate our luciferase translation data in cis (Fig. 4C). A 5-fold molar excess of free RNA corresponding to nt 442 to 739 reduced translation by 50%. However, a 20-fold molar excess of free RNAs spanning nt 539 to 739 or 549 to 739 had no inhibitory effect on cap-dependent translation. The inability of these two free RNA mutants to inhibit translation in trans is consistent with their inabilities to drive translation in cis (Fig. 4B). These observations suggest that the 5′ region of the TriMV leader needed for cap-independent translation resides roughly between nucleotides 442 and 739.
Next, we made deletions from the 3′ end of the TriMV 5′ UTR. We tested each deletion mutant with a monocistronic firefly luciferase reporter and as free RNA in an in vitro trans-inhibition assay. Deletion of 30 nt from the 3′ end (nt 1 to 709) maintained full translation of the mRNA and, in fact, resulted in a significant boost in translation (P value of 0.0078). However, further truncation (nt 1 to 601 and 1 to 550) abolished translation (Fig. 4D). We performed an in vitro trans-inhibition assay of capped RNA with a molar excess of mutant free RNAs (nt 1 to 709, 1 to 601, and 1 to 550), control BTE RNA, and control GFP RNA. Despite their inability to drive translation in cis, truncated RNAs spanning nt 1 to 601 and nt 1 to 550 inhibited the translation of capped and polyadenylated mRNA in trans as efficiently as the mutant RNA spanning nt 1 to 709 and the full-length TriMV 5′-UTR (nt 1 to 739) free RNA (Fig. 4E).
To further map the minimal TriMV 5′ UTR required for translation, we investigated whether the deletion mutant at nt 442 to 709 is able to drive cap-independent translation compared to the full-length TriMV 5′ UTR RNA (Fig. 4F). Our results show that the truncated RNA transcript spanning nt 442 to 709 can still support cap-independent translation at ∼50% of the level of the full-length 5′-UTR RNA.
Finally, to eliminate the possibility that the observed losses of translation in the deletion mutants were linked to losses in RNA stability, we estimated the functional half-lives of the full-length TriMV 5′ UTR (nt 1 to 739) and the mutant RNAs (nt 442 to 739, 549 to 739, 1 to 601, and 442 to 709) by monitoring the rate of luciferase accumulation in wheat germ extract over a 3-h course of time (Fig. 4G). This approach measures the stability of mRNAs that are actively being translated, as opposed to the measurement of the physical integrity of the transcripts (44, 45). We defined the functional half-life of the mRNA as the time of half-maximum accumulation of luciferase expression minus lag time (45). Our analysis revealed that all mutant RNAs that had a significant loss in translation activity had functional half-lives that were similar to those of the wild-type TriMV construct, which showed a half-life of 57.2 min. This supports that the observed loss of translation efficiency of RNA deletion constructs results from regulation at the translational level and not RNA instability. Interestingly, the mutant spanning nt 442 to 739 had a half-life of 78.9 min, which may have contributed to its increased translation ability (Fig. 4G). Taken together, these observations suggest that the minimal region of the TriMV leader for cap-independent translation resides roughly between nucleotides 442 and 709.
The stem-loop structure at positions 469 to 490 is required for translational activity.
To further examine the region of the TriMV 5′ UTR that is functionally involved in translation, we used mFOLD (46) to predict the secondary structure of the region spanning nt 442 to 709 (predicted ΔG = −67.33 kcal) (data not shown), which we validated using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (Fig. 5A). The structural probing results, which covered nucleotides 440 to 610 of the TriMV 5′ UTR, were superimposed on the best-fitting secondary structure using VARNA software (33). Our assay validated the presence of an 8-bp-long stem structure at nucleotide positions 469 to 490, as indicated by lowly reacting nucleotides within the stem (Fig. 5A, dark green), which could result from the pairing of the bases. The stem is capped with a UGGU tetraloop.
To examine the relevance of the stem in TriMV 5′-UTR-mediated translation, three mutations were introduced into the upper portion of the stem structure within the full-length 5′ UTR (three consecutive guanines into three cytosines at nucleotide positions 472, 473, and 474) (construct Δstem-G>C) (Fig. 5B). The mutations, predicted by mFOLD, would disrupt the stem region spanning positions 469 to 490 but preserve the overall surrounding structure. We then measured translation of the TriMV 5′-UTR stem mutant in wheat germ extract and compared it to those of the wild-type and nonfunctional TriMV reversed sequences. The disruption of the stem resulted in a significant loss of 70% of translation activity (Fig. 5B). The restoration of the stem structure with double-compensatory mutations (construct Δstem-C=G) restored translation. It significantly reestablished the translation of the mRNA in vitro (P value of <0.00001) to ∼70% of the translational activity of the wild-type TriMV 5′ UTR.
