Abstract
Autophagy, which is a mechanism for the turnover of intracellular molecules and organelles, protects cells during stress responses; however, the role of autophagy in the stages of bone fracture remains to be elucidated. The aim of the present study was to investigate the process of autophagy in bone tissue at different time-points after fracture. A femur fracture model was established in male adult Wistar rats via surgery. The protein expression of microtubule-associated protein II light chain 3 (LC3-II) was analyzed in a femur fracture (experimental) group and a sham-surgery group using immunofluorescence. The protein expression of proliferating cell nuclear antigen (PCNA) was used to investigate the cell proliferation in bone tissue following fracture via immunohistochemical analysis. The correlation between cell proliferation and autophagy was analyzed using linear regression. LC3-II protein was constitutively expressed in the sham-surgery group; however, compared with the expression in the sham-surgery group, the LC3-II expression in the experimental group was significantly increased at each time-point (P<0.05). Similarly, immunohistochemistry revealed that the number of PCNA-positive cells in each section was significantly increased following fracture injury (P<0.01). A comparison of the LC3-II- and PCNA-positive rates in the experimental group rats at each time-point revealed a linear correlation (R2=0.43, P<0.01). In conclusion, surgically induced fracture in rats is associated with an increase in LC3-II and PCNA protein expression during the initial stages of fracture injury, and a correlation exists between the expression of the two proteins. These results suggest that potential treatment aimed at improving fracture healing should target the process of autophagy.
Keywords: autophagy, rat, bone fracture
Introduction
Bone fracture can initiate a series of bio-physiological and pathological reactions, and a variety of biomechanical and biological factors that determine the outcome of fracture healing have been identified during past decades (1,2). Numerous studies have indicated that autophagy plays an important role in nutrient and energy regulation, as well as in the removal of dysfunctional or damaged organelles and molecules (3,4). Bone fracture can disrupt the cellular homeostasis and place the bone cells under considerable stress, which can result in the activation of autophagy. The involvement of autophagy in neurodegeneration, cardiomyopathies and abnormal skeletal development has been investigated in previous studies (5–7); however, the role of autophagy in the various stages of bone fracture remains to be studied.
Autophagy is a common physiological mechanism that targets altered and dysfunctional cytosolic macromolecules, membranes and organelles for delivery to lysosomes for degradation and recycling (8,9). The process of autophagy is activated when there is a shortage of energy or nutrients. At the beginning of the process, an autophagosome is created via a double membrane that forms around cellular substances, which then fuses with a lysosome. The organelles and proteins in cells are degraded to amino and fatty acids by the formed lysosome, to enable cell survival (10). Previous studies have demonstrated that autophagy is associated with the progression of numerous diseases, such as heart disease (11), and is involved in tumor suppression (12) and the removal of toxic agents (13). The role of autophagy has additionally been investigated in the field of bone science, including in the pathogenesis of osteoporosis (14) and Paget's disease (15).
Microtubule-associated protein II light chain 3 (LC3-II) is one of the major regulators of the autophagy pathway. The protein binds to the membrane of the autophagosome, and the extent of autophagosome formation is correlated with the level of LC3-II formation (16). Thus, the upregulation of LC3-II is often used to illustrate the activation of autophagy in disease models (17).
In a regenerating tissue, cell proliferation plays an important role in tissue amplification, particularly in physiological and pathological processes (18). In a previous study, Lee et al (19) quantified the proliferating cells in each of the cellular events occurring during bone fracture healing. The degree of cell proliferation varied according to the length of time that had passed since the fracture, suggesting the existence of local regulatory factors, such as growth factors and cytokines (20). Proliferating cell nuclear antigen (PCNA) is a crucial protein for the proliferation of osteoblasts, its expression and cell proliferation cycle are closely associated, and thus accurately reflect the cell proliferation. The aim of the present study, therefore, was to investigate the role of autophagy in the bone following fracture and describe the association between cell proliferation and autophagy, in order to identify a potential therapeutic target to improve fracture healing. The protein expression of PCNA was used to investigate the cell proliferation in bone tissue following fracture via immunohistochemical analysis.
