ABSTRACT
Simian foamy virus (SFV) is a ubiquitous retrovirus in nonhuman primates (NHPs) that can be transmitted to humans, mostly through severe bites. In the past few years, our laboratory has identified more than 50 hunters from central Africa infected with zoonotic SFVs. Analysis of the complete sequences of five SFVs obtained from these individuals revealed that env was the most variable gene. Furthermore, recombinant SFV strains, some of which involve sequences in the env gene, were recently identified. Here, we investigated the variability of the env genes of zoonotic SFV strains and searched for possible recombinants. We sequenced the complete env gene or its surface glycoprotein region (SU) from DNA amplified from the blood of (i) a series of 40 individuals from Cameroon or Gabon infected with a gorilla or chimpanzee foamy virus (FV) strain and (ii) 1 gorilla and 3 infected chimpanzees living in the same areas as these hunters. Phylogenetic analyses revealed the existence of two env variants among both the gorilla and chimpanzee FV strains that were present in zoonotic and NHP strains. These variants differ greatly (>30% variability) in a 753-bp-long region located in the receptor-binding domain of SU, whereas the rest of the gene is very conserved. Although the organizations of the Env protein sequences are similar, the potential glycosylation patterns differ between variants. Analysis of recombination suggests that the variants emerged through recombination between different strains, although all parental strains could not be identified.
IMPORTANCE SFV infection in humans is a great example of a zoonotic retroviral infection that has not spread among human populations, in contrast to human immunodeficiency viruses (HIVs) and human T-lymphotropic viruses (HTLVs). Recombination was a major mechanism leading to the emergence of HIV. Here, we show that two SFV molecular envelope gene variants circulate among ape populations in Central Africa and that both can be transmitted to humans. These variants differ greatly in the SU region that corresponds to the part of the Env protein in contact with the environment. These variants may have emerged through recombination between SFV strains infecting different NHP species.
INTRODUCTION
The emergence of zoonotic viruses is a multistep process involving the transmission of viruses from domestic or wild animals to humans and their subsequent spread among human populations. Such viral infections are frequent (1). In particular, several viruses originating from nonhuman primates (NHPs) have had a major impact on human health. Among them, the retroviruses simian immunodeficiency virus (SIV) and simian T-lymphotropic virus (STLV) have crossed the species barrier from NHPs to humans, leading to the emergence of human immunodeficiency virus (HIV) and human T-lymphotropic virus (HTLV), respectively, in human populations (2).
Another retrovirus that can be transmitted from NHPs to humans is simian foamy virus (SFV). SFVs are complex retroviruses from the Spumaretrovirinae subfamily that are ubiquitous in both Old World and New World NHPs, with a seroprevalence of up to 75 to 100% in adult NHPs. Phylogenetic studies suggest that SFVs have evolved by cospeciation with Old World primates over more than 85 million years (3, 4). Consequently, host-specific groups have emerged. Recombination can also be involved in the genetic diversity of SFVs. Indeed, NHPs can become coinfected with strains from either the same group (5–7) or different groups, as shown by some chimpanzees, which were found to be coinfected with SFV from chimpanzees (SFVcpz) and SFVs from colobus monkeys or Cercopithecus monkeys (5, 8). Coinfection may create recombinant strains, as suggested for the env, gag, and pol genes (5, 7, 9). Of note, both cospeciation and host switching may have contributed to the evolutionary history of SFVs infecting New World prosimians (10).
Humans are not considered to be natural SFV hosts; however, more than 100 cases of human SFV infection have been reported, mostly among individuals exposed to NHPs in either a professional context (e.g., veterinarians or zookeepers) (11–15) or natural settings (e.g., hunters in Africa and monkey temple workers or visitors, pet owners, or people living around free-ranging macaques in Asia) (16–22). The clinical significance of zoonotic SFV infection remains unknown. This may be due to the very small number of persons yet studied, with limited follow-up (23), and also a possible recruitment bias (among healthy individuals). SFV is transmitted mainly through biting (22). Indeed, in African green monkeys and macaques, the oral mucosa is a major site for viral replication (24–26), and SFV RNA accumulates to high concentrations in saliva (25, 27). In humans, SFV infection is persistent. SFV DNA is detectable in peripheral blood and saliva cells. However, cell-associated viral RNA has not been detected (21, 28), and secondary human-to-human transmission (22, 29) has not been reported. Hence, human infection with zoonotic SFVs represents a unique natural model to study the role of viral and immunological factors in the restriction of viral transmission.
In the past few years, we have undertaken a large epidemiological and molecular study involving hunters of monkeys and apes living in central Africa to obtain insight into the natural history of SFV emergence in humans. We identified a large series (>50) of hunters that had been infected with SFVs, mostly following severe bites from NHPs (17, 22). We previously showed that the virus mostly localizes to CD8+, CD4+ T, and B lymphocytes in the blood of infected humans (30). Our study also revealed natural polymorphisms in gag and bet between SFV strains originating from different chimpanzee subspecies and polymorphisms in U3 and tas at the interindividual level (31). Based on the complete sequence of five replication-competent strains, we found no evidence of viral adaptation among SFV strains isolated from humans (31).
Our previous study revealed some genetic diversity in the env genes of zoonotic strains of SFV from apes. The env gene is involved in different viral cycle steps, such as receptor binding (32, 33), fusion (34–36), and budding (37–41). The broad tropism of SFV suggests a ubiquitous cellular receptor (42, 43). While all SFVs seem to use the same cellular receptor (44–46), only heparin sulfate has been identified as an attachment factor so far (47, 48).
Here, we aimed to investigate the genetic variability of env genes among SFVs infecting humans or NHPs living in Cameroon and Gabon. We found that viral strains from both gorilla and chimpanzee foamy viruses segregate into two env variants. These variants cocirculate among humans and NHPs and may have arisen by recombination.
MATERIALS AND METHODS
Populations. (i) Human population.
In three previous studies, we identified 48 individuals from Cameroon who were infected with either a SFV from Gorilla gorilla gorilla (Ggo-FV) or a SFV from Pan troglodytes troglodytes (Ptr-FV), as defined by the analysis of a 465-bp-long pol-integrase gene fragment (17, 22, 49), In a few cases, SFV was detected by analyzing a smaller long terminal repeat (LTR) fragment. These individuals live in villages or settlements located in the rain forest of south and east Cameroon. They belong to different Bantu tribes or the Baka and Bakola Pygmy tribes. Fourteen Ggo- or Ptr-FV-infected individuals from Gabon, all of whom were Bantus, were also included in this study (20).
This study was approved by the research division of the Ministry of Public Health and the National Committee of Ethics in Cameroon, the Ministry of Health and the Ethics Committee of the CIRMF in Gabon, and the Comité de Protection des Personnes and the Commission Nationale de l'Informatique et des Libertés in France. All individuals provided written informed consent.
(ii) NHP population.
One gorilla from Cameroon and three Pan troglodytes troglodytes chimpanzees (one from Cameroon and two from Gabon) were included in this study. NHPs from Cameroon were born and caught in the wild in the southern rain forest area (50, 51). NHPs from Gabon were all born in the wild (20). Samples were collected in accordance with rules of the animal care committees.
Sampling and DNA preparation.
Blood samples were collected from all individuals and NHPs into EDTA tubes. The buffy coat (BC) was obtained after centrifugation, and genomic DNA was extracted by using the QIAamp DNA Blood minikit (Qiagen, Courtaboeuf, France). DNA samples were quantified by using a Nanodrop instrument (ThermoScientific).
PCR amplification of env from genomic DNA.
Regarding Ggo-FVs, three different overlapping env fragments (720, 1,102, and 1,490 bp long) were amplified from BC DNA by using specific primers (Table 1). Nested PCR was performed as follows: 500 ng of DNA was mixed in the enzyme buffer with the external primers (0.25 μM each), MgCl2 (3.5 mM), deoxynucleoside triphosphates (dNTPs) (200 μM each), and 0.5 μl of HotStarTaq polymerase (Qiagen) in a final volume of 50 μl. External PCR consisted of a 15-min-long denaturation step at 95°C, followed by 40 amplification cycles (45 s at 95°C, 45 s at 57°C, and 1 min per kb at 72°C) and a 7-min-long extension step at 72°C. The product (5 μl) was then used as the template for a second internal PCR under the same conditions but using the internal primers (Table 1).
TABLE 1.