To further examine the function of the stem structure, we performed an in vitro trans-inhibition assay of the capped RNA with a molar excess of the TriMV 5′-UTR RNA with the stem mutations (Δstem-G>C and Δstem-C=G) and compared their trans-inhibitory effects to those of the wild-type, control GFP, and reversed TriMV free RNAs (Fig. 5C). As noted above, the reversed TriMV free RNA behaves similarly to the control GFP RNA in trans-inhibition. The disruption of the stem resulted in a considerable loss of the competitive trans-inhibitory effect of the TriMV 5′ UTR. At least a 10-fold molar excess of mutant Δstem-G>C free RNA is required to attain 50% inhibition of cap-driven translation. However, the free RNA of the double-compensatory mutant with the restored stem structure (Δstem-C=G) inhibited translation as efficiently as the wild-type sequence (Fig. 5C). A 5-fold molar excess of the double-compensatory mutant RNA was sufficient to achieve >50% inhibition of capped-RNA translation. We conclude that the stem structure at nucleotide positions 469 to 490 is required for the TriMV 5′-UTR translational activity.
The 5′ UTR of TriMV can drive internal initiation.
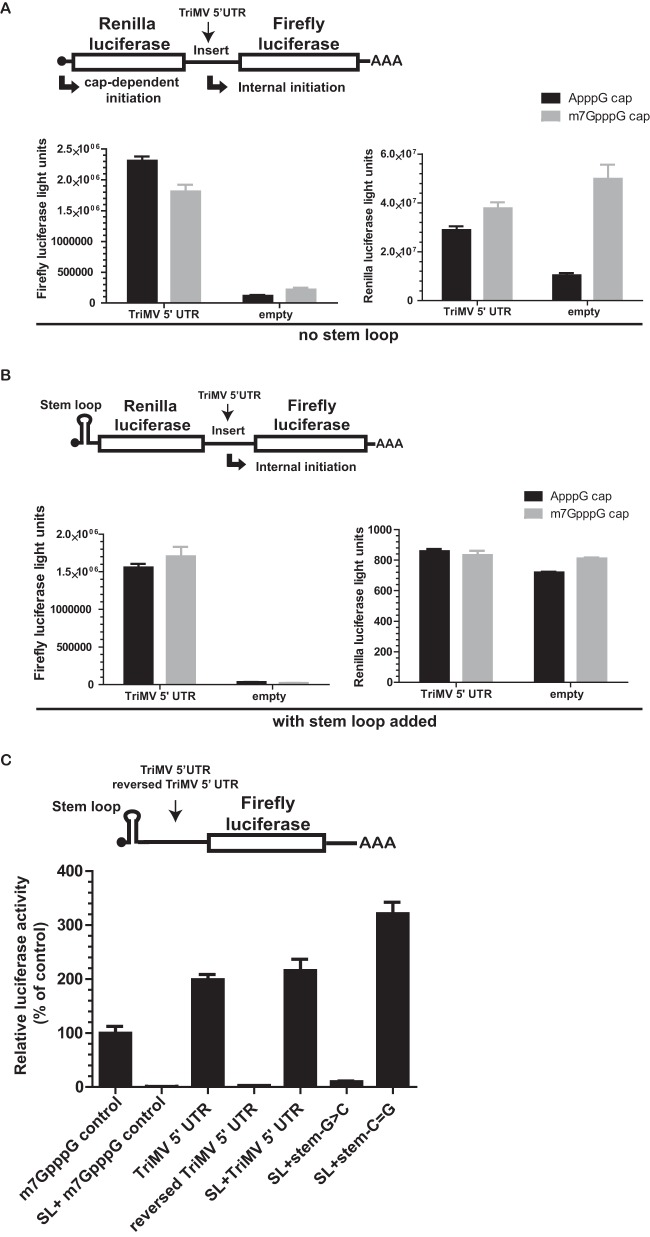
We next assessed whether the TriMV 5′ UTR can direct translation from an internal position. We inserted the TriMV 5′ UTR between the renilla luciferase and firefly luciferase reporter genes in a standard bicistronic RNA construct (Fig. 6A). Expression of the downstream, second cistron (firefly luciferase) depends on the internal initiation ability of the element placed in the intergenic region. We tested the bicistronic polyadenylated mRNA in wheat germ extract with an ApppG or an m7GpppG cap analog at its 5′ end and compared the ratio of firefly luciferase to renilla luciferase light units. We compared the translation levels of the TriMV 5′ leader sequence to those of a 30-nt vector sequence (empty control). The TriMV 5′ UTR directed internal initiation, in both ApppG- and m7GpppG-capped RNAs, and this was well above the level of the empty control (Fig. 6A).