Materials and methods
Experimental groups and surgical procedure
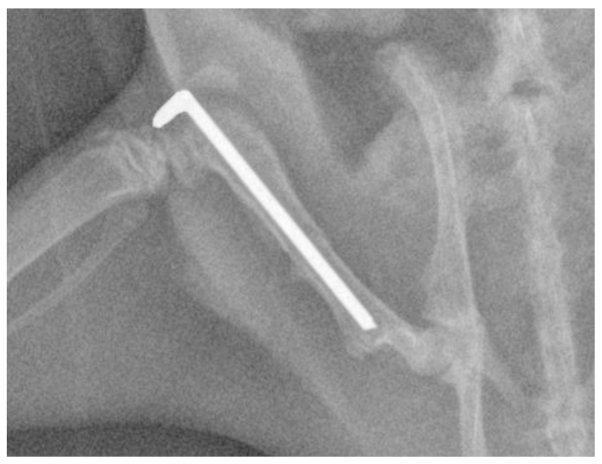
The present study was performed according to protocols approved by the local governmental Animal Care Committee and the institutional Animal Care and Use Committee at Xiamen University (Xiamen, China). Every effort was made to minimize animal suffering and to reduce the number of animals used. In total, 36 adult male Wistar rats weighing 230–260 g were obtained from the Experimental Animal Center, the Affiliated Southeast Hospital of Xiamen University (SCZZ(min)2012-0203). The rats were randomly separated into six groups (n=6/group): Five experimental groups (for examination at different time-points) and one sham-surgery group (as a control group). The rats were housed with 3 or 4 rats to a cage, 1 week before the experiments began. For the experimental procedure, in brief, the rats were anesthetized using an intraperitoneal injection of ketamine (75 mg/kg) and xylazine (25 mg/kg), and a lateral incision was then made through the shaved skin and fascia lata from the right knee to the greater trochanter. The plane between the vasti and hamstrings was opened through blunt dissection to expose the femur. The right femur of each animal was fractured using a 3-point bending device and stabilized using Kirschner wire (diameter, 1.0 mm; Shanghai Pudong Jinhuan Medical Products Co., Ltd., Pudong, Shanghai, China), as described previously (21). The fascia lata and skin were closed with polyglactin absorbable sutures (Shanghai Pudong Jinhuan Medical Products Co., Ltd.). The fracture configuration was a closed, midshaft fracture type A2-A3, according to AO classification (22), and the implant positions were documented using X-rays (Fig. 1). The sham-surgery group underwent an identical procedure without fracture.
Figure 1.
Representative X-ray of a rat femur following open fracture and stabilization by a Kirschner wire measuring 1.0 mm in diameter.
The experimental group samples were extracted near the fracture site at 6 h, 12 h, 24 h, 3 days and 7 days after fracture. The samples were then fixed in 4% formalin for 24 h and decalcified in 10% EDTA solution for 5 weeks. A total of 10 slices per animal (the results from which were averaged) were randomly selected for paraffin embedding and used for quantitative immunofluorescence and immunohistochemical analyses.
