Abstract
The aim of the present study was to obtain comprehensive microRNA (miRNA) profiles of type I [Ishikawa (ISK)] and type II (HEC-1B) human endometrial adenocarcinoma cell lines, utilizing the latest high-throughput sequencing techniques. RNA was extracted from ISK and HEC-1B cell lines. Sequencing results were obtained from a next-generation sequencing platform. Using the miRBase database and a series of software pipelines, miRNA expression was analyzed in the ISK and HEC-1B cell lines. It was revealed that the type and quantity of miRNAs in the two cell types varied significantly; 34 miRNAs were upregulated and 105 miRNAs were downregulated in HEC-1B cells compared with those of ISK cells. Furthermore, it was observed that the expression pattern of the miRNA (miR)-17-92 cluster differed between the two cell types, and the expression levels of the miR-200 family in ISK cells were markedly increased compared with those of HEC-1B cells. The present study therefore identified potential novel biomarkers, which may be useful in the differentiation between type I and type II endometrial cancer, and also revealed miRNA alterations that may be associated with endometrial cancer and its underlying pathogenic mechanisms.
Keywords: deep sequencing, endometrial cancer, HEC-1B, Ishikawa, microRNA
Introduction
MicroRNAs (miRNAs) are a class of single-stranded, non-coding RNA molecules that are 18–22 nucleotides in length, are encoded by endogenous genes and are transcribed from genomic DNA (1). However, miRNAs are not directly translated into proteins; instead, they control protein synthesis by binding their complementary sequence to the 3′ untranslated region (UTR) of the target gene messenger RNA (mRNA) molecule and inducing mRNA degradation or inhibition of translation, which subsequently inhibits protein synthesis (2). Therefore, miRNAs are capable of regulating protein-coding genes (1). Previous research has demonstrated that miRNAs participate in a series of critical physiological processes, including development, hematopoiesis, organogenesis, apoptosis and cell proliferation, and miRNAs have also been associated with the incidence of multiple diseases, including the development of tumors (3). Endometrial cancer is a common malignant tumor of the female reproductive system, which has a significant impact on the quality of life of patients and may result in mortality. Endometrial cancer may be classified as type I or type II, according to certain clinical and pathological characteristics. The initiation of type I endometrial cancer is associated with long-term exposure to increased levels of estrogen stimulation; therefore type I endometrial cancer is known as estrogen-dependent endometrial cancer (4). The initiation of type II endometrial cancer is not associated with estrogen stimulation and it is therefore referred to as estrogen-independent endometrial cancer (4). Ishikawa (ISK) cells are a highly differentiated endometrial adenocarcinoma cell line that express the estrogen receptor (ER), and are considered to be a classic ER-positive cell line, exhibiting typical type I characteristics of endometrial cancer (5). Therefore, ISK cells are frequently used for in vitro experiments examining the role of estrogen and the ER in endometrial cancer. Conversely, the HEC-1B cell line, which consists of intermediately-differentiated endometrial adenocarcinoma cells, is ER-negative or expresses the ER at reduced levels, exhibiting typical type II endometrial cancer characteristics (6). These two cell types are widely used for basic endometrial cancer research (5). Type II endometrial cancer is less frequently observed in patients compared with type I, and accounts for 20–30% of total cases (7). However, type II endometrial cancer has been revealed to be associated with a poorer prognosis, and to differ significantly from type I endometrial cancer, in terms of histological differentiation, myometrial invasion, lymph node metastasis and recurrence rate (8). In addition, ER and progesterone receptor expression has been observed to differ significantly between type I and II endometrial cancer, and is considered to be a critical marker for the assessment of prognosis (9).
Previous studies on ISK and HEC-1B endometrial cancer cell lines have mainly focused on hormone and mRNA expression levels. Previous research has revealed that miRNAs are critical regulators of the progression of cancer; therefore the abnormal miRNA expression observed in endometrial cancer has gained attention (10–12). Myatt et al (10) identified that expression levels of the tumor suppressor Forkhead box protein O1 (FOXO1) were significantly downregulated in endometrial cancer cell tumor tissues compared with normal endometrial tissues. miRNA (miR)-9, miR-27, miR-96, miR-153, miR-182 and miR-183 were predicted, using bioinformatic tools (miRNA_Targets; http://mamsap.it.deakin.edu.au/~amitkuma/mirna_targetsnew/find_gene.html), to bind to the FOXO1 3′UTR and this hypothesis was subsequently verified using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and northern blotting. These miRNAs are associated with the downregulation of FOXO1 and therefore may be involved in the development of endometrial cancer. Chung et al (11) utilized RT-qPCR to selectively screen for miR-205, which was observed to be expressed at increased levels in endometrial adenocarcinoma-like cells. Following endometrial cancer cell transfection with an miR-205 inhibitor, miR-205 expression was observed to be significantly reduced and the protein expression of its predicted target gene JPH4 was observed to be increased. This result indicated that miR-205 may be involved in the initiation and development of endometrial cancer. Snowdon et al (12) demonstrated that the expression of the miRNA-200 family was significantly increased in endometrial cancer, compared with the miRNA-200 family expression observed in atypical endometrial hyperplasia. Furthermore, compared with studies focusing on the expression of individual miRNAs, investigations into the variation in the expression of miRNA clusters and families have provided novel hypotheses for the study of miRNAs in endometrial cancer (13).