FIG 6.
The TriMV 5′ leader can drive internal initiation. (A) Schematic diagram of the bicistronic dual-luciferase reporter RNA and the position of the insertion of the RNA elements tested. Translation of the renilla luciferase gene is cap mediated, but translation of the downstream firefly luciferase gene can be directed only by internal initiation driven by the RNA sequence inserted into the intergenic region. The internal initiation ability is quantified as the ratio of firefly luciferase to renilla luciferase activities. The ratio of firefly/renilla luciferase activities in wheat germ extracts of the bicistronic mRNAs was determined by adding an ApppG or m7GpppG cap at the 5′ end and either the TriMV 5′ UTR or no insertion (empty control) in the intergenic region. (B) Schematic diagram of the bicistronic luciferase reporter RNA with a stable hairpin insertion (ΔG = −34 kcal) (25) immediately at the 5′ end of the mRNAs. The ratio of firefly/renilla luciferase activities in wheat germ extract of the bicistronic mRNAs was determined by adding an ApppG or m7GpppG cap at the 5′ end with the strong hairpin and using either the TriMV 5′ UTR or no insertion (empty control) in the intergenic region. It is worth noting that due to the presence of the 5′ hairpin, the baseline renilla luciferase activity is at its lowest, which in turn makes the value for the ratio much higher. (C) Schematic diagram of monocistronic firefly luciferase reporter RNAs containing a stable hairpin insertion (ΔG = −34 kcal) (25) immediately at the 5′ end of the mRNA. Translation is driven by the TriMV 5′ UTR, the reversed TriMV 5′ UTR, or the m7GpppG-capped vector control sequence. The relative luciferase activity in wheat germ extract of the TriMV 5′ UTR and its stem-loop-derived mutants with the strong hairpin is shown and is relativized to that of the control capped, polyadenylated mRNA.
To test that translation of the downstream firefly luciferase is independent from translation of the first cistron, we introduced a stable hairpin (ΔG = −34 kcal) (25) at the immediate 5′ end of the mRNAs to presumably block the 5′ end for ribosomal entry. This strong hairpin should impair translation of the first cistron and leave translation of the downstream cistron strictly dependent on internal initiation (47). We again compared TriMV 5′-UTR translation to that of the empty control in ApppG- and m7GpppG-capped RNA transcripts in wheat germ extract (Fig. 6B). Our data showed that despite the addition of the stable hairpin at the 5′ end of the mRNA, the TriMV 5′ UTR maintained its ability to drive internal translation, and its level of translation was well above that of the empty control (Fig. 6B).
To further examine the observed internal initiation activity, we compared translation of the monocistronic TriMV 5′-UTR luciferase reporter RNA to that of the m7GpppG-capped control mRNA with or without the strong stem-loop (ΔG = −34 kcal) at the immediate 5′ end of the mRNA to presumably block the 5′ end for ribosomal entry (Fig. 6C). As expected, cap-dependent translation was abolished with the presence of the strong hairpin (construct SL+m7GpppG control). However, the TriMV 5′ UTR fully retained its activity when the 5′ end was blocked (SL+TriMV 5′ UTR) and drove translation at the level of the TriMV RNA without the stem-loop. To determine whether the identified stem structure at nucleotide positions 469 to 490 was involved in the internal initiation ability of the TriMV 5′ UTR, we measured translation of the mutant RNAs with the strong hairpin at the 5′ end of the mRNA. The three point mutations on the stem structure were sufficient to abolish the translational advantage of the TriMV 5′ UTR. The mutant RNA failed to drive translation of the luciferase reporter when the 5′ end was obstructed with a strong hairpin (construct SL+Δstem-G>C). However, the double-compensatory mutation that restored the stem structure showed maximum translational activity (construct SL+Δstem-C=G). Together, our results suggest that the TriMV 5′ UTR can drive translation internally and clearly independently of the 5′ end. This internal initiation activity requires the identified stem structure at nucleotide positions 469 to 490 within the TriMV 5′ UTR.
