Abstract
Any tissue is made up of a heterogeneous mix of spatially distributed cell types. In response to any (patho) physiological cue, responses of each cell type in any given tissue may be unique and cannot be homogenized across cell-types and spatial co-ordinates. For example, in response to myocardial infarction, on one hand myocytes and fibroblasts of the heart tissue respond differently. On the other hand, myocytes in the infarct core respond differently compared to those in the peri-infarct zone. Therefore, isolation of pure targeted cells is an important and essential step for the molecular analysis of cells involved say in the progression of disease. Laser capture microdissection (LCM) is powerful to obtain a pure targeted cell subgroup, or even a single cell, quickly and precisely under the microscope, successfully tackling the problem of tissue heterogeneity in molecular analysis. This review presents an overview of LCM technology, the principles, advantages and limitations and its down-stream applications in the fields of proteomics, genomics and transcriptomics. With powerful technologies and appropriate applications, this technique provides unprecedented insights into cell biology from cells grown in their natural tissue habitat as opposed to those cultured in artificial petri dish conditions.
Keywords: Laser capture microdissection (LCM), Genomics, Proteomics, Transcriptomics
1. Introduction
A large component of tissue analysis approaches depends on tissue homogenization. The process of homogenization disregards (a) the spatial localization of tissue being studied with respect to the location of the focal event such as, ischemia-reperfusion or injury site, and (b) the fact that biological events in different cell types of same tissue are likely to be dissimilar. Cell heterogeneity is widespread and increasingly apparent in eukaryotic cell populations where cells differ not only in terms of function and specialization, but also in size and morphology [1]. Signaling pathways are complex and highly interconnected, have a spatially dependent nature, and rely on low abundance molecules. Because of these stochastic properties, cellular signaling pathways depend on underpinnings of heterogeneity in cell systems [2].
The molecular analysis of DNA, RNA, and protein derived from tissue specimens has revolutionized pathology and led to the identification of a broad range of diagnostic and prognostic markers influencing the clinical practice [3]. Techniques such as Southern blot analysis and polymerase chain reaction (PCR) are integral parts of the diagnostic repertoire of current pathology [3]. Next-generation sequencing technologies have reshaped our understanding of the molecular constituents of cells and their regulatory elements. The completion of the Human Genome Project has led to a surge in the use of genomic and proteomic technologies in the identification of markers for early detection of several diseases and in the discovery of molecular-targeted treatments [3–5].
The number of defined human genes and expressed sequence tags continues to grow. New tools are being developed for the interrogation of the databases which have sprung up housing these newly characterized genes [4, 5]. The majority of the mammalian genome is transcribed, generating a vast repertoire of transcripts that includes protein-coding RNAs as well as non-coding RNAs (ncRNAs). ncRNAs include transcripts that can greatly differ in size and biogenesis and whose biological activities remain largely unexplored [6–8].
When used together, technologies that isolate discrete cell types/tissues and modern sequencing platforms, genome-wide transcriptional profiling has the power to unveil new hypothesis generating information [9]. An increasing number of these observations challenge the concept of functional “ectopic” expression suggesting that proteins with defined biochemical activities may exert their biological function or acquire some new ones in previously unidentified cells and tissues [9]. Proteomic analysis, therefore, has the unique capacity to snap shot the current status and composition of cell phenotypes within a defined time frame, which is highly relevant to biological functions [2].
High-throughput screening techniques are now widely available enabling investigators to rapidly screen and confirm new genes, mRNA transcripts and proteins. The last decade has shown considerable changes in the instrumentation and analytical techniques used in the area of single cell analysis, which have led to major achievements in the field of diagnosis and treatment [2, 10–12]. Laser Microbeam Microdissection (LMM) and Laser Capture Microdissection (LCM) were the first among those new technologies, have been used in attempts to overcome the heterogeneity of the tissues. Considerable efforts are being made to apply such methodologies to a limited number of cells. However, the analytical advances in proteomics have not been as rapid or fortunate as those of single cell transcriptomics mainly because of the lack of protein amplification techniques [13].
2. Laser Capture Microdissection
Modern laser microdissection technology was first described in the early 20th century but it has been steadily advanced and modified over the years. LMM was known to use a pulsed UV laser with a small beam focus to cut out areas or cells of interest by photo-ablation of adjacent tissue [12, 14]. But it was less effective at single cell collection and technically became more challenging and time consuming process. LCM was then introduced by the National Cancer Institute of the National Institutes of Health in Bethesda, as the next generation technology for LMM. LCM has rapidly found widespread interest as an attractive addition to the repertoire of microdissection techniques [15–17]. It has allowed accurate separation of tumor, stromal and normal cells within a single biopsy specimen [15, 18, 19]. Besides that LCM technology has been used in a wide variety of applications, such as pathology [20], pre-fertilization genetic diagnosis [21], organ transplantation [22, 23], psychiatric disorders [24], single cell mutation analysis [25], analysis of keratinocytes from wounds [26], transcriptome-wide analysis of blood vessels from human skin and wound-edge tissue [27], gene expression [28, 29], tissue chimerism [22], and molecular characterization of cancer cells [30, 31]. In addition, high-precision surgical technologies coupled with LCM now make it possible to isolate targeted nuclei with single-cell precision from surgically removed tissue. This approach has also been used to successfully characterize the protein content or differential genomic profiles of a number of specific cell populations and subcellular structures [32–34]. Application of these technologies to patient samples has allowed dissection of genomic changes, expression events, and differential expression, activation, and signaling of a variety of proteins in tumor samples to be possible [35, 36].