PCR primers used to amplify the envelope genes of gorilla and chimpanzee foamy virus strains
| FV and fragment | Length (bp) | Use | Directiona | Primer | Sequence (5′→3′) | Site of hybridizationb (bp) |
|---|---|---|---|---|---|---|
| Gorilla FV | ||||||
| Fragment 1 | 720 | External PCR | F | AENVF1b | GAACTGTGGTAATTGTGGACC | 6566–6586 |
| R | AENVR1b | CCACGAGACCAAGAACAATA | 7292–7311 | |||
| Internal PCR | F | AENVF1 | GGTAATTGTGGACCATCTTGG | 6573–6593 | ||
| R | AENVR1 | GGATCCACGAGACCAAGAAC | 7288–7307 | |||
| Fragment 2 | 1,102 | External PCR | F | BF3 | CATCCACCCCTCCTGCCT | 6431–6448 |
| R | RR3 | CCTGTAAATGAAATGCCTAAT | 8228–8248 | |||
| Internal PCR | F | DF3 | CTATAATACACACGGAGAGG | 7113–7132 | ||
| R | RR3 | CCTGTAAATGAAATGCCTAAT | 8228–8248 | |||
| Fragment 3 | 1,490 | External PCR | F | AENVF2 | TACGACAACAAGATTATGAAG | 8109–8129 |
| R | AENVR3 | CTGAGTGAGCTTGTTGGTCC | 9640–9659 | |||
| Internal PCR | F | AENVF2b | ATATCAAGAATGTAAGTTGG | 8140–8159 | ||
| R | AENVR3b | TCTGCAAACTCTGAGTGAGC | 9630–9649 | |||
| Chimpanzee FV | ||||||
| Fragment 1 | 1,645 | External PCR | F | CPZENVF1b | ACTGTTGTTATTTTGGACCA | 7066–7085 |
| R | CPZENVR1 | CCTTTGTAGGCCTAGTAGAT | 8763–8782 | |||
| Internal PCR | F | CPZENVF1 | GGCAACAACAGAACTGTAAG | 7090–7109 | ||
| R | CPZENVR1b | CCTGTAAATGAAATGCCTAA | 8735–8754 | |||
| Fragment 2 | 1,802 | External PCR | F | CPZENVF2 | TTCTCTTTGTGGGAAGGAG | 8330–8349 |
| R | CPZENVR2 | CTTAGTGAGCTTGTTGGTCC | 10141–10160 | |||
| Internal PCR | F | CPZENVF2b | TCTTTGTGGGAAGGAGATTG | 8333–8352 | ||
| R | CPZENVR2b | CAGACTCTTAGTGAGCTTGT | 10135–10154 |
F, forward; R, reverse.
Based on the SFVggo or PFV genome.
Regarding Ptr-FVs, two different overlapping env fragments (1,645 and 1,802 bp) were amplified from BC DNA by using specific primers (Table 1). The PCR parameters were the same as those used for the Ggo-FV strains, except for the annealing temperature for the PCR for the second fragment (60°C instead of 57°C).
The env gene of the prototypic Pan troglodytes verus SFVpvr.SFV7 strain (52) was amplified from SFVpvr.SFV7-infected BHK-21 cells (a gift from A. Rethwilm) using primers CPZENVF1 and CPZENVR2b (Table 1) with an annealing temperature of 57°C and an extension time of 3 min 30 s.
PCR products were directly sequenced by MWG Operon (Ebersberg, Germany). Both sense and antisense sequences were obtained for each fragment and were found to be identical. To obtain complete env sequences, the different env fragments were concatenated (the overlapping regions were identical in all samples).
Phylogenetic analyses.
Multiple-sequence alignments of previously known (Table 2) and newly generated sequences were performed by using the DAMBE program (v4.2.13 [http://dambe.bio.uottawa.ca]). Modeltest v3.6 was used to select the most appropriate nucleotide substitution model, based on the Akaike information criterion (AIC). Generalized time reversible (GTR) was found to be the best-fitting model. Phylogenetic trees were constructed by using the neighbor-joining method, and bootstrap values were calculated with 1,000 replicates. Phylogenetic tree topologies were confirmed by using the maximum likelihood method with the PAUP program (v4.0 [http://www.paup.csit.fsu.edu]). Percent identity was calculated by CLC software (CLC DNA Workbench 6; CLCbio) on the alignment obtained with DAMBE.
TABLE 2.
GenBank accession numbers and origins of other SFV isolates used in this study
| Isolate(s) | Strain | Origin | Type of sequence | GenBank accession no. |
|---|---|---|---|---|
| PFV | Chimpanzee (P. troglodytes schweinfurthii) | Human | Complete genome | Y07725 |
| SFVcpz | Chimpanzee (P. troglodytes verus) | NHP | Complete genome | U04327 |
| AG15 | Chimpanzee (P. troglodytes troglodytes) | Human | Complete genome | JQ867462 |
| Bad327 | Chimpanzee (P. troglodytes troglodytes) | Human | Complete genome | JQ867463 |
| SFVggo | Gorilla (G. gorilla gorilla) | NHP | Complete genome | HM245790 |
| Bak74 | Gorilla (G. gorilla gorilla) | Human | Complete genome | JQ867464 |
| Bad468 | Gorilla (G. gorilla gorilla) | Human | Complete genome | JQ867465 |
| SFVora | Orangutan | NHP | Complete genome | AJ544579 |
| SFVAGMhu | African green monkey (Chlorocebus aethiops) | Human | Complete genome | AX575326 |
| SFV-agm3 | African green monkey (Chlorocebus aethiops) | NHP | Complete genome | NC_010820 |
| SFVka | African green monkey (Chlorocebus aethiops) | Human | Partial env | AJ244092 |
| SFV3 | African green monkey (Chlorocebus aethiops) | NHP | Partial env | AJ244094 |
| agm1–agm37 (19 isolates) | African green monkey (Chlorocebus aethiops) | NHP | Partial env | AJ244067 to AJ244091 |
| AG16 | Cercopithecus | NHP | Complete genome | JQ867466 |
| SFV-mcy1 | Macaque (Macaca cyclopis) | NHP | Complete genome | NC_010819 |
| SFV-mcy2 | Macaque (Macaca cyclopis) | NHP | Complete genome | KF026286 |
| SFVR289HybAGM | Macaque (Macaca mulatta) | NHP | Complete genome | JN801175 |
| SFVmmu-K3T | Macaque (Macaca mulatta) | NHP | env | KF026287 |
| SFVmmu-A4W | Macaque (Macaca mulatta) | NHP | env | KF026288 |
| SFVsp | Spider monkey | NHP | Complete genome | EU010385 |
| SFVmar | Marmoset | NHP | Complete genome | GU356395 |
| SFVsq | Squirrel monkey | Human | Complete genome | GU356394 |
By definition, a clade is a monophyletic group supported by a strong bootstrap value. Here, we used “group” as a host-specific clade and “subgroup” as a clade defined based on the central region of env.
Recombinant analyses.
To search for potential recombination events, similarity plot and bootscanning analyses were performed with Simplot software (v3.5.1; J. R. Simplot Co.). These analyses were performed with default parameters, except for the bootscan repetitions (set to 1,000) and the evolution model (Kimura two parameter). The similarity plot depicts a similarity score between a group of interest (query) and other groups (defined based on the phylogenetic tree) over a 200-bp-long region, and the overall env gene was analyzed in 20-bp-long steps. The bootscan analysis reflects the phylogenetic relationship (bootstrap value) between a group of interest (query) and the other groups (window, 200 bp; step, 20 bp).
The Recombination Detection Program (RDP4) (53) was also used to investigate putative recombination. Unlike Simplot-derived studies, the RDP method looks for recombination by analyzing every possible sequence triplet (instead of considering groups). A window is moved along the genome, and a percentage of identity between each of the three possible pairs is calculated at each position. A P value is calculated to determine the likelihood that the potential recombination event is due to chance.
Protein analysis.
Percent identity was calculated by using CLC software (CLC DNA Workbench 6) on the alignment obtained with DAMBE after translation of the sequences into amino acids. Putative glycosylation sites were identified by the NetGlyc 1.0 program and consisted of NXS/T sites, where X denotes any amino acid except proline (http://www.cbs.dtu.dk/services/NetNGlyc).
Nucleotide sequence accession numbers.
All complete and partial env sequences generated in this work were submitted to GenBank. Complete sequences can be found under accession numbers KT211246 to KT211269, the latter corresponding to the SFVpvr.SFV7 sequence, and partial sequences can be found under accession numbers KT211270 to KT211290.
RESULTS
We previously identified 48 SFV-infected individuals in Cameroon (17, 22, 49) and 14 SFV-infected individuals in Gabon (20). The strains belonged to the Ggo-FV or Ptr-FV group according to their pol and/or LTR sequences. In addition, we previously obtained complete SFV sequences (Bad468, Bak74, AG15, and Bad327) from four Cameroonian individuals (31). Buffy coat or DNA samples were available for 51 of the remaining 58 individuals. From these samples, 40 new env sequences were generated by PCR and direct sequencing (19 env sequences were complete, and 21 were partial). Low viral load may explain the absence of amplification for the 11 remaining samples. Thus, overall, our study population yielded 44 env sequences from zoonotic SFVs (33 Ggo-FV and 4 Ptr-FV isolates from Cameroon and 5 Ggo-FV and 2 Ptr-FV isolates from Gabon) (Table 3 and Fig. 1).
TABLE 3.
