Abstract
Caspase-8 (CASP8) is a key controller of apoptosis, and its deregulation is crucially involved in carcinogenesis. The aim of the present study was to evaluate the function of CASP8 polymorphisms in oral squamous carcinoma (OSCC) by evaluating the risk associated with three single-nucleotide polymorphisms (SNPs) in a case-control study in a Han Chinese patient population. A total of 505 individuals with clinically diagnosed OSCC and 507 healthy controls were tested for the three SNPs rs3834129, rs13016963 and rs1045485, using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and DNA sequencing analysis. After adjusting for other confounders, the genotype frequencies of CASP8 −652 6N ins/del promoter polymorphism (rs3834129) were found to be lower in patients with OSCC compared with normal subjects. No significant difference was detected in the genotype frequencies of rs13016963 between the patients and control subjects. However, the AA genotype frequency of rs1306963 was associated with OSCC as a risk factor among non-smokers and non-drinkers. For CASP8, rs1045485 was not present in any of the patients with OSCC or control subjects. These results suggest that the del allele of rs3834129 may play a protective role in the tumorigenesis of OSCC and may be useful as a genetic susceptibility marker for OSCC in the population studied.
Keywords: oral squamous carcinoma, caspase-8, single nucleotide polymorphisms, Chinese population
Introduction
Previous studies indicate that oral squamous cell carcinoma (OSCC) is the eighth most prevalent type of cancer worldwide (1), in men and women (2). Despite considerable investigation and novel therapeutic developments, the 5-year survival rate associated with OSCC is ~50%, which is in part due to the lack of effective biomarkers for early diagnosis and optimal treatment (1,3). Therefore, OSCC is a significant public health issue worldwide (4). A significant number of genetic single-nucleotide polymorphisms (SNPs) have been associated with OSCC (5–8); however, the contribution of these gene interactions in the development of OSCC remains unclear. Therefore, the identification of novel and effective biomarkers is necessary to improve the prognosis of OSCC.
Apoptosis, or programmed cell death, is a crucial mechanism against hyperproliferation and malignancy (9). Apoptosis is predominantly mediated by enzymes known as caspases (9,10). Caspase-8 (CASP8) is a key regulator of apoptosis in T lymphocytes and is encoded by the CASP8 gene. However, a previous study has suggested that CASP8 additionally serves certain non-apoptotic functions in cells, such as promoting activation nuclear factor (NF)-κB signaling, regulating autophagy, altering endosomal trafficking and enhancing cellular adhesion and migration (11). The human CASP8 gene contains at least 11 exons, spanning 30 kb on the highly polymorphic chromosome 2q33–34 (12,13). In addition to rare mutations, a number of common variants of the CASP8 gene disrupt apoptosis and enhance the risk of developing various types of cancer, including breast cancer (14,15), colorectal cancer (16), ovarian cancer (17), prostate cancer (18) and others (19,20). The ins/del polymorphism rs3834129 in the promoter region of the CASP8 gene may block the stimulatory protein 1 (Sp1) binding site, and has been associated with reduced susceptibility to various types of cancer, including lung, esophageal, gastric, colorectal, cervical and breast cancers in Chinese individuals (21). Barrett et al (22) conducted a genome-wide association study (GWAS) among European populations and identified three novel melanoma susceptibility loci, including an SNP adjacent to CASP8 (rs13016963; P=8.6×10−10). In addition, a GWAS study revealed that rs13016963 is a significant susceptibility locus for esophageal squamous cell carcinoma in Chinese individuals (23). The SNP rs1045485 (Asp302His), which is located in the CASP8 gene, has been investigated in previous studies, and the results suggest that this locus is associated with breast cancer and prostate carcinoma (18,24).
The aim of the present study was to determine whether the CASP8 −652 6 N ins/del polymorphisms rs1045485 (Asp302His) and rs13016963 are associated with an increased risk of OSCC. To test this hypothesis, the polymorphisms were genotyped and their associations with OSCC risk were assessed in a hospital-based, case-control study of Chinese patients.
Materials and methods
Patients and controls
The study population included 505 Han Chinese patients with clinically diagnosed OSCC, that were recruited at the West China Hospital of Stomatology, Sichuan University (Chengdu, China) between December 2009 and January 2013, in addition to 507 healthy Han Chinese that visited the general health check-up division of the West China Hospital of Stomatology, Sichuan University. OSCC was diagnosed according to the 1997 World Health Organization criteria, and clinical stage (Tumor Node Metastasis) was determined according to the 2002 American Joint Committee on Cancer criteria (25,26).
All subjects were informed of the detailed study protocol, required to sign consent forms and instructed to complete a standardized questionnaire that was conducted by two trained interviewers. The detailed standardized questionnaire completed by the patients was described previously (6). The ethics committee of Sichuan University and all of the participating patients signed written consent forms prior to the collection of samples. The characteristics of the case group and control group are shown in Table I and clinicopathological characteristics of the case group are detailed in Table II.
Table I.
Characteristics of case group patients and control group subjects.
| Characteristic | Case, n=505 | Control, n=507 | P-valuea |
|---|---|---|---|
| Age | |||
| Mean ± SD, years | 60.01±11.67 | 58.95±12.71 | 0.166 |
| ≤45 years, n | 77 | 56 | 0.091 |
| 45<age≤65 years, n | 264 | 292 | |
| >65 years, n | 164 | 159 | |
| Gender, n (%) | |||
| Male | 330 (65.3) | 348 (68.6) | |
| Female | 175 (34.7) | 159 (31.4) | 0.265 |
| Smoking, n (%) | |||
| Yes | 304 (60.2) | 165 (32.5) | |
| No | 201 (39.8) | 342 (67.5) | <0.001 |
| Drinking, n (%) | |||
| Yes | 275 (54.5) | 209 (41.2) | |
| No | 230 (45.5) | 298 (58.8) | <0.001 |
Student's t-test or χ2 test. SD, standard deviation.
Table II.