Until recently, studies of miRNA expression in endometrial cancer utilized conventional microarrays, which mainly rely on sequence hybridization and qPCR. Next-generation sequencing technology is capable of detecting novel miRNAs, and additionally demonstrates increased sensitivity and a broad dynamic range (14). A significant advantage of deep sequencing is that the final sequencing results do not contain saturation effects, which frequently occur during the analysis of microarray results (14).
Although fluorescent qPCR possesses the advantages of being sensitive, reproducible, quantitatively accurate and rapid, it has additional disadvantages. Similarly to microarray technology, fluorescent qPCR is only capable of detecting known miRNAs. A significant disadvantage of fluorescent qPCR is that it does not support high throughput detection of miRNAs. By contrast, next-generation sequencing has successfully overcome these disadvantages and is able to simultaneously achieve high throughput analysis with increased sensitivity (15).
Materials and methods
Sample preparation and RNA extraction
ISK and HEC-1B endometrial cancer cell lines were obtained from the Obstetrics and Gynecology Research Institute of Fudan University Women's Hospital (Shanghai, P.R. China). Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum, 5 µg/ml bovine insulin, 100 U/ml penicillin and 100 µg/ml streptomycin (all Solarbio Science & Technology Co.,Ltd, Beijing, China) in a 5% CO2 atmosphere at 37°C. Total RNA was extracted from cells using the mirVana™ miRNA isolation kit (Ambion Life Technologies, Carlsbad, CA, USA). RNA concentrations were measured using a NanoDrop® ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
Small RNA library generation and sequencing
Small RNA libraries for deep sequencing analysis were prepared from total RNA using the Applied Biosystems Small RNA Expression kit (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, ~500 ng total RNA was hybridized and ligated with the 5′ and 3′ adapter A and adapter B overnight. Subsequently, template bead preparation and emulsion PCR were performed using the Applied Biosystems SOLiD emulsion PCR and SOLiD buffer kits (Thermo Fisher Scientific), according to the manufacturer's instructions. Briefly, the oil phase was prepared: A total of 400 µl Tween 80 (Sigma-Aldrich, St. Louis, MO, USA) and 1.8 ml SP80 (Sigma-Aldrich) were added to 37.8 ml mineral oil (Sigma-Aldrich) and mixed thoroughly. Next, 80 µl P1 primer-labeled beads (DNA sequence, 5′-TTTTTTCCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT-3′; Applied Biosystems; Thermo Fisher Scientific) were added to the aqueous phase [consisting of 10X PCR Buffer (280 µl), 100 mM dNTP Mix (392 µl), 1M MgCl2 (70 µl), 10 µM ePCR Primer 1 (11.2 µl), 500 µM ePCR Primer 2 (16.8 µl), 500 pM prepared library (2.8 µl), nuclease-free water (1647.2 µl) and 5 U/µl AmpliTaq Gold® DNA Polymerase, UP (300 µl)] to a final volume of 2,800 µl. The DNA primer sequences of the forward ePCR Primer 1 and reverse ePCR Primer 2 were 5′-CCACTACGCCTCCGCTTTCCTCTCTATG-3′ and 5′-CTGCCCCGGGTTCCTCATTCT-3′, respectively. The emulsion was produced using the ULTRA-TURRAX®Tube Drive (IKA®-Werke GmbH & Co. KG, Staufen, Germany). The aqueous phase (2,800 µl) was added to 9 ml oil phase gradually, and 100 µl emulsion was gently dispensed into each well of a 96-well PCR plate. ePCR was performed using the Applied Biosystems GeneAmp® PCR System 9700 (Thermo Fisher Scientific) under the following conditions: Initial denaturation step at 95°C for 5 min, followed by 60 cycles of 93°C for 15 sec, 62°C for 30 sec and 72°C for 75 sec, with a final extension step at 72°C for 7 min. Following PCR, the emulsion was broken by the addition of two volumes of 2-butanol (294810-1L; Sigma-Aldrich) and the template beads were washed three times with wash solution (10 mM Tris-HCl, 50 mM KCl, 2 mM EDTA and 0.01% (v/v) Triton X-100) and suspended in Applied Biosystems TE buffer (10 mM Tris-HCl and 1 mM EDTA; Thermo Fisher Scientific) for further sequencing. Finally, deposition was performed according to the standard protocol.