Epidemiological data for the 44 individuals infected with simian foamy virus included in this studya
| Country of isolation | Ethnicity | Individual | Sex | Age at contact/age at sampling (yr) | FV strain | Type of env FV sequence | FV variant | FV isolate (GenBank accession no.) |
|---|---|---|---|---|---|---|---|---|
| Cameroon | Pygmy | Pyl106 | M | 15/60 | Chimpanzee | Complete | SFV-CpzI | SFVptr-hu.Pyl106 (KT211263) |
| BAK46 | M | 26/50 | Gorilla | Complete | SFV-GorI | SFVggo-hu.Bak46 (KT211255) | ||
| BAK74 | M | 26/47 | Gorilla | Complete | SFV-GorII | Bak74 (JQ867464) | ||
| BAK82 | M | 46/50 | Gorilla | Complete | SFV-GorI | SFVggo-hu.Bak82 (KT211247) | ||
| BAK228 | M | 29/70 | Gorilla | Complete | SFV-GorII | SFVggo-hu.Bak228 (KT211246) | ||
| BAK242 | M | 30/49 | Gorilla | Complete | SFV-GorI | SFVggo-hu.Bak242 (KT211254) | ||
| Sabak36 | M | 40/68 | Gorilla | Complete | SFV-GorII | SFVggo-hu.Sabak36 (KT211249) | ||
| BAK33 | M | 25/45 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bak33 (KT211277) | ||
| Bak55 | M | 30/65 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bak55 (KT211273) | ||
| BAK56 | M | 40/65 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bak56 (KT211274) | ||
| BAK132 | M | 30/64 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bak132 (KT211276) | ||
| BAK177 | M | 26/36 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bak177 (KT211281) | ||
| BAK224 | M | 19/38 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bak224 (KT211280) | ||
| BAK232 | M | 40/60 | Gorilla | Partial | SFV-GorII | SFVggo-hu.Bak232 (KT211278) | ||
| BAK270 | M | 25/60 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bak270 (KT211272) | ||
| Bobak153 | M | 53/59 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bobak153 (KT211279) | ||
| Bobak237 | M | ?/68 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bobak237 (KT211275) | ||
| Lobak2 | M | 37/57 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Lobak2 (KT211282) | ||
| Lobak89 | M | 20/50 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Lobak89 (KT211287) | ||
| Mebak65 | M | 20/40 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Mebak65 (KT211283) | ||
| 801001 | M | 35/60 | Gorilla | Partial | SFV-GorI | SFVggo-hu.801001 (KT211284) | ||
| CH29 | M | 49/50 | Gorilla | Partial | SFV-GorI | SFVggo-hu.CH29 (KT211289) | ||
| Bantu | AG15 | M | 28/71 | Chimpanzee | Complete | SFV-CpzII | AG15 (JQ867462) | |
| Bad316 | M | 35/51 | Chimpanzee | Complete | SFV-CpzII | SFVptr-hu.Bad316 (KT211262) | ||
| Bad327 | M | 30/33 | Chimpanzee | Complete | SFV-CpzII | Bad327 (JQ867463) | ||
| BAD348 | M | 19/27 | Gorilla | Complete | SFV-GorI | SFVggo-hu.Bad348 (KT211252) | ||
| BAD456 | M | 24/30 | Gorilla | Complete | SFV-GorI | SFVggo-hu.Bad456 (KT211251) | ||
| BAD463 | M | 37/43 | Gorilla | Complete | SFV-GorI | SFVggo-hu.Bad463 (KT211253) | ||
| BAD468 | M | 25/35 | Gorilla | Complete | SFV-GorI | Bad468 (JQ867465) | ||
| BAD551 | M | 37/38 | Gorilla | Complete | SFV-GorII | SFVggo-hu.Bad551 (KT211248) | ||
| CH101 | M | 65/76 | Gorilla | Complete | SFV-GorII | SFVggo-hu.CH101 (KT211256) | ||
| BAD332 | M | 25/37 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bad332 (KT211288) | ||
| BAD349 | M | 32/40 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bad349 (KT211270) | ||
| BAD350 | M | 40/68 | Gorilla | Partial | SFV-GorII | SFVggo-hu.Bad350 (KT211290) | ||
| BAD447 | M | 40/56 | Gorilla | Partial | SFV-GorI | SFVggo-hu.Bad447 (KT211271) | ||
| AKO394 | M | 53/53 | Gorilla | Partial | SFV-GorII | SFVggo-hu.AKO394 (KT211286) | ||
| CH61 | M | 52/65 | Gorilla | Partial | SFV-GorI | SFVggo-hu.CH61 (KT211285) | ||
| Gabon | Bantu | H3Gab56 | M | 47/48 | Chimpanzee | Complete | SFV-CpzII | SFVptr-hu.H3Gab56 (KT211267) |
| H4Gab59 | M | 50/51 | Chimpanzee | Complete | SFV-CpzII | SFVptr-hu.H4Gab59 (KT211268) | ||
| H2Gab54 | M | 52/53 | Gorilla | Complete | SFV-GorI | SFVggo-hu.H2Gab54 (KT211257) | ||
| H5Gab27 | M | 53/80 | Gorilla | Complete | SFV-GorI | SFVggo-hu.H5Gab27 (KT211258) | ||
| H6Gab51 | M | 28/56 | Gorilla | Complete | SFV-GorI | SFVggo-hu.H6Gab51 (KT211259) | ||
| H7Gab42 | M | 20/65 | Gorilla | Complete | SFV-GorII | SFVggo-hu.H7Gab42 (KT211261) | ||
| H12Gab69 | M | 22/38 | Gorilla | Complete | SFV-GorI | SFVggo-hu.H12Gab69 (KT211260) |
The env genes of FVs from individuals in boldface type (BAK74, AG15, BAD327, and BAD468) were sequenced by our laboratory in a previous study (31). M, male; F, female.
FIG 1.
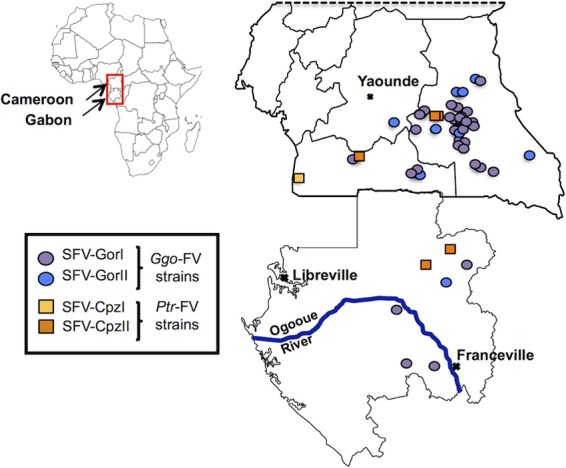
Geographic locations of the 44 individuals infected with Ggo- or Ptr-FV strains. Individuals infected with Ggo-FV strains are indicated by circles. Individuals infected with a Ptr-FV strain are indicated by squares. These colors are used in all figures.
In addition, we also obtained complete env sequences from four wild-born NHPs living in the same region as the infected individuals (one gorilla and three chimpanzees), and we sequenced the env gene of a prototypic Pan troglodytes verus strain, SFVpvr.SFV7 (Table 4).
TABLE 4.
Epidemiological data for four nonhuman primates infected with simian foamy virus included in this study
| Country | Species | Animal | Sex | Age at sampling (yr) | Situation | Type of env FV sequence | FV variant | Strain (GenBank accession no.) |
|---|---|---|---|---|---|---|---|---|
| Cameroon | Gorilla | GgoCam7SFV | F | 7 | Wild born, zoo | Complete | SFV-GorI | SFVggo.Cam7 (KT211250) |
| Chimpanzee | CpzCam15SFV | M | 6 | Wild born, zoo | Complete | SFV-CpzI | SFVptr.Cam15 (KT211264) | |
| Gabon | Chimpanzee | CpzJudWd | F | adult | Wild born, zoo | Complete | SFV-CpzII | SFVptr.JudWd (KT211266) |
| Cpz133Wd | NAa | adult | Wild born, pet | Complete | SFV-CpzII | SFVptr.133Wd (KT211265) |
NA, not available.
All newly obtained sequences contained an open reading frame of 2,958 to 2,970 bp and were unique.
Complete env sequences define host-specific SFV groups.