Clinicopathological features of the patients in the case group (n=505).
| Characteristic | Case, n (%) |
|---|---|
| Site | |
| Tongue | 161 (31.9) |
| Buccal mucosa | 131 (25.9) |
| Gingiva | 84 (6.6) |
| Floor of mouth | 61 (12.1) |
| Palate | 35 (6.9) |
| Lip | 24 (4.8) |
| Maxillary sinus | 9 (1.8) |
| Tumor size | |
| T1+T2 | 304 (60.2) |
| T3+T4 | 201 (39.8) |
| Lymph node metastases | |
| N0 | 332 (65.7) |
| N+ | 173 (34.3) |
| Clinical stage | |
| Stage I + II | 244 (48.3) |
| Stage III + IV | 261 (51.7) |
| Pathological stage | |
| SCCI (highly differentiated) | 358 (70.9) |
| SCCII (moderately differentiated) | 128 (25.3) |
| SCCIII (poorly differentiated) | 19 (3.8) |
SCC, squamous cell carcinoma.
DNA extraction and genotyping
Blood samples were extracted from all of the participants by peripheral antecubital venous puncture and stored at −20°C until required for analysis.
Genomic DNA was extracted from the samples using a previously described salting-out method (27). The DNA concentration of each individual sample was determined using a NanoDrop ND 1000 spectrophotometer and NanoDrop software, version 2.4.7c (NanoDrop Technologies Inc., Wilmington, DE, USA). The details of the genotyping method used for rs1045485 and rs3834129 are described in previous studies (28,29). The CASP8 genotype rs13016963 was determined via polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The PCR primers were designed on the basis of the National Center for Biotechnology Information dbSNP database reference sequence (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=13016963). The PCR reaction mixture consisted of 2.5 µl 10X PCR buffer, 0.1 mM/l dNTPs, 1 mM/l MgCl2, 2 U Taq polymerase [Tiangen Biotech (Beijing) Co., Ltd., Beijing China], 10 ng genomic DNA, 0.4 pM/l of each primer (forward, 5′-GTG CCG AGG CTC AGG CTA GAG GAA GGA AAC ATC CGC-3′; and reverse, 5′-TTT CCC CAC TAT TGA GGT AA-3′) and sufficient ddH2O to increase the volume to a total of 25 µl per reaction.
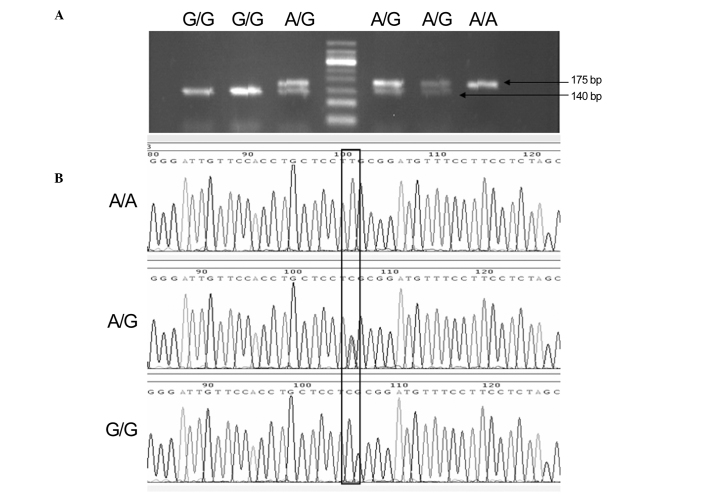
PCR products were each digested with 3 units of the specific endonuclease Bsh1236I for 2 h at 37°C. PCR and digestion products were analyzed via electrophoresis in 2% agarose gels using TAE 1X buffer in runs of 20 min at 120 V; the gels were subsequently visualized by staining with ethidium bromide and images were captured using Tanon 1600 Gel Imaging system (Tanon Science & Technology Co., Ltd., Shanghai, China). To confirm the genotyping results, Sanger sequencing was conducted by Sangon Biotech Co., Ltd. (Shanghai, China), and the results were 100% concordant. Product sizes were 175 bp for the A allele and 140/35 bp for the G allele (Fig. 1). After each genetic polymorphism had been genotyped, 10–15% of the samples in each genotype group were randomly selected for Sanger sequencing to validate the results (Fig. 1). Sanger sequencing results were consistent with those of the RFLP analysis.
Figure 1.
(A) PCR products of the rs13016963 polymorphism of the CASP8 gene as determined by restriction fragment length polymorphism and (B) Sanger sequencing. PCR, polymerase chain reaction; CASP8. caspase-8.
Statistical analysis
The genotype frequencies of rs3834129 and rs13016963 in the patients with OSCC and control subjects were compared using the χ2 test. Odds ratio (OR) with a 95% confidence interval (CI) was estimated using unconditional logistic regression. The adjusted odds ratios (AORs) with 95% CIs of the association between genotype frequencies and OSCC susceptibility were estimated using multiple logistic regression models after other covariates, such as age, gender, smoking and drinking, were controlled. The observed genotype frequencies of CASP8 polymorphisms in the control subjects were analyzed to determine deviation from Hardy-Weinberg equilibrium using Power-Stats software (Promega Corporation, Madison, WI, USA). P<0.05 was considered to indicate a statistically significant difference. SPSS software, version 11.5 (SPSS, Inc., Chicago, IL, USA) was used to perform all statistical analyses. SHEsis software (http://analysis.bio-x.cn/SHEsisMain.htm) was used to analyze linkage disequilibrium (LD) (30). Lewontin's D' (31) and r2 (32) were calculated between each pair of SNPs.
Results
Study groups
The frequency distributions of the selected characteristics of the case and control groups are presented in Table I. Statistical data indicated no significant differences in gender and age between the patient and control groups, but showed significant differences in smoking (P<0.001) and drinking (P<0.001; Table I). Increased numbers of smokers were present among the case group patients (60.2%) compared with the control group subjects (32.5%; P<0.001). In addition, more drinkers were included in the case group (54.5%) compared with the control group (41.2%; P<0.001).
Regression analysis
Multiple logistic regression analyses were conducted for rs3834129 (additive, ins/del vs. ins/ins and del/del vs. ins/ins; dominant, ins/del + del/del vs. ins/ins; recessive, del/del vs. ins/ins + ins/del) and rs13016963 (additive, AG vs. GG and AA vs. GG; dominant, AG/AA vs. GG; recessive, AA vs. GG/GA). rs1045485 was not detected in any of the patients with OSCC or control subjects. Therefore, this SNP was excluded from subsequent analyses. The AORs and 95% CIs of the genotype distributions of rs3834129 and rs13016963 associated with the susceptibility of OSCC between the case and control groups are shown in Table III. No significant association was observed between genetic polymorphisms of rs1306963 and oral cancer in additive, dominant and recessive models (Table III). However, the AA genotype frequencies of rs1306963 were significantly associated with OSCC in additive (AA vs. GG, 95% CI, 1.045–3.052) and recessive models (AA vs. GG + AG, 95% CI, 1.199–3.089) in non-smokers (Table IV). In additive and recessive models, the AA genotype was associated with a significantly higher risk of 1.834-fold (AA vs. GG, 95% CI, 1.052–3.199) and 2.127-fold (AA vs. GG/AG, 95% CI, 1.298–3.487) among non-drinkers (Table V).
