Abstract
Moderate consumption of wine seems to produce positive health effects derived from the occurrence of bioactive polyphenols. The gut microbiota is involved in the metabolism of phenolic compounds, and these compounds and/or their metabolites may modulate gut microbiota through the stimulation of the growth of beneficial bacteria and the inhibition of pathogenic bacteria. The characterization of bacterial metabolites derived from polyphenols is essential in order to understand their effects, including microbial modulation, and therefore to associate dietary intake with particular health effects. This review aims to summarize the current information about the two-way “wine polyphenols–gut microbiota” interaction, from a perspective based on the experimental and analytical designs used. The availability of advanced methods for monitoring bacterial communities, along with the combination of in vitro and in vivo models, could help to assess the metabolism of polyphenols in the human body and to monitor total bacterial communities, and, therefore, to elucidate the implications of diet on the modulation of microbiota for delivering health benefits.
Keywords: wine, polyphenols, gut microbiota, probiotics, modulation, batch culture fermentation, gastrointestinal simulators, animal models, human studies
1. Introduction
Wine is considered to be a high bioactive polyphenol content source. Many studies have revealed the key role played by phenolic compounds from grapes and wine on human health; cardiovascular diseases being the pathologies that have received much attention [1,2].
Several studies confirm the importance of the intestinal microbiota to the health of the host, including mental health [3]. Gut bacteria not only help to maximize the absorption of nutrients and energy, but also are essential in the body’s defense mechanisms [4]. Although polyphenol metabolism starts in the mouth and proceeds along the gastrointestinal tract, most of the dietary polyphenols reach the colon, where they are subjected to the action of the gut microbiota, thus releasing aglycones that might, to a certain extent, be absorbed and degraded to simpler phenolic derivatives and other metabolites which could present higher activity at a physiological level than the corresponding food precursors [5]. These metabolites could also be absorbed, increasing polyphenol bioavailability [6]. Non-absorbed polyphenols and/or the resultant phenolic metabolites could affect the growth of gut microbiota, thereby modifying their diversity and metabolic activity. In fact, modulation of gut microbiota by polyphenols has been a topic that has gained increasing attention from the scientific community in recent years, as can be seen from several reviews [7,8,9,10,11,12]. Among the major wine phenolic compounds that may reach the gut, special attention has been given to polymeric flavan-3-ols or proanthocyanidins (also known as condensed tannins) since there is evidence that these polyphenols promote the growth of beneficial bacteria and the inhibition of pathogenic bacteria while they are extensively metabolized by gut microbiota to produce a great range of active metabolites. As relevant reference, an intervention study of cocoa flavan-3-ols in healthy volunteers has shown that they enhance the growth of Lactobacillus spp. and Bifidobacterium spp. and limit the growth of the Clostridium histolyticum group [13]. On the other hand, proanthocyanidins have been found to be largely metabolized into phenylvalerolactones and phenolic acids after cocoa intake in humans and rats [14].
The aim of this review was to summarize the information available on the action of gut microbiota on wine polyphenols (metabolism), as well as the effect of phenolic compounds on the growth of gut bacteria (modulation). This two-way polyphenols–gut microbiota interaction will be assessed from a perspective based on the experimental designs used, from isolated cultures to omic approaches in the case of microbiota analysis, and advanced analytical techniques in the case of metabolite analysis.
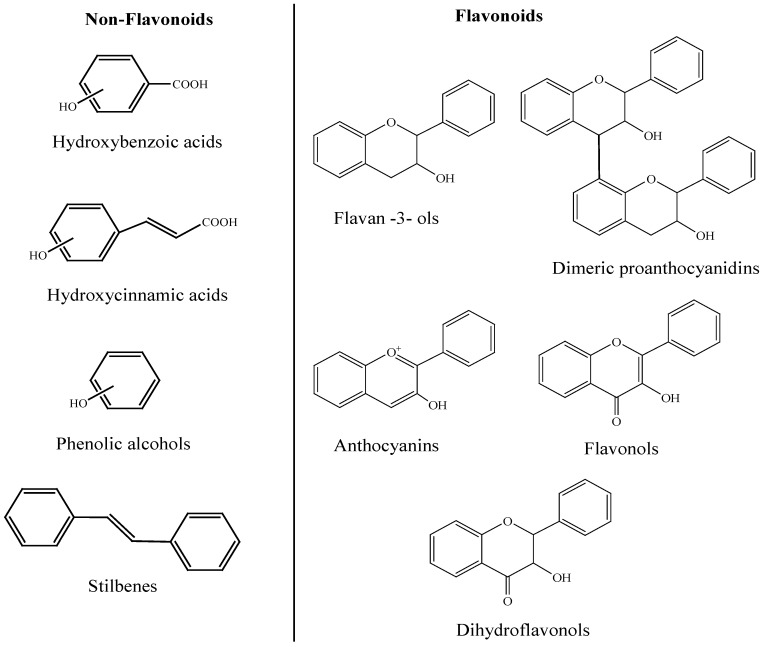
2. Phenolic Compounds in Wine
The term “phenolic” describes those compounds that possess a benzenic ring substituted by one or several hydroxyl groups (−OH). Polyphenols are secondary metabolites of plants and play an important role in the plant’s defense mechanism against external agents, such as animals or microbial infections, they facilitate pollination and seed dispersion through signals that attract insects and animals, and participate in protection mechanisms against ultraviolet radiation and/or oxidant agents. Phenolic compounds presented in grapes, located in the solid parts of the fruit (skins and seeds), have a wide diversity of chemical structures, including flavonoid compounds (flavan-3-ols (monomers and oligomeric and polymeric proanthocyanidins), anthocyanins, flavonols, and dihydroflavonols) and non-flavonoid compounds (hydroxybenzoic and hydroxycinnamic acids, phenolic alcohols, and stilbenes) (Figure 1). Although the concentration of phenolic compounds in wine is conditioned by several factors related to the grape (variety, soil, geography, climate, etc.) and by enological practices, the total polyphenol content is around 50–400 mg/L for white wines, and 900–1400 mg/L for young red wines. Therefore, moderate consumption of wine (250 mL/day) corresponds to an intake of 60 mg of polyphenols for white wines and 210 mg for young red wines [15]. With respect to their distribution by compound groups, acids and hydroxybenzoic derivatives represent approximately 6% of the total; acids and hydroxycinnamic derivatives, 1.1%; stilbenes, 0.5%; alcohols, 3.8%; flavanols, 15%; flavonols, 3.6%; and anthocyanins, 70% in young red wines [15]. Other anthocyanin derivatives such as pyranoanthocyanins present much lower proportions.
Figure 1.
Common phenolic compounds in wine.
Over the last few decades, the role of polyphenols has been the focus of considerable research in the field of nutrition. Several epidemiological studies have shown that the intake of these compounds is inversely associated with the risk of various chronic diseases, such as coronary heart disease, specific cancers, and neurodegenerative disorders [16,17]. Indeed, potential beneficial effects have been demonstrated for different phenolic compounds (especially flavonoids) through in vitro assays. In particular, these compounds act as powerful inhibitors of low-density lipoprotein (LDL) oxidation, one of the main mechanisms responsible for the development of atherosclerosis. However, it is currently believed that the physiological activities and mechanisms of these compounds are more diverse and complex. Thus, phenolic compounds are able to inhibit the growth of human cancer cell lines, cholesterol-related processes, and the activity of enzymes, such as telomerase, lipoxygenase and cyclooxygenase involved in inflammatory processes [18,19]. They also interact in different signal transduction pathways, and can affect the cell-cycle regulation, platelet function, and prevent endothelium dysfunction [20,21].
