ABSTRACT
Positive-strand RNA [(+) RNA] viruses remodel cellular membranes to facilitate virus replication and assembly. In the case of turnip mosaic virus (TuMV), the viral membrane protein 6K2 plays an essential role in endomembrane alterations. Although 6K2-induced membrane dynamics have been widely studied by confocal microscopy, the ultrastructure of this remodeling has not been extensively examined. In this study, we investigated the formation of TuMV-induced membrane changes by chemical fixation and high-pressure freezing/freeze substitution (HPF/FS) for transmission electron microscopy at different times of infection. We observed the formation of convoluted membranes connected to rough endoplasmic reticulum (rER) early in the infection process, followed by the production of single-membrane vesicle-like (SMVL) structures at the midstage of infection. Both SMVL and double-membrane vesicle-like structures with electron-dense cores, as well as electron-dense bodies, were found late in the infection process. Immunogold labeling results showed that the vesicle-like structures were 6K2 tagged and suggested that only the SMVL structures were viral RNA replication sites. Electron tomography (ET) was used to regenerate a three-dimensional model of these vesicle-like structures, which showed that they were, in fact, tubules. Late in infection, we observed filamentous particle bundles associated with electron-dense bodies, which suggests that these are sites for viral particle assembly. In addition, TuMV particles were observed to accumulate in the central vacuole as membrane-associated linear arrays. Our work thus unravels the sequential appearance of distinct TuMV-induced membrane structures for viral RNA replication, viral particle assembly, and accumulation.
IMPORTANCE Positive-strand RNA viruses remodel cellular membranes for different stages of the infection process, such as protein translation and processing, viral RNA synthesis, particle assembly, and virus transmission. The ultrastructure of turnip mosaic virus (TuMV)-induced membrane remodeling was investigated over several days of infection. The first change that was observed involved endoplasmic reticulum-connected convoluted membrane accumulation. This was followed by the formation of single-membrane tubules, which were shown to be viral RNA replication sites. Later in the infection process, double-membrane tubular structures were observed and were associated with viral particle bundles. In addition, TuMV particles were observed to accumulate in the central vacuole as membrane-associated linear arrays. This work thus unravels the sequential appearance of distinct TuMV-induced membrane structures for viral RNA replication, viral particle assembly, and accumulation.
INTRODUCTION
Positive-strand RNA [(+) RNA] viruses remodel cellular membranes for different stages of the infection process, such as protein translation and processing, viral RNA (vRNA) synthesis, particle assembly, and virus transmission. Ultrastructural studies showed that distinct membrane structures could be simultaneously generated in one cell that was infected with animal (+) RNA viruses. For instance, severe acute respiratory syndrome (SARS) coronavirus-induced membrane structures are a unique reticulovesicular network of modified endoplasmic reticulum (ER) that integrates convoluted membranes (CMs) for polyprotein synthesis and processing, numerous interconnected double-membrane vesicles (DMVs) for vRNA synthesis, and vesicle packets (VPs) for virus assembly and budding (1). (+) RNA virus-induced distinct membrane structures can also be observed during the time course of infection. Coxsackievirus B3 (CVB3) induces single-membrane tubules (SMTs) at an early stage of infection for vRNA synthesis, followed by the formation of DMVs and multilamellar structures late in infection (2).
Plant (+) RNA viruses also manipulate host membranes for their replication (reviewed in reference 3). However, limited transmission electron microscopy (TEM) data on the remodeling of the cellular membrane during vRNA replication and encapsidation have been generated. Several plant (+) RNA viruses have been observed to produce spherule-like structures on different membrane-bound organelles, such as the ER (brome mosaic virus [BMV] [4]), vacuole (cucumber mosaic virus [CMV] [5]), peroxisome (tomato bushy stunt virus [TBSV] [6, 7]), mitochondrion (carnation Italian ringspot virus [CIRV] [8, 9]), and chloroplast (turnip yellow mosaic virus [TYMV; Tymovirus, Tymoviridae] [10, 11]). These spherule-like structures are similar to those induced by some animal (+) RNA viruses for vRNA synthesis, as they are all formed by membrane invagination of the outer organelle membrane and have a pore-like opening allowing the exchange of materials between the lumen of the spherule and the cytoplasm. These spherule-like structures are static and reside inside the different membrane-bound organelles. Conversely, other types of plant (+) RNA virus-induced membrane structures are ER-derived vesicles, which are highly motile and morphologically dynamic, such as in cells infected with cowpea mosaic virus (CPMV) (12), grapevine fanleaf virus (GFLV) (13), potato virus X (PVX) (14, 15), and turnip mosaic virus (TuMV) (16). However, not much is known about the ultrastructure of these ER-derived vesicles.
TuMV is a (+) RNA virus that belongs to the genus Potyvirus in the family Potyviridae. Potyviruses are the largest genera of plant viruses and are responsible for more than half of the viral crop damage in the world (reviewed in reference 17). Potyviral particles are flexuous rods of approximately 680 to 900 nm in length and 11 to 15 nm in diameter. The viral genome is a single RNA molecule of approximately 10 kb. The 5′ end is covalently linked to a protein, VPg (for viral protein genome-linked), and the 3′ end has a poly(A) tail. The vRNA codes for a polyprotein that is processed by three viral proteinases into at least 12 proteins.
TuMV infection leads to the formation of numerous vesicles that originate from the ER (16, 18, 19). The viral membrane protein 6K2 is responsible for vesicle formation (18). 6K2-induced vesicles contain vRNA and several replication-related viral and host proteins (18–21) and are thus considered to be the site of vRNA replication. These vesicles also participate in the cell-to-cell movement of the vRNA. They are motile (19), traffic through the secretory pathway, and use myosin motors (22). Ultimately, the vesicles associate with plasmodesmata (PD) and are then released into neighboring cells (23) for a new round of infection. They are also involved in long-distance virus movement, as they are present in phloem sieve elements and xylem vessels (24).
The studies cited above relied mainly on observations made by light microscopy. TEM images of virus-induced vesicles have also been reported for TuMV-infected (16) and potato virus Y (PVY)-infected cells (25). These images showed the presence of numerous single-membrane vesicles (SMVs) or what appeared to be vesicles, and on some occasions, the vesicles were found to be in direct continuity with the rough endoplasmic reticulum (rER). However, no study has looked at the biogenesis of these apparent vesicles over the time course of infection and their relationship with viral particle assembly.
In this study, we investigated the formation and maturation of the TuMV-induced membrane remodeling that took place over several days of infection. The first change that was observed involved rER-connected convoluted membrane accumulation. This was followed by the formation of SMV-like (SMVL) structures, which were shown to be vRNA replication sites. Later in the infection process, double-membrane vesicle-like (DMVL) structures having an electron-dense core, along with electron-dense bodies associated with viral particle bundles, were observed. Electron tomography (ET) showed that the vesicle-like structures were in fact tubules. In addition, TuMV particles were observed to accumulate in the central vacuole as membrane-associated linear arrays.
MATERIALS AND METHODS
Plasmid DNA and plant inoculation.
The infectious clone pCambiaTuMV/6K2:GFP has been described previously (26). The plasmid pCambiaTuMV/6K2:GFP:HA was constructed as follows. The green fluorescent protein (GFP) fragment was amplified by PCR from pGreen/PABP:GFP (20) by using the forward primer 5′-CGGGATCCATGGTGAGCAAGGGCGA-3′ and the reverse primer 5′-CGGAATTCTTACTTGTACAGCTCGTCCA-3′ (the restriction sites are underlined) and digested with BamHI and EcoRI. The digested GFP fragment was then used to replace the mCherry fragment of pCambia/6K2:mCherry (23), and the resulting construct was named pCambia/6K2:GFP. The 6K2:GFP:hemagglutinin (HA)-coding region was amplified by PCR from pCambia/6K2:GFP by using the forward primer 5′-GCTCTAGAATGAACACCAGCGACATGAGC-3′ and the reverse primer 5′-CGGAATTCTTAAGCGTAGTCTGGAACGTCGTATGGGTACTTGTACAGCTCGTCCATGCC-3′ (the restriction sites are underlined, the HA tag-coding sequence is highlighted in bold italics) and digested with XbaI and EcoRI. The digested 6K2:GFP:HA fragment was then used to replace the 6K2:GFP fragment of pCambia/6K2:GFP, and the resulting construct was named pCambia/6K2:GFP:HA. The 6K2:GFP:HA-coding region was again amplified by PCR from pCambia/6K2:GFP:HA by using the forward primer 5′-TCCCCGCGGGAAACACCAGCGACATGAGC-3′ and the reverse primer 5′-TCCCCGCGGCTGCCTGGTGATAGACACAAGCAGCGTAGTCTGGAACGTC-3′ (the restriction sites are underlined, the proteinase cleavage site-coding sequence is highlighted in bold italics). The PCR product was digested with SacII and introduced into the SacII site of p35Tunos/SacII (27) to obtain p35Tunos/6K2:GFP:HA. This plasmid was then cut with SmaI and KpnI and ligated in pCambiaTunos (23), which was cut with the same enzymes. Kanamycin-resistant Escherichia coli colonies were screened for plasmids containing the fragment encoding 6K2:GFP:HA, and the resulting construct was pCambiaTuMV/6K2:GFP:HA. All plasmid constructs were verified by sequencing.
TuMV infectious clones and the mock-infected empty vector pCambia 0390 were electroporated into Agrobacterium tumefaciens strain AGL1 and selected on Luria broth (LB) plates containing ampicillin and kanamycin. The pellet of an overnight culture was resuspended in water supplemented with 10 mM MgCl2 and 150 μM acetosyringone and kept at room temperature (RT) for 2 to 4 h. The solution was then diluted to an optical density at 600 nm of 0.3. Agroinfiltration was performed with 4-week-old Nicotiana benthamiana plants, which were grown under a 16-h light and 8-h dark photoperiod, at temperatures of 22°C during the day and 20°C at night.
Histological preparation and confocal microscopy.
The midrib area of N. benthamiana leaves systemically infected with TuMV/6K2:GFP was cut and fixed in 4% (wt/vol) paraformaldehyde plus 0.5% (wt/vol) glutaraldehyde at different days postinfection (dpi) for more than 24 h at 4°C. The air between the intercellular spaces was removed by infiltration with the fixative before the leaves were cut. The fixed samples were treated with 100 mM glycine in phosphate-buffered saline (PBS) for 1 h to reduce the background fluorescence, followed by sucrose gradient infiltration and cryosectioning as described previously (28). The sections were observed using a 20× objective (numerical aperture = 0.8) on an LSM-780 confocal microscope (Carl Zeiss Microscopy). Zen 2011 software (Carl Zeiss Microscopy) was used for image acquisition. Excitation and emission wavelengths were 405 and 410 to 440 nm, respectively, for Fluorescent Brightener 28 and 488 and 490 to 560 nm, respectively, for GFP.