Sequence processing and annotation
The SOLiD System Small RNA Analysis Pipeline tool (RNA2MAP, version 0.5.0; Life Technologies, Carlsbad, CA, USA) was used to analyze the miRNA sequencing data, according to the following protocol. Initially, two mismatches for the sequencing data were defined, the data from the present study was compared with the available known RNA library (www.mirbase.org) and subsequently, known RNA sequences were eliminated to prevent interference with the comparison with the miRNA library. Processed reads resulting from the above method were compared with the known miRNA precursor library (miRBase 20.0; www.mirbase.org) (16). Comparison data for each miRNA was collected and a statistical analysis of the length of the reads that matched the precursors was undertaken. Finally, the remaining reads were matched to the genome to identify potential novel miRNAs.
Normalization of read counts
It was assumed that unique sequence reads of <10 copies may represent sequencing errors; therefore, these reads were eliminated from the total sequencing data. To simplify the comparison of miRNA expression variation in various samples, the total copy number in each sample was standardized and uniformly set to 1,000,000 for each sample.
Statistical analysis
The characteristics of study subjects were compared using Student's t-test. To show the differential expression level of specific miRNA between the ISK and HEC-1B. The data were normalized and log2-transformed. GraphPad Prism 5.0 software (GraphPad Software Inc., La Jolla, CA, USA) was used for all statistical analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
Overview of sequencing data
The raw sequencing data was classified into the following six databases: Ribosomal RNA (rRNA), small nucleolar RNA, transfer RNA (tRNA), small nuclear RNA, miRNA and genome. The results indicated that the proportion of miRNAs among the total non-coding small RNAs was similar for HEC-1B and ISK cells (77 vs. 78%, respectively). Excluding miRNA, rRNA was the most abundant RNA observed among the non-coding small RNAs, and was revealed to represent 62 and 64% of non-coding small RNAs in HEC-1B and ISK cells, respectively. The distribution of small RNAs in each sample is illustrated in Fig. 1A. In conclusion, small RNA sequencing results demonstrated that miRNAs and rRNAs were the most predominant non-coding small RNAs detected in the HEC-1B and ISK cell lines and the proportions of these RNAs, compared with the total observed non-coding small RNA population, were comparable between the two cell lines.
Figure 1.
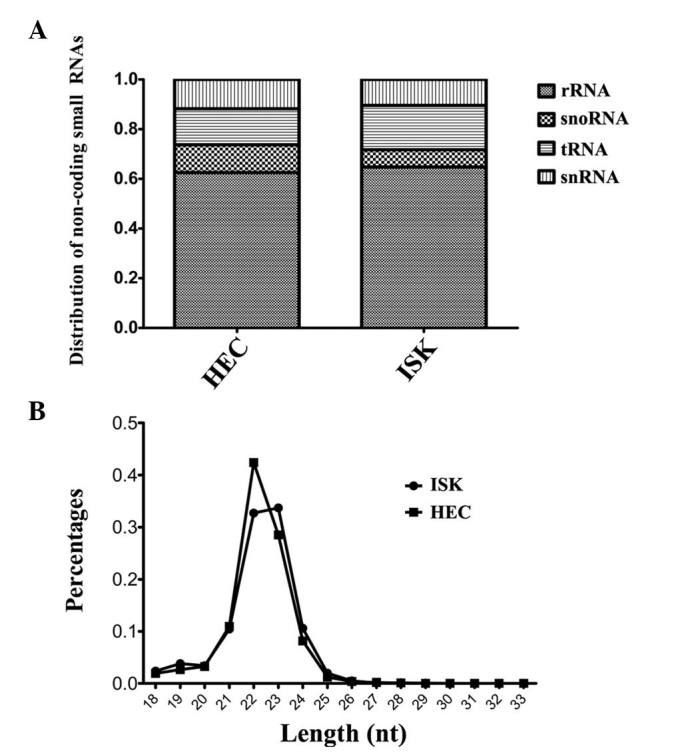
Overview of sequencing data. (A) Distribution of non-coding RNA in the samples analyzed (proportion of RNA was analyzed from data in which miRNAs were already excluded). (B) Distribution of miRNA length in HEC and ISK cell lines. miRNA, microRNA; ISK, Ishikawa; nt, nucleotides; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; tRNA, transfer RNA; snRNA, small nuclear RNA.
The length distribution of the miRNAs in the sequencing data was analyzed. It was revealed that in the HEC-1B and ISK cell lines, miRNAs with lengths of 22 and 23 nucleotides (nt) appeared as the two highest peaks. The overall length distribution indicated that the authenticity of the miRNA expression profile was maximally preserved during the process of sequencing library preparation (Fig. 1B). However, in the ISK cell line, the proportion of miRNAs with lengths of 22 and 23 nt was similar, whereas the proportion of 22 nt miRNAs was greater than that of 23 nt miRNAs in the HEC-1B cell line.