DISCUSSION
RNA elements have evolved functions as translational enhancers that perform under various conditions (10, 48). Viral RNAs of the large Potyviridae family lack a 5′ m7GpppG cap, and many of them reportedly rely on their 5′ leader sequences to initiate translation (6). Diverse translation enhancers that employ unique functional mechanisms have been identified within this family (11). Here, we report the function of the 5′ translation element identified in a recently emerged member of the Potyviridae family, Triticum mosaic virus (TriMV). Our results reveal a uniquely regulated translation enhancer that may provide new insights into the large functional diversity of translation mechanisms among plant-infecting viruses.
While no clear consensus sequence or structure has been reported for the potyviral translation elements, these elements demonstrate a similar function: they are able to drive cap-independent translation at no less than the level of a 5′-methylated capped control RNA (11). Here, we show that the noncapped TriMV leader sequence shares similar functions with other members of the Potyviridae family. The TriMV 5′ UTR is able to drive cap-independent translation both in vivo and in vitro (Fig. 1). Similarly to other reported potyviral translation elements (11), the TriMV 5′ UTR initiated translation internally. Standard approaches used to measure the internal initiation activity of the potyviral elements test their ability to either (i) initiate the translation of a downstream gene when placed in the intergenic region of a bicistronic construct, or (ii) conduct translation in a monocistronic mRNA with a stable secondary structure placed at the immediate 5′ end of the mRNA (11). A unique characteristic of the TriMV translation element compared to those of other potyviruses is that its translation mechanism is clearly driven in a 5′-end-independent manner (11). When a large stem-loop structure was placed at the 5′ end of the uncapped Turnip mosaic virus (TuMV) 5′ UTR, the potyviral 5′-UTR element sustained translation; however, it was detected at <30% of the level observed without the stem-loop (13). A similar observation was reported previously for tobacco etch virus (TEV) cap-independent regulatory elements (CIREs), which show internal initiation activity (5, 18). The presence of a 24-nt stem-loop structure added upstream of the TEV leader in a monocistronic mRNA decreased CIRE-1-driven translation by ∼30% and CIRE-2-driven translation by ∼70% (18). Although these translation levels were higher than that of an uncapped control RNA, these observations indicate that these leaders appear to require an accessible 5′ end for optimal translation; however, the exact mechanism of ribosomal recruitment remains unknown. It is worth noting that the ability to drive cap-independent translation does not necessarily correlate with an ability to drive internal initiation. For example, several noncapped and nonadenylated plant viruses rely on 3′ cap-independent translation elements (3′ CITEs) for translation (10). These 3′ CITEs are located at the 3′ end of the viral RNAs and mediate translation of the 5′-end-proximal AUG. However, these elements cannot recruit the ribosomes from an internal position to mediate downstream translation, and their translation mechanism requires ribosomal scanning from the 5′ end of the mRNA (10). The ability of TriMV to mediate translation both cap independently and 5′-end independently seems novel among plant viruses.