Immunofluorescence
Paraffin-embedded samples for immunofluorescence were deparaffinized in the xylene substitute Pro-Par Clearant (Anatech, Ltd., Battle Creek, MI, USA) and rehydrated in graded ethanol and water. After washing with phosphate-buffered saline (PBS), the sections (5-µm) were blocked with 5% goat serum for 1 h at room temperature and then incubated at 4°C with rabbit polyclonal anti-LC3II (1:50; NB910-40435; Sigma-Aldrich, St. Louis, MO, USA) overnight. After further washing with PBS, the sections were incubated with the Alexa Fluor® 488 anti-rabbit IgG secondary antibody (Abcam, Cambridge, MA, USA) for 30 min. Finally, the sections were washed and observed using fluorescence microscopy
Immunohistochemistry
For immunohistochemistry, 6-µm sections from paraffin-embedded samples were deparaffinized and rehydrated. For antigen unmasking, the sections were immersed in 10 mM sodium citrate buffer (pH 6.0), boiled in a microwave oven and kept at 92–95°C for 2 min. Following antigen unmasking, the slides were cooled for 20 min at room temperature and then washed with PBS. The sections were subsequently blocked with 0.03% hydrogen peroxide sodium azide (supplied by the Affiliated Southeast Hospital of Xiamen University) for 5 min at room temperature. Purified rabbit anti-rat PCNA monoclonal antibody (1:100; GTX12496; Dako North America, Inc., Carpinteria, CA, USA) was incubated for 45 min at room temperature. After washing with PBS, the secondary purified goat anti-rabbit/mouse biotinylated antibody (1:100; Dako North America, Inc.) was applied to the sections and incubated for 30 min at room temperature. The sections were then washed gently with PBS and incubated with streptavidin-peroxidase (1:50; Dako North America, Inc.) for 30 min at room temperature. Diaminobenzidine substrate chromagen was added to the sections, which were subsequently incubated at room temperature for >5 min and washed. The tissue sections were counterstained with hematoxylin (Sigma-Aldrich) for 5 min, washed with distilled water and dehydrated through a series of graded ethanol solutions to xylene, mounted in DPX (Olympus Corporation, Tokyo, Japan) and examined using light microscopy.
Statistical analysis
The percentage of PCNA-positive cells and the number of the cells with punctuate LC3-II fluorescence were calculated in 10 randomly selected slices per animal and averaged, and the results were then quantitatively analyzed using Image-Pro Plus software (Media Cybernetics, Inc., Silver Spring, MD, USA). Results are presented as the mean ± standard deviation. Significant differences between animals at each time-point were assessed using analysis of variance, and correlation analysis was conducted using linear regression. Statistical analysis was performed using SPSS software for Windows (version 13.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
LC3-II is upregulated in bone tissue following fracture injury
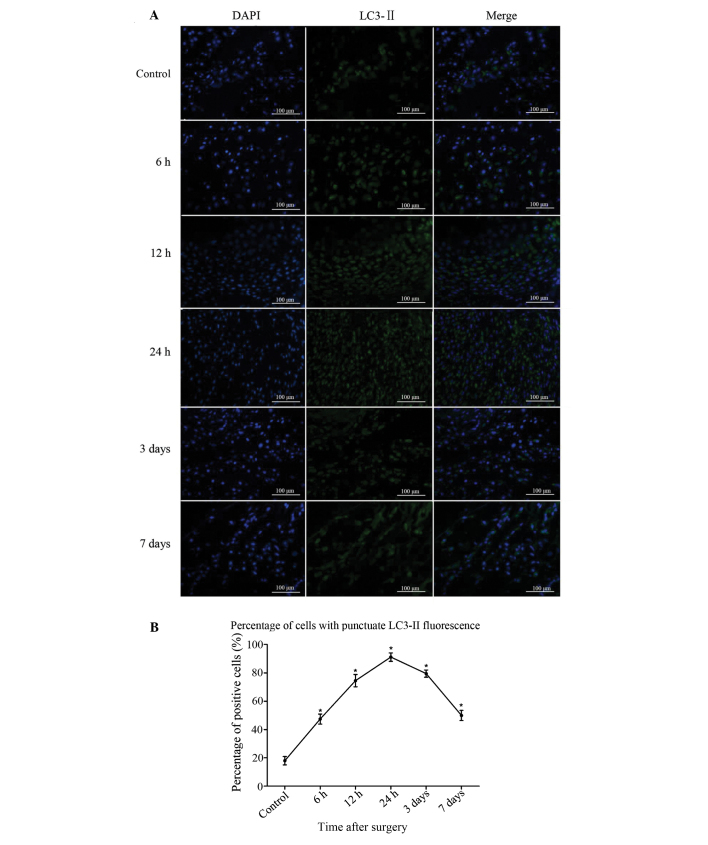
LC3-II-positive cells were sparse in the bone of the sham-surgery group. At 6 h after injury, the number of LC3-II-positive cells began to increase, peaked at 24 h and then reduced gradually from 3 days after bone fracture (Fig. 2A). The difference between the experimental and sham-surgery groups was significant at all time-points (P<0.05). Significant differences were also found among the data from the 5 experimental groups (P<0.05). The results demonstrate that the process of autophagy was induced in the bone tissue following fracture injury (Fig. 2B).