However, the health effects of these compounds depend on their bioavailability, and therefore it is important to understand how they are absorbed, metabolized and eliminated from the body, in order to ascertain their in vivo actions.
3. General Metabolism of Polyphenols in the Human Body
Polyphenols are considered as xenobiotics by the human organism and therefore are extensively metabolized and finally eliminated, mainly in the bile but also in the urine. The first step in their metabolism is likely to be deglycosylation before absorption in the small intestine. Hydrolysis of some flavonoid glycosides might have already occurred in the oral cavity, as both saliva and oral microbiota show β-glucosidase activity, giving rise to the corresponding aglycones. The hydrolytic activity begins in the mouth, and continues through the digestive tract into the stomach, where the size of food particles is reduced, which prompts the release of phenolic compounds. It has been estimated that 5%–10% of ingested polyphenols are absorbed in the small intestine, while 90%–95% reach the colon where they are intensively degraded by microbiota into a diversity of bioactive phenolic metabolites, lactones and phenolic acids that are then further absorbed [22].
In particular, the glycosylated polyphenols, such as anthocyanins, flavonol glycosides and glycosides of resveratrol, can be hydrolyzed by intestinal β-glucosidases. In contrast, monomeric flavanols, and dimer procyanidins can be absorbed directly into the small intestine. Once absorbed, the resulting aglycones would enter enterocyte by passive diffusion. Thus, the resulting aglycone is rapidly biotransformed by phase II enzymes into conjugated metabolites (i.e., glucuronides, O-methylethers and/or sulfates) within the enterocyte and again in the liver [23]. Other wine polyphenols, mainly oligomeric flavan-3-ols with a degree of polymerization (mDP) >3 and polymeric flavanols (proantocyanidins and condensed tannins), esters of hydroxycinnamic acids, and flavonols conjugated with rhamnose, such as rutin, are not absorbed in their native forms. These compounds reach the colon, where compounds are subjected to the action of the colonic microflora and transformed into various phenolic acids and other metabolites [24]. The methylated, glucuronide and sulfate conjugates (phase II metabolites) can reach the colon via enterohepatic circulation and are also susceptible to degradation by the intestinal microbiota. Finally, the phenolic metabolites are excreted via urine and feces.
4. Gut Microbiota
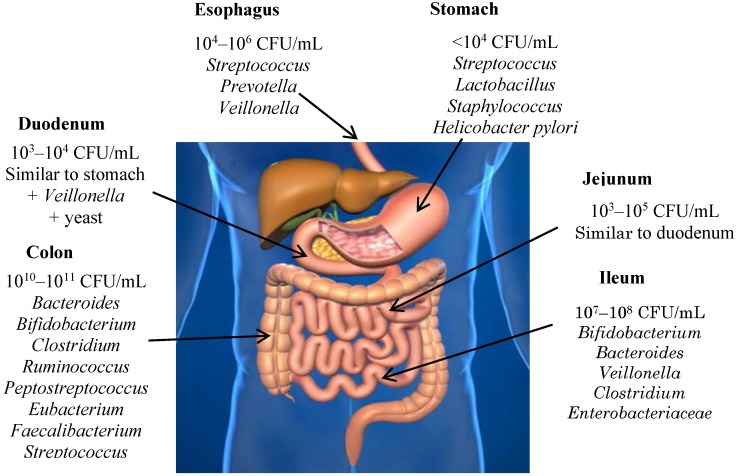
The orogastrointestinal tract of humans has an abundant microbiota dominated by anaerobic bacteria. The number of bacteria in the oral cavity is about 1011 bacteria/g in dental plaque and 108–109 bacteria/mL in saliva, whereas in feces the corresponding numbers are 1011–1012 bacteria/g [25]. More precisely, the gut microbial ecosystem includes native species that permanently colonize the gastrointestinal tract, and a variable number of live microorganisms that temporarily pass through the digestive tract [4]. Native bacteria are mainly acquired at birth and during the first year of life, whereas transient bacteria are continuously being ingested from food, drinks and the environment.
Among the human gastrointestinal microbiota, the majority of the species belong to the phyla Firmicutes, Bacteroides, Actinobacteria and Proteobacteria [26] (Figure 2). The phylogenetic composition of the gut microbiota is considered specific and stable over time for each individual. The species vary greatly between individuals. In fact, interindividual variation in gut microbiota may, in part, reflect differences in dietary intake, although the response of the gut microbiota to dietary change can also differ among individuals. The composition of the individual’s microbiota can fluctuate under some circumstances, for instance acute diarrheal illnesses, antibiotic treatment, or to a lesser extent when being induced by dietary interventions, but the individual flora composition patterns usually remain constant [27]. Among the microbiota associated with the esophagus, are included microorganisms belonging to the genera Streptococcus, Prevotella and Veillonella, which also appear in the oral cavity. The density of colonization is increased about eight times from proximal regions of the small intestine (103 bacterias/g) until the colon. In the stomach and duodenum, the number of microorganisms is reduced due to acid, bile and pancreatic secretions; as advances in the small intestine, the acidity decreases due to the dilution of the acid, which facilitates bacterial colonization, reaching 1011 CFU (colony forming units)/mL in the colon. The most frequently found bacteria in this area are members of the genus Bacteroides, Bifidobacterium, Eubacterium, Clostridium, Lactobacillus, and Gram-positive cocci [28], while Enterococci and representatives of the Enterobacteriaceae family are found to a lesser extent [29].
Figure 2.
Distribution and composition of bacterial species in the gastrointestinal tract. CFU: colony forming units.
Currently, there is evidence that confirms the importance of the gut microbiota in host health is associated to bacterial groups that colonize the intestine. The large intestine contains a complex and dynamic microbial ecosystem composed of commensal bacteria, potentially harmful opportunistic bacteria and others that can have both effects [30]. In particular, having a well-balanced gut microbiota composition is essential for human health. Undesirable bacteria include species of the genus Clostridium, Staphylococcus and Veillonella. These species can produce potentially harmful products, such as toxins and carcinogens, which are associated with intestinal disorders such as chronic inflammatory bowel diseases and other immune-related disorders. However, the use of antibiotics can disrupt the ecological balance and allow the overgrowth of species with potential pathogenicity, such as Clostridium difficile, associated with pseudomembranous colitis [31]. With regards to beneficial bacteria, among which are mainly included species of the genus Lactobacillus and Bifidobacterium, these play a key role in nutritional and disease-prevention functions, so they are used as probiotics. The main functions of these bacteria are: decreasing gas production, the production of short-chain fatty acids (SCFA), immunostimulating and antitumor activity [32,33].
Although the investigations concerning the influence of polyphenols on the gut microbiota and their mechanisms of action in humans are scarce, nevertheless drastic changes in fecal- and mucosa-associated microbiota could be related to determined diseases, such as colon cancer, short bowel syndrome and obesity [34]. It is important, therefore, to investigate gut microbiota composition and the influence of dietary intake on the human microbiome, to elucidate the implications of diet on the modulation of microbiota for delivering health benefits.