Clearly, some characteristics of the TriMV 5′ UTR are quite unique, which broadens the divergence among the potyviral translation enhancers. First, the observed cap-independent translation activity of most of the reported potyviral 5′ translation elements relies on a 60- to 190-nt-long 5′-UTR sequence (11). Most potyviruses require only a minimal portion of their genome for translation. The TriMV 5′ UTR is considerably longer, as the minimally derived region of the 739-nt-long TriMV leader sequence is ∼300 nt long. Second, uncommon to 5′ leader sequences of most plant RNA viruses, the TriMV 5′ UTR contains 12 AUG start codons upstream of the presumed correct initiation site of the viral polyprotein at position 740 (Fig. 3). While it remains possible that translation initiates at one or more of the alternative positions, it is clear that primary initiation occurs at the furthest downstream AUG codon (Fig. 3). This translation strategy must differ from the canonical scanning mechanism where the ribosomal complex enters from an accessible 5′ end of the mRNA and scans in the 5′-to-3′ direction in search and recognition of the 5′-proximal AUG (1). We cannot dismiss, however, whether any of the upstream AUGs play a regulatory role in the recognition and/or regulation of the main initiation site, as observed for the uncapped plum pox virus (PPV) 5′ leader (19). Whether the PPV 5′ UTR was m7GpppG capped or not, the same level of translation was reported in vitro and in vivo, which supports its ability to drive translation cap independently (19). This 145-nt-long potyviral 5′ UTR contains an in-frame upstream AUG at position 36 preceding the authentic initiator AUG at position 147 (19). The PPV leader failed to drive internal initiation in the context of a bicistronic construct. However, when the first AUG codon was placed in a good Kozak sequence context, it became the preferred initiation site (19). This suggests that the correct AUG codon is recognized through a leaky scanning mechanism. In internal initiation, the ribosomal complex is recruited within the vicinity of the correct initiation site without scanning from the 5′ end of the mRNA (47). The fact that TriMV-mediated translation initiates at the 13th AUG codon could be supported by its ability to recruit the translation initiation machinery internally (Fig. 6). Our remaining question is how TriMV directs the assembly of the translation machinery at the correct AUG codon. Mechanistically, in picornaviruses, the position of a polypyrimidine CUUU-rich tract determines the location of the landing site of the ribosomes for the correct AUG and contributes to internal initiation (49). For encephalomyocarditis virus (EMCV)-type internal ribosome entry sites, the ribosomes bind closely to the AUG used for translation initiation, which is 25 nt downstream of the polypyrimidine tract (50, 51). For other picornaviral serotypes, including poliovirus, the initiation event occurs at the following AUG, ∼100 nt downstream of the polypyrimidine tract, which the ribosome reaches by scanning (52, 53). Our sequence analysis of the TriMV 5′ UTR revealed a CUUU(4) tract that is 11 nucleotides upstream of the correct AUG. However, deletion of the TriMV leader sequence (Fig. 4) shows that without the last 30 nt of the viral 5′ UTR [which includes the CUUU(4) tract] (nt 1 to 709), the TriMV 5′ UTR maintains its ability to drive optimal cap-independent translation.
The RNA structure and/or sequence requirements of most potyviral translation elements remain to be investigated. The well-characterized TEV translation element relies on a 45-nt-long pseudoknot (17). Mutations that disrupt the stems or loops on the pseudoknot have a 60 to 92% loss in translational activity compared to that of the wild-type construct (17). On the other hand, the translation activity of the 131-nt-long TuMV 5′ UTR seems to be both structure and sequence independent. With a GC content of 29.2%, this potyviral 5′ leader remains translationally functional in the antisense orientation and retains its ability to trans-inhibit cap-dependent translation (13). While the entire structure of the full-length TriMV 5′ UTR has yet to be analyzed, an overlapping, yet different, requirement for the dual TriMV 5′-UTR functions for cap-independent translation activity and internal initiation ability is an 8-bp-long hairpin positioned ∼270 bases upstream of the initiation site.
While the exact requirement for the host translation machinery for TriMV 5′-UTR-mediated translation is yet to be dissected, our findings suggest a largely eIF4E-independent translation mechanism (Fig. 2). It remains possible that TriMV translation requires eIF4E, but (i) the affinity of the TriMV 5′-UTR mRNA for eIF4E is much stronger than that of eIF4E for the m7GpppG cap analog or (ii) the TriMV-eIF4E interaction domain is not in conflict with the cap analog-eIF4E interaction. Clearly, eIF4E-independent translation could confer a translational advantage to the virus for competing for the limited number of host factors available.
In summary, with their similar genome architecture with the animal Picornaviridae family, it has been largely assumed that members of the plant Potyviridae family share similar translation mechanisms with their animal counterparts. However, it is clear that the potyviruses show remarkably diverse translation elements unseen in animal viruses. Here, we have heightened knowledge of such diversity with the discovery of a uniquely controlled translation enhancer that may provide new insights into mechanisms of plant virus translational regulation. It remains unclear why TriMV would evolve such a unique mechanism of translation to ensure efficient expression. Future studies in the context of the viral life cycle and its synergistic interaction with other wheat viruses and/or with its vector could provide new insights.
ACKNOWLEDGMENTS
This work was supported by HATCH-ACT Formula funds (WIS01654 and WIS01796 to A.R.) and by the National Science Foundation (MCB1052530 and Arabidopsis 2010 S-0000335 to K.S.B.).