Table III.
Comparison of genotype frequencies of the CASP8 gene polymorphisms between oral squamous cell carcinoma patients and control subjects.
| SNP | Model/allele | Genotype | Case (%) n=505 | Control (%) n=507 | P-valuea | OR (95% CI)b | Pac | AOR (95% CI)d |
|---|---|---|---|---|---|---|---|---|
| Rs3834129 | Additive | ii | 328 (65.0) | 276 (54.4) | 1.00 (Ref) | |||
| id | 159 (31.5) | 197 (38.9) | 0.04 | 0.679 (0.522–0.884) | 0.014 | 0.447 (0.235–0.850) | ||
| dd | 18 (3.5) | 34 (6.7) | 0.006 | 0.445 (0.246–0.806) | 0.002 | 0.640 (0.482–0.851) | ||
| Dominant | ii | 328 (65.0) | 276 (54.4) | 1.00 (Ref) | ||||
| id/dd | 177 (35.0) | 231 (45.6) | 0.001 | 0.645 (0.501–0.830) | <0.001 | 0.613 (0.467–0.806) | ||
| Recessive | ii/id | 487 (96.5) | 473 (93.3) | |||||
| dd | 18 (3.5) | 34 (6.7) | 0.024 | 0.514 (0.286–0.923) | 0.048 | 0.528 (0.281–0.993) | ||
| Rs13016963 | Additive | GG | 162 (32.1) | 164 (32.3) | 1.00 (Ref) | |||
| AG | 243 (48.1) | 264 (52.1) | 0.619 | 0.932 (0.705–1.231) | 0.765 | 0.955 (0.706–1.291) | ||
| AA | 100 (19.8) | 79 (15.6) | 0.184 | 1.281 (0.888–1.848) | 0.156 | 1.332 (0.896–1.981) | ||
| Dominant | GG | 162 (32.1) | 164 (32.3) | 1.00 (Ref) | ||||
| AG/AA | 343 (67.9) | 343 (67.7) | 0.927 | 1.012 (0.778–1.318) | 0.787 | 1.040 (0.781–1.385) | ||
| Recessive | GG/AG | 405 (80.2) | 428 (84.4) | 1.00 (Ref) | ||||
| AA | 100 (19.8) | 79 (15.6) | 0.079 | 1.338 (0. 967–1.851) | 0.077 | 1.1371 (0.967–1.945) |
P-value and
OR with 95% CIs estimated by unconditional logistic regression.
P-value and
AOR with 95% CIs estimated by multiple logistic regression models after controlling age, gender, smoking and drinking. CASP8, caspase-8; SNP, single nucleotide polymorphism; ref, reference; OR, odds ratio; AOR, adjusted odds ratio; CI, confidence interval; ii, ins/ins; id, ins/del; dd, del/del.
Table IV.
Comparison of genotype frequencies of CASP8 gene polymorphisms in smokers and non-smokers.
| Smoking status | SNP | Model/allele | Genotype | Case, n (%) | Control, n (%) | P-valuea | OR (95% CI)b | Pac | AOR (95% CI)d |
|---|---|---|---|---|---|---|---|---|---|
| Smokere | rs3834129 | Additive | ii | 194 (63.8) | 87 (52.7) | 1.00 (Ref) | |||
| id | 100 (32.9) | 70 (42.4) | 0.027 | 0.641 (0.431–0.952) | 0.057 | 0.660 (0.430–1.013) | |||
| dd | 10 (3.3) | 8 (4.9) | 0.234 | 0.561 (0.214–1.469) | 0.076 | 0.395 (0.142–1.102) | |||
| Dominant | ii | 194 (63.8) | 87 (52.7) | 1.00 (Ref) | |||||
| id/dd | 110 (36.2) | 78 (47.3) | 0.019 | 0.632 (0.430–0.929) | 0.027 | 0.627 (0.415–0.949) | |||
| Recessive | ii/id | 294 (96.7) | 157 (95.1) | 1.00 (Ref) | |||||
| dd | 10 (3.3) | 8 (4.9) | 0.401 | 0.668 (0.258–1.725) | 0.132 | 0.459 (0.167–1.263) | |||
| rs13016963 | Additive | GG | 99 (32.6) | 54 (32.7) | 1.00 (Ref) | ||||
| AG | 150 (49.3) | 83 (50.3) | 0.947 | 0.986 (0.644–1.510) | 0.916 | 1.025 (0.646–1.627) | |||
| AA | 55 (18.1) | 28 (17.0) | 0.810 | 1.071 (0.610–1.881) | 0.897 | 1.041 (0.569–1.902) | |||
| Dominant | GG | 99 (32.6) | 54 (32.7) | 1.00 (Ref) | |||||
| AG/AA | 205 (67.4) | 111 (67.3) | 0.972 | 1.007 (0.673–1.509) | 0.897 | 1.029 (0.665–1.593) | |||
| Recessive | GG/AG | 249 (81.9) | 137 (83.0) | 1.00 (Ref) | |||||
| AA | 55 (18.1) | 28 (17.0) | 0.761 | 1.081 (0.655–1.783) | 0.928 | 1.025 (0.600–1.752) | |||
| Non-smokerf | rs3834129 | Additive | ii | 134 (66.7) | 189 (55.3) | 1.00 (Ref) | |||
| id | 59 (29.3) | 127 (37.1) | 0.029 | 0.655 (0.448–0.958) | 0.036 | 0.655 (0.440–0.973) | |||
| dd | 8 (4.0) | 26 (7.6) | 0.042 | 0.434 (0.191–0.988) | 0.032 | 0.395 (0.169–0.922) | |||
| Dominant | ii | 134 (66.7) | 189 (55.3) | 1.00 (Ref) | |||||
| id/dd | 67 (33.3) | 153 (44.7) | 0.009 | 0.618 (0.430–0.888) | 0.010 | 0.608 (0.417–0.887) | |||
| Recessive | ii/id | 193 (96) | 316 (92.4) | 1.00 (Ref) | |||||
| dd | 8 (4.0) | 26 (7.6) | 0.093 | 0.504 (0.224–1.135) | 0.069 | 0.460 (0.199–1.061) | |||
| rs13016963 | Additive | GG | 63 (31.3) | 110 (32.2) | 1.00 (Ref) | ||||
| AG | 93 (46.3) | 181 (52.9) | 0.593 | 0.897 (0.603–1.336) | 0.563 | 0.885 (0.584–1.340) | |||
| AA | 45 (22.4) | 51 (14.9) | 0.094 | 1.541 (0.928–2.557) | 0.034 | 1.785 (1.045–3.052) | |||
| Dominant | GG | 63 (31.3) | 110 (32.2) | 1.00 (Ref) | |||||
| AG/AA | 138 (68.7) | 232 (67.8) | 0.843 | 1.039 (0.714–1.510) | 0.758 | 1.063 (0.720–1.571) | |||
| Recessive | GG/AG | 156 (77.6) | 291 (85.1) | 1.00 (Ref) | |||||
| AA | 45 (22.4) | 51 (14.9) | 0.027 | 1.646 (1.054–2.570) | 0.007 | 1.924 (1.199–3.089) |
P-value and
OR with their 95% CIs were estimated using unconditional logistic regression.