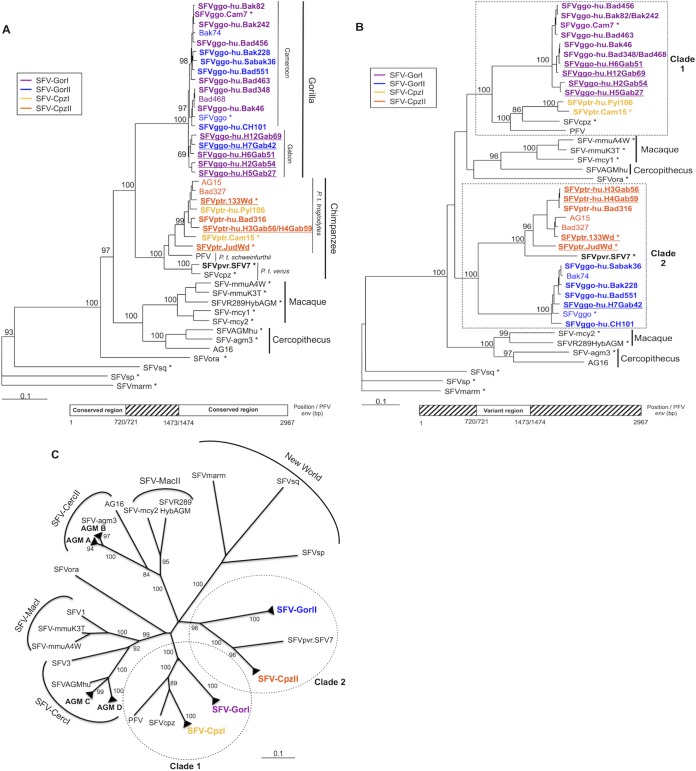
We analyzed phylogenetically the complete env sequences by using the neighbor-joining (Fig. 2) and maximum likelihood (data not shown) methods. The newly generated complete env sequences were included together with the SFV env sequences available in GenBank (Table 2). The topologies of both phylogenetic trees were comparable. Host-specific groups (gorilla, chimpanzee, cercopithecus, and macaque) were identifiable and supported by strong bootstrap values (>90). Moreover, among the strains from the chimpanzee-specific FV group, we identified three different clades corresponding to different subspecies: Pan troglodytes troglodytes, P. troglodytes schweinfurthii, and P. troglodytes verus.
FIG 2.
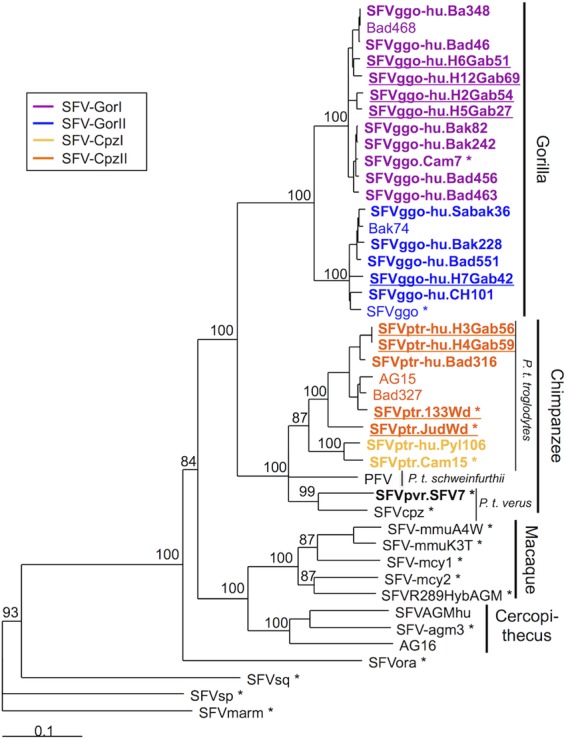
Phylogenetic analysis of complete simian foamy virus envelope gene sequences. Phylogenetic analysis of complete env sequences (2,967 bp long for the PFV strain) from 43 SFV isolates, including the 24 complete sequences generated in this study (in boldface type) and 19 previously reported sequences, was performed. SFVmarm, a marmoset strain from South America, was the outgroup. Gabon isolates are underlined, and asterisks indicate FVs isolated from nonhuman primates. The phylogenic tree was derived by the neighbor-joining method using the GTR model (gamma = 0.6749). Horizontal branch lengths are drawn to scale, with the bar indicating 0.1 nucleotide replacements per site. Numbers on each node indicate the bootstrap value (calculated for 1,000 replicates) supporting the group.
Interestingly, the gorilla-specific FV group was composed of two monophyletic subgroups supported by strong bootstrap values. These subgroups did not correspond to geographic segregation. Indeed, each subgroup contained sequences from both Cameroon and Gabon (underlined strains on the tree in Fig. 2) as well as both zoonotic and NHP strains (SFVggo or SFVggo.Cam7). We named the subgroup containing the previously described Bad468 env FV sequence SFV-GorI and the one containing the Bak74 env FV and prototypic SFVggo sequences SFV-GorII.
Similarly, two phylogenetic subgroups were identifiable within the Ptr-FV group. We named the one containing the previously described Bad327 and AG15 env sequences SFV-CpzII and the other one SFV-CpzI.
Definition of a central variant region within the surface env sequence.
We performed sequence alignments to better understand the origins of the two subgroups present in the Ggo-FV and Ptr-FV groups.
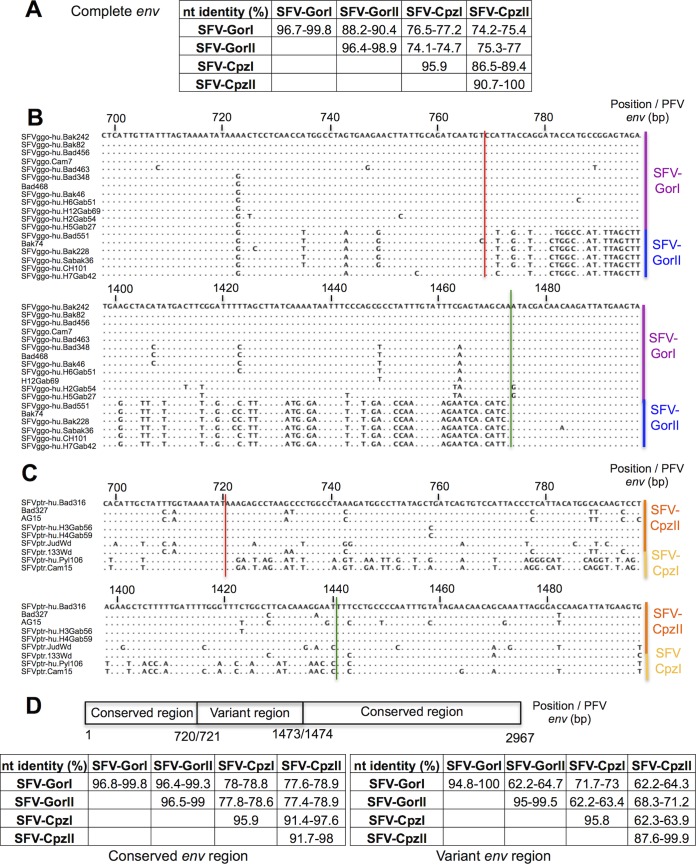
Concerning the Ggo-FV strains, SFV-GorI env sequences were 2,970 bp long, whereas SFV-GorII sequences were 2,964 bp long. Complete sequences within a subgroup were 96.4 to 99.8% identical, whereas those belonging to different subgroups were only 88.2 to 90.4% identical (Fig. 3A). SFV-GorI and SFV-GorII sequences differed the most in a central region beginning at position 769 (Fig. 3B, red line) and ending at position 1473 (the positions were defined on the env prototype foamy virus (PFV) strain) (Fig. 3B, green line). In the Ptr-FV strains, SFV-CpzI sequences were 2,967 bp long, whereas SFV-CpzII sequences were 2,958 bp long. These two Ptr-FV subgroups differed the most between positions 721 (Fig. 3C, red line) and 1440 (green line). Interestingly, this corresponds closely to the same variable region defined for Ggo-FV strains.
FIG 3.
Definition of conserved and variant regions in the envelope genes of Ggo- and Ptr-FVs. (A and D) Percent nucleotide identity between the different subgroups of Ggo- and Ptr-FVs in the complete (A), conserved (D), or variant (D) region of the envelope gene. Percent identity was determined after alignment with the DAMBE program and by using CLC software. nt, nucleotides. (B and C) Gorilla and chimpanzee foamy virus sequences were aligned by using DAMBE software. The regions from bp 698 to 798 and from bp 1398 to 1498 (positions defined according to the PFV env sequence) are shown for Ggo-FV sequences (B) and Ptr-FV sequences (C), respectively. Dots indicate identity. The red vertical lines indicate the position between two codons where the env sequences diverge into separate variants (defined at the right of the alignment), and the green vertical lines show the position where the variants become very similar again.
Thus, we defined two regions in the env sequence (Fig 3D): a conserved region, which comprises nucleotides 1 to 720 and 1474 to 2967 (positions defined in the PFV strain), and a variant region, from nucleotides 721 to 1473. We confirmed a high sequence similarity (96.4 to 99.3% identity) in the conserved region between the SFV-GorI and SFV-GorII subgroups and a very important divergence (62.2 to 64.7% identity) in the variant region (Fig. 3D). The level of similarity of the variant region was very high between strains from the same subgroup (94.8 to 100% identity). This was also true for the two Ptr-FV subgroups (SFV-CpzI and SFV-CpzII). Strikingly, in the variant region, SFV-GorI and SFV-CpzI showed 71.7 to 73% identity, which is higher than that between SFV-GorI and SFV-GorII (62.2 to 64.7%) (Fig. 3D).