Differential expression of miRNAs between the HEC-1B and ISK cell lines
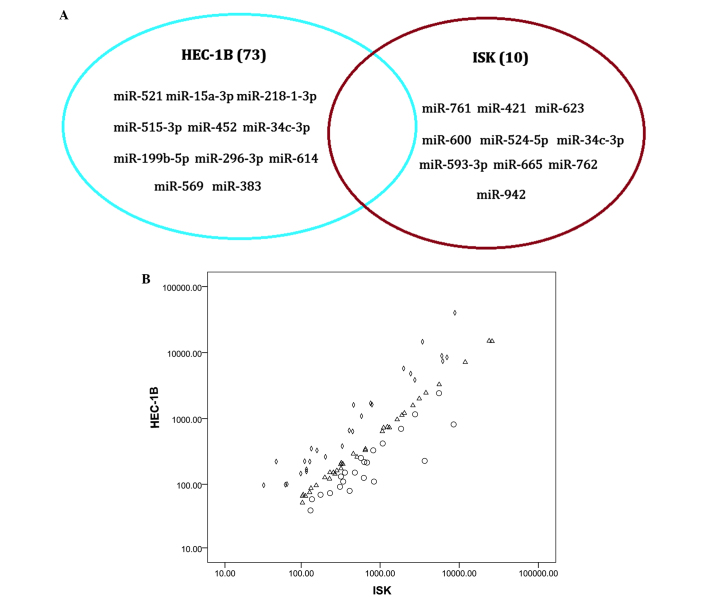
The primary focus of the present study was to examine the differential expression of miRNAs between an estrogen-dependent (ISK) and an estrogen-independent endometrial cancer cell line (HEC-1B). Mature miRNAs with copy numbers >10 were considered to be representative of a valid read. According to this definition, 381 miRNAs in the HEC-1B cell line and 318 miRNAs in the ISK cell line were detected. Furthermore, 73 unique miRNAs were identified to be present in the HEC-1B cell line and absent in ISK cell line, whereas 10 miRNAs were expressed in the ISK cell line and absent in HEC-1B cells (Fig. 2A).
Figure 2.
miRNA expression in HEC-1B and ISK cell lines. (A) Unique sets of miRNAs in the HEC-1B and ISK cell lines. (B) Differential expression of total miRNA in the ISK and HEC-1B cell lines. Diamonds (◊) represent miRNAs with expression levels in HEC-1B cells that are >2-fold greater than those in ISK cells; circles (o) represent miRNAs with expression levels in ISK cells >2-fold greater than those in HEC-1B cells and triangles (∆) represent miRNAs with a differential expression between ISK and HEC-1B cells that was <2-fold. miRNA, microRNA; ISK, Ishikawa.
The ten most abundant miRNAs in the HEC-1B and ISK cell lines were identified (Table I) and those that were included in the lists from the two cell types included Homo sapiens (hsa)-miR-19b, hsa-miR-23b, hsa-miR-17, hsa-let-7a, hsa-miR-18a and hsa-miR-30d. However, expression of these miRNAs varied significantly between the two samples; miR-141 was observed to be highly expressed in the HEC-1B cell line but was not abundantly expressed in the ISK cell line.
Table I.
Ten most abundantly expressed miRNAs in HEC and ISK cell lines.
| A, HEC | |
|---|---|
| Most abundantly expressed | All isomiRs |
| hsa-miR-141 | hsa-miR-141 |
| hsa-miR-17 | hsa-miR-19b |
| hsa-miR-130a | hsa-miR-23b |
| hsa-miR-18a | hsa-miR-17 |
| hsa-miR-200a | hsa-let-7a |
| hsa-miR-93 | hsa-miR-200a |
| hsa-miR-19b | hsa-miR-191 |
| hsa-miR-30d | hsa-miR-18a |
| hsa-miR-210 | hsa-miR-130a |
| hsa-miR-23b | hsa-miR-30d |
| B, ISK | |
| Most abundantly expressed | All isomiRs |
| hsa-miR-18a | hsa-miR-19b |
| hsa-miR-17 | hsa-miR-18a |
| hsa-miR-30d | hsa-miR-191 |
| hsa-miR-19b | hsa-miR-17 |
| hsa-miR-93 | hsa-miR-23b |
| hsa-miR-130a | hsa-miR-93 |
| hsa-miR-191 | hsa-miR-103 |
| hsa-let-7g | hsa-let-7a |
| hsa-miR-23b | hsa-miR-20a |
miRNA, microRNA; ISK, Ishikawa; hsa, Homo sapiens.
In addition, to understand the differential expression of miRNAs between the HEC-1B and ISK cell lines, the miRNA reads were normalized and the log2 values of each miRNA in the ISK and HEC-1B cells were calculated and compared (Fig. 2B).