Figure 2.
Immunofluorescence staining of LC3-II in the bone tissue near the fracture site. (A) In the control group, LC3-II-positive cells were sparse; in the fracture group, the population of LC3-II-positive cells increased after 6 h, peaked at 24 h and then gradually decreased at 3 days. (B) Analysis of the percentage of cells in fracture-site tissue with punctuate LC3-II fluorescence. Results are presented as the mean ± standard deviation. *P<0.05 vs. control. Scale bar, 100 µm; magnification, ×400. LC3-II, microtubule-associated protein II light chain 3.
An increase in the number of PCNA-positive cells is observed in bones from rats with surgically induced fracture
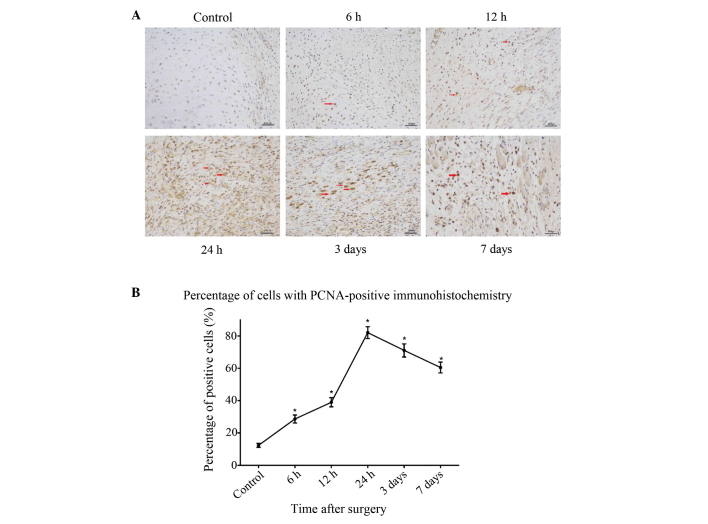
To further investigate the association between autophagy and cell proliferation following fracture injury, immunohistochemical staining for the cell proliferation marker PCNA was performed on the same samples that were analyzed for the autophagy marker in the preceding experiments. At 6 h after fracture, an increased number of PCNA-positive cells was observed compared with the sham-surgery group, and this increase lasted for 24 h after the surgery (P<0.05) (Fig. 3). After 3 days, the pathological changes began to decrease, which coincided with the recovery of blood flow and nutrition.
Figure 3.
Immunohistochemical staining of PCNA in controls and at each time-point after fracture injury. (A) In the control group, PCNA-positive cells were constitutively expressed; in the fracture group, the population of PCNA-positive cells increased after 6 h, peaked at 24 h and then gradually decreased at 3 days, as a result of the recovery of blood flow and nutrition. Arrows show the PCNA-positive cells. (B) Analysis of the percentage of cells in fracture-site tissue with PCNA-positive innmunohistochemistry. Results are presented as the mean ± standard deviation. *P<0.05 vs. control. Scale bar, 400 µm; magnification, ×200. PCNA, proliferating cell nuclear antigen.