P-value and
AOR with 95% CIs estimated using multiple logistic regression models after controlling age, gender and drinking.
Case, n=304; control, n=165.
Case, n=201; control, n=342. CASP, caspase-8; SNP, single nucleotide polymorphism; ref, reference; OR, odds ratio; AOR, adjusted odds ratio; CI, confidence interval; ii, ins/ins; id, ins/del; dd, del/del.
Table V.
Comparison of genotype frequencies of CASP8 gene polymorphisms in drinkers and non-drinkers.
| Drinking status | SNP | Model/allele | Genotype | Case, n (%) | Control, n (%) | P-valuea | OR (95% CI)b | Pac | AOR (95% CI)d |
|---|---|---|---|---|---|---|---|---|---|
| Drinkere | rs3834129 | Additive | ii | 175 (63.6) | 120 (57.4) | 1.00 (Ref) | |||
| id | 90 (32.7) | 76 (36.4) | 0.287 | 0.812 (0.553–1.192) | 0.149 | 0.713 (0.451–1.128) | |||
| dd | 10 (3.6) | 13 (6.2) | 0.138 | 0.527 (0.224–1.242) | 0.139 | 0.471 (0.174–1.277) | |||
| Dominant | ii | 175 (63.6) | 120 (57.4) | 1.00 (Ref) | |||||
| id/dd | 100 (36.4) | 89 (42.6) | 0.165 | 0.770 (0.533–1.113) | 0.083 | 0.678 (0.436–1.052) | |||
| Recessive | ii/id | 265 (96.4) | 196 (93.8) | 1.00 (Ref) | |||||
| dd | 10 (3.6) | 13 (6.2) | 0.186 | 0.569 (0.244–1.324) | 0.210 | 0.534 (0.201–1.423) | |||
| rs13016963 | Additive | GG | 88 (32.0) | 65 (31.1) | 1.00 (Ref) | ||||
| AG | 140 (50.9) | 100 (47.8) | 0.873 | 1.034 (0.686–1.559) | 0.768 | 1.077 (0.657–1.767) | |||
| AA | 47 (17.1) | 44 (21.1) | 0.373 | 0.789 (0.468–1.329) | 0.440 | 0.782 (0.419–1.459) | |||
| Dominant | GG | 88 (32.0) | 65 (31.1) | 1.00 (Ref) | |||||
| AG/AA | 187 (68.0) | 144 (68.9) | 0.833 | 0.959 (0.651–1.413) | 0.954 | 0.986 (0.618–1.573) | |||
| Recessive | GG/AG | 228 (82.9) | 165 (78.9) | 1.00 (Ref) | |||||
| AA | 47 (17.1) | 44 (21.1) | 0.269 | 0.773 (0.489–1.221) | 0.292 | 0.747 (0.434–1.285) | |||
| Non-drinkerf | rs3834129 | Additive | ii | 153 (66.5) | 156 (52.3) | 1.00 (Ref) | |||
| id | 69 (30.0) | 121 (40.6) | 0.004 | 0.581 (0.401–0.842) | 0.010 | 0.596 (0.401–0.886) | |||
| dd | 8 (3.5) | 21 (7.1) | 0.024 | 0.388 (0.167–0.904) | 0.016 | 0.336 (0.138–0.817) | |||
| Dominant | ii | 153 (66.5) | 156 (52.3) | 1.00 (Ref) | |||||
| id/dd | 77 (33.5) | 142 (47.7) | 0.001 | 0.553 (0.287–0.789) | 0.002 | 0.553 (0.378–0.809) | |||
| Recessive | ii/id | 222 (96.5) | 277 (92.9) | 1.00 (Ref) | |||||
| dd | 8 (3.5) | 21 (7.1) | 0.074 | 0.475 (0.207–1.094) | 0.069 | 0.460 (0.199–1.061) | |||
| rs13016963 | Additive | GG | 74 (32.2) | 99 (33.2) | 1.00 (Ref) | ||||
| AG | 103 (44.8) | 164 (55.0) | 0.380 | 0.840 (0.569–1.240) | 0.258 | 0.786 (0.517–1.193) | |||
| AA | 53 (23.0) | 35 (11.8) | 0.008 | 2.026 (1.201–3.416) | 0.033 | 1.834 (1.052–3.199) | |||
| Dominant | GG | 74 (32.2) | 99 (33.2) | 1.00 (Ref) | |||||
| AG/AA | 156 (67.8) | 199 (66.8) | 0.799 | 1.049 (0.727–1.514) | 0.893 | 0.973 (0.656–1.445) | |||
| Recessive | GG/AG | 177 (77.0) | 263 (88.2) | 1.00 (Ref) | |||||
| AA | 53 (23.0) | 35 (11.8) | 0.001 | 2.250 (1.410–3.591) | 0.003 | 2.127 (1.298–3.487) |
P-value and
OR with 95% CIs were estimated using unconditional logistic regression.