As miRNAs with copy numbers <10 reads may represent sequencing errors, the differential expression of miRNAs with copy numbers >10 reads was compared. It was observed that following normalization, there were 34 miRNAs in the HEC-1B cell line which differed >1.5-fold compared with those in the ISK cell line, and there were 105 miRNAs in the ISK cell line which differed >1.5-fold compared with those in the HEC-1B cell line.
Notably, when TAM online software (17) (http://cmbi.bjmu.edu.cn/tam) was used to analyze miRNAs with expression levels that were significantly different between the ISK and HEC-1B cell lines, it was revealed that there were 18 upregulated miRNAs in the ISK cell line that were correlated with hormone regulation (P=1.25×10−3, where P<0.01 was considered to represent a significant correlation). However, in the HEC-1B cell line, there were only 4 upregulated miRNAs that were correlated with hormone regulation (P=0.36). These results may indicate that there are a number of miRNAs associated with hormone regulation in the ISK cell line. These results support the findings of previous studies, as the ISK cell line is associated with ER expression, which is known to possess a critical role in hormonal regulation (18).
Expression patterns of miRNA variants in HEC-1B and ISK cells
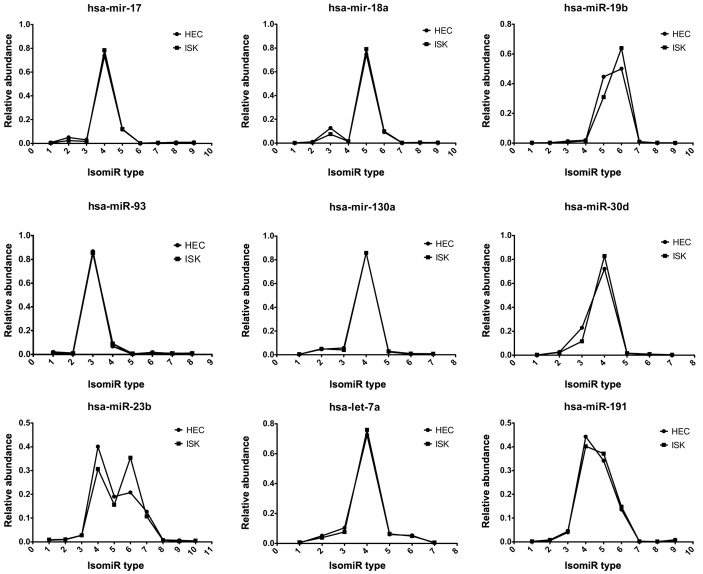
Multiple isomiRs, including isomiRs with 3′ additions, were detected from a given miRNA locus (Fig. 3). These nine abundant miRNAs demonstrated similar numbers of miRNA variants with comparable expression levels. Furthermore, the isomiR expression patterns of hsa-miR-17, hsa-mir-18a and hsa-miR-19b, which are encoded by the miR-17 cluster in HEC-1B and ISK cells, were highly consistent (Fig. 3).
Figure 3.
Comparison of isomiR distribution patterns of nine abundant miRNAs between HEC-1B and ISK cells. The x-axis indicates type of isomiRs from a specific miRNA locus and the y-axis indicates percentage expression. Of these, hsa-mir-17, hsa-mir-18a and hsa-mir-19b were associated with the mir-17 cluster, while hsa-mir-17, hsa-mir-18a and hsa-mir-93 were part of the mir-17 family. miRNA, microRNA; ISK, Ishikawa; hsa, Homo sapiens.
Differential expression of miRNA clusters and families
Previous studies have demonstrated that miRNA genes are arranged as clusters on the chromosome (19,20). The members of an miRNA cluster typically share sequence similarities (21) and coordinate the regulation of similar biological processes (22). When members of a gene cluster exhibit sequence similarity and homologous regulation, they form miRNA gene families, which further increase the diversity of miRNA gene cluster distribution (23). Previous studies have generally focused on the expression and regulation of single miRNAs, while studies investigating the expression and function of clustered miRNAs are infrequent and studies examining the expression profile of miRNA clusters at the genome-wide level are particularly limited. However, the expression patterns of homologous miRNAs arranged as clusters are typically coordinated (22).
miRNAs exhibiting significantly different expression patterns between the ISK and HEC-1B cell lines were classified into clusters and families. miRNA clusters and families were considered distinct if there were ≥2 miRNA members in the cluster or family that exhibited significantly different expression levels between ISK and HEC-1B cells.