The pLGMS2 plasmid was gifted to us by the laboratory of Marvin Wickens at the University of Wisconsin—Madison. The pDluc plasmid was gifted to us by the laboratory of W. Allen Miller at Iowa State University, Ames, IA.
We claim that we have no conflicts of interest.
REFERENCES
- 1.Merrick WC. 2004. Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 2.Gallie DR, Browning KS. 2001. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem 276:36951–36960. doi: 10.1074/jbc.M103869200. [DOI] [PubMed] [Google Scholar]
- 3.Mayberry LK, Allen ML, Dennis MD, Browning KS. 2009. Evidence for variation in the optimal translation initiation complex: plant eIF4B, eIF4F, and eIF(iso)4F differentially promote translation of mRNAs. Plant Physiol 150:1844–1854. doi: 10.1104/pp.109.138438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elroy-Stein O, Fuerst TR, Moss B. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci U S A 86:6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrington JC, Freed DD. 1990. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol 64:1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kneller EL, Rakotondrafara AM, Miller WA. 2006. Cap-independent translation of plant viral RNAs. Virus Res 119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reineke LC, Lloyd RE. 2011. Animal virus schemes for translation dominance. Curr Opin Virol 1:363–372. doi: 10.1016/j.coviro.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller WA, Wang Z, Treder K. 2007. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem Soc Trans 35:1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson BL, White KA. 2011. 3′ cap-independent translation enhancers of positive-strand RNA plant viruses. Curr Opin Virol 1:373–380. doi: 10.1016/j.coviro.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Simon AE, Miller WA. 2013. 3′ cap-independent translation enhancers of plant viruses. Annu Rev Microbiol 67:21–42. doi: 10.1146/annurev-micro-092412-155609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Roberts R, Rakotondrafara AM. 2015. The role of the 5′ untranslated regions of Potyviridae in translation. Virus Res 206:74–81. doi: 10.1016/j.virusres.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Gallie DR, Tanguay RL, Leathers V. 1995. The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene 165:233–238. doi: 10.1016/0378-1119(95)00521-7. [DOI] [PubMed] [Google Scholar]
- 13.Basso J, Dallaire P, Charest PJ, Devantier Y, Laliberte JF. 1994. Evidence for an internal ribosome entry site within the 5′ non-translated region of turnip mosaic potyvirus RNA. J Gen Virol 75(Part 11):3157–3165. [DOI] [PubMed] [Google Scholar]
- 14.Yang LJ, Hidaka M, Sonoda J, Masaki H, Uozumi T. 1997. Mutational analysis of the potato virus Y 5′ untranslated region for alteration in translational enhancement in tobacco protoplasts. Biosci Biotechnol Biochem 61:2131–2133. doi: 10.1271/bbb.61.2131. [DOI] [PubMed] [Google Scholar]
- 15.Adams MJ, Antoniw JF, Beaudoin F. 2005. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol Plant Pathol 6:471–487. doi: 10.1111/j.1364-3703.2005.00296.x. [DOI] [PubMed] [Google Scholar]
- 16.Khan MA, Yumak H, Gallie DR, Goss DJ. 2008. Effects of poly(A)-binding protein on the interactions of translation initiation factor eIF4F and eIF4F.4B with internal ribosome entry site (IRES) of tobacco etch virus RNA. Biochim Biophys Acta 1779:622–627. doi: 10.1016/j.bbagrm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Zeenko V, Gallie DR. 2005. Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5′-leader. J Biol Chem 280:26813–26824. doi: 10.1074/jbc.M503576200. [DOI] [PubMed] [Google Scholar]
- 18.Niepel M, Gallie DR. 1999. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J Virol 73:9080–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simón-Buela L, Guo HS, García JA. 1997. Cap-independent leaky scanning as the mechanism of translation initiation of a plant viral genomic RNA. J Gen Virol 78(Part 10):2691–2699. [DOI] [PubMed] [Google Scholar]
- 20.Tatineni S, Ziems AD, Wegulo SN, French R. 2009. Triticum mosaic virus: a distinct member of the family Potyviridae with an unusually long leader sequence. Phytopathology 99:943–950. doi: 10.1094/PHYTO-99-8-0943. [DOI] [PubMed] [Google Scholar]
- 21.Seifers DL, Martin TJ, Harvey TL, Fellers JP, Stack JP, Ryba-White M, Haber S, Krokhin O, Spicer V, Lovat N, Yamchuk A, Standing KG. 2008. Triticum mosaic virus: a new virus isolated from wheat in Kansas. Plant Dis 92:808–817. doi: 10.1094/PDIS-92-5-0808. [DOI] [PubMed] [Google Scholar]