2.1. Overview of LCM technology
LCM is a state-of-the-art technology for isolating pure cell populations from a heterogeneous tissue specimen. It can precisely target and capture the cells of interest for a wide range of downstream assays [37]. In 1976, Isenberg et al were among the first to use primitive UV laser technology in the surgery, but their approach required massive space-occupying instruments to dissect subpopulations of cell types from a heterogeneous background [38]. LCM was devised at the NIH by Lance Liotta, Emmert-Buck and their team who recognized a need to develop a microscope-based microdissection system for accurate and efficient dissection of cells from histological tissue sections of solid tumors to fully exploit emerging molecular analytical technologies [38]. That system was rapidly moved into commercial production by Arcturus Engineering (Mountainview, CA) and offers one of several laser-assisted dissection strategies that allow direct selection of cell types without the need for enzymatic processing or growth in culture [37–40]. The PALM Microbeam (Carl Zeiss MicroImaging GmbH, Bernried, Germany) and Leica LMD6000 (Leica Microsystems Inc., Bannockburn, IL, USA) laser microdissection systems were also developed promptly and broadened its applications internationally [37]. At present, thousands of researchers’ worldwide benefit from this technology, and thousands of publications involving LCM have appeared. As a result, approaches to molecular analysis of pathologic processes have been enhanced significantly.
2.2. Types and devices of LCM
There are two general classes of laser capture microdissection systems: infrared (IR LCM) and ultraviolet (UV LCM) [37, 41, 42]. LCM instruments exist in a form of manual and automated (robotic) platforms [18]. The system is based on an inverted light microscope (with or without a fluorescent module), fitted with a laser device to facilitate the visualization and procurement of cells. This platform consists of an inverted microscope, a solid state near infrared laser diode, a laser control unit, a joy stick controlled microscope stage with a vacuum chuck for slide immobilization, a CCD camera, and a color monitor. The LCM microscope is usually connected to a personal computer for additional laser control and image archiving [43]. The minimum diameter of the laser beam of the LCM microscope is 7.5 μm and the maximum diameter is 30 μm. In this system most of the energy is absorbed by the membrane, the maximum temperatures reached by the tissue upon laser activation are in the range of 90°C for several milliseconds; thus leaving biological macromolecules of interest intact [43, 44]. The low energy of the infrared laser also avoids potentially damaging photochemical effects. All commercially available laser microdissection systems are essentially based on one of these two platforms, with the main variations concerning system configuration and intended applications.
2.2.1. Infrared LCM (IR LCM)
In 1996, Emmert-Buck and coworkers at the National Institutes of Health introduced the infrared (IR) laser capture microdissection system [38]. This system became commercially available by Arcturus Engineering as the PixCell system a year after the first publication describing its use was reported [38]. The PixCell platform is based on the placement of a thin transparent thermoplastic film over a tissue section. Consequently, the tissue is visualized microscopically. Cells of interest are selectively adhered to the film with a fixed-position, short duration, focused pulse from an IR laser [38]. The adherence of the cells to the film exceeds the adhesion to the glass slide, which allows selective removal of the cells of interest [45]. Removed cells are detached by lifting of the film, which is then transferred to a microcentrifuge tube containing buffer solutions required for the isolation of DNA or RNA [43, 46].
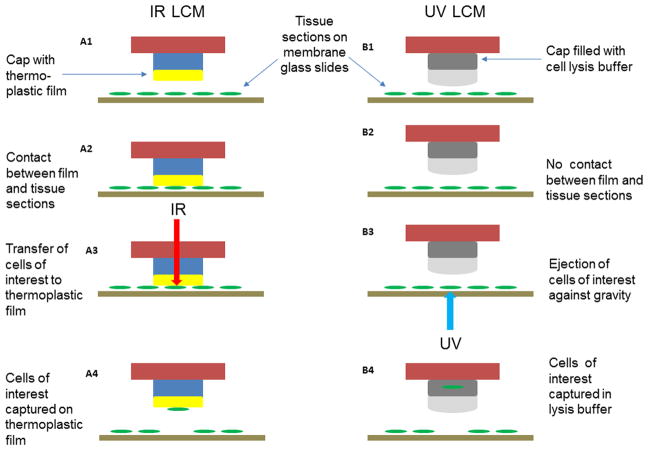
Fig. 1. Mechanism of tissue capture using the PALM and Arcturus LCM systems.