Correlation between the number of LC3-II- and PCNA-positive cells
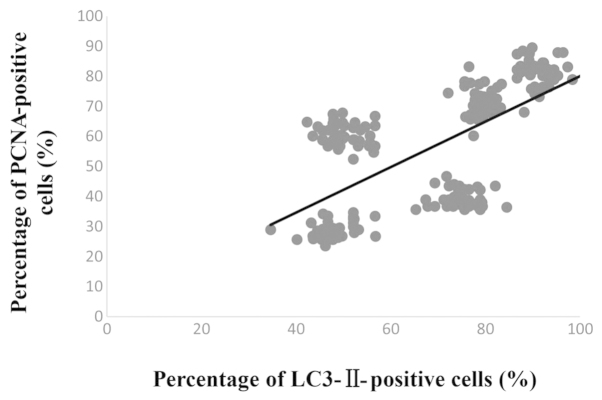
Simple linear regression was used to analyze the correlation between the number of LC3-II- and PCNA-positive cells. The result showed that a significant correlation existed between the degree of LC3-II and PCNA expression following fracture injury (R2=0.43, P<0.01) (Fig. 4). These findings demonstrate that surgically induced femur fracture is associated with the activation of autophagy (LC3-II expression) along with an increase in cell proliferation (PCNA expression).
Figure 4.
A significant correlation between the percentage of LC3-II- and PCNA-positive cells was observed following fracture injury (R2=0.43, P<0.01). LC3-II, microtubule-associated protein II light chain 3; PCNA, proliferating cell nuclear antigen.
Discussion
In the present study, the occurrence of certain cellular events, i.e. autophagy and proliferation, was analyzed in the various phases of bone fracture healing following internal fixation in a rat femoral fracture model. Autophagy, type II programmed cell death, is a cellular mechanism of self-cycling for survival in eukaryotes. The study of autophagy is an emerging field within bone research. To investigate the degree of autophagy in bone tissue following fracture injury, immunofluorescence was used with an LC3-II polyclonal antibody, a method that has already been described and used previously (23). In the present study, the protein expression of LC3-II increased in the bone tissue following fracture, peaking at 24 h, and then exhibited a gradual decline from 3 days. Significant differences were observed at each time-point (P<0.05). A few LC3-II-positive cells were additionally observed in the sham-surgery group, indicating that autophagy exists within a narrow homeostatic range and in delicate balance with other cellular systems that regulate protein homeostasis and cell survival. This theory has been described in a previous study (24). Once the balance is disrupted, autophagy can be induced as a survival mechanism in normal cells in response to hypoxic conditions or stress (25). There is growing evidence that energy status and nutrient levels are key modulators of autophagy, and autophagy is defined as a catabolic, energy-generating mechanism for the cell (26,27). In the present study it was hypothesized that autophagy would be activated in bone tissue following fracture injury due to the sudden reduction or interruption of the nutrient supply.
Cell proliferation appears to be a major cellular response at the beginning of a cellular event, such as inflammation, chondrogenesis and endochondral or intramembranous ossification. PCNA is a key marker for bone cell proliferation and plays an important role in both pathological and physiological activity (28). In the present study, PCNA-positive cells were detected at 6 h near the fracture site, concurring with the findings of Iwaki et al (29), which suggested that fracture healing began with cell proliferation. In addition, the present results showed that the percentage of PCNA- and LC3-II-positive cells exhibited similar trends in variation following fracture injury at each experimental time-point. Linear regression analysis demonstrated that a significant correlation was present between PCNA and LC3-II protein expression following fracture injury (R2=0.43, P<0.01). In a previous study, Morrow et al (30) demonstrated that rapamycin caused the activation of the nuclear translocation of PCNA in CD4+ T cells. It has been reported that one of the key suppressors of autophagy is mammalian target of rapamycin (mTOR); rapamycin exerts significant inhibitory effects on mTOR, with no off-target effects on other enzymes (31,32). Based on the findings of Morrow et al (30), we propose that the activation of the nuclear translocation of PCNA may be associated with the induction of autophagy; however, the potential mechanism underlying the association between the process of autophagy and PCNA expression has not yet been elucidated in the context of fracture injury and healing, although it may involve the self-regulation pathway of the cell, among others. An inhibitor or enhancer of autophagy, such as 3-methyladenine or rapamycin, respectively, should be used in further studies to attempt to illustrate the specific role of autophagy in bone following fracture injury.