Two variants are present within SFV groups.
Given that a conserved region and a variant region were defined, we performed phylogenetic analyses on each segment separately.
In the conserved region (Fig. 4A), sequences segregated according to host species and subspecies. The four major monophyletic and highly supported groups corresponded to SFV strains of gorilla, chimpanzee, macaque, or Cercopithecus origin. Moreover, Ggo-FV strains were now subdivided into groups reflecting geographic origin (two Cameroonian clades and one Gabonese clade) (Fig. 4A, underlining). This distribution resembles that of the pol-based phylogenetic tree established in previous studies for the same individuals (20, 22).
FIG 4.
Phylogenetic analysis of the envelope gene conserved and variant regions. (A and B) Phylogenetic trees corresponding to the conserved region (2,214 bp for PFV) (A) or the variant region (753 bp for PFV) (B) were derived by the neighbor-joining method (GTR) (gamma = 0.6749). Horizontal branch lengths are drawn to scale, with the bar indicating 0.1 nucleotide replacements per site. Bootstrap values were calculated for 1,000 replicates. The SFVmarm strain was used as an outgroup; Gabon isolates are underlined, and asterisks indicate FVs isolated from nonhuman primates. (C) Unrooted phylogenetic tree of the variant region. Closely related strains that formed a robust cluster (as shown in panel B) are represented with black triangles. AGM A, B, C, and D are clusters of African green monkey (Cercopithecus) sequences previously defined by Schweizer et al. (54).
The phylogenetic tree based on the variant region (Fig. 4B) was quite different. Ggo-FVs and Ptr-FVs segregated according to the subgroups identified previously (SFV-GorI, SFV-GorII, SFV-CpzI, and SFV-CpzII). In this phylogenetic tree, we were able to define two clades for African ape SFVs (clade 1 and clade 2), each comprising both Ggo-FV and Ptr-FV strains. Of note, P. troglodytes verus FV sequences (SFV7 and SFVcpz) were also split: the SFV7 env sequence was closer to the SFV-CpzII sequences, whereas the SFVcpz env sequence was closer to the P. troglodytes schweinfurthii PFV and SFV-CpzI strains. Finally, macaque and cercopithecus FV isolates were also divided into two distinct subgroups.
To confirm these findings, in the analysis of the variant region, we included the 21 partial env sequences obtained from other individuals infected by a Ggo-FV strain (Table 3) and 21 previously reported African green monkey (AGM) (cercopithecus group) env surface glycoprotein (SU) sequences (Table 2) (54). Phylogenetic analysis of the variant region (Fig. 4C) confirmed the robustness of the two major African ape clades (clade 1 and clade 2) and the different subgroups SFV-GorI, SFV-GorII, SFV-CpzI, and SFV-CpzII. Interestingly, cercopithecus strains separated into two major subgroups: SFV-CercI and SFV-CercII. Macaque FV strains were also split into two distinct subgroups (SFV-MacI and SFV-MacII). Of note, the unrooted phylogenetic tree was star shaped (Fig. 4C), suggesting that although SFV-GorI and SFV-CpzI are the closest groups, the sequences are still genetically distant and do not share a recent common ancestor.
Thus, the strains from four simian species (gorilla, chimpanzee, macaque, and cercopithecus) systematically segregate into two subgroups that differ in the env central region.
Genomic and functional organization of the envelope gene.
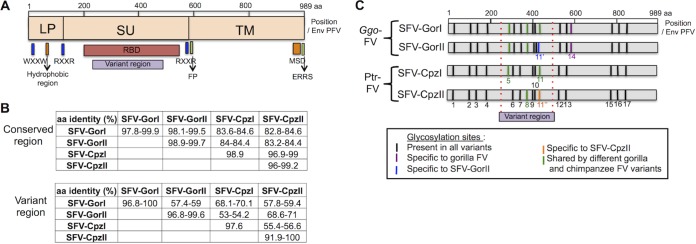
All protein sequences generated from the new env sequences showed typical Env organization, with the leader peptide (LP), SU, and transmembrane glycoprotein (TM) regions separated by RXXR cleavage signals (data not shown). Other important domains, such as the fusion peptide (FP) or the membrane-spanning domain (MSD), were also present (Fig. 5A and data not shown). Every cysteine residue (essential for proper folding) described for PFV Env (32) was also conserved.
FIG 5.
Comparison of Env proteins of Ggo-FV and Ptr-FV strains. (A) Structure and important domains of Env FV protein present in all sequences in this study. WXXW motif, Trp-X-X-Trp motif (site of interaction with Gag protein); RXXR, Arg-X-X-Arg cleavage sites; RBD, receptor-binding domain; FP, fusion peptide (i, i + 3/4, i + 7 pattern); MSD, membrane-spanning domain; ERRS, endoplasmic reticulum retrieval signal (Lys at position −3 and Lys or Arg at positions −4 and/or −5 relative to the C terminus). (B) Amino acid identity in the conserved and variant regions was investigated by using CLC software based on the alignment performed with DAMBE. (C) Location of N-glycosylation sites of the different Ggo- and Ptr-FV subgroups.
The variant region was located within the SU domain, more specifically within the receptor-binding domain (RBD) identified on PFV (32). Given that glycosylation is important for RBD function, we examined the conservation of the putative glycosylation sites. Although amino acid sequences diverge in the central region between variants (Fig. 5B), 13 of the potential glycosylation sites (previously defined for PFV) were conserved among Ggo- and Ptr-FV strains (55) (Fig. 5C). Ggo-FV subgroups differed at position N11, and chimpanzee FV subgroups differed at positions N11, N5, and N8. Thus, the different subgroups might have different glycosylation patterns.
Could variants be generated by recombination?
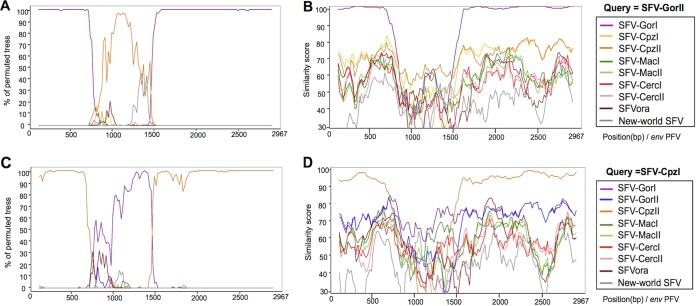
We performed bootscan analysis of the env sequences grouped by host-specific subgroups to search for potential recombinants among Ggo- and Ptr-FV strains (Fig. 6). Consistent with the phylogenetic trees, the conserved region of SFV-GorII env sequences was closely related to that of SFV-GorI (>99% of permuted trees grouped them together), whereas the variable region was closer to sequences from SFV-CpzII (Fig. 6A). Similarly, the conserved region of SFV-CpzI strains was closer to that of SFV-CpzII strains, whereas the variable region of SFV-CpzI strains was closer to that of SFV-GorI strains (Fig. 6C).
FIG 6.
Recombination analysis of the SFV-GorII and SFV-CpzI envelope gene sequences. Bootscan analyses (A and C) and Simplot analyses (B and D) using the SFV-GorII (A and B) or SFV-CpzI (C and D) env sequences as a query against the other groups of SFV isolates were performed.
Such a pattern suggests that strains from the SFV-GorII subgroup originated from a recombination event between SFV-GorI and SFV-CpzII strains. However, data from similarity plot analyses argue against this conclusion. Indeed, the similarity scores for SFV-GorII and SFV-CpzII strains are low throughout the whole gene and are lowest in the variant region (between 52 and 62% similarity) (Fig. 6B). Thus, if the strains from the SFV-GorII subgroup indeed appeared by recombination, one of the parental strains remains unknown.
To investigate this hypothesis further, we performed Recombinant Detection Program (RDP) analysis to detect recombinant sequences. This analysis strongly suggests that the strains from the SFV-GorI and SFV-GorII subgroups diverged upon recombination (P value of 4.745 × 10−12). Similarly, the SFV-CpzI and SFV-CpzII subgroups probably recombined (P value of 2.074 × 10−12). However, in both cases, the parental strain from which the central region was obtained could not be identified.
We conclude that the two variants observed among Ggo- and Ptr-FV strains probably arose by a recombination event and that one of the parental strains is still unknown.
DISCUSSION
Here, we studied the variability of the envelope genes of SFVs infecting apes (gorillas and chimpanzees) and humans living in Cameroon or Gabon. We demonstrate the cocirculation of two SFV env molecular variants in both the gorilla and chimpanzee FV groups. In a given group, the complete nucleotide sequence of env differs by >10% between the variants. These differences are located mostly in a central region of the envelope gene that we have called the “variant” region, in contrast with the “conserved” region that corresponds to the rest of the env gene. The genetic diversity of env may have arisen from recombination.
These findings raise the following several important issues and questions.
Genetic variability in the conserved region reflects host species origin and geographical clustering.