The availability and feasibility of advanced methods for monitoring total bacterial communities is a prerequisite to be considered when designing a study to assess gut microbiota. Over recent decades, the introduction of culture-independent techniques like terminal restriction fragment length polymorphism (T-RFLP), fluorescence in situ hybridation (FISH), denaturing gradient gel electrophoresis (DGGE), and quantitative polymerase chain reaction (qPCR) have improved the analysis of gut microbiota. At present, next-generation sequencing (NGS) techniques have promoted the emergence of new, high-throughput technologies, such as genomics, metagenomics, transcriptomics, metatranscriptomics, etc. The development of these techniques has provided the opportunity to explore the taxonomic, protein-coding gene or expression diversity by applying more comprehensive and less biased measurements to all systems involved (i.e., diet, microbiota and host) [35]. However, the enormous amount of data generated becomes cumbersome to analyze, requiring much dedicated time as well as expertise to manage data in such quantity [36]. The link between high-throughput qPCR and next generation sequencing technologies provides manageable data with valuable quantitative and taxonomic information. NGS platforms involve many different technologies [37] all of which generate large, genome-scale datasets. Examples of current commercial platforms are the 454 (Roche), Solexa (Illumina), SOLiD and Ion Torrent (Life Technologies), and PacBio (Pacific Biosciences) systems.
5. Interaction between Wine Polyphenols and Gut Microbiota
Taking the above-mentioned points into account, it is evident that the gut microbiome is implicated in the metabolism of wine polyphenols, and it also suggests that phenolic compounds and their metabolites may modulate the gut microbiota to a certain extent. The so-called two-way interaction between wine polyphenols and gut microbiota [7] can be subjected to various factors related to the phenolic compounds concentration and structure as well as the characteristics of bacterial strains, microbial environment, food matrix, etc. Prior to the specific sections about wine polyphenol metabolism and bacterial modulation by polyphenols, a preliminary section about experimental and analytical approaches used in these studies is included.
5.1. Experimental Designs and Analytical Techniques
5.1.1. In Vitro Gastrointestinal Tract Simulators
Studies regarding polyphenol metabolism and microbial modulation have been carried out using different experimental designs, from simple approaches involving fermentation experiments in batch cultures and in continuous simulators, to intervention trials in animals and humans. Although in vivo trials are most relevant physiologically, in vitro tools have been designed to simulate intestinal conditions and the transformations undergone in foods during transit through the gastrointestinal tract.
Simply, static gut models, also known as batch-type cultures, are generally closed systems using sealed bottles or reactors containing suspensions of fecal material, which are maintained under anaerobic conditions. This model approach is primarily used when assessing the stability of polyphenols in the presence of human-derived gut microbiota and evaluating which environmental conditions favor or limit polyphenol bioconversion.
In contrast to short-duration experiments with static gut models, longer-term experiments are required when the adaptation of the gut microbial community to dietary polyphenols needs to be assessed. To that end, simulators of the gastrointestinal tract comprise stomach and small intestinal sections for the pre-digestion of food as well as vessels stimulating the ascending, transverse and descending sections of the human colon, allowing the assessment of changes in colonic areas that are very challenging to access in a human intervention, as well as the physiological variables (i.e., retention time, pH variations, gastrointestinal fluids, microbiota).
In the past few years, dynamic in vitro gut models, such as SHIME (simulator of the human intestinal microbial ecosystem) (University of Ghent, Belgium) [38], the TIM-1 and TIM-2 (developed by TNO, Netherlands Organization for Applied Scientific Research, The Netherlands) [39] and the recent SIMGI (dynamic gastrointestinal tract simulator) (CIAL, CSIC-UAM, Spain), have been developed to simulate physiological conditions that can have an influence on the gut microbiota and their metabolic activity. All these systems contain different compartments representing the stomach, small intestine and colon.
5.1.2. Analytical Approaches to the Analysis of Phenolic Microbial-Derived Metabolites
Rapid, sensitive, and reliable analytical methods are needed for profiling microbe-derived phenolic metabolites in biological fluids in order to determine their contribution to the overall bioavailability of polyphenols and to allow identification of biomarkers of polyphenol exposure that could be further used to correlate dietary intake with particular health effects, including microbial modulation.
Gas chromatography (GC) coupled to mass spectrometry techniques (MS) and liquid chromatography (LC) coupled to a diode array detector (DAD), electrochemical (ECD) and, in particular, tandem mass spectrometry (ESI-MS/MS) are the most widely used analytical techniques for the analysis of microbial phenolic metabolites in biological fluids, which can be found in micromolar concentration. GC methodologies provide higher resolution and sensitivity than LC methodologies but a laborious sample preparation stage is required, since they usually involve the isolation of metabolites by liquid-liquid extraction (LLE) or other extraction procedures followed by further derivatization [40]. In contrast, for LC analysis the sample preparation step is more simple and faster and provides a very sensitive method for the quantification of selected phenolic metabolites when coupled to MS/MS [41]. The efficiency of separation and the sensitivity of LC–MS/MS methodologies can be largely improved by the use of ultra performance liquid chromatography (UPLC), which operates with smaller particle size (<2 μm) sorbent materials and at very high pressures (up to 15,000 psi). This technique coupled to MS detection is considered a good application in the analysis of phenolic compounds in biological fluids and food matrixes [41].
Metabolomics approaches have been used in clinical, pharmaceutical, and toxicological applications and have also recently emerged as fields of increasing importance in food and nutrition sciences [42]. The metabolome can be affected by different external and internal factors. Among them, diet is a very important external factor affecting the urinary metabolome because it produces notable changes in its composition [43]. Metabolomics studies of the polyphenols may lead to the discovery of new phytochemical metabolites and new biomarkers of intake that could allow the intake of dietary phytochemicals to be monitored, and eventually relate them to the expected biological effects [44].
5.2. Biotransformation of Wine Polyphenols by Gut Microbiota
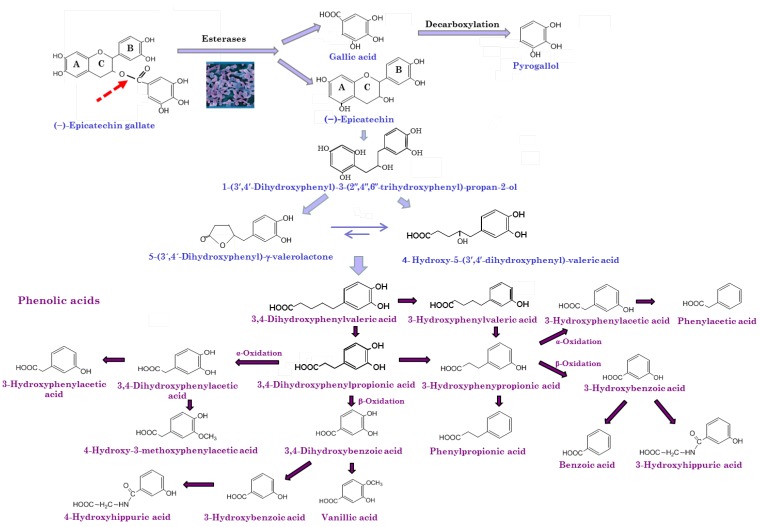
The microbial biotransformation of wine polyphenols is widely influenced by their chemical structure. Oligomers and polymers of flavan-3-ols are the major phenolic compounds present in the wine that reaches the colon. The colonic metabolism of dimeric procyanidins involves C-ring opening, followed by lactonization, decarboxylation, dehydroxylation, and oxidation reactions, among others [45] (Figure 3). In the case of galloylated monomeric flavan-3-ols, the microbial metabolism usually starts with the rapid cleavage of the gallic acid ester moiety by microbial esterases, giving rise to gallic acid which is further decarboxylated into pyrogallol. The C-ring is subsequently opened, giving rise to 1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol, which is later converted into 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone in the case of (epi)catechin or 5-(3′,4′,5′-trihydroxyphenyl)-γ-valerolactone in the case of (epi)gallocatechin. The valerolactone ring later breaks, giving rise to 5-(3′,4′-dihydroxyphenyl)valeric acid and/or 4-hydroxy-5-(3′,4′-dihydroxyphenyl) valeric acid. Subsequent biotransformations of these valeric acids give rise to hydroxyphenylpropionic and hydroxybenzoic acids by successive loss of carbon atoms from the side chain through β-oxidation (Figure 3).