P-value and
AOR with 95% CIs were estimated using multiple logistic regression models after controlling age, gender and smoking.
Case, n=275; control, n=209. eCase, n=230; control, n=298. CASP, caspase-8; SNP, single nucleotide polymorphism; ref, reference; OR, odds ratio; AOR, adjusted odds ratio; CI, confidence interval; ii, ins/ins; id, ins/del; dd, del/del.
The genotype frequencies of rs3834129 were found to be significantly associated with OSCC in the additive, dominant and recessive models (Table III). The frequencies of the rs3834129 genotypes containing the del allele exhibited significant differences of 0.613-fold between patients and control subjects in the dominant model (ins/del + del/del vs. ins/ins, 95% CI, 0.467–0.806). In the additive model, ins/del and del/del genotypes were associated with a significantly lower risk of 0.447-fold (ins/del vs. ins/ins, 95% CI, 0.235–0.850) and 0.640-fold (del/del vs. ins/ins, 95% CI, 0.482–0.851). In the recessive model, del/del genotype was associated with a 0.528-fold lower risk (del/del vs. ins/del + ins/ins, 95% CI, 0.281–0.993).
The genotype frequencies of rs3834129 were significantly associated with OSCC in men and women in the additive and dominant models (Table VI). In the additive model, the del/del genotype was associated with a significantly reduced risk of 0.687-fold (del/del vs. ins/ins, 95% CI, 0.477–0.989) in men and 0.582-fold (ins/del vs. ins/ins, 95% CI, 0.325–0.961) in women. In the dominant model, the frequencies of the genotypes containing the del allele in the OSCC case group showed significant differences of 0.652-fold (ins/del + del/del vs. ins/ins, 95% CI, 0.458–0.928) in men and 0.549-fold (ins/del + del/del vs. ins/ins, 95% CI, 0.341–0.883) in women compared with he control group (Table VI). No significant association between rs13016963 and OSCC was observed in either gender. In addition, no significant association was identified for rs3834129 and OSCC in the recessive model in women (Table VI).
Table VI.
Comparison of genotype frequencies of CASP8 gene polymorphisms in men and women.
| Gender | SNP | Model/allele | Genotype | Case, n (%) | Control, n (%) | P-valuea | OR (95% CI)b | Pac | AOR (95% CI)d |
|---|---|---|---|---|---|---|---|---|---|
| Malee | rs3834129 | Additive | ii | 212 (64.2) | 195 (56.0) | 1.00 (Ref) | |||
| id | 109 (33.0) | 133 (38.2) | 0.082 | 0.754 (0.548–1.037) | 0.056 | 0.409 (0.164–1.022) | |||
| dd | 9 (2.8) | 20 (5.8) | 0.028 | 0.414 (0.184–0.931) | 0.044 | 0.687 (0.477–0.989) | |||
| Dominant | ii | 212 (64.2) | 195 (56.0) | 1.00 (Ref) | |||||
| id/dd | 118 (35.8) | 153 (44) | 0.029 | 0.709 (0.521–0.966) | 0.017 | 0.652 (0.458–0.928) | |||
| Recessive | ii/id | 321 (97.2) | 328 (94.2) | 1.00 (Ref) | |||||
| dd | 9 (2.8) | 20 (5.8) | 0.052 | 0.460 (0.206–1.025) | 0.100 | 0.467 (0.189–1.155) | |||
| rs13016963 | Additive | GG | 101 (30.6) | 120 (34.5) | 1.00 (Ref) | ||||
| AG | 166 (50.3) | 176 (50.6) | 0.510 | 1.121 (0.798–1.573) | 0.651 | 1.094 (0.742–1.611) | |||
| AA | 63 (19.1) | 52 (14.9) | 0.114 | 1.439 (0.915–2.264) | 0.262 | 1.342 (0.803–2.241) | |||
| Dominant | GG | 101 (30.6) | 120 (34.5) | 1.00 (Ref) | |||||
| AG/AA | 229 (69.4) | 228 (65.5) | 0.282 | 1.193 (0.865–1.647) | 0.453 | 1.152 (0.797–1.665) | |||
| Recessive | GG/AG | 267 (80.9) | 296 (85.1) | 1.00 (Ref) | |||||
| AA | 63 (19.1) | 52 (14.9) | 0.150 | 1.343 (0.898–2.009) | 0.303 | 1.269 (0.806–1.997) | |||
| Femalef | rs3834129 | Additive | ii | 116 (66.3) | 81 (50.9) | 1.00 (Ref) | |||
| id | 50 (28.6) | 64 (40.3) | 0.010 | 0.546 (0.342–0.870) | 0.034 | 0.582 (0.352–0.961) | |||
| dd | 9 (5.1) | 14 (8.8) | 0.070 | 0.449 (0.185–1.087) | 0.064 | 0.408 (0.158–1.054) | |||
| Dominant | ii | 116 (66.3) | 81 (50.9) | 1.00 (Ref) | |||||
| id/dd | 59 (33.7) | 78 (49.1) | 0.004 | 0.528 (0.340–0.821) | 0.013 | 0.549 (0.341–0.883) | |||
| Recessive | ii/id | 166 (94.9) | 145 (91.2) | 1.00 (Ref) | |||||
| dd | 9 (5.1) | 14 (8.8) | 0.187 | 0.562 (0.236–1.336) | 0.171 | 0.527 (0.210–1.320) | |||
| Rs13016963 | Additive | GG | 61 (34.9) | 44 (27.7) | 1.00 (Ref) | ||||
| AG | 77 (44.0) | 88 (55.3) | 0.067 | 0.631 (0.385–1.034) | 0.082 | 0.619 (0.360–1.062) | |||
| AA | 37 (21.1) | 27 (17.0) | 0.971 | 0.988 (0.527–1.855) | 0.980 | 0.991 (0.503–1.955) | |||
| Dominant | GG | 61 (34.9) | 44 (27.7) | 1.00 (Ref) | |||||
| AG/AA | 114 (65.1) | 115 (72.3) | 0.158 | 0.715 (0.449–1.140) | 0.182 | 0.706 (0.423–1.178) | |||
| Recessive | GG/AG | 138 (78.9) | 132 (83.0) | 1.00 (Ref) | |||||
| AA | 37 (21.1) | 27 (17.0) | 0.334 | 1.311 (0.756–2.273) | 0.317 | 1.346 (0.752–2.411) |
P-value and
OR with 95% CIs estimated by unconditional logistic regression.