In conclusion, this is the first study, to the best of our knowledge, to report the activation of autophagy in bone tissue following fracture. This phenomenon may indicate that autophagy is associated with the occurrence of trauma and could be used as a target to improve bone fracture healing. Future studies should investigate the role of autophagy in bone fracture, in order to facilitate the development of treatments aimed at improving fracture healing.
References
- 1.Megas P. Classification of non-union. Injury. 2005;36(Suppl 4):S30–S37. doi: 10.1016/j.injury.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Rueff-Barroso CR, Milagres D, Valle J, Casimiro-Lopes G, Nogueira-Neto JF, Zanier JF, Porto LC. Bone healing in rats submitted to weight-bearing and non-weight-bearing exercises. Med Sci Monit. 2008;14:BR231–BR236. [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y, Tang C, He H, Duan R. Activation of mTOR ameliorates fragile X premutation rCGG repeat-mediated neurodegeneration. PloS One. 2013;8:e62572. doi: 10.1371/journal.pone.0062572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su H, Li J, Osinska H, Li F, Robbins J, Liu J, Wei N, Wang X. The COP9 signalosome is required for autophagy, proteasome-mediated proteolysis, and cardiomyocyte survival in adult mice. Circ Heart Fail. 2013;6:1049–1057. doi: 10.1161/CIRCHEARTFAILURE.113.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caramés B, Hasegaa A, Taniguchi N, Miyaki S, Blanco FJ, Lotz M. Autophagy activation by rapamycin reduces severity of experimental osteoarthritis. Ann Rheum Dis. 2012;71:575–581. doi: 10.1136/annrheumdis-2011-200557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vellai T. Autophagy genes and ageing. Cell Death Differ. 2009;16:94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- 9.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorburn A. Apoptosis and autophagy: Regulatory connections between two supposedly different processes. Apoptosis. 2008;13:1–9. doi: 10.1007/s10495-007-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao XH, Naomoto Y, Hao HF, Watanabe N, Sakurama K, Noma K, Motoki T, Tomono Y, Fukazawa T, Shirakawa Y, et al. Autophagy: Can it become a potential therapeutic target? Int J Mol Med. 2010;25:493–503. doi: 10.3892/ijmm_00000369. [DOI] [PubMed] [Google Scholar]
- 12.Beau I, Mehrpour M, Codogno P. Autophagosomes and human diseases. Int J Biochem Cell Biol. 2011;43:460–464. doi: 10.1016/j.biocel.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Mijaljica D, Prescott M, Devenish RJ. Autophagy in disease. Methods Mol Biol. 2010;648:79–92. doi: 10.1007/978-1-60761-756-3_5. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Guo YF, Liu YZ, Liu YJ, Xiong DH, Liu XG, Wang L, Yang TL, Lei SF, Guo Y, et al. Pathway-based genome-wide association analysis identified the importance of regulation-of-autophagy pathway for ultradistal radius BMD. J Bone Miner Res. 2010;25:1572–1580. doi: 10.1002/jbmr.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daroszewska A, van't Hof RJ, Rojas JA, Layfield R, Landao-Basonga E, Rose L, Rose K, Ralston SH. A point mutation in the ubiquitin-associated domain of SQSMT1 is sufficient to cause a Paget's disease-like disorder in mice. Hum Mol Genet. 2011;20:2734–2744. doi: 10.1093/hmg/ddr172. [DOI] [PubMed] [Google Scholar]