Figure 3.
Catabolism of monomeric flavan-3-ols by gut microbiota.
With regard to microbial catabolism of flavonols, other flavonoid compounds present in the wine are directly transformed into 3,4- or 3,5-dihydroxylated phenylacetic acids [46]. In the case of anthocyanins, they are converted into 3,4-dihydroxy-, 4-hydroxy-, 3,4-dimethoxy- or 3-methoxyl-4-hydroxyl benzoic acids according to the substitution pattern of the B-ring of the precursor anthocyanin molecule [46]. However, in spite of the fact that anthocyanins are abundant in wine, their circulating levels in plasma are very low, which has been attributed to anthocyanin instability under neutral pH [47], their extensive metabolism in vivo [48], and their probable catabolism by intestinal microbiota [5]. Thus, it has been described that less than 1% of the ingested anthocyanins are typically absorbed and excreted in the urine [49], which might be found in low concentrations in serum, feces, urine (in the nanomolar concentrations) [50].
In the case of non-flavonoid compounds present in wine, hydroxycinnamic esters (i.e., caffeic acid derivatives) are mainly transformed into 3-hydroxyphenylpropionic acid, benzoic acid and 4-ethylcatechol. Once absorbed, the microbial metabolites are mainly metabolized in the liver by phase II enzymes as conjugated metabolites (glucuronides and sulfates). Other reactions that can take place are glycination, dehydrogenation, hydroxylation and methylation. In fact, different studies have proved the presence of phase II and microbial-derived phenolic metabolites in plasma and urine after ingestion of wine and wine polyphenols (see for review [51,52]). Some previous studies have confirmed that a moderate intake of red wine effectively led to a significant increase of phenolic metabolites in human feces [53,54].
Despite the advances recently made in the knowledge of the identification of phenolic metabolites, the specific bacterial species able to metabolize most wine polyphenols in the gastrointestinal tract and the anaerobic degradation pathways have still not been identified. So far, only a few bacterial species, some of them belonging to the class Clostridiales, such as Eubacterium sp. and Flavonifractor sp., and Eggerthella spp., have been reported to be able to initiate the metabolism of flavanol-3-ols [55,56,57].
5.3. Modulation of Gut Microbiota by Wine
Table 1 and Table 2 report different studies of modulation of gut microbiota by wine and grape polyphenols using different experimental approaches: batch culture fermentations, human gastrointestinal simulators, animal model studies and human interventions. For all of them, details about experimental conditions and/or study design, microbial techniques used and main effects on bacteria groups (growth enhancement, growth inhibition or no effect) are included.
Table 1.
Studies regarding modulation of gut microbiota by wine polyphenols using batch culture fermentations and gastrointestinal tract simulator.
| Studies Using Batch Culture Fermentation | ||||||||
|---|---|---|---|---|---|---|---|---|
| Reference | Fecal Concentration | Phenolic Compound/Food | Dose | Time of Incubation | Microbial Technique | Growth Enhancement | Growth Inhibition | No Effect |
| [58] | 10%, w/v | (+)-Catechin | 150 mg/L, 1000 mg/L | <48 h | FISH | Lactobacillus–Enterococcus spp.; Bifidobacterium spp.; C. coccoides–E. Rectale group E. coli | C. histolyticum group | |
| [59] | 10%, w/v | Malvidin-3-O-glucoside Anthocyanidins mixture | 20 mg/L and 200 mg/L 4850 mg/L and 48,500 mg/L |
<24 h | FISH | Lactobacillus–Enterococcus spp.; Bifidobacterium spp.; C. coccoides–E. rectale group | ||
| [60] | 10%, w/v | Grape seed extract fractions | 300–450 mg/L | <48 h | FISH | Lactobacillus–Enterococcus spp. | C. histolyticum group | |
| [61] | 1% w/v | Red wine extract | 600 mg/L | 48 h | FISH | C. histolyticum group | Lactobacillus–Enterococcus spp. | |
| [62] | Red wine extract | 500 mg/L | 48 h | qPCR | Lactobacillus spp.; Bifidobacterium spp.; Bacteroides spp.; Ruminococcus spp. | |||
| [63] | 20% w/v | Red wine/grape extract | 500–1000 mg/L | 72 h | HITChip | |||
| Studies Using a Gastrointestinal Simulator | ||||||||
| Reference | Simulator | Phenolic Compound/Food | Dose | Time | Microbial Technique | Population Increase | Population Decrease | No Effect |
| [64] | Twin-SHIME | Red wine-grape extract | 3 × daily dosing (1000 mg polyphenols as total daily dose) | 2 weeks | Plate count qPCR PCR-DGGE; Pyrosequencing | Klebsiella spp.; Alistipes spp.; Cloacibacillus spp.; Victivallis spp.; Akkermansia spp. | Bifidobacteria; Blautia coccoides group; Anaeroglobus spp.; Subdoligranulum spp. Bacteroides | |
Table 2.
Studies regarding modulation of gut microbiota by wine polyphenols in studies with animals and humans.
| Animal Model Studies | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reference | Simulator | Phenolic Compound/Food | Dose | Time | Microbial Technique | Population Increase | Population Decrease | No Effect | |
| [65] | Rats | Red wine polyphenols powder | 50 mg/kg | 16 weeks | Plate count | Lactobacilli; Bifidobacteria | Propionibacteria; Bacteroides; Clostridia | ||
| [66] | Broiler chicks | Grape seed extract (GSE) | 7.2 g/kg diet (GSE) (free access) | 21 days | Plate count T-RFLP | E. coli; Enterococcus spp.; Lactobacillus spp. | |||
| [67] | Pigs | Grape seed extract | 1% (free access) | 4 weeks | qPCR | Streptococcus spp.; Clostridium Cluster XIVa | Lactobacillus spp; Bifidobacterium spp. | ||
| [68] | Pigs | Grape seed extract | 1% w/w | 6 days | Ilumina MiSeq platform | Lachnospiraceae, Clostridales, Lactobacillu, Ruminococcacceae | |||
| Human Intervention Studies | |||||||||
| Reference | Volunteer Numbers | Phenolic Compound/Food | Dose | Treatment Duration | Microbial Technique | Population Increase | Population Decrease | No Effect | |
| [69] | 9 | Proantocyanidin-rich extract from grape seeds | 0.5 g/day | 6 weeks | Plate count | Bifidobacterium spp. | Enterobacteriaceae | ||
| [70] | 10 | Red wine | 272 mL/day | 20 days | qPCR | Enterococcus spp.; Prevotella spp.; Bacteroides Bifidobacterium spp.; Bacteroides uniformis Eggerthella lenta Blautia coccoides–E. rectale group | Clostridium spp.; C. histolyticum group | Actinobacteria | |
5.3.1. Studies Using Batch Culture Fermentations
One of the first relevant experiments using batch culture fermentation and a standard compound present in grape seeds and wine was carried out by Tzounis et al. [58] who found that the flavan-3-ol monomers ((−)-epicatechin and (+)-catechin) promoted the growth of the Clostridium coccoides–Eubacterium rectale group, which is known to produce large amounts of butyrate, a short-chain fatty acid (SCFA) with anti-inflammatory and antineoplastic properties (Table 1). Compared to (−)-epicatechin, (+)-catechin exposure resulted in a greater modification of the growth of the bacterial groups; (+)-catechin also increased the growth of Lactobacillus–Enterococcus spp., Bifidobacterium spp. and Escherichia coli but decreased the growth of Clostridium histolyticum. In contrast, the effect of (−)-epicatechin only significantly increased the growth of the C. coccoides–E. rectale group. Also using standard compounds of anthocyanins (i.e., malvidin-3-glucoside and a mixture of anthocyanins), Hidalgo et al. [59] found a significant increase in the growth of Lactobacillus–Enterococcus spp. and Bifidobacterium spp.