P-value and
AOR with 95% CIs estimated by multiple logistic regression models after controlling age, smoking and drinking.
Case, n=330; control, n=348.
Case, n=175; control, n=159. CASP, caspase-8; SNP, single-nucleotide polymorphism; Ref, reference; OR, odds ratio; AOR, adjusted odds ratio; CI, confidence interval; ii, ins/ins; id, ins/del; dd, del/del.
The genotype frequencies of rs3834129 were significantly associated with OSCC in smokers in the dominant model (Table IV). The genotypes containing the del allele were associated with a significantly lower risk of 0.627-fold (del/del + del/ins vs. ins/ins, 95% CI, 0.415–0.949). In additive and dominant models, the genotype frequencies of rs3834129 were significantly associated with OSCC in non-smokers (del/ins vs. ins/ins, 95% CI, 0.440–0.973; del/del vs. ins/ins, 95% CI, 0.169–0.922; del/del + del/ins vs. ins/ ins, 95% CI, 0.417–0.887).
The genotype frequencies of rs3834129 were significantly associated with OSCC in non-drinkers in the additive and dominant models (Table V). In the additive model, the del/del genotype was associated with a significantly lower risk of 0.336-fold (del/del vs. ins/ins, 95% CI, 0.138–0.817) and 0.596-fold (ins/del vs. ins/ins, 95% CI, 0.401–0.886) in non-drinkers. In the dominant model, the frequencies of the genotypes containing the del allele were significantly different by 0.553-fold (ins/del and del/del vs. ins/ins, 95% CI, 0.378–0.809) in non-drinkers in the OSCC case group, as compared with the control group in non-drinkers. Furthermore, the genotype frequencies of rs13016963 were significantly associated with OSCC in non-drinkers in the additive and recessive models (Table V). Among non-drinkers, the AA genotype was associated with a significantly higher risk of 1.834-fold (AA vs. GG, 95% CI, 1.052–3.199) in the additive model and 2.127-fold (AA vs. GG and AG, 95% CI, 1.298–3.487) in the recessive model (Table V).
The LD values of rs3834129 and 13016963 were evaluated using SHEsis software. No significant LD was observed for the two loci rs3834129 and 13016963; the r2 value was 0.003 and the D' value was 0.107. Analysis using PowerStats software revealed no marked deviations (P>0.05) from the Hardy-Weinberg equilibrium for the two loci (rs3834129, P=0.9228; and rs13016963, P=0.1225). The statistical power in the present study was >80%, with an estimated OR of 2.0 (rs3834129, 83%; rs13016963, 87%).
Discussion
OSCC is among the most common malignancies of the oral cavity (33). To date, no reliable methods have been developed to indicate the OSCC risk of an individual. However, until now, there are no solid methods to warn against OSCC risk. Genetic diagnosis has attracted more attention in recent years (34). The aim of the present study was to identify new suitable biomarkers or genes that are able to indicate the physiological state and alterations in cells prior to or during the pathogenesis of OSCC, in order to provide early and accurate prediction and diagnosis for patients with OSCC, particularly in the early stages (35).
Apoptosis is an crucial physiological mechanism that eliminates cells with unrepairable DNA damage, and thus sustains homeostasis. Apoptosis occurs via two mechanisms: The death receptor Fas/FasL (also known as extrinsic) pathway and the mitochondrial (DNA damage-induced and p53-mediated, also known as intrinsic) pathway (36). CASP8 participates in the FAS-FAS ligand mediated extrinsic (death-receptor) pathway (37,38), while additionally interacting with the BH3 interacting-domain death agonist protein to influence the intrinsic (mitochondrial) pathway, which functions through caspases (39). However, CASP8 has also been observed to serve a number of non-apoptotic functions in cells, such as promoting the activation of NF-κB signaling, regulating autophagy, altering endosomal trafficking and enhancing cellular adhesion and migration (11). Therefore, depending on the specific cellular context, CASP8 may potentiate or suppress tumor malignancy.
Molecular epidemiological studies have suggested that SNPs may contribute to the susceptibility of an individual to OSCC by affecting enzyme expression levels or enzyme activities (40–42). However, the association between SNPs and major apoptosis regulatory caspase genes in OSCC remains unknown (43). The CASP8 promoter −652 6 N del allele substantially affects the promoter activity of the CASP8 gene, as this allele destroys a binding element for Sp1, resulting in reduced apoptotic reactivity in T lymphocytes following stimulation by cancer cells or phytohemagglutinin in an ex vivo model (44). Alternatively, reduced cell apoptosis involved in the antitumor process may provide protection against cancer (44). A previous study reported that −652 6 N ins/del polymorphism is able to influence the risk of multiple cancer types, including cancer of the lungs, esophagus, stomach, colorectum, breast, and cervix in Chinese populations and cutaneous melanoma in Caucasian populations (11). Li et al (44) reported that the six-nucleotide deletion variant in the CASP8 promoter region is inversely associated with the risk of squamous cell carcinoma of the head and neck in non-Hispanic Caucasian populations. However, studies on breast cancer in Europe and colorectal cancer in the United Kingdom have not yet confirmed such an association with cancer risk (45,46). Furthermore, Haiman et al were unable to replicate the results of the CASP8 polymorphism associated with the risk of cancer of the prostate, breast, and colorectum in multiple US populations (47). However, the results of the present study indicate that the CASP8 −652 6 N ins/del polymorphism is reversely associated with OSCC risk in a Chinese population, which is consistent with the findings reported by Sun et al (21) and Li et al (44) in Chinese populations. rs13016963 is a rarely studied locus adjacent to CASP8 (22). A GWAS indicated that rs13016963 was significantly associated with the risk of melanoma in French and UK populations (P=8.6×10−10; OR, 1.14; 95% CI, 1.09–1.19). Using GWAS data, Abnet et al (23) identified an association between esophageal squamous cell carcinoma and 2q33 that achieved genome-wide significance. The strongest signal was for rs13016963, with a combined OR (95% CI) of 1.29 (1.19–1.40) and P=7.63×10−10 in a Chinese population. Abnet et al further suggested that future studies of esophageal cancer and other cancers should focus on the comprehensive sequencing of this 2q33 locus and functional analysis of rs13016963 and other strongly correlated variants.