Chemical fixation.
Small pieces (1.5 mm by 2 mm) of mock- and TuMV-infected leaf midrib were cut and fixed in 2.5% (wt/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, for 24 h at 4°C. After rinsing 3 times for 10 min each time in washing buffer at RT, the samples were postfixed in 1% (wt/vol) osmium tetroxide with 1.5% (wt/vol) potassium ferrocyanide in sodium cacodylate buffer for 2 h at 4°C. The samples were then rinsed in washing buffer at RT (3 times for 10 min each time) and stained with 1% (wt/vol) tannic acid for 1 h at 4°C. After rinsing 3 times in water at RT, the samples were dehydrated in a graded acetone series (30, 50, 70, 80, 90, and 100%) for 20 min at each step at RT. A rinse with 100% acetone was repeated two more times. The samples were then gradually infiltrated with increasing concentrations (50, 66, 75, and 100%) of Epon 812 resin mixed with acetone for a minimum of 8 h for each step. A vacuum of 25 lb/in2 was applied when the samples were in pure Epon 812 resin. The samples were finally embedded in pure, fresh Epon 812 resin and polymerized at 60°C for 48 h. After polymerization, 90- to 100-nm ultrathin sections were obtained and stained with 4% (wt/vol) uranyl acetate for 8 min and Reynolds lead citrate for 5 min. The sections were examined in a Tecnai T12 transmission electron microscope (FEI) operating at 120 kV. Images were recorded using an AMT XR80C charge-coupled-device (CCD) camera system (Advanced Microscopy Techniques Corp.).
Immunogold labeling.
Large pieces (1.5 mm by 5 mm) of mock- and TuMV-infected leaf midrib were cut and fixed in 4% (wt/vol) formaldehyde and 0.25% (wt/vol) glutaraldehyde in 0.1 M Sorensen's phosphate buffer, pH 7.4, for 4 h at 4°C. The samples were then rinsed 3 times for 10 min each time in washing buffer at RT. For anti-double-stranded RNA (anti-dsRNA) antibody-treated samples, the samples were postfixed in 0.1% (wt/vol) reduced osmium tetroxide for 15 min at 4°C (29). The samples were then rinsed in water at RT (3 times for 10 min each time) and dehydrated in a graded alcohol series (30, 50, 70, 80, 90, 95, and 100%) for 20 min at each step at 4°C on a rotator. Rinsing in 100% alcohol was repeated one more time. The samples were then gradually infiltrated with increasing concentrations of LR White resin (50, 75, and 100%) mixed with alcohol for a minimum of 8 h for each step at 4°C on a rotator. The samples were finally embedded in pure LR White resin and polymerized at 50°C for 48 h. Sections of 90 to 100 nm were incubated in 20 mM glycine in Dulbecco's phosphate-buffered saline (DPBS; 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 6.5 mM Na2HPO4, 1 mM CaCl2, 0.5 mM MgCl2, pH 7.4) for 10 min to inactivate residual aldehyde groups and then in a blocking solution consisting of 2% (wt/vol) bovine serum albumin (BSA), 2% (wt/vol) casein, and 0.5% (wt/vol) ovalbumin in DPBS (DPBS-BCO) for 5 min. Sections were then incubated with primary antibody diluted in DPBS-BCO for 1 h at RT. After 6 washings (5 min each time) in DPBS, the sections were incubated with secondary antibodies diluted in DPBS-BCO. After washing with DPBS and distilled water, grids were stained with 4% (wt/vol) uranyl acetate for 5 min and Reynolds lead citrate for 3 min. Background labeling was determined using mock-infected leaf cross sections. The number of gold particles per square micrometer and the relative labeling distribution over mock-infected and TuMV-infected sections were determined as described previously (30).
For the primary antibodies, the mouse monoclonal anti-dsRNA antibody J2 (stock solution concentration, 1 mg ml−1; English and Scientific Consulting Bt.) was diluted in DPBS-BCO to 1:40, the rat monoclonal anti-HA antibody 3F10 (stock solution concentration, 0.1 mg ml−1; Roche) was diluted in DPBS-BCO to 1:10, and rabbit polyclonal anti-RNA-dependent RNA polymerase (anti-RdRp) antibodies (21) were diluted in DPBS-BCO to 1:20. For the secondary antibodies, goat anti-mouse and anti-rat antibodies conjugated to 10-nm gold particles (Sigma-Aldrich), as well as goat anti-rabbit antibodies conjugated to 12-nm gold particles (Jackson ImmunoResearch), were used at a dilution of 1:20.
HPF/FS.
For high-pressure freezing (HPF), discs of mock- and TuMV-infected leaf tissues (rib tissue was avoided) with a diameter of 1.2 mm were punched out with a punching device for flat specimen carriers (Leica Microsystems) in a drop of 1-hexadecene on a soft piece of rubber. Subsequently, the samples were transferred into the cavity (diameter, 1.2 mm) of gold-plated flat specimen carriers that were 200 μm in depth and that had been prefilled with 1-hexadecene as a cryoprotectant. The samples were immediately frozen in a high-pressure freezer (Leica EM PACT2; Leica Microsystems). Platelets containing frozen samples were then transferred and stored under liquid nitrogen conditions in transfer boxes.
Freeze substitution (FS) was carried out using an automatic FS system (Leica EM AFS2; Leica Microsystems) at −80°C (72 h), −65°C (24 h), −40°C (12 h), −20°C (12 h), 0°C (6 h), and RT (2 h) in anhydrous acetone containing 2% osmium tetroxide and 0.2% uranyl acetate. After the samples were rinsed three times in anhydrous acetone for 15 min each time, they were infiltrated with increasing concentrations of Epon 812 resin (5% [4 h], 10% [8 h], 25% [8 h], 50% [18 h], 75% [24 h], 100% [24 h]) mixed with acetone at RT. The embedded samples were then polymerized in pure, fresh Epon 812 resin for 48 h at 60°C. Ultrathin sections were cut, poststained as described above, and observed in the TEM.
Electron tomography.
Chemical-fixed sections 90 nm thick and 200 nm thick were collected on Formvar-coated copper grids. A series of single-axis-tilt images was collected with a Tecnai G2 F20 cryo-S/TEM (FEI) operated at an accelerating voltage of 200 kV, and these images were recorded with a Gatan Ultrascan 4000 4k × 4k digital CCD camera system (model 895). Images were captured over a tilt range of ±4° (1° increments) on the 90-nm-thick section, and the resulting images had a pixel size of 0.5 nm. Images were captured over a tilt range of ±60° (2° increments in low tilts [up to a tilt angle of ±30°] and 1° increments at high tilts [from tilt angles of ±30° to ±60°]) on the 200-nm-thick sections, and the resulting images had pixel sizes of 1.0 nm (see Fig. 6) and 0.78 nm (see Fig. 7). The images from the tilt series were aligned and reconstructed into a series of tomographic slices using the IMOD software package (31). The three-dimensional (3-D) surface models were created with the Amira (version 6.0) visualization package (FEI Visualization Sciences Group) by manually selecting areas of interest.
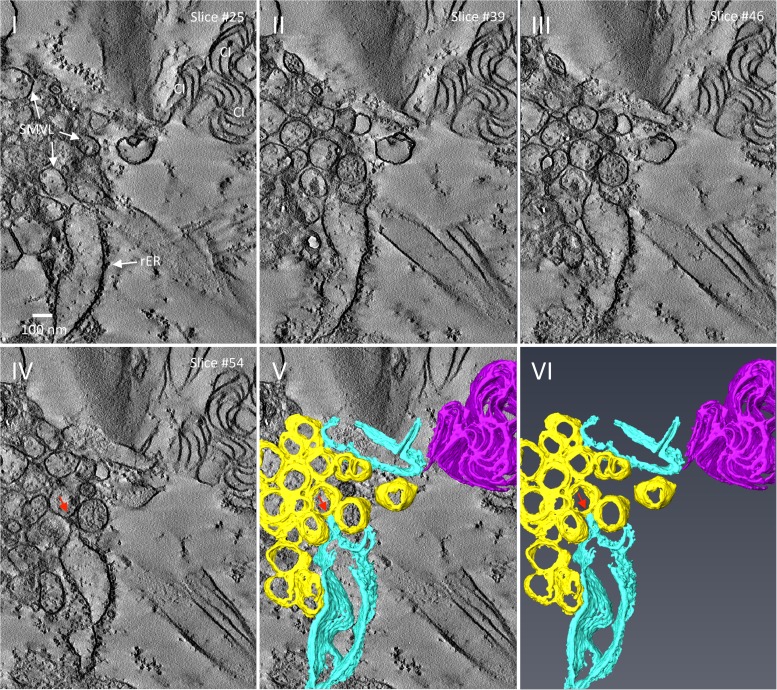
FIG 6.
3-D reconstruction of TuMV-induced membrane rearrangement at midstage of infection. (I to IV) Representative tomogram slices generated on a 200-nm-thick section from TuMV-infected vascular parenchymal cell at 6 dpi. The SMVL structures in close proximity to dilated rER and a cytoplasmic inclusion body are shown. (V and VI) A 3-D surface rendering of the closely packed SMTs, dilated rER, and cytoplasmic inclusion body. Red arrows, connection between a SMT and the rER membrane; yellow, SMTs, sky blue, rER; magenta, cytoplasmic inclusion body. CI, cytoplasmic inclusion body; rER, rough endoplasmic reticulum; SMVL, single-membrane vesicle-like structure.
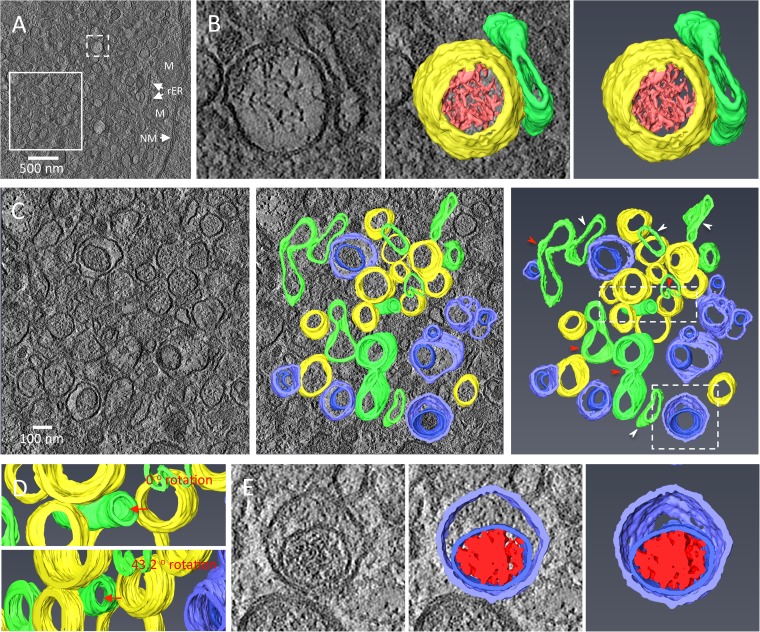
FIG 7.