Recently, our research group carried out several batch culture fermentations of two flavan-3-ol fractions with different degrees of polymerization from grape seed extract and of wine polyphenols, with fecal microbiota from three healthy volunteers [60,61]. Both flavan-3-ol fractions induced the growth of Lactobacillus/Enterococcus spp. and inhibited the C. histolyticum group during fermentation, although the effects were only statistically significant with the less polymerized fraction. However, wine polyphenols only showed a slight inhibition in the C. histolyticum group, probably due to their lower content in flavan-3-ols and the time of exposure. Additionally, this type of fermentation has also been used to assess the contribution of certain probiotic strains to the colonic metabolism of polyphenols. With this in mind, Barroso et al. [62] carried out fermentations of a red wine extract inoculated with human microbiota obtained from the colonic compartments of a dynamic simulator, in the presence and absence of the probiotic strain Lactobacillus plantarum IFPL935. Microbial analysis by qPCR indicated that red wine polyphenols induced greater variations among in vitro batches harboring different colon-region (ascending colon, descending colon and effluent) microbiota than those found when L. plantarum IFPL935 was added. Batches inoculated with microbiota from the ascending colon were shown to harbor the major proportion of saccharolytic bacteria (Bacteroides, Bifidobacterium, Prevotella) whereas Clostridium groups were found in major numbers in the batches inoculated with microbiota simulating the distal regions [62].
Gross et al. [63] carried out in vitro fermentation of a red wine/grape and black tea polyphenols with fecal samples from 10 human volunteers. Microbial composition of fecal samples using HITChip analysis indicated different bacterial populations present in the individual fecal microbial communities. This inter-individual variability was related to the production of different metabolites, suggesting that each individual generates a specific microbial metabolome.
5.3.2. Studies Using Human Gastrointestinal Simulators
As an example of the versatility and potential of human gastrointestinal simulators, Table 1 also reports a study concerning the modulation of gut microbiota by polyphenols using the SHIME [38]. This validated model comprises stomach and small intestinal sections for predigestion of food as well as vessels stimulating the ascending, transverse and descending parts of the human colon, allowing assessment of changes in the different colonic areas that are very challenging to access in a human intervention.
The use of the twin-SHIME to investigate the effects of a red wine extract on the colonic microbiota was the aim of a study recently carried out by Kemperman et al. [64]. These authors characterized microbial community changes using a combination of analyses including cultivation, PCR-denaturing gradient gel electrophoresis (DGGE), quantitative PCR and high-throughput pyrosequencing of the 16S ribosomal RNA gene. They observed that the continuous administration of red wine extracts for two weeks could modulate select members of the human gut microbiota, revealing novel targets potentially involved in polyphenol metabolism and/or resistant microbes.
5.3.3. Animal Model Studies
It is widely known that preliminary evidence should be warranted in animal models before human intervention trials. Animal models contribute to a better understanding of the mechanisms and biological effects that could be likely to happen in the human body. Caution is required in extrapolating results to humans because culture-independent comparisons have revealed that most bacterial genera and species found in mice are not seen in humans, although the distal gut microbiota of mice and humans harbor the same bacterial phyla. Nevertheless, the use of germ-free mice inoculated with human fecal microbiota is of great relevance. Such humanized mice provide a model system for controlling host genotype, gut microbiota composition, diet, and housing conditions. To date, no studies have followed the composition of the intestinal microbiota in humanized rodents fed a diet rich in wine polyphenols [71].
In this section, studies performed on animals in order to assess the effects of wine polyphenols on the modulation of intestinal microbiota are summarized (Table 2).
Dolara et al. [65] showed that treatment with wine polyphenols in carcinogen-treated F344 rats was associated with a strong variation in the colonic microbiota, compared to the control-fed rats. Although the total bacterial counts and anaerobe/aerobe ratio of microorganisms in the feces from polyphenol-treated rats were similar to those from control rats, Propionibacteria, Bacteroides and Clostridia decreased while Lactobacilli and Bifidobacteria increased. Based on additional experiments, these authors concluded that reduction of oxidative damage, modulation of colonic flora and variation in gene expression may all be connected in the action of wine polyphenols on the intestinal function and carcinogenesis.
Another animal experiment was conducted to study the effect of the inclusion of grape seed extracts in the diet of broiler chicks [66] on intestinal microbiota. It was observed that, for the cecum, birds fed grape extracts had higher populations of E. coli, and Lactobacillus and Enterococcus species than birds in control groups. These authors concluded that polyphenol-rich grape products modified the gut morphology and intestinal microbiota and increased the degree of biodiversity in intestinal bacteria of broiler chicks.
Animal studies performed in pigs [67,68] demonstrated that grape seed extract administration caused an ecological shift in the microbiome. Thus, the administration of grape seed extracts produced lower counts of Streptococcus spp and Clostridium cluster XIVa [67], and increased Lachnospiraceae, Clostridales, Lactobacillus and Ruminococcus during the intervention period [68].
5.3.4. Human Intervention Studies
Investigations carried out with humans potentially provide the best models for studying the interactions of food components (e.g., polyphenols) with microbiota, although in vivo intervention trials hold inevitable practical and ethical limitations [8]. The use of crossover designs, where volunteers serve as their own control, permit multilevel analysis schemes that increase power, but as with other human intervention studies, they require a relevant number of volunteers to allow for statistically significant multivariate models [72]. Up to now, only a few studies have examined the in vivo impact of dietary polyphenols on the human gut microbiota, and most of them were focused on single polyphenol molecules and selected bacterial populations (Table 2).
In a study with a reduced number of subjects (n = 9), Yamakoski et al. [69] reported that administration of a proanthocyanidin-rich extract from grape seed significantly increased the fecal number of Bifidobacterium spp., whereas a reduction in the level of putrefactive bacteria, such as Enterobacteriaceae was observed.
Queipo-Ortuño et al. [70] performed a randomized, crossover and controlled trial (n = 10), which studied the effect of the intake of red wine, de-alcoholized red wine and gin over three consecutive periods. After the red wine period, the bacterial concentrations of Proteobacteria, Fusobacteria, Firmicutes and Bacterioidetes markedly increased compared with the washout period; significant increases in the number of Bifidobacterium spp. and Prevotella spp. were also observed. In contrast, Clostridium spp. and C. histolyticum group concentrations decreased after the red wine period.
6. Conclusions
Assessment of microbial metabolism of wine polyphenols is necessary to clarify the potential physiological effects on human health associated with moderate consumption of wine. There is scientific evidence that polyphenols may contribute to the maintenance of gut health by the modulation of the gut microbial balance through the stimulation of the growth of beneficial bacteria and the inhibition of pathogen bacteria, exerting prebiotic-like effects. Although genetic and environmental factors are the main determinants of gut microbiota composition, it is well established that diet influences microbial fermentation and total bacteria in the intestine. In fact, inter-individual variation in gut microbiota may, in part, reflect differences in dietary intake, although the response of the gut microbiota to dietary change can also differ among individuals. Therefore, the availability of advanced methods for monitoring bacterial communities could provide a better understanding of the underlying mechanisms in the polyphenols–microbiota–host triangle, and elucidate the implications of polyphenols on host health.