In the present study, common SNPs for CASP8, namely rs3834129, rs13016963 and rs1045485, were selected to evaluate the susceptibility of patients with OSCC and healthy control subjects. To the best of our knowledge, the present study is the first to investigate rs13016963 polymorphisms without available genotype data from the SNP database of the National Institute of Health (http://www.ncbi.nlm.nih.gov/projects/SNP/). In addition, none of these polymorphisms was identified in the Han population of Sichuan province that was included in this study.
In the present study, no significant difference was observed in the genotype and allele frequencies of rs13016963 SNPs between patient the and control groups. However, the AA genotype frequencies of rs1306963 were significantly associated with OSCC in non-smokers and non-drinkers. These results corresponded well with those of previous GWAS studies (22). The del allele frequency of rs3834129 was significantly lower in OSCC patients compared with control subjects in the dominant model (ins/del + del/del vs. ins/ins, AOR=0.613, 95% CI, 0.467–0.806). The del allele of rs3834129 was significantly reduced in smoking and non-smoking OSCC patients compared with the control group subjects. Furthermore, the genotypes containing the del allele were associated with a significantly reduced risk of OSCC compared with the ins/ins genotype among non-drinkers (ins/del + del/del vs. ins/ins, AOR=0.553, 95% CI, 0.378–0.809; ins/del vs. ins/ins, AOR=0.596, 95% CI, 0.401–0.886; del/del vs. ins/ins, AOR=0.336, 95% CI, 0.138–0.817). Stratification of OSCC patients on the basis of gender showed that the del allele of the rs3834129 polymorphism exhibited a protective effect in men and women.
In the present study, rs3834129 was found to be associated with a significantly decreased risk of OSCC. This result suggests that the CASP8 −652 6 N ins/del polymorphism may affect patient susceptibility to OSCC and may be used as a biomarker for this disease. Future studies involving a larger sample size and various expression studies are required to evaluate the association between these polymorphisms and OSCC risk.
Acknowledgements
This study was financially supported by the ‘Support for the Recruitment of Under-Represented Faculty’, State Key Laboratory of Oral Diseases, Sichuan University (no. SKLODYISF2012-14 and SKLODSCU20130045).
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Kessler P, Grabenbauer G, Leher A, Bloch-Birkholz A, Vairaktaris E, Neukam FW. Neoadjuvant and adjuvant therapy in patients with oral squamous cell carcinoma long-term survival in a prospective, non-randomized study. Br J Oral Maxillofac Surg. 2008;46:1–5. doi: 10.1016/j.bjoms.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Sankaranarayanan R, Masuyer E, Swaminathan R, Ferlay J, Whelan S. Head and neck cancer: A global perspective on epidemiology and prognosis. Anticancer Res. 1998;18:4779–4786. [PubMed] [Google Scholar]
- 5.de Maria S, Lo Muzio L, Braca A, et al. Survivin promoter-31 G/C polymorphism in oral cancer cell lines. Oncol Lett. 2011;2:935–939. doi: 10.3892/ol.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Zha L, Li B, Zhang L, Yu T, Li L. Correlation between superoxide dismutase 1 and 2 polymorphisms and susceptibility to oral squamous cell carcinoma. Exp Ther Med. 2014;7:171–178. doi: 10.3892/etm.2013.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Long L, Li T, et al. Polymorphisms of microRNA-binding sites in integrin genes are associated with oral squamous cell carcinoma susceptibility and progression. Tohoku J Exp Med. 2014;233:33–41. doi: 10.1620/tjem.233.33. [DOI] [PubMed] [Google Scholar]
- 8.Zhong F, Yang XC, Bu LX, Li NY, Chen WT. Single nucleotide polymorphisms in the u-PA gene are related to susceptibility to oral tongue squamous cell carcinoma in the Northern Chinese Han population. Asian Pac J Cancer Prev. 2013;14:781–784. doi: 10.7314/APJCP.2013.14.2.781. [DOI] [PubMed] [Google Scholar]
- 9.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 10.Siegel RM. Caspases at the crossroads of immune-cell life and death. Nat Rev Immunol. 2006;6:308–317. doi: 10.1038/nri1809. [DOI] [PubMed] [Google Scholar]
- 11.Stupack DG. Caspase-8 as a therapeutic target in cancer. Cancer Lett. 2013;332:133–140. doi: 10.1016/j.canlet.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenet J, Teitz T, Wei T, Valentine V, Kidd VJ. Structure and chromosome localization of the human CASP8 gene. Gene. 1999;226:225–232. doi: 10.1016/S0378-1119(98)00565-4. [DOI] [PubMed] [Google Scholar]
- 13.Nunez G, Benedict MA, Hu Y, Inohara N. Caspases: The proteases of the apoptotic pathway. Oncogene. 1998;17:3237–3245. doi: 10.1038/sj.onc.1202581. [DOI] [PubMed] [Google Scholar]
- 14.Frank B, Hemminki K, Wappenschmidt B, et al. Association of the CASP10 V410I variant with reduced familial breast cancer risk and interaction with the CASP8 D302H variant. Carcinogenesis. 2006;27:606–609. doi: 10.1093/carcin/bgi248. [DOI] [PubMed] [Google Scholar]
- 15.Palanca Suela S, Esteban Cardenosa E, Barragán González E, et al. CASP8 D302H polymorphism delays the age of onset of breast cancer in BRCA1 and BRCA2 carriers. Breast Cancer Res Treat. 2010;119:87–93. doi: 10.1007/s10549-009-0316-2. [DOI] [PubMed] [Google Scholar]
- 16.Yang C, Liu HZ, Fu ZX. PEG-liposomal oxaliplatin induces apoptosis in human colorectal cancer cells via Fas/FasL and caspase-8. Cell Biol Int. 2012;36:289–296. doi: 10.1042/CBI20100825. [DOI] [PubMed] [Google Scholar]