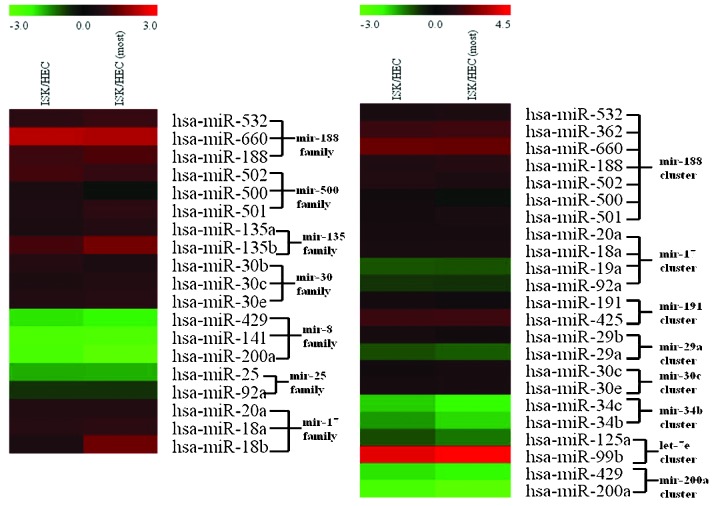
Based on the cluster comparison definition used in the present study, it was observed that there were 5 miRNA clusters in the ISK cell line in which expression levels were significantly upregulated compared with those of the HEC-1B cell line, and 3 miRNA clusters in which expression levels were significantly downregulated in ISK cells as compared with HEC-1B cells. In addition, there were also 5 miRNA families in the ISK cell line for which expression levels were significantly upregulated compared with the HEC-1B cell line and 2 miRNA families for which expression levels were significantly downregulated. Notably, expression levels of all 7 miRNA members in the hsa-mir-188 cluster were significantly upregulated (P=1.12×10−6), which demonstrated that these 7 miRNAs exhibited expression patterns consistent with each other. The presence of similar expression levels of miRNA members within an miRNA cluster indicated a co-transcription process and suggested that clustered miRNAs possess identical expression levels. Notably, the hsa-mir-17 cluster exhibited an inconsistent expression pattern; in the ISK cell line, hsa-mir-20a and hsa-mir-18a were upregulated, while hsa-mir-19a and hsa-mir-92a were downregulated. This suggested that clustered miRNAs may additionally possess an independent transcription process and subsequent post-transcriptional regulation may induce miRNA species within the same miRNA cluster to exhibit differential expression patterns. It was also hypothesized that the observed inconsistent expression patterns among members of miRNA clusters may partially result from the increased sensitivity of high-throughput technology.
Furthermore, members within miRNA families typically demonstrated similar expression levels (Fig. 4) compared with members of miRNA clusters. Inconsistent expression patterns were absent from miRNA members within each evaluated miRNA family, for example the mir-17 family. Due to the fact that the miRNA species within an miRNA gene family were homologous, their similar expression levels may be indicative of their closely associated functions.
Figure 4.
Differential miRNA gene cluster and family expression. The right panel indicates differential expression of miRNAs based on cluster analysis. The left panel indicates differential expression of miRNAs based on family analysis. miRNA/miR, microRNA; hsa, Homo sapiens.
The miR-17-92 gene cluster is widely expressed in humans and mice, and represents a potentially oncogenic miRNA cluster. This gene cluster may induce tumors by inhibiting the expression of genes that regulate oncogenes and the cell cycle (24). It is of note that certain miRNA sequences in the miR-17-92 gene cluster (hsa-mir-20a and hsa-mir-18a) were observed to be highly similar to the miRNA sequences in the miR-106a gene cluster, which may be grouped into the hsa-mir-17 family. These miRNAs may therefore possess similar biological functions and may regulate identical target genes. The sequences of the additional members of the miR-17-92 gene cluster (hsa-mir-19a and hsa-mir-92a) exhibited variation compared with miRNA sequences within the hsa-mir-17 family.
Castellano et al (25) reported that the expression levels of pri-mir-17-92 in ERα-positive breast cancer cells were significantly upregulated compared with those of ERα-negative breast cancer cells. The miRNA that is transcribed from this miRNA gene cluster downregulates the expression of the target gene ERα and is also able to inhibit the protein expression of AIBI, a p160 family member and co-activator that is required for transcription. The results of the present study exhibited a similar phenomenon; in the ERα-positive ISK cell line, the expression of certain mature miRNA species within the miR-17-92 cluster was upregulated, however the expression of a small number of miRNA species in the miR-17-92 cluster was downregulated. Therefore, the results of the present study appeared to indicate that the non-homologous miRNA species in the miR-17-92 cluster not only had differential expression levels, induced by additional post-transcriptional regulation, but additionally may perform distinct biological functions.
It was also revealed that for the miR-200 family in particular, the expression levels of miR-141 in the HEC-1B cell line were significantly increased (Fig. 4). Notably, miR-141 is highly expressed in a number of common epithelial cancers (26). This suggests that the miR-200 family, which was highly expressed in the HEC-1B cell line, may promote cancer cell invasion and metastasis and may have the potential to be used as a biomarker for the differentiation of endometrial cancer subtypes.
Discussion
In the present study, high-throughput sequencing technology was used to screen for the differential expression of miRNAs in ISK and HEC-1B cells. The results of the present study suggested that although the two cell types investigated were endometrial cancer cell lines, their miRNA expression profiles varied significantly. Of the known human miRNAs, there were 139 miRNAs that were differentially expressed between these two types of cell, and a number of these miRNAs have previously been demonstrated to be associated with tumors (27).