Acknowledgments
Authors of this review were funded by the Spanish MINECO through different projects (AGL2012-04172-C02-01, AGL2010-17499, BFU2012-35228), the CONSOLIDER INGENIO 2010 Program (project FUN-C-FOOD, CSD2007-063), as well by the CSIC (“Intramural” project 201270E065) and the “Comunidad de Madrid” (project ALIBIRD P2009/AGR-1469). Author M.D. thanks the Spanish “Ramón y Cajal” Programme for a contract.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pozo-Bayón M.A., Monagas M., Bartolome B., Moreno-Arribas M.V. Wine features related to safety and consumer health: An integrated perspective. Crit. Rev. Food Sci. Nutr. 2012;52:31–54. doi: 10.1080/10408398.2010.489398. [DOI] [PubMed] [Google Scholar]
- 2.Arranz S., Chiva-Blanch G., Valderas-Martínez P., Medina-Remón A., Lamuela-Raventós R.M., Estruch R. Wine, beer, alcohol and polyphenols on cardiovascular disease and cancer. Nutrients. 2012;4:759–781. doi: 10.3390/nu4070759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthoud H.R., Shin A.C., Zheng H. Obesity surgery and gut-brain communication. Physiol. Behav. 2011;105:106–119. doi: 10.1016/j.physbeh.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guarner F., Malagelada J.R. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 5.Williamson G., Clifford M.N. Colonic metabolites of berry polyphenols: The missing link to biological activity? Br. J. Nutr. 2010;104:S48–S66. doi: 10.1017/S0007114510003946. [DOI] [PubMed] [Google Scholar]
- 6.Selma M.V., Espín J.C., Tomás-Barberán F.A. Interaction between phenolics and gut microbiota: Role in human health. J. Agric. Food Chem. 2009;57:6485–6501. doi: 10.1021/jf902107d. [DOI] [PubMed] [Google Scholar]
- 7.Requena T., Monagas M., Pozo-Bayón M.A., Martín-Alvárez P.J., Bartolomé B., del Campo R., Ávila M., Martínez-Cuesta M.C., Peláez C., Moreno-Arribas M.V. Perspectives of the potential implications of wine polyphenols on human oral and gut microbiota. Trends Food Sci. Tech. 2010;21:332–344. doi: 10.1016/j.tifs.2010.04.004. [DOI] [Google Scholar]
- 8.Hervert-Hernández D., Goñi I. Dietary polyphenols and human gut microbiota: A review. Food Rev. Int. 2011;27:154–169. doi: 10.1080/87559129.2010.535233. [DOI] [Google Scholar]
- 9.Tuohy K.M., Conterno L., Gasperotti M., Viola R. Up-regulating the human intestinal microbiome using whole plant foods, polyphenols, and/or fiber. J. Agric. Food Chem. 2012;60:8776–8782. doi: 10.1021/jf2053959. [DOI] [PubMed] [Google Scholar]
- 10.Etxebarria U., Fernández-Quintela A., Milagro F.I., Aguirre L., Martinez J.A., Portillo M.P. Impact of polyphenols and polyphenol-rich dietary sources on gut microbiota composition. J. Agric. Food Chem. 2013;61:9517–9533. doi: 10.1021/jf402506c. [DOI] [PubMed] [Google Scholar]
- 11.Van Duynhoven J., Vaughan E.E., van Drosten F., Gomez-Roldan V., de Vos R., Vervoort J., van der Hooft J.J.J., Roger L., Draijer R., Jacobs D.M. Interactions of black tea polyphenols with human gut microbiota: Implications for gut and cardiovascular health. Am. J. Clin. Nutr. 2013;98:1631S–1641S. doi: 10.3945/ajcn.113.058263. [DOI] [PubMed] [Google Scholar]
- 12.Cardona F., Andrés-Lacueva C., Tulipani S., Tinahones F.J., Queipo-Ortuño M.I. Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 2013;24:1415–1422. doi: 10.1016/j.jnutbio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Tzounis X., Rodriguez-Mateos A., Vulevic J., Gibson G.R., Kwik-Uribe C., Spencer J.P.E. Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, double-blind, crossover intervention study. Am. J. Clin. Nutr. 2011;93:62–72. doi: 10.3945/ajcn.110.000075. [DOI] [PubMed] [Google Scholar]
- 14.Urpi-Sarda M., Monagas M., Khan N., Lamuela-Raventos R.M., Santos-Buelga C., Sacanella E., Castell M., Permanyer J., Andrés-Lacueva C. Epicatechin, procyanidins, and phenolic microbial metabolites after cocoa intake in humans and rats. Anal. Bioanal. Chem. 2009;394:1545–1556. doi: 10.1007/s00216-009-2676-1. [DOI] [PubMed] [Google Scholar]
- 15.García-Ruiz A., Bartolomé B., Martínez-Rodríguez A.J., Pueyo E., Martín-Alvárez P.J., Moreno-Arribas M.V. Potential of phenolic compounds for controlling lactic acid bacteria growth in wine. Food Control. 2008;19:835–841. doi: 10.1016/j.foodcont.2007.08.018. [DOI] [Google Scholar]
- 16.Hertog M.G.L., Feskens E.J.M., Hollman P.C.H., Katan M.B., Kromhout D. Dietary antioxidant flavonoids and risk of coronary heart disease: The Zutphen Elderly Study. Lancet. 1993;342:1007–1011. doi: 10.1016/0140-6736(93)92876-U. [DOI] [PubMed] [Google Scholar]
- 17.Del Rio D., Rodriguez-Mateos A., Spencer J.P.E., Tognolini M., Borges G., Crozier A. Dietary (poly)phenolics in human health: structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid. Redox Sign. 2013;18:1818–1892. doi: 10.1089/ars.2012.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vison J.A., Teufel K., Wu N. Red wine, dealcoholized red wine, and especially grape juice, inhibit atherosclerosis in a harmster model. Atherosclerosis. 2001;156:67–72. doi: 10.1016/S0021-9150(00)00625-0. [DOI] [PubMed] [Google Scholar]
- 19.Khan N., Adhami V.M., Mukhtar H. Apoptosis by dietary agents for prevention and treatment of prostate cancer. Endocr. Relat. Cancer. 2010;17:R39–R52. doi: 10.1677/ERC-09-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo P., Tedesco I., Russo M., Russo G.L., Venezia A., Cicala C. Effects of de-alcoholated red wine and its phenolic fractions on platelet aggregation. Nutr. Metab. Cardiovasc. Dis. 2001;11:25–29. [PubMed] [Google Scholar]
- 21.Rechner A.R., Kroner C. Anthocyanins and colonic metabolites of dietary polyphenols inhibit platelet function. Thromb. Res. 2005;116:327–334. doi: 10.1016/j.thromres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Clifford M.N. Diet-derived phenols in plasma and tissues and their implications for health. Planta Medica. 2004;70:1103–1114. doi: 10.1055/s-2004-835835. [DOI] [PubMed] [Google Scholar]
- 23.Scalbert A., Williamson G. Dietary intake and bioavailability of polyphenols. J. Nutr. 2000;130:2073S–2085S. doi: 10.1093/jn/130.8.2073S. [DOI] [PubMed] [Google Scholar]
- 24.Manach C., Scalbert A., Morand C., Rémésy C., Jiménez L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004;79:727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 25.Maukonen J., Motto J., Suihko M.L., Saarela M. Intra-individual diversity and similarity of salivary and faecal microbiota. J. Med. Microbiol. 2008;57:1560–1568. doi: 10.1099/jmm.0.47352-0. [DOI] [PubMed] [Google Scholar]