- 17.Engel C, Versmold B, Wappenschmidt B, et al. Association of the variants CASP8 D302H and CASP10 V410I with breast and ovarian cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol Biomarkers Prev. 2010;19:2859–2868. doi: 10.1158/1055-9965.EPI-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lubahn J, Berndt SI, Jin CH, et al. Association of CASP8 D302H polymorphism with reduced risk of aggressive prostate carcinoma. Prostate. 2010;70:646–653. doi: 10.1002/pros.21098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bethke L, Sullivan K, Webb E, et al. CASP8 D302H and meningioma risk: An analysis of five case-control series. Cancer Lett. 2009;273:312–315. doi: 10.1016/j.canlet.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Bethke L, Sullivan K, Webb E, et al. The common D302H variant of CASP8 is associated with risk of glioma. Cancer Epidemiol Biomarkers Prev. 2008;17:987–989. doi: 10.1158/1055-9965.EPI-07-2807. [DOI] [PubMed] [Google Scholar]
- 21.Sun T, Gao Y, Tan W, et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605–613. doi: 10.1038/ng2030. [DOI] [PubMed] [Google Scholar]
- 22.Barrett JH, Iles MM, Harland M, et al. GenoMel Consortium: Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011;43:1108–1113. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abnet CC, Wang Z, Song X, et al. Genotypic variants at 2q33 and risk of esophageal squamous cell carcinoma in China: A meta-analysis of genome-wide association studies. Hum Mol Genet. 2012;21:2132–2141. doi: 10.1093/hmg/dds029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sergentanis TN, Economopoulos KP. Association of two CASP8 polymorphisms with breast cancer risk: A meta-analysis. Breast Cancer Res Treat. 2010;120:229–234. doi: 10.1007/s10549-009-0471-5. [DOI] [PubMed] [Google Scholar]
- 25.Pindborg JJ, Reichart PA, Smith CJ, van der Waal I, editors. Histological Typing of Cancer and Precancer of the Oral Mucosa. 2nd. Berlin: Springer-Verlag; 1997. [DOI] [Google Scholar]
- 26.Green FL. AJCC Cancer Staging Manual. 6th. Berlin Heidelberg: Springer-Verlag; 2002. [DOI] [Google Scholar]
- 27.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashemi M, Eskandari-Nasab E, Fazaeli A, et al. Bi-directional PCR allele-specific amplification (bi-PASA) for detection of caspase-8 −652 6 N ins/del promoter polymorphism (rs3834129) in breast cancer. Gene. 2012;505:176–179. doi: 10.1016/j.gene.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 29.Hu Z, Li C, Chen K, et al. Single nucleotide polymorphisms in selected apoptotic genes and BPDE-induced apoptotic capacity in apparently normal primary lymphocytes: A genotype-phenotype correlation analysis. J Cancer Epidemiol. 2008;2008:147905. doi: 10.1155/2008/147905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 31.Lewontin R. On measures of gametic disequilibrium. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson RR. The sampling distribution of linkage disequilibrium under an infinite allele model without selection. Genetics. 1985;109:611–631. doi: 10.1093/genetics/109.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jemal A. Global burden of cancer: Opportunities for prevention. Lancet. 2012;380:1797–1799. doi: 10.1016/S0140-6736(12)61688-2. [DOI] [PubMed] [Google Scholar]
- 34.Chawla JP, Iyer N, Soodan KS, Sharma A, Khurana SK, Priyadarshni P. Role of miRNA in cancer diagnosis, prognosis, therapy and regulation of its expression by Epstein-Barr virus and human papillomaviruses: With special reference to oral cancer. Oral Oncol. 2015;51:731–737. doi: 10.1016/j.oraloncology.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- 36.Wang M, Zhang Z, Tian Y, Shao J, Zhang Z. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter associated with risk and progression of bladder cancer. Clin Cancer Res. 2009;15:2567–2572. doi: 10.1158/1078-0432.CCR-08-2829. [DOI] [PubMed] [Google Scholar]
- 37.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 38.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485–495. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 39.Andersen MH, Becker JC, Straten P. Regulators of apoptosis: suitable targets for immune therapy of cancer. Nat Rev Drug Discov. 2005;4:399–409. doi: 10.1038/nrd1717. [DOI] [PubMed] [Google Scholar]
- 40.Hsing AW, Sakoda LC, Rashid A, et al. Variants in inflammation genes and the risk of biliary tract cancers and stones: A population-based study in China. Cancer Res. 2008;68:6442–6452. doi: 10.1158/0008-5472.CAN-08-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava A, Srivastava K, Pandey SN, Choudhuri G, Mittal B. Single-nucleotide polymorphisms of DNA repair genes OGG1 and XRCC1: Association with gallbladder cancer in North Indian population. Ann Surg Oncol. 2009;16:1695–1703. doi: 10.1245/s10434-009-0354-3. [DOI] [PubMed] [Google Scholar]
- 42.Srivastava K, Srivastava A, Pandey SN, Kumar A, Mittal B. Functional polymorphisms of the cyclooxygenase (PTGS2) gene and risk for gallbladder cancer in a North Indian population. J Gastroenterol. 2009;44:774–780. doi: 10.1007/s00535-009-0071-5. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava K, Srivastava A, Mittal B. Caspase-8 polymorphisms and risk of gallbladder cancer in a northern Indian population. Mol Carcinog. 2010;49:684–692. doi: 10.1002/mc.20641. [DOI] [PubMed] [Google Scholar]
- 44.Li C, Lu J, Liu Z, et al. The six-nucleotide deletion/insertion variant in the CASP8 promoter region is inversely associated with risk of squamous cell carcinoma of the head and neck. Cancer Prev Res (Phila) 2010;3:246–253. doi: 10.1158/1940-6207.CAPR-08-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank B, Rigas SH, Bermejo JL, et al. The CASP8 −652 6 N del promoter polymorphism and breast cancer risk: A multicenter study. Breast Cancer Res Treat. 2008;111:139–144. doi: 10.1007/s10549-007-9752-z. [DOI] [PubMed] [Google Scholar]
- 46.De Vecchi G, Verderio P, Pizzamiglio S, et al. Evidences for association of the CASP8 −652 6 N del promoter polymorphism with age at diagnosis in familial breast cancer cases. Breast Cancer Res Treat. 2009;113:607–608. doi: 10.1007/s10549-008-9963-y. [DOI] [PubMed] [Google Scholar]
- 47.Haiman CA, Garcia RR, Kolonel LN, Henderson BE, Wu AH, Le Marchand L. A promoter polymorphism in the CASP8 gene is not associated with cancer risk. Nat Genet. 2008;40:259–260. doi: 10.1038/ng0308-259. [DOI] [PubMed] [Google Scholar]