3-D architecture of TuMV-induced complex membrane structures at late stage of infection. (A) Overview of a single slice of a tomogram generated on a 200-nm-thick section in the perinuclear region of a TuMV-infected vascular parenchymal cell at 7 dpi. (B) A higher-magnification image (left) of the area in the dashed white square of panel A is shown, and the 3-D model (middle and right) shows an SMT with fibrillar material inside and with an adjacent intermediate tubular structure. (C) A higher-magnification image (left) of the area in the solid white square in panel A is shown, and the 3-D model (middle and right) shows SMTs, intermediate tubular structures, and DMTs. White arrowheads, intermediate SMTs that paired into flattened cisternae; red arrowheads, intermediate SMTs with a slightly negative curvature that may result in the wrapping of the cytoplasm to form DMTs. (D) A higher-magnification image of the area in the top dashed white rectangle in the rightmost panel of panel C. Red arrow in the upper panel, a slightly bent, intermediate tubular structure (green) located among several SMTs (yellow) with similar orientations; red arrow in the lower panel, a view rotated 43.2° around the y axis, highlighting its tubular nature. (E) A higher-magnification image of the area in the bottom dashed white square in the rightmost panel of panel C. (Left) A DMVL structure and its electron-dense materials; (middle and right) the 3-D model of the DMT with a core of electron-dense materials. Yellow, SMTs; light red, electron-dense materials; green, intermediate tubular structures; light blue, outer membranes of DMTs; dark blue, inner membranes of DMTs; dark red, the electron-dense materials inside DMTs. rER, rough endoplasmic reticulum; M, mitochondrion; NM, nuclear membrane.
Vacuole isolation.
The vacuole isolation was done as described previously (32) with some modifications. Four-week-old N. benthamiana plants were agroinfiltrated with the infectious clone TuMV/6K2:GFP or the mock-infected empty vector, pCambia 0390. Leaves systemically infected with TuMV or mock-infected young leaves were collected at 9 dpi and sliced into 1-mm strips using a razor blade. The processed leaves were placed in protoplast enzyme solution (0.4 M mannitol, 20 mM MES [morpholineethanesulfonic acid], pH 5.7, 20 mM KCl, 1.5% [wt/vol] Cellulase R-10, 0.2% [wt/vol] Macerozyme R-10 macerating enzyme, 0.1% [wt/vol] BSA, 10 mM CaCl2). A vacuum was applied for 20 min to remove the air within the leaf tissues, and then the vacuum was slowly released for about 10 min to allow the protoplast enzyme solution to enter into the leaf tissues. The leaf strips were kept in the dark at RT for another 3.5 h. The released protoplasts were filtered with a 41-μm-pore-size filter and centrifuged at 100 × g for 3 min at 4°C. Protoplasts were washed two times in washing buffer (0.4 M mannitol, 20 mM MES, pH 5.7) and then resuspended in 10 ml prewarmed (37°C) lysis buffer (0.2 M mannitol, 10% [wt/vol] Ficoll, 10 mM EDTA, pH 8.0, 5 mM sodium phosphate, pH 8.0). After 5 min, 5 ml of the solution was overlaid with 3 ml 4% (wt/vol) Ficoll solution and 1 ml ice-cold vacuole buffer (0.2 M mannitol, 2 mM EDTA, pH 8.0, 5 mM sodium phosphate, pH 7.5). The gradient was centrifuged at 1,500 × g for 20 min at 10°C, and the vacuoles were located at the interface between 0 and 4% Ficoll.
RESULTS
TEM protocol for improved membrane contrast.
To obtain a well-defined description of TuMV-induced cellular reorganization, we optimized several steps in sample preparation to enhance the contrast of the membrane structure in TEM. Different fixatives (2.5% glutaraldehyde or 4% paraformaldehyde plus 2% glutaraldehyde), postfixation solutions (1% osmium tetroxide or reduced osmium–1% osmium tetroxide plus 1.5% potassium ferrocyanide), dehydration solutions (ethanol or acetone), and embedding media (Epon or Spurr) were tested. Tannic acid was also added before dehydration, since tannic acid-treated samples showed increased contrast and the greater delineation of cell membranes (33). Epon tended to give a higher image contrast than Spurr, but its penetration into the sample was inhibited because of its higher viscosity (34). To solve this problem, the time period for each Epon embedding step was increased and vacuum was applied during pure Epon embedding to improve solute penetration into the dense cell wall (see Materials and Methods).
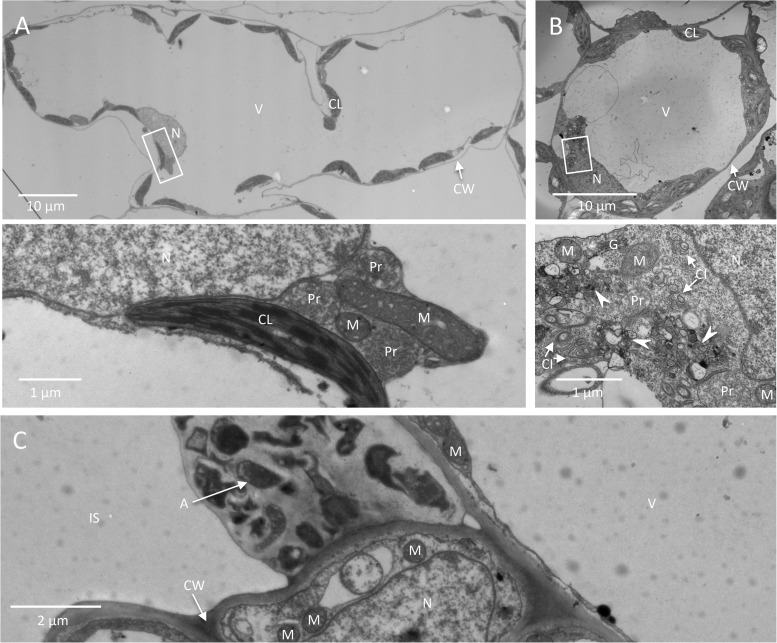
Cells from a leaf infiltrated with an A. tumefaciens suspension that contained pCambiaTuMV/6K2:GFP or the resulting upper leaf systemically infected with TuMV were analyzed by TEM. The cytoplasm of mesophyll cells from an agroinfiltrated leaf was pushed to the periphery of the cell owing to the presence of the large central vacuole (Fig. 1A, top). Although it was easy to find organelles, such as the nucleus, chloroplasts, mitochondria, and peroxisomes, TuMV cytoplasmic inclusions and virus-induced membrane structures were more difficult to observe, likely due to the compacted cytosol (Fig. 1A, bottom). On the other hand, TuMV cytoplasmic inclusions (arrows) and virus-induced membrane structures (arrowheads) (Fig. 1B, bottom) were more apparent in the relatively more abundant cytoplasm of younger mesophyll cells from the systemically infected leaves (Fig. 1B, top). Finally, choosing systemically infected leaves over agroinfiltrated leaves for TEM characterization avoided unknown bacterium-induced negative effects, since agrobacteria were frequently found in the intercellular space of the infiltrated leaf (Fig. 1C, arrow).
FIG 1.
Comparison of agroinfiltrated leaves and systemically TuMV-infected leaves. Cross sections of TuMV-agroinfiltrated (A, C) and systemically TuMV-infected (B) N. benthamiana leaves were collected and observed by TEM. (A and B) Overview of mesophyll cells in TuMV-agroinfiltrated (A) and systemically TuMV-infected (B) leaves. The areas delineated by rectangles in the top panels are shown at higher magnification in the bottom panels, where organelles and TuMV-induced membranous aggregates and cytoplasmic inclusion bodies are highlighted. Arrowheads, TuMV-induced membranous aggregates. (C) Agrobacteria in the intercellular space of the TuMV-agroinfiltrated leaf. N, nucleus; V, vacuole; CL, chloroplast; CW, cell wall; Pr, peroxisome; M, mitochondrion; G, Golgi apparatus; CI, cytoplasmic inclusion body; IS, intercellular space; A, Agrobacterium.
Time course analysis of TuMV-induced cellular reorganization.
The midrib area of systemically infected leaves was collected at different days postinfection with the infectious clone TuMV/6K2:GFP, which produces GFP-fluorescing 6K2-tagged vesicles during infection (Fig. 2A, white rectangle). No 6K2:GFP signal was observed by confocal microscopy in the cross sections of the midrib area at 4 dpi (n = 8), suggesting that infection had not yet reached the upper noninfiltrated leaves (Fig. 2B, left). Infection of vascular tissues, however, was noted at 5 dpi (Fig. 2B, middle), which then spread to all the other tissues (e.g., angular collenchyma cells, epidermal cells, and mesophyll cells) at 6 dpi (Fig. 2B, right). This time frame was thus chosen to observe by TEM the progression of TuMV cellular reorganization.
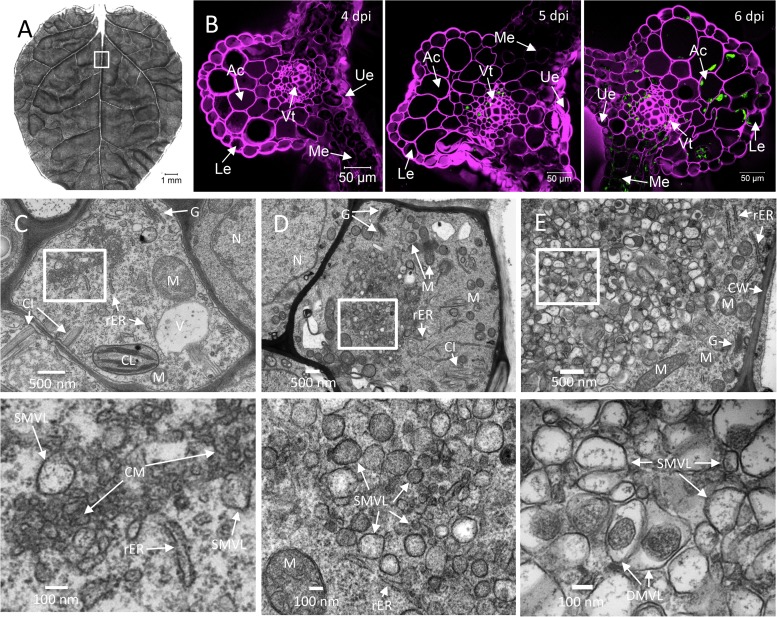
FIG 2.