Clustered miRNA genes may coordinate the regulation of numerous critical biological processes, including embryonic development, the cell cycle and cell proliferation (28). As these miRNAs may inhibit the expression of numerous proteins involved in tumor development, they possess crucial roles during organogenesis, as well as in the initiation and development of human diseases, including cancer (29). miRNA clusters possess a more complex regulatory network, associated with expression, compared with the regulation of individual miRNAs. The miRNA members of certain gene clusters are functionally associated and may target or regulate identical genes or the same gene family, or target various protein components of the same signaling pathway (30). The sequences of members in the miR-17-92 gene cluster are highly conserved across vertebrates, and the miR-17-92 gene cluster has received extensive attention as has been suggested to be involved in mammalian organ development, which is a function thought to be closely associated with the initiation of solid tumor development (24). Members of the miR-17-92 gene cluster are expressed at high levels in numerous tumor cells, including those of B-cell lymphoma, as well as lung, liver, bladder, colon, stomach and pancreatic cancer. Furthermore, members of the miR-17-92 gene cluster are thought to induce tumorigenesis mainly by inhibiting the expression of genes that regulate oncogenes and the cell cycle (31).
The unique distribution and expression pattern of miRNA gene clusters has suggested that they may possess important regulatory functions. As numerous miRNAs are distributed in clusters, the majority of these gene clusters are transcribed in a single transcript and co-regulate a number of biological processes. Thus, the investigation of miRNA gene clusters, rather than the study of the expression of single miRNAs, may present a novel research strategy. As high-throughput sequencing technology has been widely used in miRNA research, it is possible that studies regarding the expression and function of gene clusters will further improve miRNA research.
In the present study, it was revealed that the hsa-miR-17 cluster exhibited an inconsistent pattern of expression, and it was also identified that the expression of the miR-200 family was significantly upregulated, particularly the expression of miR-141, in the HEC-1B cell line. These findings not only provide novel potential biomarkers for differentiating between type I and type II endometrial cancer, but also reveal the miRNA alterations associated with the underlying mechanisms of endometrial cancer.
Following a review of previous literature regarding endometrial cancer and miRNA expression (Table II), it was also identified that expression of the miR-200 family was significantly increased in endometrial cancer. It was observed that the expression levels of hsa-miR-200a, hsa-miR-429 and hsa-miR-141 in the miR-200 family were significantly increased in the HEC-1B cell line compared with the ISK cell line. This finding was also verified by sequencing (Table II).
Table II.
Comparison of upregulated miRNAs identified in previous studies with the results of the present study.
| Study | Increased miRNA expression | Tissue | Technique | Ref |
|---|---|---|---|---|
| Chung et al | miR-103, miR-106a, miR-107, miR-10a, miR-130b, miR-141, miR-151, miR-155, miR-17-5p, miR-182, miR-183, miR-184, miR-191, miR-194, miR-200a, miR-200c | EEC compared with normal endometrium | RT-PCR | (11) |
| Snowdon et al | miR-141, miR-146a, miR-18a, miR-200a, miR-200b, miR-200c, miR-203, miR-205, miR-210, miR-421, miR-429 | EEC compared with normal endometrium | Microarray & RT-PCR | (12) |
| Present study | miR-141, miR-200a, miR-92a, let-7b, miR-429, miR-125a miR-25, miR-124, miR-185 miR-29a, miR-34c, miR-519a | HEC cell line compared with ISK cell line | Deep sequencing |
miRNA/miR, microRNA; RT-PCR, reverse transcription-polymerase chain reaction; EEC, endometrial epithelial cells; ISK, Ishikawa.
Notably, the miR-200 family possesses the potential to be used as a biomarker for differentiating between the HEC-1B and ISK cell lines, and may also be closely associated with the distinct pathogenic mechanisms of these two cell types. A previous study demonstrated that the expression of the miR-200 family may be highly correlated with the epithelial phenotype of cancer cells (32). The study used two markers to classify 60 types of cell line into two groups: One group consisted of cells with an epithelial phenotype (marked by E-cadherin) and the other consisted of cells with a mesenchymal phenotype (indicated by vimentin). This previous study revealed that the expression of miR-200 family members was positively correlated with an epithelial phenotype and that these miRNAs demonstrated high expression levels in epithelial-type tumor cells. Recent studies also demonstrated that the overexpression of miR-200 family members in mouse breast cancer cell lines stimulated macroscopic metastases (33,34). Based on the characteristics of the HEC-1B and ISK cell lines, the miR-200 family may be used as a biomarker for differentiating between endometrial cancer subtypes. In addition, miR-200 family members that were expressed at increased levels in HEC-1B cells, particularly miR-141, may also be involved in myometrial invasion and lymph node metastasis (33).