- 16.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su JC, Tseng PH, Hsu CY, Tai WT, Huang JW, Ko CH, Lin MW, Liu CY, Chen KF, Shiau CW. RFX1-dependent activation of SHP-1 induces autophagy by a novel obatoclax derivative in hepatocellular carcinoma cells. Oncotarget. 2014;5:4909–4919. doi: 10.18632/oncotarget.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manolagas SC. Birth and death of bone cells: Basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev. 2000;21:115–137. doi: 10.1210/edrv.21.2.0395. [DOI] [PubMed] [Google Scholar]
- 19.Lee FY, Choi YN, Beherns FF, Defoun DO, Einhorn TA. Programmed removal of chondrocytes during endochondral fracture healing. J Orthop Res. 1998;16:144–150. doi: 10.1002/jor.1100160124. [DOI] [PubMed] [Google Scholar]
- 20.Einhorn TA. The cell and molecular biology of fracture dealing. Clin Orthop Relat Res. 1998;(355 Suppl):S7–S21. doi: 10.1097/00003086-199810001-00003. [DOI] [PubMed] [Google Scholar]
- 21.Holstein JH, Menger MD, Culemann U, Meier C, Pohlemann T. Development of a locking femur nail for mice. J Biomech. 2007;40:215–219. doi: 10.1016/j.jbiomech.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 22.Müller E, Koch P, Nazarian S, Schatzker J, editors. The Comprehensive Classification of Fractures of Long Bones. 1st. New York, NY: Springer; 1994. [Google Scholar]
- 23.Hou H, Zhang L, Zhang L, Liu D, Xiong Q, Du H, Tang P. Acute spinal cord injury could cause activation of autophagy in dorsal root ganglia. Spinal Cord. 2013;51:679–682. doi: 10.1038/sc.2013.52. [DOI] [PubMed] [Google Scholar]
- 24.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dosenko VE, Nagibin VS, Tumanovska LV, Moibenko AA. Protective effect of autophagy in anoxia-reoxygenation of isolated cardiomyocyte? Autophagy. 2006;2:305–306. doi: 10.4161/auto.2946. [DOI] [PubMed] [Google Scholar]
- 26.Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174–176. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- 27.Quan W, Jung HS, Lee MS. Role of autophagy in the progression from obesity to diabetes and in the control of energy balance. Arch Pharm Res. 2013;36:223–229. doi: 10.1007/s12272-013-0024-7. [DOI] [PubMed] [Google Scholar]
- 28.Barnouti ZP, Owtad P, Shen G, Petocz P, Darendeliler MA. The biological mechanisms of PCNA and BMP in TMJ adaptive remodeling. Angle Orthod. 2011;81:91–99. doi: 10.2319/091609-522.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwaki A, Jingushi S, Oda Y, Izumi T, Shida JI, Tsuneyoshi M, Sugioka Y. Localization and quantification of proliferating cells during rat fracture repair: Detection of proliferating cell nuclear antigen by immunohistochemistry. J Bone Miner Res. 1997;12:96–102. doi: 10.1359/jbmr.1997.12.1.96. [DOI] [PubMed] [Google Scholar]
- 30.Morrow PW, Tung HY, Hemmings HC., Jr Rapamycin causes activation of protein phosphatase-2A1 and nuclear translocation of PCNA in CD4+ T cells. Biochem Biophys Res Commun. 2004;323:645–651. doi: 10.1016/j.bbrc.2004.08.147. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi S, Kishimoto T, Kamata S, Otsuka M, Miyazaki M, Ishikura H. Rapamycin, a specific inhibitor of the mammalian target of rapamycin, suppresses lymphangiogenesis and lymphatic metastasis. Cancer Sci. 2007;98:726–733. doi: 10.1111/j.1349-7006.2007.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawahara T, Asthana S, Kneteman NM. m-TOR inhibitors: What role in liver transplantation? J Hepatol. 2011;55:1441–1451. doi: 10.1016/j.jhep.2011.06.015. [DOI] [PubMed] [Google Scholar]