In the conserved region, the nucleotide variability among Ggo-FV strains was low, ranging from 0.2% to 3.6%. This genetic stability is consistent with the low in vivo variation rate, estimated to be ∼1.7 × 10−8 substitutions per site per year (3). The genetic variability of the conserved regions of the different Ggo-FV strains reflects geographic location. Indeed, we defined two clusters of Cameroonian sequences and a cluster from strains isolated in Gabon by phylogenetic analysis. Interestingly, these clusters were observed previously when the same strains were divided according to their pol sequences (20, 22). Similarly, geographic segregation was also observed for SFV strains in macaque populations in Bangladesh (7) and mandrill populations in Gabon (56). Indeed, geographic segregation and consequent isolation lead to the accumulation of distinct mutations and speciation.
In contrast, Cameroonian and Gabonese Ptr-FV sequences did not segregate together, consistent with the findings of studies on the pol-integrase gene (20, 22). This may suggest that, unlike gorillas, P. troglodytes troglodytes populations are not genetically or virologically isolated despite being geographically distant. However, such results are based on few sequences.
For the central variant region, we found two subgroups for each FV host-specific group with >35% nucleotide variability. This variability did not reflect geographic location. Strains from every Ggo- and Ptr-FV subgroup were present in both humans and NHPs.
What is the mechanism leading to the emergence of env diversity among the SFV variants?
We first examined whether recombination, which is a frequent evolutionary event in retroviruses, could explain the diversity of env sequences (57–60). Indeed, recombination is also frequent among SFVs. In vitro studies show that it can occur between PFV-based vectors and reveal that the probability of a template-switching event within a 1-kb-long region is 27% (61).
Our analyses suggested that a Ggo-FV variant originated from recombination between Ggo-FV strains and unknown FV strains (the closest known strain being the chimpanzee strains). Similarly, one of the Ptr-FV variants may have arisen from recombination between chimpanzee- and gorilla-like FV strains. Until the parental strains are found, the recombination origin of these sequences might be debated. However, this hypothesis is the most realistic.
Such a scenario implies that NHPs can be coinfected with SFV strains of different host species and that these strains have recombined. Coinfection has been documented for chimpanzees (5, 6) and rhesus macaques (7). However, in these cases, coinfection occurred with strains from the same host specific group. Interestingly, in these populations, recombination in FVs has been observed in the pol or gag gene (5, 7). Furthermore, coinfection with strains belonging to different NHP-related groups has been documented for wild chimpanzees. These animals were infected with both Colobus monkey- and chimpanzee-related FVs (5, 8), but no recombinant strain was detected. Recently, recombination between macaque and Cercopithecus FV strains has been reported (9, 62). However, macaques and Cercopithecus live in different continents, suggesting that the recombination event occurred in captivity. Indeed, animals from various species and/or geographical areas are frequently mixed in primate centers. The variants that we report would be the first detected recombinants between SFVs of different primate hosts found in a natural setting.
Interestingly, the putative recombination sites between the gorilla and the chimpanzee FV variants were quite similar. This site also corresponds closely to the same region described in macaque (9) and African green monkey (54) FV sequences. A similar variant region, with similar recombination points, has been described for feline FVs (63). Together, this suggests that a recombination hot spot may exist in the env region of foamy viruses.
Our data suggest the different env-based subgroups arose by recombination, but many questions remain unsolved. First, the identity of one of the parental strains is still undetermined. This second parental strain, which purportedly introduced the central variable region into the env gene, may be either an existing but as-yet-undescribed FV or a strain that has disappeared. Second, the date of the recombination events has not been addressed. If the recombination event was ancient, genetic drift would have segregated SFV-GorI and SFV-GorII even in the conserved region. Given that the conserved regions of SFV-GorI and SFV-GorII strains are very similar and that they do not form separate monophyletic groups in the conserved region, the recombination event probably occurred fairly recently.
Why are only two env genetic variants present?
Strikingly, we observed two env molecular variants for each NHP species studied (i.e., gorilla, chimpanzee, macaque, and Cercopithecus). If the defined sites were hot spots of recombination, we would expect many more variants to be generated. Strong functional constraints may be an effective source of purifying selection that would allow for the emergence of only two subgroups per host.
The variant central region is located in the putative receptor-binding domain of the SU domain, as identified for PFV (32). SFV uses heparin sulfate to attach to target cells (47, 48), but the receptor for SFV is still unknown (64). Interference studies showed that one cellular receptor is used by all SFVs (44–46). Binding studies of recombinant envelope proteins suggest that SFVcpz interacts with two receptors, one with low affinity and one with high affinity (65). Although most glycosylation sites are conserved, we found that the pattern of glycosylation may differ between the Ggo- and Ptr-FV subgroups. Consequently, it is possible that the two FV variants correspond to viruses that use different receptor complexes, as is the case for murine leukemia virus (66). Alternatively, the SFV receptor may be expressed in different conformations at the cell surface, allowing two modes of engagement by the two Env variants. Indeed, such conformational heterogeneity was described for the CCR5 molecule and its interaction with the envelope of HIV and chemokines (67).
Finally, env molecular variants will probably induce distinct immune responses. The Env SU is indeed the target of neutralizing antibodies (65). Two env genotypes corresponding to two serotypes have been described for feline FVs (63, 68). The two genotypes differ in a central region nearly corresponding to the one described in our study. SFV strains from macaques and chimpanzees also segregate into two serotypes as defined by neutralization assays with immune plasma or sera: serotypes 1 and 2 for macaques (69) and serotypes 6 and 7 for chimpanzees (52, 70, 71). We show here two cases in which strains belonging to different serotypes belong to distinct genetic subgroups. Indeed, SFV-mcy1 and SFV-mcy2 are from serotypes 1 and 2, respectively, while belonging to SFV-MacI and SFV-MacII, respectively (Fig. 4C); PFV/SFVcpz and SFVpvr.SFV7 belong to serotypes 6 and 7, respectively, while segregating with SFV-CpzI or SFV-CpzII, respectively (Fig. 4C). Preliminary results on neutralizing antibodies present in plasma of infected persons support the correspondence between serotype and genotype for SFV from the chimpanzee group (72). Studies are currently ongoing to characterize immune responses induced by SFV strains from the gorilla group.
ACKNOWLEDGMENTS
We thank the Institut de Recherche pour le Développement (IRD) and the Centre Pasteur du Cameroun for collaborating with us concerning the field work in Cameroon. We thank Axel Rethwilm for providing SFV7-infected BHK cells and Arifa Khan for helpful discussions about foamy virus taxonomy.