As the function and targets of numerous miRNAs remain unclear, the alteration of cell phenotypes due to the differential expression of miRNAs remains to be fully elucidated. Thus, further research may be required in this area. The roles of the miR-17-92 cluster and the miR-200 family in the regulation of signaling pathways in the HEC-1B and ISK cell lines remain to be elucidated by further experiments.
Furthermore, based on high-throughput sequencing data, the protocol for how to combine the characteristics of miRNAs that are distributed throughout the genome in clusters remains to be elucidated. However, the analysis of the expression profiles of specific types of miRNAs, including sense/antisense miRNAs located in specific positions in the genome or miRNAs possessing homologous sequences remains a notable direction for future research.
The present study identified potential novel biomarkers as certain representative members of hsa-mir-17 cluster and the miR-200 family: has-miR-20a, has-miR-18a, hsa-miR-19a, has-miR-20a in the hsa-mir-17 cluster and hsa-miR-429, hsa-miR-141, hsa-miR-200a in the miR-200 family.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 61271055).
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2006;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 4.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 5.Treeck O, Diedrich K, Ortmann O. The activation of an extracellular signal-regulated kinase by oestradiol interferes with the effects of trastuzumab on HER2 signalling in endometrial adenocarcinoma cell lines. Eur J Cancer. 2003;39:1302–1309. doi: 10.1016/S0959-8049(02)00777-3. [DOI] [PubMed] [Google Scholar]
- 6.Boggess JF, Zhou C, Bae-Jump VL, Gehrig PA, Whang YE. Estrogen-receptor-dependent regulation of telomerase activity in human endometrial cancer cell lines. Gynecol Oncol. 2006;103:417–424. doi: 10.1016/j.ygyno.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 7.Doll A, Abal M, Rigau M, Monge M, Gonzalez M, Demajo S, Reventos J. Novel molecular profiles of endometrial cancer-new light through old windows. J Steroid Biochem Mol Biol. 2008;108:221–229. doi: 10.1016/j.jsbmb.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Gründker C, Günthert AR, Emons G. Hormonal heterogeneity of endometrial cancer. Adv Exp Med Biol. 2008;630:166–188. doi: 10.1007/978-0-387-78818-0_11. [DOI] [PubMed] [Google Scholar]
- 9.Uharcek P. Prognostic factors in endometrial carcinoma. J Obstet Gynaecol Res. 2008;34:776–783. doi: 10.1111/j.1447-0756.2008.00796.x. [DOI] [PubMed] [Google Scholar]
- 10.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung TK, Cheung TH, Huen NY, Wong KW, Lo KW, Yim SF, Siu NS, Wong YM, Tsang PT, Pang MW, et al. Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int J Cancer. 2009;124:1358–1365. doi: 10.1002/ijc.24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snowdon J, Zhang X, Childs T, Tron VA, Feilotter H. The microRNA-200 family is upregulated in endometrial carcinoma. PLoS One. 2011;6:e22828. doi: 10.1371/journal.pone.0022828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo L, Yang Q, Lu J, Li H, Ge Q, Gu W, Bai Y, Lu Z. A comprehensive survey of miRNA repertoire and 3′ addition events in the placentas of patients with pre-eclampsia from high-throughput sequencing. PLoS One. 2011;6:e21072. doi: 10.1371/journal.pone.0021072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coppée JY. Do DNA microarrays have their future behind them? Microbes Infect. 2008;10:1067–1071. doi: 10.1016/j.micinf.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu M, Shi B, Wang J, Cao Q, Cui Q. TAM: A method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinformatics. 2010;11:419. doi: 10.1186/1471-2105-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhat KP, Pezzuto JM. Resveratrol exhibits cytostatic and antiestrogenic properties with human endometrial adenocarcinoma (Ishikawa) cells. Cancer Research. 2001;61:6137–6144. [PubMed] [Google Scholar]
- 19.Guo L, Sun B, Sang F, Wang W, Lu Z. Haplotype distribution and evolutionary pattern of miR-17 and miR-124 families based on population analysis. PLoS One. 2009;4:e7944. doi: 10.1371/journal.pone.0007944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo L, Yang S, Zhao Y, Zhang H, Wu Q, Chen F. Global analysis of miRNA gene clusters and gene families reveals dynamic and coordinated expression. Biomed Res Int. 2014;2014:782490. doi: 10.1155/2014/782490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/S1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, Zhang JW. Human microRNA clusters: Genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashita YI, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Rese. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 25.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: from functions to targets. PLoS One. 2010;5:e13067. doi: 10.1371/journal.pone.0013067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altuvia YI, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;3:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Wong C. A computational screen for mouse signaling pathways targeted by microRNA clusters. RNA. 2008;14:1276–1283. doi: 10.1261/rna.997708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ventura AI, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory PAI, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 33.Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pecot CVI, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]