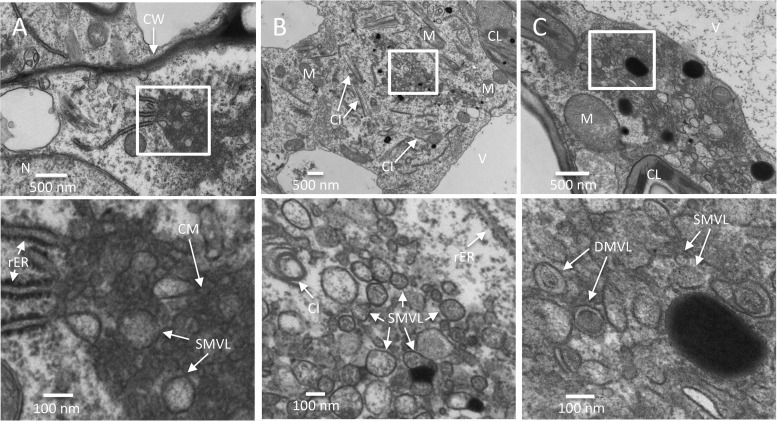
Time course analysis of TuMV-induced membranous aggregates in N. benthamiana leaf midrib. (A) An upper young leaf of an N. benthamiana plant was imaged by tile scanning with a Zeiss LSM-780 confocal microscope using a 10× objective. The tile scan was carried out by assembling images of 11 by 13 tiles. (B) Cross sections of a leaf midrib area systemically infected with TuMV/6K2:GFP (marked with a rectangle in panel A) were imaged by confocal microscopy using a 20× objective on the day postinfection indicated in the upper right. The Fluorescent Brightener 28-stained cell wall is shown in false magenta color. 6K2:GFP is shown in green. All images are single optical slices. (C to E) Time course analysis of TuMV-induced membranous aggregates in vascular parenchymal cells. N. benthamiana leaf midribs systemically infected with TuMV/6K2:GFP were chemically fixed, processed, and observed by TEM. The lower panels show higher-magnification images of the areas in the rectangles in the upper panels. (C) TuMV-induced CM structures, which were associated with SMVL structures, were located close to the rER in a vascular parenchymal cell at 5 dpi. (D) TuMV-induced heterogeneous SMVL structures in a vascular parenchymal cell at 6 dpi. (E) TuMV-induced aggregates contain both SMVL structures and DMVL structures with an electron-dense core in a vascular parenchymal cell at 7 dpi. Ue, upper epidermis; Le, lower epidermis; Ac, angular collenchyma cells; Vt, vascular tissue; Me, mesophyll cells; N, nucleus; G, Golgi apparatus; M, mitochondrion; V, vacuole; CL, chloroplast; CW, cell wall; rER, rough endoplasmic reticulum; CI, cytoplasmic inclusion body; CM, convoluted membranes; SMVL, single-membrane vesicle-like structure; DMVL, double-membrane vesicle-like structure.
Since TuMV reached the vascular tissues first and then moved to the rest of the systemically infected leaf tissues, we initially focused on vascular parenchymal cells. Cells were scored positive for TuMV infection by the presence of cytoplasmic inclusions of different morphologies (e.g., pinwheels, bundles, and short curved laminated aggregates or scrolls) (35). Infected cells were characterized by the accumulation of CM structures that were adjacent to the rER at 5 dpi (Fig. 2C). Few SMVL structures were present at the periphery of CM amalgams (Fig. 2C, bottom). We then observed the presence of numerous heterogeneously sized SMVL structures in most of the vascular parenchymal cells at 6 dpi (Fig. 2D). The shape of the SMVL structures was round or oval, and the size was estimated to be 113 nm ± 38 nm (n = 56) in diameter or in length. Finally, we observed both SMVL structures and DMVL structures with an electron-dense core in most of the cells at 7 dpi (Fig. 2E). The shape of DMVL structures was also round or oval, with the outer membrane diameter or length being 244 nm ± 60 nm (n = 52) and the inner membrane diameter or length being 162 nm ± 48 nm (n = 52).
As expected, a 1-day delay in membrane modification took place in mesophyll cells compared to the time of modification in vascular parenchymal cells. At 6 dpi, the CM structures were either located near or connected with the rER, and some SMVL structures were associated with the CM structures (Fig. 3A). At 7 dpi, aggregations of heterogeneously sized SMVL structures of 111 nm ± 42 nm (n = 72) in diameter or in length were observed (Fig. 3B). At 8 dpi, both SMVL structures and DMVL structures with an electron-dense core were observed (Fig. 3C). DMVL structures had an outer membrane diameter or length of 172 nm ± 56 nm (n = 40) and an electron-dense core diameter or length of 114 nm ± 45 nm (n = 40).
FIG 3.
Time course analysis of TuMV-induced membranous aggregates in mesophyll cells. (A to C) N. benthamiana leaf midribs systemically infected with TuMV/6K2:GFP were chemically fixed, processed, and observed by TEM. Higher-magnification images of the areas in the white rectangles are shown in the panels below. (A) TuMV-induced CM structures amid SMVL structures were connected with several rERs in a mesophyll cell at 6 dpi. (B) TuMV-induced heterogeneous SMVL structures close to the rER in a mesophyll cell at 7 dpi. (C) TuMV-induced aggregates contain both SMVL structures and DMVL structures with an electron-dense core in a mesophyll cell at 8 dpi. CW, cell wall; M, mitochondrion; CL, chloroplast; V, vacuole; CI, cytoplasmic inclusion body; rER, rough endoplasmic reticulum; CM, convoluted membranes; SMVL, single-membrane vesicle-like structure; DMVL, double-membrane vesicle-like structure.
SMVL structures are RNA replication sites of TuMV.
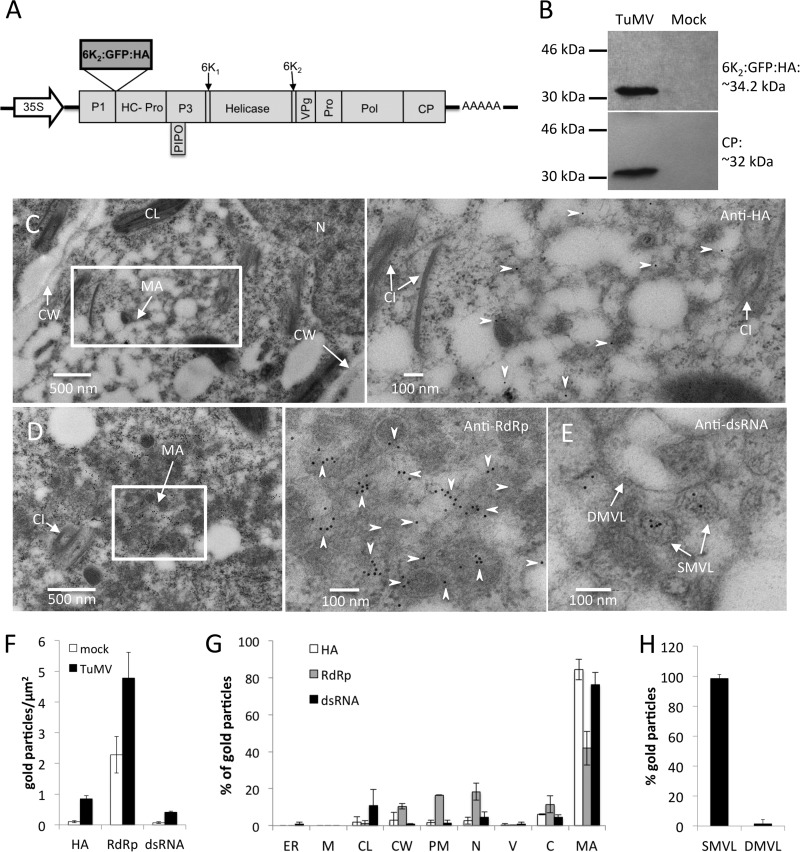
To confirm if TuMV-induced membranous aggregates were 6K2-containing structures (Fig. 2 and 3), an HA tag was fused to the C terminus of 6K2:GFP (6K2:GFP:HA). The 6K2:GFP:HA-coding sequence was thus introduced into the TuMV infectious clone pCambiaTuMV to produce pCambiaTuMV/6K2:GFP:HA (Fig. 4A). Introduction of the 6K2:GFP:HA fragment into the polyprotein produced the expected protein with the correct molecular mass, as shown by Western blot analysis using an anti-HA monoclonal antibody (Fig. 4B). TuMV infection was not compromised, as analyzed by Western blot analysis using a rabbit serum raised against the coat protein (CP) of TuMV (Fig. 4B), nor was the morphology of 6K2 vesicles compromised when they were observed under a confocal microscope (data not shown). Immunogold labeling using an anti-HA monoclonal antibody confirmed that the TuMV-induced membranous aggregates were truly tagged with 6K2 (Fig. 4C, arrowheads). Immunogold labeling using anti-RNA-dependent RNA polymerase (anti-RdRp) polyclonal antibodies also showed the vesicle aggregates contained RdRp (Fig. 4D, arrowheads).
FIG 4.