- 26.Gerritsen J., Smidt H., Rijkers G.T., de Vos W.M. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6:209–240. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore W.E., Moore L.H. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 1995;61:3200–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson G.R., Roberfroid M.B. Dietary modulation of the human colonic microbiota-Introducing the concept of prebiotics. J. Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 29.Guarner F. Enteric flora in health and disease. Digestion. 2006;73:5–12. doi: 10.1159/000089775. [DOI] [PubMed] [Google Scholar]
- 30.Gibson G.R. Dietary modulation of the human gut microflora using prebiotics. Br. J. Nutr. 1998;80:S209–S212. [PubMed] [Google Scholar]
- 31.Van der Waaij L.A., Limburg P.C., Mesander G., van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996;38:348–354. doi: 10.1136/gut.38.3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salminen S., Wright A., Morelli L., Marteau P., Brassart D., Vos W.M., Fondem R., Saxelin M., Collins K., Mogensen G., et al. Demostration of safety of probiotic—A review. Int. J. Food Microbiol. 1998;44:93–106. doi: 10.1016/S0168-1605(98)00128-7. [DOI] [PubMed] [Google Scholar]
- 33.Rastall R.A., Gibson G.R., Gill H.S., Guarner F., Klaenhammer T.R., Pot B., Reid G., Rowland I.R., Sanders M.E. Modulation of the microbial ecology of the human colon by probiotic, prebiotics and synbiotics to enhance human health: An overview of enabling science and potential applications. FEMS Microbiol. Ecol. 2005;52:145–152. doi: 10.1016/j.femsec.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Joly F., Mayeur C., Bruneau A., Noordine M.L., Meylheuc T., Langella P., Messing B., Duée P.H., Cherbuy C., Thomas M. Drastic changes in fecal and mucosa-associated microbiota in adult patients with short bowel syndrome. Biochimie. 2010;92:753–761. doi: 10.1016/j.biochi.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Kemperman R.A., Bolca S., Roger L.C., Vaughan E.E. Novel approaches for analysing gut microbes and dietary polyphenols: Challeges and opportunities. Microbiology. 2010;156:3224–3231. doi: 10.1099/mic.0.042127-0. [DOI] [PubMed] [Google Scholar]
- 36.Fraher M.H., O’Toole P.W., Quigley E.M.M. Techniques used to characterize the gut microbiota: A guide for the clinician. Nat. Rev. Gastroentero. 2012;9:312–322. doi: 10.1038/nrgastro.2012.44. [DOI] [PubMed] [Google Scholar]
- 37.Glenn T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resources. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 38.De Boever P., Deplancke B., Verstraete W. Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem is improved by supplementing a soygerm powder. J. Nutr. 2000;130:2599–2606. doi: 10.1093/jn/130.10.2599. [DOI] [PubMed] [Google Scholar]
- 39.Molly K., Vande Woestyne M., Verstraete W. Development of a 5 step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993;39:254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- 40.Muñoz-González C., Moreno-Arribas M.V., Rodríguez-Bencomo J.J., Cueva C., Martín-Álvarez P.J., Pozo-Bayón M.A. Feasibility and application of liquid-liquid extraction combined with gas chromatography-mass spectrometry for the analysis of phenolic acids from grape polyphenols degraded by human faecal microbiota. Food Chem. 2012;133:526–535. doi: 10.1016/j.foodchem.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 41.Sánchez-Patán F., Monagas M., Moreno-Arribas M.V., Bartolomé B. Determination of microbial phenolic acids in human faeces by UPLC-ESI-TQ MS. J. Agric. Food Chem. 2011;59:2241–2247. doi: 10.1021/jf104574z. [DOI] [PubMed] [Google Scholar]
- 42.García-Cañas V., Simó C., León C., Cifuentes A. Advances in nutrigenomics research: Novel and future analytical approaches to invesigate the biological activity of natural compounds and food functions. J. Pharmaceut. Biomed. 2010;51:290–304. doi: 10.1016/j.jpba.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 43.Brennan L. Metabolomics in nutrition research: current status and perspectives. Biochem. Soc. Trans. 2013;41:670–673. doi: 10.1042/BST20120350. [DOI] [PubMed] [Google Scholar]
- 44.Llorach R., Garrido I., Monagas M., Urpi-Sarda M., Tulipani S., Bartolomé B., Andrés-Lacueva C. Metabolomics study of human urinary metabolome modifications after intake of almond (Prunus. dulcis (Mill.) D.A. Webb) skin polyphenols. J. Proteome Res. 2010;9:5859–5867. doi: 10.1021/pr100639v. [DOI] [PubMed] [Google Scholar]
- 45.Monagas M., Urpi-Sarda M., Sánchez-Patán F., Llorach R., Garrido I., Gómez-Cordovés C., Andrés-Lacueva C., Bartolomé B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010;1:233–253. doi: 10.1039/c0fo00132e. [DOI] [PubMed] [Google Scholar]
- 46.Aura A.M. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem. Rev. 2008;7:407–429. doi: 10.1007/s11101-008-9095-3. [DOI] [Google Scholar]
- 47.Fossen T., Cabrita L., Andersen O.M. Colour and stability of pure anthocyanins influenced by pH including the alkaline region. Food Chem. 1998;63:435–440. doi: 10.1016/S0308-8146(98)00065-X. [DOI] [Google Scholar]
- 48.Mullen W., Edwards C.A., Serafini M., Crozier A. Bioavailability of pelargonidin-3-O-glucoside and its metabolites in humans following the ingestion of strawberries with and without cream. J. Agric. Food Chem. 2008;56:713–719. doi: 10.1021/jf072000p. [DOI] [PubMed] [Google Scholar]
- 49.McGhie T.K., Walton M.C. The bioavailability and absorption of anthocyanins: Towards a better understanding. Mol. Nutr. Food Res. 2007;51:702–713. doi: 10.1002/mnfr.200700092. [DOI] [PubMed] [Google Scholar]
- 50.De Ferrars R.M., Czank C., Saha S., Needs P.W., Zhang Q., Raheem K.S., Botting N.P., Kroon P.A., Kay C.D. Methods for isolating, identifying, and quantifying anthocyanin metabolites in clinical samples. Anal. Chem. 2014;86:10052–10058. doi: 10.1021/ac500565a. [DOI] [PubMed] [Google Scholar]
- 51.Stockley C., Teissedre P.L., Boban M., di Lorenzo C., Restani P. Bioavailability of wine-derived phenolic compounds in humans: A review. Food Funct. 2012;3:995–1007. doi: 10.1039/c2fo10208k. [DOI] [PubMed] [Google Scholar]
- 52.Boto-Ordoñez M., Rothwell J.A., Andres-Lacueva C., Manach C., Scalbert A., Urpi-Sarda M. Prediction of the wine polyphenol metabolic space: An application of the Phenol-Explorer database. Mol. Nutr. Food Res. 2014;3:466–477. doi: 10.1002/mnfr.201300411. [DOI] [PubMed] [Google Scholar]