- 22.Gallie DR. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 23.Thoma C, Bergamini G, Galy B, Hundsdoerfer P, Hentze MW. 2004. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol Cell 15:925–935. doi: 10.1016/j.molcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 24.Cooke A, Prigge A, Wickens M. 2010. Translational repression by deadenylases. J Biol Chem 285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol 9:5134–5142. doi: 10.1128/MCB.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijayapalani P, Maeshima M, Nagasaki-Takekuchi N, Miller WA. 2012. Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog 8:e1002639. doi: 10.1371/journal.ppat.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft JJ, Hoy JA, Miller WA. 2011. Crystallization and preliminary X-ray diffraction analysis of the barley yellow dwarf virus cap-independent translation element. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:561–564. doi: 10.1107/S1744309111007196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo L, Allen E, Miller WA. 2000. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA 6:1808–1820. doi: 10.1017/S1355838200001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lax SR, Lauer SJ, Browning KS, Ravel JM. 1986. Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol 118:109–128. doi: 10.1016/0076-6879(86)18068-2. [DOI] [PubMed] [Google Scholar]
- 30.Rakotondrafara A, Jackson RJ, Kneller EP, Miller WA. 2007. Preparation and electroporation of oat protoplasts from cell suspension culture. Curr Protoc Microbiol Chapter 16:Unit 16D.3. doi: 10.1002/9780471729259.mc16d03s05. [DOI] [PubMed] [Google Scholar]
- 31.Mayberry LK, Dennis MD, Leah Allen M, Ruud Nitka K, Murphy PA, Campbell L, Browning KS. 2007. Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F, and eIF5. Methods Enzymol 430:397–408. doi: 10.1016/S0076-6879(07)30015-3. [DOI] [PubMed] [Google Scholar]
- 32.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. 2013. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 19:63–73. doi: 10.1261/rna.036327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darty K, Denise A, Ponty Y. 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2:135–140. doi: 10.1016/S1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 35.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treder K, Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA. 2008. The 3′ cap-independent translation element of barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 14:134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallie DR. 2001. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J Virol 75:12141–12152. doi: 10.1128/JVI.75.24.12141-12152.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gazo BM, Murphy P, Gatchel JR, Browning KS. 2004. A novel interaction of cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J Biol Chem 279:13584–13592. doi: 10.1074/jbc.M311361200. [DOI] [PubMed] [Google Scholar]
- 39.Wang S, Guo L, Allen E, Miller WA. 1999. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans. RNA 5:728–738. doi: 10.1017/S1355838299981979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Kraft JJ, Hui AY, Miller WA. 2010. Structural plasticity of barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology 402:177–186. doi: 10.1016/j.virol.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Browning KS, Miller WA. 1997. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J 16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K. 2008. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res 36:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA. 1987. Selection of AUG initiation codons differs in plants and animals. EMBO J 6:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuda D, Dreher TW. 2005. In vivo translation studies of plant viral unit RNAs using reporter genes. Curr Protoc Microbiol Chapter 16:Unit 16K.2. doi: 10.1002/9780471729259.mc16k02s00. [DOI] [PubMed] [Google Scholar]
- 45.Meulewaeter F, Van Montagu M, Cornelissen M. 1998. Features of the autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA 4:1347–1356. doi: 10.1017/S135583829898092X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson SR. 2012. So you want to know if your message has an IRES? Wiley Interdiscip Rev RNA 3:697–705. doi: 10.1002/wrna.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plank TD, Kieft JS. 2012. The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip Rev RNA 3:195–212. doi: 10.1002/wrna.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolupaeva VG, Hellen CU, Shatsky IN. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 50.Kaminski A, Belsham GJ, Jackson RJ. 1994. Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J 13:1673–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaminski A, Howell MT, Jackson RJ. 1990. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J 9:3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belsham GJ. 1992. Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J 11:1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borman AM, Le Mercier P, Girard M, Kean KM. 1997. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res 25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]