Subcellular localization of TuMV RNA replication sites. (A) Schematic representation of the infectious clone TuMV/6K2:GFP:HA that coexpresses 6K2 as a GFP:HA protein fusion. Black lines, the plasmid backbone; arrow, the cauliflower mosaic virus 35S promoter; AAAAA, the position of the polyadenylated tail; rectangles, TuMV proteins. 6K2:GFP:HA is inserted between P1 and HCpro. PIPO, pretty interesting Potyviridae ORF. (B) Western blot analysis of 6K2:GFP:HA and CP expression in N. benthamiana plants after TuMV/6K2:GFP:HA infection. (C to E) Immunogold labeling was performed on the cross sections of mock- and TuMV-infected N. benthamiana leaf tissues by using anti-HA, anti-RdRp, and anti-dsRNA antibodies. Higher-magnification images of the areas in the white rectangles are shown on the right of each panel. Arrowheads, HA-specific (C) and RdRp-specific (D) gold particles, which are located in TuMV-induced membranous aggregates. (E) The dsRNA-specific gold particles are mainly localized to TuMV-induced SMVL structures. (F to H) The number of gold particles per square micrometer in mock-infected versus TuMV-infected cells (F), the relative labeling distribution in infected cells (G), and the relative labeling distribution in TuMV-induced membranous aggregates (H) are shown. The results of two different labeling experiments were considered, and 200 gold particles were counted for each experiment. N, nucleus; CL, chloroplast; CW, cell wall; MA, membranous aggregate; CI, cytoplasmic inclusion body; SMVL, single-membrane vesicle-like structure; DMVL, double-membrane vesicle-like structure; ER, endoplasmic reticulum; M, mitochondrion; PM, plasma membrane; C, cytosol; V, vacuole.
To better differentiate between the distinctive TuMV-induced membrane structures (i.e., the CM, SMVL, and DMVL structures), a 15-min postfixation incubation period with 0.1% reduced osmium tetroxide was added before the dehydration step. This step increased the membrane contrast but also decreased the antigenicity of the tested proteins. For instance, anti-HA and anti-RdRp labeling was weak with osmium-treated samples, since only a few gold particles were found on the sections (data not shown). However, the anti-dsRNA monoclonal antibody J2 worked well on the reduced osmium-treated samples (Fig. 4E). In mesophyll cells at 8 dpi, when SMVL structures were distinguishable from DMVL structures on osmium-treated samples, anti-dsRNA-specific gold particles mainly decorated the SMVL structures (Fig. 4E).
The number of gold particles per square micrometer and the relative labeling distribution over mock-infected and TuMV-infected sections were determined. For each of the three different antibodies, at least 10 sections were observed. Practically no labeling with the anti-HA and anti-dsRNA monoclonal antibodies was observed in mock-infected samples (Fig. 4F). On the other hand, anti-HA and anti-dsRNA labeling was specifically associated with TuMV-induced membranous aggregates and was essentially absent from the rest of the cell (Fig. 4G). Background labeling was detected with the polyclonal anti-RdRp antiserum in mock-infected samples (Fig. 4F), but a larger amount of labeling was observed on TuMV-induced membranous aggregates (Fig. 4G). Quantification of the anti-dsRNA gold particle distribution on SMVL and DMVL structures in mesophyll cells on TuMV-infected sections was performed at 8 dpi. The results showed that the gold particles were mainly associated with SMVL structures (98.5%), with few gold particles being found on the DMVL structures (Fig. 4H). No gold particles decorated CM structures (data not shown). These data suggest that SMVL structures are TuMV RNA replication sites.
Comparison of TuMV-induced cellular reorganization in chemically fixed and HPF/FS-prepared samples.
Chemical fixation may induce ultrastructural artifacts owing to the slow diffusion of chemical fixatives that results in the nonsynchronized immobilization of cellular macromolecular components and owing to the selective cross-linking reactions of chemical fixatives, such as aqueous aldehydes and osmium tetroxide. High-pressure freezing (HPF) is widely regarded as the optimal fixation method for TEM (36). HPF is complete within milliseconds and ensures the simultaneous immobilization of all macromolecular components (37). Subsequent processing for morphological studies is predominantly achieved by freeze substitution (FS), where ice is removed by organic solvents at about −90°C. Freeze-substituted samples appear similar to materials prepared for TEM by conventional chemical fixation methods, but they have a greater likelihood of displaying structures in their native state (38). Since HPF/FS leads to an improved ultrastructural preservation, we also conducted HPF/FS for sample preparation.
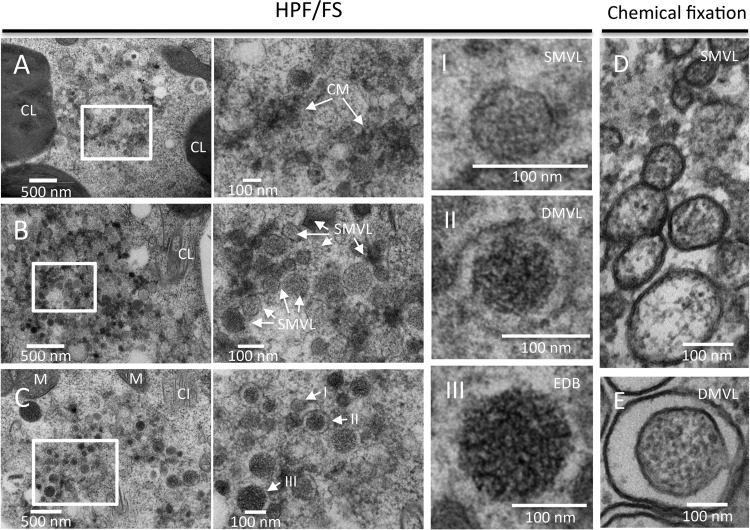
Currently, HPF allows a depth of 200 μm thick without detectable ice crystallization damage (39). In systemically infected N. benthamiana leaves, the thickness of the midrib is 356 μm ± 64 μm (n = 35), while it is 97 μm ± 23 μm (n = 35) for the other leaf area. To avoid damage from ice crystallization, the leaf area without the rib tissue was consequently chosen. Consistent with what we observed in chemically fixed samples, HPF/FS-prepared mesophyll cells showed similar membrane remodeling. CM structures were observed at 6 dpi (Fig. 5A) and SMVL structures of 74 nm ± 22 nm (n = 48) in diameter were observed at 7 dpi (Fig. 5B) in mesophyll cells. However, instead of only SMVL structures and DMVL structures with an electron-dense core being observed in chemically fixed mesophyll cells (Fig. 5D and E), electron-dense bodies of 172 nm ± 59 nm (n = 20) in diameter were also frequently observed in HPF/FS-prepared mesophyll cells at 8 dpi (Fig. 5C, arrows labeled I to III). DMVL structures had an outer vesicle diameter of 138 nm ± 20 nm (n = 48) and an electron-dense core diameter of 93 nm ± 19 nm (n = 48). Thus, the sizes of SMVL and DMVL structures in HPF/FS-prepared samples were smaller than those in chemically fixed samples. Another noticeable difference was that HPF/FS-prepared vesicles were largely rounded and had more electron-dense materials within them (Fig. 5C to E). We have no explanation for the different sizes observed between the HPF/HS and chemically fixed samples. Likewise, Royer and Kinnamon (40) found that T-cell tubules were more uniform and somewhat smaller in diameter in HPF/FS-prepared samples than in chemically fixed samples, but no reason for this was given.
FIG 5.
Comparison of TuMV-induced membranous aggregates in HPF/FS-prepared and chemically fixed samples. (A to C) Time course analysis of TuMV-induced membranous aggregates in mesophyll cells. TuMV/6K2:GFP systemically infected N. benthamiana leaf without rib tissues were fixed by HPF, processed by FS, and observed by TEM. Higher-magnification images of the areas in the white rectangles are shown on the right of each panel. (A) TuMV-induced CM structures in a mesophyll cell at 6 dpi. (B) TuMV-induced heterogeneous SMVL structures in a mesophyll cell at 7 dpi. (C) TuMV-induced aggregates contain SMVL structures (I), DMVL structures with an electron-dense core (II), and electron-dense bodies (III) in a mesophyll cell at 8 dpi. The images in panels I to III are higher-magnification images of the structures identified by the arrows labeled I to III, respectively, in panel C. (D and E) TuMV-induced heterogeneous SMVL structures (D) and a DMVL structure (E) in chemically fixed mesophyll cells. CL, chloroplast; M, mitochondrion; CM, convoluted membranes; SMVL, single-membrane vesicle-like structure; DMVL, double-membrane vesicle-like structure; EDB, electron-dense body.
3-D architecture of TuMV-induced membrane reorganization.
We performed ET on semithin (200-nm) sections to generate the three-dimensional (3-D) representation of the numerous heterogeneously sized SMVL structures that were observed in vascular parenchymal cells at 6 dpi (Fig. 2D). Figures 6I to IV show the representative tomograms that were generated from a single-axis-tilt series. SMVL structures associated with the rER, along with three closely associated cytoplasmic inclusions having a pinwheel configuration, were clearly observed. Figures 6V and VI show the 3-D surface rendering generated from tilted images over a range of ±60° (1° increments in high tilts and 2° increments in low tilts) and indicate that the SMVL structures were actually SMTs (represented in yellow), similar to those observed in CVB3-infected (2) and poliovirus-infected (41) cells (see also Movie S1 in the supplemental material). The 3-D surface rendering shows that most SMTs are closely packed together and have a similar orientation. The SMTs were touching one another, but no holes or connections were observed. Cytoplasmic inclusions (represented in magenta in Fig. 6V and VI) and rER were often found in close proximity to TuMV-modified membranes, and some dilated rER (represented in sky blue in Fig. 6V and VI), including ER membranes and the associated ribosomes, was tightly associated with the SMTs (Fig. 6IV to VI, red arrows).
We next investigated the 3-D architecture of the membrane structures in TuMV-infected vascular parenchymal cells at 7 dpi (Fig. 7; see also Movie S2 in the supplemental material). At this stage, the membrane structures had various shapes and complexities (Fig. 7A), and 3-D renderings were generated from selected membrane structures (boxed areas in Fig. 7A). Figure 7B is a close-up view of an SMVL structure (left) along with its 3-D surface rendering (right two panels), which shows its tubular nature (represented in yellow). The 3-D rendering of the electron-dense fibrillar materials inside this SMT (represented in light red) was also regenerated. These fibrillar materials could be the replication complexes containing vRNA, as proposed for beet black scorch virus (BBSV)-induced replication factories (42), which is consistent with our observation that SMVL structures contain dsRNA (Fig. 4E). Adjacent to the SMT is an irregularly shaped tubule (represented in green) that might represent an intermediate structure leading to the formation of DMTs (see below).
The left panel of Fig. 7C is a close-up of the area identified by the solid square depicted in Fig. 7A, and the two right panels are 3-D surface renderings for some of the membrane structures. These membrane structures are SMTs (in yellow) along with double-membrane tubules (DMTs) (the outer membrane is represented in light blue, and the inner membrane is represented in dark blue) interspersed between them with irregularly shaped tubules (represented in green) with frequent membrane pairing (Fig. 7C, white arrowheads) or curving (Fig. 7C, red arrowheads) (see also Movie S2 in the supplemental material). Figure 7D also shows tubule bending. Finally, the left panel of Fig. 7E is a close-up view of a DMVL structure (left), along with its 3-D surface rendering (right two panels). The DMT contents (represented in dark red) did not have a fibrillar profile but almost fill the inner core of the DMT.