- 53.Jiménez-Girón A., Queipo-Ortuno M.I., Boto-Ordoñez M., Muñoz-González I., Sánchez-Patán F., Monagas M., Martín-Álvarez P.J., Murri M., Tinahones F.J., Andres-Lacueva C., et al. More comparative study of microbial-derived phenolic metabolites in human feces after intake of gin, red wine, and dealcoholized red wine. J. Agric. Food Chem. 2012;61:3909–3915. doi: 10.1021/jf400678d. [DOI] [PubMed] [Google Scholar]
- 54.Muñoz-González I., Jiménez-Girón A., Martín-Álvarez P.J., Bartolomé B., Moreno-Arribas M.V. Profiling of microbial-derived phenolic metabolites in human feces after moderate red wine intake. J. Agric. Food Chem. 2013;61:9470–9479. doi: 10.1021/jf4025135. [DOI] [PubMed] [Google Scholar]
- 55.Wang L.Q., Meselhy M.R., Li Y., Nakamura N., Min B.S., Qin G.W., Hattori M. The heterocyclic ring fission and dehydroxylation of catechins and related compounds by Eubacterium sp. strain SDG-2, a human intestinal. Chem. Pharm. Bull. 2001;49:1640–1643. doi: 10.1248/cpb.49.1640. [DOI] [PubMed] [Google Scholar]
- 56.Kutschera M., Engst W., Blaut M., Braune A. Isolation of catechin-converting human intestinal bacteria. J. Appl. Microbiol. 2011;111:165–175. doi: 10.1111/j.1365-2672.2011.05025.x. [DOI] [PubMed] [Google Scholar]
- 57.Jin J.S., Hattori M. Isolation and characterization of a human intestinal bacterium Eggerthella sp. CAT-1 capable of cleaving the C-Ring of (+)-catechin and (−)-epicatechin, followed by p-dehydroxylation of the B-Ring. Biol. Pharm. Bull. 2012;35:2252–2256. doi: 10.1248/bpb.b12-00726. [DOI] [PubMed] [Google Scholar]
- 58.Tzounis X., Vulevic J., Kuhnle G.G.C., George T., Leonczak J., Gibson G.R., Kwik-Uribe C., Spencer J.P.E. Flavanol monomer-induced changes to the human faecal microflora. Br. J. Nutr. 2008;99:782–792. doi: 10.1017/S0007114507853384. [DOI] [PubMed] [Google Scholar]
- 59.Hidalgo M., Oruna-Concha M.J., Kolida S., Walton G.E., Kallithraka S., Spencer S.P.E., Gibson G.R., Pascual-Teresa S. Metabolism of antocyanins by human gut microflora and their influence on gut bacterial growth. J. Agric. Food Chem. 2012;60:3882–3890. doi: 10.1021/jf3002153. [DOI] [PubMed] [Google Scholar]
- 60.Cueva C., Sánchez-Patán F., Monagas M., Walton G.E., Gibson G.R., Martin-Alvarez P.J., Bartolome B., Moreno-Arribas M.V. In vitro fermentation of grape seed flavan-3-ol fractions by human faecal microbiota: Changes in microbial groups and phenolic metabolites. FEMS Microbiol. Ecol. 2013;83:729–805. doi: 10.1111/1574-6941.12037. [DOI] [PubMed] [Google Scholar]
- 61.Sánchez-Patán F., Cueva C., Monagas M., Walton G.E., Gibson G.R., Quintanilla-Lopez J.E., Lebron-Aguilar R., Martin-Alvarez P.J., Moreno-Arribas M.V., Bartolome B. In vitro fermentation of a red wine extract by human gut microbiota; changes in microbial groups and formation and phenolic metabolites. J. Agric. Food Chem. 2012;60:2136–2147. doi: 10.1021/jf2040115. [DOI] [PubMed] [Google Scholar]
- 62.Barroso E., Sánchez-Patán F., Martin-Alvarez P.J., Bartolome B., Moreno-Arribas M.V., Pelaez C., Requena T., van de Wiele T., Martinez-Cuesta M.C. Lactobacillus plantarum IFPL935 favors the initial metabolism of red wine polyphenols when added to a colonic microbiota. J. Agric. Food Chem. 2013;61:10163–10172. doi: 10.1021/jf402816r. [DOI] [PubMed] [Google Scholar]
- 63.Gross G., Jacobs D.M., Peters S., Possemiers S., van Duynhoven J., Vaughan E.E., van de Wiele T. In vitro bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays strong interindividual variability. J. Agric. Food Chem. 2010;58:10236–10246. doi: 10.1021/jf101475m. [DOI] [PubMed] [Google Scholar]
- 64.Kemperman R.A., Gross G., Mondot S., Possemiers S., Marzorati M., van de Wiele T., Dore J., Vaughan E.E. Impact of polyphenols from black tea and red wine/grape juice on a gut model microbiome. Food Res. Int. 2013;53:659–669. doi: 10.1016/j.foodres.2013.01.034. [DOI] [Google Scholar]
- 65.Dolara P., Luceri C., de Filippo C., Femia A.P., Giovannelli L., Caderni G., Cecchini C., Silvi S., Orpianesi C., Cresci A. Red wine polyphenols influence carcinogenesis, intetinalmicroflora, oxidative damage and gene expression profile of colonic mucosa in F344 rats. Mutat. Res. 2005;591:237–246. doi: 10.1016/j.mrfmmm.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 66.Viveros A., Chamorro S., Pizarro M., Arija I., Centeno C., Brenes A. Effects of dietary polyphenols-rich grape productus on intestinal microflora and gut morphology in broiler chicks. Poultry Sci. 2011;90:566–578. doi: 10.3382/ps.2010-00889. [DOI] [PubMed] [Google Scholar]
- 67.Fiesel A., Gessner D.K., Most E., Eder K. Effects of dietary polyphenol-rich plant products from grape or hop on pro-inflammatory gene expression in the intestine, nutrient digestibility and faecal microbiota of weaned pigs. BMC Vet. Res. 2014;10:196. doi: 10.1186/s12917-014-0196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choy Y.Y., Quifer-Rada F., Holstege D.M., Frese S.A., Calvert C.C., Mills D.A., Lamuela-Raventos R.M., Waterhouse A.L. Phenolic metabolites and substantial microbiome changes in pig feces by ingesting grape seed proantocyanidins. Food Funct. 2014;5:2298–2308. doi: 10.1039/C4FO00325J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamakoshi J., Tokutake S., Kikuchi M., Kubota Y., Konishi H., Mitsuoka T. Effect of proantocyanidin-rich extract from grape seed on human fecal flora and fecalodor. Microb. Ecol. Health Dis. 2001;13:25–31. [Google Scholar]
- 70.Queipo-Ortuño M.I., Boto-Ordoñez M., Murri M., Gómez-Zumaquero J.M., Clemente-Postigo M., Estruch R., Diaz F.C., Andres-Lacueva C., Tinahones F.J. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am. J. Clin. Nutr. 2012;95:1323–1334. doi: 10.3945/ajcn.111.027847. [DOI] [PubMed] [Google Scholar]
- 71.Van Duynhoven J., Vaughan E.E., Jacobs D.M., Kemperman R.A., van Velzen E.J.J., Gross G., Roger L.C., Possemiers S., Smilde A.K., Dore J., et al. Metabolic fate of polyphenols in the human superorganism. Proc. Natl. Acad. Sci. USA. 2011;108:4531–4538. doi: 10.1073/pnas.1000098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Velzen E.J.J., Wasterhuis J.A., van Duynhoven J.P.M., van Drosten F.A., Hoefsloot H.C.J., Jacobs D.M., Smit S., Draijer R., Kroner C.I., Smilde A.K. Multilevel data analysis of a crossover designed human nutritional intervention study. J. Proteome Res. 2008;7:4483–4491. doi: 10.1021/pr800145j. [DOI] [PubMed] [Google Scholar]