L.R. was personally supported by the Bourse de l'Ecole Normale Supérieure, Faculté Paris Diderot. E.B. was supported by the Virus Cancer Prevention Association and by the Institut National pour le Cancer. This work was supported by the Institut Pasteur in Paris, France, by Programme Transversal de Recherche 437 from the Institut Pasteur, and by the French government program Investissement d'Avenir (grant ANR-10-LABX-62-IBEID).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Wolfe ND, Dunavan CP, Diamond J. 2007. Origins of major human infectious diseases. Nature 447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peeters M, D'Arc M, Delaporte E. 2014. Origin and diversity of human retroviruses. AIDS Rev 16:23–34. [PMC free article] [PubMed] [Google Scholar]
- 3.Switzer WM, Salemi M, Shanmugam V, Gao F, Cong ME, Kuiken C, Bhullar V, Beer BE, Vallet D, Gautier-Hion A, Tooze Z, Villinger F, Holmes EC, Heneine W. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376–380. doi: 10.1038/nature03341. [DOI] [PubMed] [Google Scholar]
- 4.Rethwilm A, Bodem J. 2013. Evolution of foamy viruses: the most ancient of all retroviruses. Viruses 5:2349–2374. doi: 10.3390/v5102349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu W, Worobey M, Li Y, Keele BF, Bibollet-Ruche F, Guo Y, Goepfert PA, Santiago ML, Ndjango JB, Neel C, Clifford SL, Sanz C, Kamenya S, Wilson ML, Pusey AE, Gross-Camp N, Boesch C, Smith V, Zamma K, Huffman MA, Mitani JC, Watts DP, Peeters M, Shaw GM, Switzer WM, Sharp PM, Hahn BH. 2008. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog 4:e1000097. doi: 10.1371/journal.ppat.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasse A, Calvignac-Spencer S, Merkel K, Goffe AS, Boesch C, Mundry R, Leendertz FH. 2013. Mother-offspring transmission and age-dependent accumulation of simian foamy virus in wild chimpanzees. J Virol 87:5193–5204. doi: 10.1128/JVI.02743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feeroz MM, Soliven K, Small CT, Engel GA, Pacheco MA, Yee JL, Wang X, Hasan MK, Oh G, Levine KL, Alam SR, Craig KL, Jackson DL, Lee E, Barry PA, Lerche NW, Escalante AA, Matsen FA IV, Linial ML, Jones-Engel L. 2013. Population dynamics of rhesus macaques and associated foamy virus in Bangladesh. Emerg Microbes Infect 2:e29. doi: 10.1038/emi.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leendertz FH, Zirkel F, Couacy-Hymann E, Ellerbrok H, Morozov VA, Pauli G, Hedemann C, Formenty P, Jensen SA, Boesch C, Junglen S. 2008. Interspecies transmission of simian foamy virus in a natural predator-prey system. J Virol 82:7741–7744. doi: 10.1128/JVI.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galvin TA, Ahmed IA, Shahabuddin M, Bryan T, Khan AS. 2013. Identification of recombination in the envelope gene of simian foamy virus serotype 2 isolated from Macaca cyclopis. J Virol 87:8792–8797. doi: 10.1128/JVI.03555-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katzourakis A, Aiewsakun P, Jia H, Wolfe ND, LeBreton M, Yoder AD, Switzer WM. 2014. Discovery of prosimian and afrotherian foamy viruses and potential cross species transmissions amidst stable and ancient mammalian co-evolution. Retrovirology 11:61. doi: 10.1186/1742-4690-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schweizer M, Turek R, Hahn H, Schliephake A, Netzer KO, Eder G, Reinhardt M, Rethwilm A, Neumann-Haefelin D. 1995. Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res Hum Retroviruses 11:161–170. doi: 10.1089/aid.1995.11.161. [DOI] [PubMed] [Google Scholar]
- 12.Heneine W, Switzer WM, Sandstrom P, Brown J, Vedapuri S, Schable CA, Khan AS, Lerche NW, Schweizer M, Neumann-Haefelin D, Chapman LE, Folks TM. 1998. Identification of a human population infected with simian foamy viruses. Nat Med 4:403–407. doi: 10.1038/nm0498-403. [DOI] [PubMed] [Google Scholar]
- 13.Brooks JI, Rud EW, Pilon RG, Smith JM, Switzer WM, Sandstrom PA. 2002. Cross-species retroviral transmission from macaques to human beings. Lancet 360:387–388. doi: 10.1016/S0140-6736(02)09597-1. [DOI] [PubMed] [Google Scholar]
- 14.Sandstrom PA, Phan KO, Switzer WM, Fredeking T, Chapman L, Heneine W, Folks TM. 2000. Simian foamy virus infection among zoo keepers. Lancet 355:551–552. doi: 10.1016/S0140-6736(99)05292-7. [DOI] [PubMed] [Google Scholar]
- 15.Switzer WM, Bhullar V, Shanmugam V, Cong ME, Parekh B, Lerche NW, Yee JL, Ely JJ, Boneva R, Chapman LE, Folks TM, Heneine W. 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J Virol 78:2780–2789. doi: 10.1128/JVI.78.6.2780-2789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Prosser AT, Torimiro JN, Wright A, Mpoudi-Ngole E, McCutchan FE, Birx DL, Folks TM, Burke DS, Heneine W. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363:932–937. doi: 10.1016/S0140-6736(04)15787-5. [DOI] [PubMed] [Google Scholar]
- 17.Calattini S, Betsem EB, Froment A, Mauclere P, Tortevoye P, Schmitt C, Njouom R, Saib A, Gessain A. 2007. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg Infect Dis 13:1314–1320. doi: 10.3201/eid1309.061162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones-Engel L, Engel GA, Schillaci MA, Rompis A, Putra A, Suaryana KG, Fuentes A, Beer B, Hicks S, White R, Wilson B, Allan JS. 2005. Primate-to-human retroviral transmission in Asia. Emerg Infect Dis 11:1028–1035. doi: 10.3201/eid1107.040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones-Engel L, May CC, Engel GA, Steinkraus KA, Schillaci MA, Fuentes A, Rompis A, Chalise MK, Aggimarangsee N, Feeroz MM, Grant R, Allan JS, Putra A, Wandia IN, Watanabe R, Kuller L, Thongsawat S, Chaiwarith R, Kyes RC, Linial ML. 2008. Diverse contexts of zoonotic transmission of simian foamy viruses in Asia. Emerg Infect Dis 14:1200–1208. doi: 10.3201/eid1408.071430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mouinga-Ondeme A, Caron M, Nkoghe D, Telfer P, Marx P, Saib A, Leroy E, Gonzalez JP, Gessain A, Kazanji M. 2012. Cross-species transmission of simian foamy virus to humans in rural Gabon, Central Africa. J Virol 86:1255–1260. doi: 10.1128/JVI.06016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engel GA, Small CT, Soliven K, Feeroz MM, Wang X, Hasan MK, Oh G, Alam SR, Craig KL, Jackson DL, Matsen FA IV, Linial ML, Jones-Engel L. 2013. Zoonotic simian foamy virus in Bangladesh reflects diverse patterns of transmission and co-infection. Emerg Microbes Infect 2:e58. doi: 10.1038/emi.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betsem E, Rua R, Tortevoye P, Froment A, Gessain A. 2011. Frequent and recent human acquisition of simian foamy viruses through apes' bites in central Africa. PLoS Pathog 7:e1002306. doi: 10.1371/journal.ppat.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boneva RS, Switzer WM, Spira TJ, Bhullar VB, Shanmugam V, Cong ME, Lam L, Heneine W, Folks TM, Chapman LE. 2007. Clinical and virological characterization of persistent human infection with simian foamy viruses. AIDS Res Hum Retroviruses 23:1330–1337. doi: 10.1089/aid.2007.0104. [DOI] [PubMed] [Google Scholar]
- 24.Falcone V, Leupold J, Clotten J, Urbanyi E, Herchenroder O, Spatz W, Volk B, Bohm N, Toniolo A, Neumann-Haefelin D, Schweizer M. 1999. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 257:7–14. doi: 10.1006/viro.1999.9634. [DOI] [PubMed] [Google Scholar]
- 25.Murray SM, Picker LJ, Axthelm MK, Linial ML. 2006. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J Virol 80:663–670. doi: 10.1128/JVI.80.2.663-670.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray SM, Picker LJ, Axthelm MK, Hudkins K, Alpers CE, Linial ML. 2008. Replication in a superficial epithelial cell niche explains the lack of pathogenicity of primate foamy virus infections. J Virol 82:5981–5985. doi: 10.1128/JVI.00367-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soliven K, Wang X, Small CT, Feeroz MM, Lee EG, Craig KL, Hasan K, Engel GA, Jones-Engel L, Matsen FA IV, Linial ML. 2013. Simian foamy virus infection of rhesus macaques in Bangladesh: relationship of latent proviruses and transcriptionally active viruses. J Virol 87:13628–13639. doi: 10.1128/JVI.01989-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rua R, Betsem E, Gessain A. 2013. Viral latency in blood and saliva of simian foamy virus-infected humans. PLoS One 8:e77072. doi: 10.1371/journal.pone.0077072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gessain A, Rua R, Betsem E, Turpin J, Mahieux R. 2013. HTLV-3/4 and simian foamy retroviruses in humans: discovery, epidemiology, cross-species transmission and molecular virology. Virology 435:187–199. doi: 10.1016/j.virol.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rua R, Betsem E, Montange T, Buseyne F, Gessain A. 2014. In vivo cellular tropism of gorilla simian foamy virus in blood of infected humans. J Virol 88:13429–13435. doi: 10.1128/JVI.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rua R, Betsem E, Calattini S, Saib A, Gessain A. 2012. Genetic characterization of simian foamy viruses infecting humans. J Virol 86:13350–13359. doi: 10.1128/JVI.01715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duda A, Luftenegger D, Pietschmann T, Lindemann D. 2006. Characterization of the prototype foamy virus envelope glycoprotein receptor-binding domain. J Virol 80:8158–8167. doi: 10.1128/JVI.00460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stirnnagel K, Luftenegger D, Stange A, Swiersy A, Mullers E, Reh J, Stanke N, Grosse A, Chiantia S, Keller H, Schwille P, Hanenberg H, Zentgraf H, Lindemann D. 2010. Analysis of prototype foamy virus particle-host cell interaction with autofluorescent retroviral particles. Retrovirology 7:45. doi: 10.1186/1742-4690-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pietschmann T, Zentgraf H, Rethwilm A, Lindemann D. 2000. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J Virol 74:4474–4482. doi: 10.1128/JVI.74.10.4474-4482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picard-Maureau M, Jarmy G, Berg A, Rethwilm A, Lindemann D. 2003. Foamy virus envelope glycoprotein-mediated entry involves a pH-dependent fusion process. J Virol 77:4722–4730. doi: 10.1128/JVI.77.8.4722-4730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stirnnagel K, Schupp D, Dupont A, Kudryavtsev V, Reh J, Mullers E, Lamb DC, Lindemann D. 2012. Differential pH-dependent cellular uptake pathways among foamy viruses elucidated using dual-colored fluorescent particles. Retrovirology 9:71. doi: 10.1186/1742-4690-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindemann D, Pietschmann T, Picard-Maureau M, Berg A, Heinkelein M, Thurow J, Knaus P, Zentgraf H, Rethwilm A. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J Virol 75:5762–5771. doi: 10.1128/JVI.75.13.5762-5771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietschmann T, Heinkelein M, Heldmann M, Zentgraf H, Rethwilm A, Lindemann D. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J Virol 73:2613–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swiersy A, Wiek C, Zentgraf H, Lindemann D. 2013. Characterization and manipulation of foamy virus membrane interactions. Cell Microbiol 15:227–236. doi: 10.1111/cmi.12042. [DOI] [PubMed] [Google Scholar]
- 40.Goepfert PA, Shaw K, Wang G, Bansal A, Edwards BH, Mulligan MJ. 1999. An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J Virol 73:7210–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goepfert PA, Shaw KL, Ritter GD Jr, Mulligan MJ. 1997. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol 71:778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mergia A, Leung NJ, Blackwell J. 1996. Cell tropism of the simian foamy virus type 1 (SFV-1). J Med Primatol 25:2–7. doi: 10.1111/j.1600-0684.1996.tb00185.x. [DOI] [PubMed] [Google Scholar]
- 43.Mikovits JA, Hoffman PM, Rethwilm A, Ruscetti FW. 1996. In vitro infection of primary and retrovirus-infected human leukocytes by human foamy virus. J Virol 70:2774–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moebes A, Enssle J, Bieniasz PD, Heinkelein M, Lindemann D, Bock M, McClure MO, Rethwilm A. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J Virol 71:7305–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill CL, Bieniasz PD, McClure MO. 1999. Properties of human foamy virus relevant to its development as a vector for gene therapy. J Gen Virol 80(Part 8):2003–2009. [DOI] [PubMed] [Google Scholar]
- 46.Berg A, Pietschmann T, Rethwilm A, Lindemann D. 2003. Determinants of foamy virus envelope glycoprotein mediated resistance to superinfection. Virology 314:243–252. doi: 10.1016/S0042-6822(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 47.Nasimuzzaman M, Persons DA. 2012. Cell membrane-associated heparan sulfate is a receptor for prototype foamy virus in human, monkey, and rodent cells. Mol Ther 20:1158–1166. doi: 10.1038/mt.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plochmann K, Horn A, Gschmack E, Armbruster N, Krieg J, Wiktorowicz T, Weber C, Stirnnagel K, Lindemann D, Rethwilm A, Scheller C. 2012. Heparan sulfate is an attachment factor for foamy virus entry. J Virol 86:10028–10035. doi: 10.1128/JVI.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calattini S, Betsem E, Bassot S, Chevalier SA, Tortevoye P, Njouom R, Mahieux R, Froment A, Gessain A. 2011. Multiple retroviral infection by HTLV type 1, 2, 3 and simian foamy virus in a family of Pygmies from Cameroon. Virology 410:48–55. doi: 10.1016/j.virol.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 50.Calattini S, Nerrienet E, Mauclere P, Georges-Courbot MC, Saib A, Gessain A. 2006. Detection and molecular characterization of foamy viruses in Central African chimpanzees of the Pan troglodytes troglodytes and Pan troglodytes vellerosus subspecies. J Med Primatol 35:59–66. doi: 10.1111/j.1600-0684.2006.00149.x. [DOI] [PubMed] [Google Scholar]
- 51.Calattini S, Nerrienet E, Mauclere P, Georges-Courbot MC, Saib A, Gessain A. 2004. Natural simian foamy virus infection in wild-caught gorillas, mandrills and drills from Cameroon and Gabon. J Gen Virol 85:3313–3317. doi: 10.1099/vir.0.80241-0. [DOI] [PubMed] [Google Scholar]
- 52.Rogers NG, Basnight M, Gibbs CJ, Gajdusek DC. 1967. Latent viruses in chimpanzees with experimental kuru. Nature 216:446–449. doi: 10.1038/216446a0. [DOI] [PubMed] [Google Scholar]
- 53.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweizer M, Schleer H, Pietrek M, Liegibel J, Falcone V, Neumann-Haefelin D. 1999. Genetic stability of foamy viruses: long-term study in an African green monkey population. J Virol 73:9256–9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luftenegger D, Picard-Maureau M, Stanke N, Rethwilm A, Lindemann D. 2005. Analysis and function of prototype foamy virus envelope N glycosylation. J Virol 79:7664–7672. doi: 10.1128/JVI.79.12.7664-7672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mouinga-Ondeme A, Betsem E, Caron M, Makuwa M, Salle B, Renault N, Saib A, Telfer P, Marx P, Gessain A, Kazanji M. 2010. Two distinct variants of simian foamy virus in naturally infected mandrills (Mandrillus sphinx) and cross-species transmission to humans. Retrovirology 7:105. doi: 10.1186/1742-4690-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elder JH, Gautsch JW, Jensen FC, Lerner RA, Hartley JW, Rowe WP. 1977. Biochemical evidence that MCF murine leukemia viruses are envelope (env) gene recombinants. Proc Natl Acad Sci U S A 74:4676–4680. doi: 10.1073/pnas.74.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cloyd MW, Hartley JW, Rowe WP. 1980. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med 151:542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robertson DL, Sharp PM, McCutchan FE, Hahn BH. 1995. Recombination in HIV-1. Nature 374:124–126. [DOI] [PubMed] [Google Scholar]
- 60.Desrames A, Cassar O, Gout O, Hermine O, Taylor GP, Afonso PV, Gessain A. 2014. Northern African strains of human T-lymphotropic virus type 1 arose from a recombination event. J Virol 88:9782–9788. doi: 10.1128/JVI.01591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gartner K, Wiktorowicz T, Park J, Mergia A, Rethwilm A, Scheller C. 2009. Accuracy estimation of foamy virus genome copying. Retrovirology 6:32. doi: 10.1186/1742-4690-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blochmann R, Curths C, Coulibaly C, Cichutek K, Kurth R, Norley S, Bannert N, Fiebig U. 2014. A novel small animal model to study the replication of simian foamy virus in vivo. Virology 448:65–73. doi: 10.1016/j.virol.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 63.Winkler IG, Flugel RM, Lochelt M, Flower RL. 1998. Detection and molecular characterisation of feline foamy virus serotypes in naturally infected cats. Virology 247:144–151. doi: 10.1006/viro.1998.9232. [DOI] [PubMed] [Google Scholar]
- 64.Berka U, Hamann MV, Lindemann D. 2013. Early events in foamy virus-host interaction and intracellular trafficking. Viruses 5:1055–1074. doi: 10.3390/v5041055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herchenroder O, Moosmayer D, Bock M, Pietschmann T, Rethwilm A, Bieniasz PD, McClure MO, Weis R, Schneider J. 1999. Specific binding of recombinant foamy virus envelope protein to host cells correlates with susceptibility to infection. Virology 255:228–236. doi: 10.1006/viro.1998.9570. [DOI] [PubMed] [Google Scholar]
- 66.Battini JL, Heard JM, Danos O. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol 66:1468–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colin P, Benureau Y, Staropoli I, Wang Y, Gonzalez N, Alcami J, Hartley O, Brelot A, Arenzana-Seisdedos F, Lagane B. 2013. HIV-1 exploits CCR5 conformational heterogeneity to escape inhibition by chemokines. Proc Natl Acad Sci U S A 110:9475–9480. doi: 10.1073/pnas.1222205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zemba M, Alke A, Bodem J, Winkler IG, Flower RL, Pfrepper K, Delius H, Flugel RM, Lochelt M. 2000. Construction of infectious feline foamy virus genomes: cat antisera do not cross-neutralize feline foamy virus chimera with serotype-specific Env sequences. Virology 266:150–156. doi: 10.1006/viro.1999.0037. [DOI] [PubMed] [Google Scholar]
- 69.Johnston PB. 1961. A second immunologic type of simian foamy virus: monkey throat infections and unmasking by both types. J Infect Dis 109:1–9. doi: 10.1093/infdis/109.1.1. [DOI] [PubMed] [Google Scholar]
- 70.Hooks JJ, Gibbs CJ Jr, Cutchins EC, Rogers NG, Lampert P, Gajdusek DC. 1972. Characterization and distribution of two new foamy viruses isolated from chimpanzees. Arch Gesamte Virusforsch 38:38–55. doi: 10.1007/BF01241354. [DOI] [PubMed] [Google Scholar]
- 71.Bieniasz PD, Rethwilm A, Pitman R, Daniel MD, Chrystie I, McClure MO. 1995. A comparative study of higher primate foamy viruses, including a new virus from a gorilla. Virology 207:217–228. doi: 10.1006/viro.1995.1068. [DOI] [PubMed] [Google Scholar]
- 72.Materniak M, Kubis P, Rola-Luszczak M, Khan AS, Buseyne F, Lindemann D, Lochelt M, Kuzmak J. 2015. Tenth International Foamy Virus Conference 2014—achievements and perspectives. Viruses 7:1651–1666. doi: 10.3390/v7041651. [DOI] [PMC free article] [PubMed] [Google Scholar]