In conclusion, the SMVL and DMVL structures that were observed in the two-dimensional (2-D) TEM images of TuMV-infected cells were, in fact, tubules. These SMTs and DMTs were also interspersed with irregularly shaped tubules, which might be intermediate forms leading to the transformation of SMTs into DMTs.
Viral particle bundles associated with electron-dense bodies.
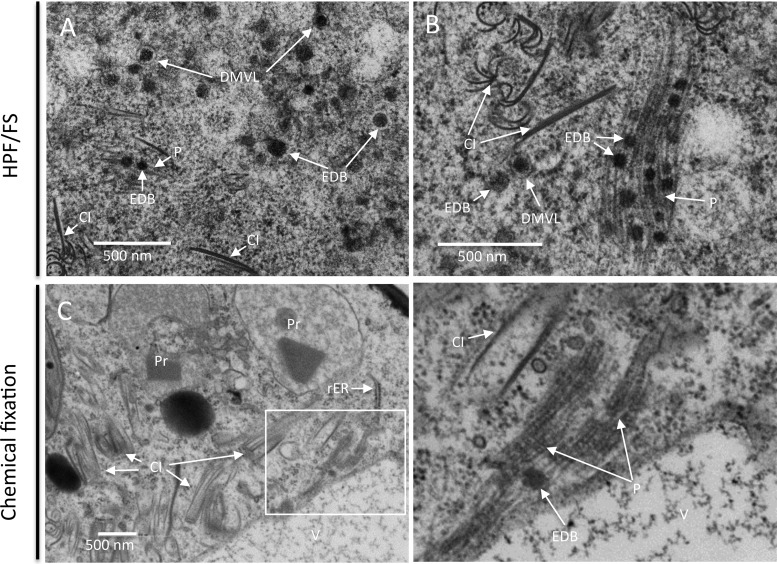
At 8 dpi, filament bundles in association with electron-dense bodies in the vicinity of TuMV-induced membranous aggregates were frequently observed in HPF/FS-prepared mesophyll cells (Fig. 8A). Individual filaments were 13 nm ± 1 nm (n = 30) in width, which corresponds to the thickness of TuMV particles. The electron-dense bodies had diameters of 64 nm ± 13 nm (n = 44), and DMVL structures with an electron-dense content of a similar intensity were found close to them (Fig. 8A and B). These structures were also seen in chemically fixed samples, although less frequently (Fig. 8C). These data suggest that these bodies are involved in viral particle assembly.
FIG 8.
TuMV particles are associated with electron-dense bodies. (A and B) In HPF/FS-prepared samples, virus-like particles are found as filament bundles associated with electron-dense bodies in the vicinity of TuMV-induced membranous aggregates at a late stage of infection (A), and the DMVL structures are found close to these filament bundle structures (B). (C) In chemically fixed samples, the filament bundles associated with electron-dense bodies are found in the cytoplasm near the tonoplast. CI, cytoplasmic inclusion body; DMVL, double-membrane vesicle-like structure; EDB, electron-dense body; P, particles; Pr, peroxisome; V, vacuole; rER, rough endoplasmic reticulum.
TuMV particles accumulate in vacuoles.
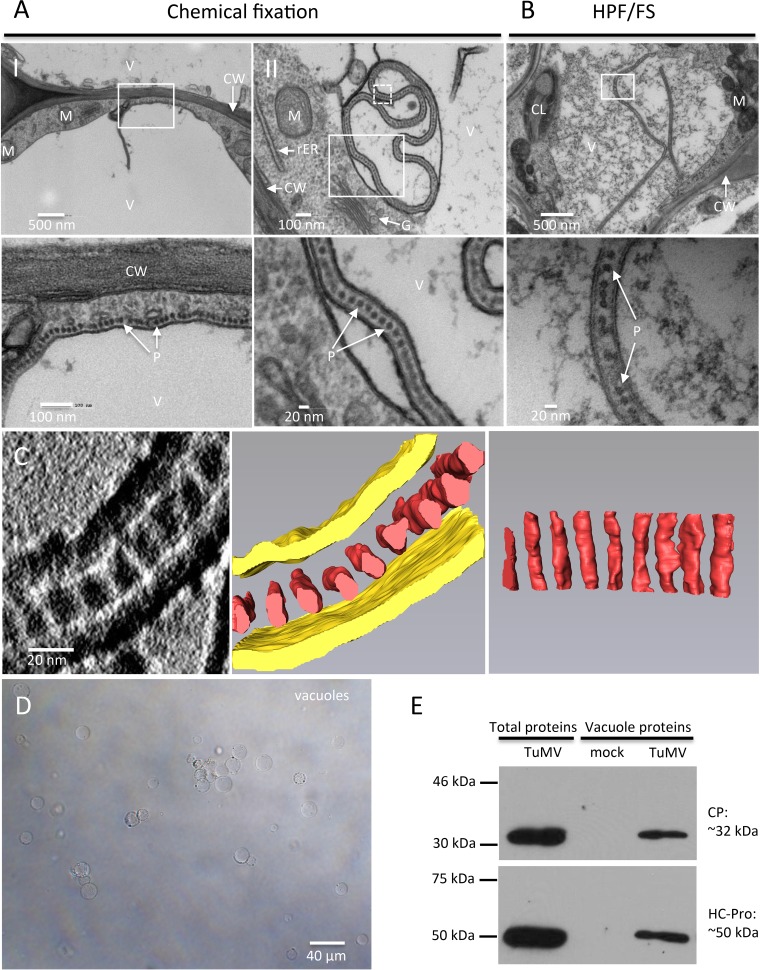
In addition to the above-described vesicle-like structures, linear arrays of dot-like structures with a diameter of 15 nm ± 1 nm (n = 30) juxtaposing the cytoplasmic face of the tonoplast and frequently protruding into the vacuole were seen (Fig. 9A, panel I). Figure 9A (panel II) shows that the aligned dots were enclosed in a membrane sac within the central vacuole. These structures were also found in HPF/FS-prepared samples, although the vacuole showed more electron-dense material (Fig. 9B).
FIG 9.
TuMV acquires an envelope by hijacking the tonoplast. (A and B) N. benthamiana leaf midribs systemically infected with TuMV/6K2:GFP were fixed by chemical fixation (A) or HPF (B), processed, and observed by TEM. A higher-magnification image of the area in the white rectangle is shown at the bottom. Monolayer dot-like structures are aligned along the tonoplast (A, panel I) and loaded into the vacuole from the cytoplasm (A, panel II, and B). (C) 3-D reconstruction of enveloped TuMV particles by ET. (Left) A higher-magnification image of the area in the white square in panel A (panel II) with a 180° rotation showing a single slice of the tomogram generated from the 90-nm-thick section and enveloped dot-like structures in the vacuole; (middle) the 3-D model generated from the whole tomogram of the left panel; (right) the 90° rotation of the dot-like structures in the middle panel. Yellow, tonoplast; red, dot-like structures. (D, E) CP and HCpro are present in purified vacuoles. (D) Vacuoles were isolated by Ficoll gradient centrifugation from N. benthamiana leaves systemically infected with TuMV. (E) Western blot analysis of viral proteins CP and HCpro in vacuoles purified from mock-infected N. benthamiana leaves and N. benthamiana leaves systemically infected with TuMV. The total proteins of N. benthamiana leaves systemically infected with TuMV were used as a positive control. V, vacuole; CW, cell wall; M, mitochondrion; rER, rough endoplasmic reticulum; G, Golgi apparatus; CL, chloroplast; P, particles.
The diameter of the dot-like structures is consistent with the diameter of viral particles. These structures may then represent the top view of vertically aligned viral particles. To confirm that this was the case, a 3-D representation was generated by ET. A series of single-axis-tilt 2-D images was collected on a 90-nm-thick ultrathin section (Fig. 9C; see also Movie S3 in the supplemental material) over a tilt range of ±64° (in 1° increments). Images were aligned and reconstructed to generate a 3-D representation of this structure. The middle and right panels of Fig. 9C show the 3-D reconstruction of the complete tomogram of the image of the left panel. A 3-D surface rendering of the aligned dot-like structures clearly showed that they were a cross section of a monolayer of TuMV particles aligned along the same axis and that they were enclosed with a membrane envelope, likely derived from the tonoplast.
Vacuoles were isolated from mock-infected leaves and leaves systemically infected with TuMV to confirm that viral particles accumulated in these organelles. Vacuoles were released from protoplasts and isolated by Ficoll gradient centrifugation (Fig. 9D). We then performed Western blot analysis with anti-CP and anti-helper component proteinase (anti-HCpro) polyclonal antibodies. Purified vacuoles from mock- and TuMV-infected leaves were visualized by phase-contrast microscopy to ascertain that similar amounts of vacuoles were loaded. The Western blot analysis results showed that the vacuoles isolated from TuMV-infected leaves contained CP (Fig. 9E), supporting the tomography data showing that TuMV particles are loaded into the vacuoles. HCpro, which is involved in aphid transmission of viral particles, was also detected in purified vacuoles (Fig. 9E).
DISCUSSION
In this study, we looked at the cellular remodeling that takes place during TuMV infection using a time course ultrastructural analysis. The very first event that was observed in the infected cells was the accumulation of CM structures close to or connected to the rER (Fig. 2C and 3A). These CM collections are reminiscent of those induced during infection by dengue virus (DENV; a flavivirus) (43) and the SARS coronavirus (1). CM structures have been proposed to be sites of viral polyprotein processing and storage for proteins and lipids involved in vRNA replication (43). Based on the morphological similarities between the CM structures induced by TuMV and by flavivirus and their observation early in the infection process, we propose that TuMV-induced CM structures are also sites for polyprotein processing and/or for storage of the proteins and lipids required for vesicle-like structure biogenesis. Indeed, the accumulation of large amounts of lipids has been observed in association with TuMV replication complexes (24) and for other plant viruses as well (44).
Few SMVL structures were observed amid CM structures in vascular parenchymal and mesophyll cells (Fig. 2C and 3A), but SMVL structures became prevalent 1 day later in these cells (Fig. 2D and 3B). The SMVL structures scored positive for 6K2 and RdRp, as well as for dsRNA, by immunogold labeling (Fig. 4C to E), suggesting that these structures are the sites for vRNA replication. The TuMV-induced SMVL structures are similar to the membrane structures induced during infection by the enteroviruses CVB3 (2) and poliovirus (45). Conventional 2-D TEM images of enterovirus CVB3- and poliovirus-induced membrane structures have been described to be either SMVs (46, 47) or DMVs (48–50), whereas for both viruses, ET-generated 3-D models revealed that the SMVs observed by TEM are SMTs (2, 41). Both potyviruses and enteroviruses belong to the picornavirus-like virus superfamily, suggesting that they might share similar membrane modification mechanisms. Similarly, in this study, the 3-D model showed that the TuMV-induced SMVL structures are SMTs (Fig. 6 and 7). Polioviral replication proteins were localized on the external cytoplasmic surface of the single-membrane tubule/vesicle. This apparently was not the case for TuMV. vRNA replication probably occurs inside the SMVL structures, since dsRNA-specific gold particles were located within the SMVL structures (Fig. 4E). In addition, the electron-dense fibrillar materials inside the SMT that were generated by ET might represent replication complexes (Fig. 7B). The intratubular localization of the replication complex is further supported by previous membrane fractionation data that showed that the replication-associated soluble viral and host proteins (VPg-Pro, RdRp, and poly(A) binding protein 2 [PABP2]) were trapped within the lumen of 6K2 vesicles (20).
DMVL structures were observed rather late in the infection cycle (Fig. 2E and 3C) and were characterized by having an electron-dense core, which was particularly apparent when samples were processed by HPF/FS (Fig. 5C). Poliovirus-induced DMTs may result from a transformation process of membrane apposition, enwrapping, and fusion of SMTs (41). The 3-D model generated at the late stage of TuMV infection also suggested that the DMVL structures are DMTs and that they were formed by enwrapping of the SMTs, which resulted in the engulfment of the cytoplasm (Fig. 7). Electron-dense bodies with a size and intensity similar to those of the electron-dense core found within DMVL structures were associated with filament bundles (Fig. 8). These bodies may derive from DMVL structures and may be associated with TuMV particle assembly. It is important to note that no SMVL structures were found near these assembly sites. This fits our confocal microscopy observations that CP did not colocalize with membrane-bound viral replication complexes (19, 24), suggesting that vRNA encapsidation occurs at a site adjacent to membrane-bound vRNA replication complexes. These tubules are likely the ultrastructure underlying the perinuclear ER amalgam observed by confocal microscopy during TuMV infection (16). However, the ultrastructure of the TuMV-induced small motile vesicles still needs to be defined.
We observed the accumulation of TuMV particles as a linear array in the central vacuole, with the tonoplast forming an envelope around them (Fig. 9A to C). Similar aligned dot-like structures were observed for cells infected with the potyviruses pokeweed mosaic virus (51) and carnation vein mottle virus (52), but their exact nature was not explained. The tonoplast has also been shown to be remodeled during several plant virus infections. TEM images showed that tonoplast-associated vesicles protruded into the vacuole in cells infected with the cucumoviruses cucumber mosaic virus (CMV) and tomato aspermy virus (TAV) (5) and the alfamovirus alfalfa mosaic virus (AMV) (53). Owing to the presence of replicase proteins, the authors concluded that these were sites of vRNA synthesis (54). On the other hand, particles of the icosahedral sobemovirus rice yellow mottle virus (RYMV) have been found in vacuoles as crystalline arrays (55). The authors proposed that the acidic pH and the presence of Ca2+ could facilitate crystal formation and thus stabilize RYMV particles, which may allow the virus to accumulate to high levels without having deleterious effects on cellular viability and integrity. The highly stable and compact RYMV particles would ultimately be released from the vacuole during xylem vessel differentiation and trafficking along the water flow for long-distance movement (55). The membrane envelope around TuMV particles would thus provide a protective shield against the harsh environmental conditions prevalent in the central vacuole for subsequent long-distance movement during xylem vessel differentiation (24).
Potyviruses are transmitted from plant to plant by aphids in a nonpersistent manner (56). The interaction between CP and the receptor in the cuticle of the aphid stylet tip is mediated by the viral protein HCpro (57). HCpro was detected in purified vacuoles from TuMV-infected cells (Fig. 9E). The accumulation of TuMV particles in the vacuole may have something to do with plant-to-plant virus transmission by aphids. Aphids are phloem feeders, since phloem sap is rich in key nutrients, such as carbohydrates (sucrose), amino acids, and minerals (58). Before the phloem nutrient source is sampled, the aphid performs host plant selection by probing the peripheral plant tissues. Potyviruses have been suggested to be acquired and inoculated during brief (<1 min) and superficial stylet penetrations in the peripheral plant tissues (epidermal and mesophyll cells) (59). Once the suitable host plant is detected, the aphid ultimately inserts the stylet along the cell wall of parenchymatous cells into the phloem sieve element for nutrient acquisition. The aphid recognizes the phloem sap by sensing the pH (7.0 to 7.5) and a high sucrose concentration (about 400 mmol−1) (60). During the search for phloem sap, the stylet tip is observed to be inserted into the vacuole of parenchymatous cells, and owing to the acidic pH (5.0 to 5.5) and low sucrose concentration, the stylet is withdrawn and changes direction until the sieve element is reached (60). This brief probing of the aphid stylet in the central vacuole may be sufficient to acquire the membrane-bound TuMV.
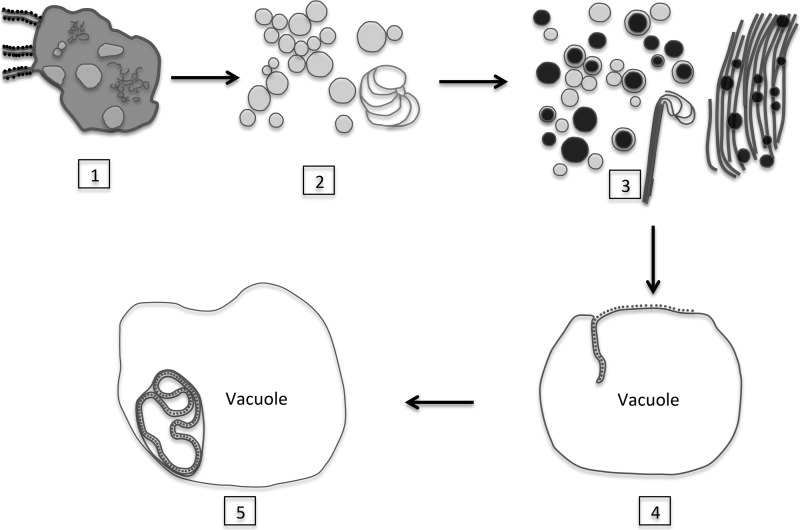
On the basis of the observations described above, we propose the following model that links the TuMV-induced membrane remodeling for vRNA replication with virus storage in the central vacuole (Fig. 10). Membrane remodeling starts with the accumulation of ER-connected CM structures. This leads to the formation of SMTs that are involved in vRNA replication. As infection proceeds, the number of SMTs decreases, and DMTs with an electron-dense core and electron-dense bodies are generated. Possibly, at this stage, vRNA and CP have been produced in sufficient quantities for particle assembly to take place in the vicinity of DMTs and electron-dense bodies. Finally, TuMV particles accumulate in a linear array along the same axis in the central vacuole. Additional steps are likely to take place between steps 4 and 5 in order to release and close the continuous double-membrane-enveloped ring of particles. These steps would involve membrane fusion events, requiring the participation of host factors, notably, secretory pathway factors, that might have been co-opted with the SMTs and DMTs. There is also the possibility of additional unseen membrane connections that increase the complexity of the relationship of the membrane structures that have been observed.
FIG 10.
Model for TuMV-induced membrane structure formation. The accumulation of ER-connected CM structures which may be involved in polyprotein processing is observed at an early stage of TuMV infection (step 1). At the midstage of TuMV infection, SMTs are produced. SMTs are transformed from CM structures and are involved in vRNA replication (step 2). In addition to SMTs, DMTs and electron-dense bodies are produced at the late stage of TuMV infection. Electron-dense bodies associated with virus-like particle filament bundles are present in the vicinity of this mixed aggregate. Possibly, at this point, vRNA and CP have been produced in sufficient quantities for particle assembly to take place (step 3). Finally, the assembled viral particles are loaded in the central vacuole (step 4) and enclosed by a modified tonoplast (step 5).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jessy Tremblay (INRS-Institut Armand-Frappier) for assistance with the Zeiss LSM 780 confocal microscope. We thank Olivier Voinnet (ETH Zürich) for the anti-HCpro serum.
This research was supported by grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada and from le Fonds de Recherche du Québec sur la Nature et les Technologies (FRQNT) to H.Z. and J.-F.L.
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02138-15.
REFERENCES
- 1.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Limpens RW, van der Schaar HM, Kumar D, Koster AJ, Snijder EJ, van Kuppeveld FJ, Barcena M. 2011. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. mBio 2:e00166–11. doi: 10.1128/mBio.00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laliberté J-F, Zheng H. 2014. Viral manipulation of plant host membranes. Annu Rev Virol 1:237–259. doi: 10.1146/annurev-virology-031413-085532. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 9:505–514. doi: 10.1016/S1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 5.Hatta T, Francki RIB. 1981. Cytopathic structures associated with tonoplasts of plant cells infected with cucumber mosaic and tomato aspermy viruses. J Gen Virol 53:343–346. doi: 10.1099/0022-1317-53-2-343. [DOI] [Google Scholar]
- 6.Scholthof KB, Scholthof HB, Jackson AO. 1995. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology 208:365–369. doi: 10.1006/viro.1995.1162. [DOI] [PubMed] [Google Scholar]
- 7.McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Franco A, Russo M, Martelli GP. 1984. Ultrastructure and origin of cytoplasmic multivesicular bodies induced by carnation Italian ringspot virus. J Gen Virol 65:1233–1237. doi: 10.1099/0022-1317-65-7-1233. [DOI] [Google Scholar]
- 9.Hwang YT, McCartney AW, Gidda SK, Mullen RT. 2008. Localization of the carnation Italian ringspot virus replication protein p36 to the mitochondrial outer membrane is mediated by an internal targeting signal and the TOM complex. BMC Cell Biol 9:54. doi: 10.1186/1471-2121-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatta T, Bullivant S, Matthews RE. 1973. Fine structure of vesicles induced in chloroplasts of Chinese cabbage leaves by infection with turnip yellow mosaic virus. J Gen Virol 20:37–50. doi: 10.1099/0022-1317-20-1-37. [DOI] [PubMed] [Google Scholar]
- 11.Hatta T, Matthews RE. 1974. The sequence of early cytological changes in Chinese cabbage leaf cells following systemic infection with turnip yellow mosaic virus. Virology 59:383–396. doi: 10.1016/0042-6822(74)90452-8. [DOI] [PubMed] [Google Scholar]
- 12.Carette JE, Stuiver M, Van Lent J, Wellink J, Van Kammen A. 2000. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J Virol 74:6556–6563. doi: 10.1128/JVI.74.14.6556-6563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritzenthaler C, Laporte C, Gaire F, Dunoyer P, Schmitt C, Duval S, Piequet A, Loudes AM, Rohfritsch O, Stussi-Garaud C, Pfeiffer P. 2002. Grapevine fanleaf virus replication occurs on endoplasmic reticulum-derived membranes. J Virol 76:8808–8819. doi: 10.1128/JVI.76.17.8808-8819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra R, Krishnamurthy K, Blancaflor E, Payton M, Nelson RS, Verchot-Lubicz J. 2003. The potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral cell-to-cell movement. Virology 312:35–48. doi: 10.1016/S0042-6822(03)00180-6. [DOI] [PubMed] [Google Scholar]
- 15.Bamunusinghe D, Hemenway CL, Nelson RS, Sanderfoot AA, Ye CM, Silva MA, Payton M, Verchot-Lubicz J. 2009. Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology 393:272–285. doi: 10.1016/j.virol.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Grangeon R, Agbeci M, Chen J, Grondin G, Zheng H, Laliberte JF. 2012. Impact on the endoplasmic reticulum and Golgi apparatus of turnip mosaic virus infection. J Virol 86:9255–9265. doi: 10.1128/JVI.01146-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Revers F, Garcia JA. 2015. Molecular biology of potyviruses. Adv Virus Res 92:101–199. doi: 10.1016/bs.aivir.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Beauchemin C, Boutet N, Laliberté J-F. 2007. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J Virol 81:775–782. doi: 10.1128/JVI.01277-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotton S, Grangeon R, Thivierge K, Mathieu I, Ide C, Wei T, Wang A, Laliberté J-F. 2009. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J Virol 83:10460–10471. doi: 10.1128/JVI.00819-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beauchemin C, Laliberte JF. 2007. The poly(A) binding protein is internalized in virus-induced vesicles or redistributed to the nucleolus during turnip mosaic virus infection. J Virol 81:10905–10913. doi: 10.1128/JVI.01243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dufresne PJ, Thivierge K, Cotton S, Beauchemin C, Ide C, Ubalijoro E, Laliberté J-F, Fortin MG. 2008. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374:217–227. doi: 10.1016/j.virol.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Agbeci M, Grangeon R, Nelson RS, Zheng H, Laliberte JF. 2013. Contribution of host intracellular transport machineries to intercellular movement of turnip mosaic virus. PLoS Pathog 9:e1003683. doi: 10.1371/journal.ppat.1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grangeon R, Jiang J, Wan J, Agbeci M, Zheng H, Laliberte JF. 2013. 6K2-induced vesicles can move cell to cell during turnip mosaic virus infection. Front Microbiol 4:351. doi: 10.3389/fmicb.2013.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan J, Cabanillas DG, Zheng H, Laliberte JF. 2015. Turnip mosaic virus moves systemically through both phloem and xylem as membrane-associated complexes. Plant Physiol 167:1374–1388. doi: 10.1104/pp.15.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otulak K, Garbaczewska G. 2012. Cytopathological Potato virus Y structures during Solanaceous plants infection. Micron 43:839–850. doi: 10.1016/j.micron.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Thivierge K, Cotton S, Dufresne PJ, Mathieu I, Beauchemin C, Ide C, Fortin MG, Laliberte JF. 2008. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377:216–225. doi: 10.1016/j.virol.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Beauchemin C, Bougie V, Laliberte JF. 2005. Simultaneous production of two foreign proteins from a polyvirus-based vector. Virus Res 112:1–8. doi: 10.1016/j.virusres.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Knapp E, Flores R, Scheiblin D, Scheiblin D, Modla S, Czymmek K, Czymmek K, Yusibov V. 2012. A cryohistological protocol for preparation of large plant tissue sections for screening intracellular fluorescent protein expression. Biotechniques 52:31–37. doi: 10.2144/000113778. [DOI] [PubMed] [Google Scholar]
- 29.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol 5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucocq JM, Habermann A, Watt S, Backer JM, Mayhew TM, Griffiths G. 2004. A rapid method for assessing the distribution of gold labeling on thin sections. J Histochem Cytochem 52:991–1000. doi: 10.1369/jhc.3A6178.2004. [DOI] [PubMed] [Google Scholar]
- 31.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 32.Robert S, Zouhar J, Carter C, Raikhel N. 2007. Isolation of intact vacuoles from Arabidopsis rosette leaf-derived protoplasts. Nat Protoc 2:259–262. doi: 10.1038/nprot.2007.26. [DOI] [PubMed] [Google Scholar]
- 33.Kalina M, Pease DC. 1977. The probable role of phosphatidyl cholines in the tannic acid enhancement of cytomembrane electron contrast. J Cell Biol 74:742–746. doi: 10.1083/jcb.74.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S, Baskin TI, Gallagher KL. 2012. Mechanical fixation techniques for processing and orienting delicate samples, such as the root of Arabidopsis thaliana, for light or electron microscopy. Nat Protoc 7:1113–1124. doi: 10.1038/nprot.2012.056. [DOI] [PubMed] [Google Scholar]
- 35.Edwardson JR, Christie RG, Ko NJ. 1984. Potyvirus cylindrical inclusions-subdivision-IV. Phytopathology 74:1111–1114. doi: 10.1094/Phyto-74-1111. [DOI] [Google Scholar]
- 36.Studer D, Michel M, Muller M. 1989. High pressure freezing comes of age. Scanning Microsc Suppl 3:253–268. [PubMed] [Google Scholar]
- 37.Gilkey JC, Staehelin LA. 1986. Advances in ultrarapid freezing for the preservation of cellular ultrastructure. J Electron Microsc Tech 3:177–210. doi: 10.1002/jemt.1060030206. [DOI] [Google Scholar]
- 38.McDonald K, Morphew MK. 1993. Improved preservation of ultrastructure in difficult-to-fix organisms by high pressure freezing and freeze substitution. I. Drosophila melanogaster and Strongylocentrotus purpuratus embryos. Microsc Res Tech 24:465–473. [DOI] [PubMed] [Google Scholar]
- 39.Vanhecke D, Graber W, Studer D. 2008. Close-to-native ultrastructural preservation by high pressure freezing. Methods Cell Biol 88:151–164. doi: 10.1016/S0091-679X(08)00409-3. [DOI] [PubMed] [Google Scholar]
- 40.Royer SM, Kinnamon JC. 1996. Comparison of high-pressure freezing/freeze substitution and chemical fixation of catfish barbel taste buds. Microsc Res Tech 35:385–412. [DOI] [PubMed] [Google Scholar]
- 41.Belov GA, Nair V, Hansen BT, Hoyt FH, Fischer ER, Ehrenfeld E. 2012. Complex dynamic development of poliovirus membranous replication complexes. J Virol 86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao X, Jin X, Zhang X, Li Y, Wang C, Wang X, Hong J, Wang X, Li D, Zhang Y. 2015. Morphogenesis of the endoplasmic reticulum membrane-invaginated vesicles during beet black scorch virus infection: the role of auxiliary replication protein and new implications of three-dimensional architecture. J Virol 89:6184–6195. doi: 10.1128/JVI.00401-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee WM, Ahlquist P. 2003. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J Virol 77:12819–12828. doi: 10.1128/JVI.77.23.12819-12828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bienz K, Egger D, Pfister T, Troxler M. 1992. Structural and functional characterization of the poliovirus replication complex. J Virol 66:2740–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bienz K, Egger D, Rasser Y, Bossart W. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 131:39–48. doi: 10.1016/0042-6822(83)90531-7. [DOI] [PubMed] [Google Scholar]
- 47.Bienz K, Egger D, Pasamontes L. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- 48.Schlegel A, Giddings TH Jr, Ladinsky MS, Kirkegaard K. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol 70:6576–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol 82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, Whitton JL. 2010. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol 84:12110–12124. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim KS, Fulton JP. 1969. Electron microscopy of pokeweed leaf cells infected with pokeweed mosaic virus. Virology 37:297–308. doi: 10.1016/0042-6822(69)90213-X. [DOI] [PubMed] [Google Scholar]
- 52.Weintraub M, Ragetli HW. 1970. Distribution of viruslike particles in leaf cells of Dianthus barbatus infected with carnation vein mottle virus. Virology 40:868–881. doi: 10.1016/0042-6822(70)90133-9. [DOI] [PubMed] [Google Scholar]
- 53.Martelli G, Russo M. 1985. Virus-host relationships, p 163–205. In Francki RIB. (ed), The plant viruses. Springer, New York, NY. [Google Scholar]
- 54.Ibrahim A, Hutchens HM, Berg RH, Loesch-Fries LS. 2012. Alfalfa mosaic virus replicase proteins, P1 and P2, localize to the tonoplast in the presence of virus RNA. Virology 433:449–461. doi: 10.1016/j.virol.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 55.Brugidou C, Opalka N, Yeager M, Beachy RN, Fauquet C. 2002. Stability of rice yellow mottle virus and cellular compartmentalization during the infection process in Oryza sativa (L.). Virology 297:98–108. doi: 10.1006/viro.2002.1398. [DOI] [PubMed] [Google Scholar]
- 56.Ng JC, Falk BW. 2006. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu Rev Phytopathol 44:183–212. doi: 10.1146/annurev.phyto.44.070505.143325. [DOI] [PubMed] [Google Scholar]
- 57.Syller J. 2005. The roles and mechanisms of helper component proteins encoded by potyviruses and caulimoviruses. Physiol Mol Plant 67:119–130. doi: 10.1016/j.pmpp.2005.12.005. [DOI] [Google Scholar]
- 58.Dinant S, Bonnemain JL, Girousse C, Kehr J. 2010. Phloem sap intricacy and interplay with aphid feeding. C R Biol 333:504–515. doi: 10.1016/j.crvi.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 59.Powell G. 1991. Cell membrane punctures during epidermal penetrations by aphids: consequences for the transmission of two potyviruses. Ann Appl Biol 119:313–321. doi: 10.1111/j.1744-7348.1991.tb04870.x. [DOI] [Google Scholar]
- 60.Hewer A, Becker A, van Bel AJ. 2011. An aphid's odyssey—the cortical quest for the vascular bundle. J Exp Biol 214:3868–3879. doi: 10.1242/jeb.060913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.