When isolating cells with the Arcturus LCM system (A1–A4), tissue sections are prepared on a conventional microscope slide (A1) and a ‘cap’ that contains an infrared-sensitive transfer film is placed physically onto cells of interest (A2). An infrared laser is fired through the cap over the cells of interest, activating the membrane, which infiltrates into the underlying tissue (A3). When the cap is removed, captured tissue is removed, leaving behind unwanted cells (A4). When isolating cells with the PALM–LCM system (B1–B4), tissue sections are firstly prepared on membrane-coated microscope slides and collecting caps are filled with lysis buffer (B1). In this system there is no contact between film and tissue sections (B2). A UV laser is focused onto the focal plane of the section and used to cut around cells of interest by laser ablation (B3), physically detaching the cells of interest and the underlying membrane from the surrounding tissue. The laser is then focused directly below the cells of interest, and a single laser pulse is fired, catapulting them vertically into the overlying eppendorf cap (B4).
2.2.2. Ultraviolet LCM (UV LCM)
In 1998, Schütze and Lahr developed an UV-based method for LCM, which operated on a quite different principle than IR LCM [12]. Currently, this platforms use tissue that has been mounted on a 6 μm membrane and placed on a glass slide, onto which the operator directs an UV laser beam under direct visualization [38, 39, 47]. The narrow-beam UV laser is used to draw around the cell or cells of interest leaving the desired cell population intact while simultaneously ablating away unwanted tissue[47]. By increasing the power of the laser, the desired cells were subsequently catapulted against gravity into an overhanging cap. This system was commercialized by PALM Zeiss Microlaser Technologies [37, 41, 42]. There are two major advantages of this method; first, it avoids any intricate operator dependent step, and second, by ablating the adjacent rim of unwanted tissue, non-specific adherence of tissue to the cap is avoided. An example of a combined IR/UV system is the automated Arcturus Veritas™ instrument (Arcturus, Mountain View, CA) [37].
2.3. Sample sources and applications for LCM
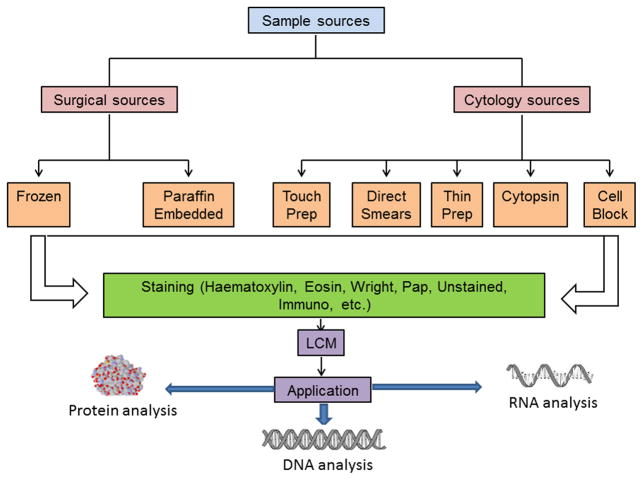
LCM techniques can be applied to histological specimens, living cells and cell cultures, plant material, chromosome spreads, forensic preparations, formalin-fixed paraffin-embedded (FFPE) or fresh-frozen tissues and stained or unstained tissues. The process of LCM is straightforward, and there are a considerable number of commercially available kits that have aided in simplifying the process. Sample preparation in a proper way is very important for successful capture. Optimal cutting temperature (OCT) compound-embedded frozen tissue or formalin-fixed paraffin-embedded (FFPE) tissues are sectioned by cryostat or microtome [37]. The optimal laser capture microdissection is achieved with tissue sections cut at thickness of 5–15 uM. Tissue sections thinner than 5 uM may not provide full cell thickness and sections greater than 15 uM may not microdissect completely. The sections are collected onto membrane slides and undergo staining protocol followed by dehydration after tissue staining with hematoxylin and eosin. Adequate dehydration of tissue section is crucial to minimize upward adhesive force between slide and the tissue. After microdissection is completed, the cap with target cells can be used for any molecular analytical methods [42] and the quality of the isolated DNA, RNA, and protein can be monitored and assessed with a bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA quality assessment is generally recommended prior to capture as high RNA integrity numbers (RIN) represent successful RNA sample preparation [37]. Frozen tissue offers excellent preservation of RNA, DNA, and proteins and making it is optimal for downstream analysis, however, it lacks histologic differentiation and often inconvenient for handling and storage. The yield of RNA from frozen tissues is affected primarily by quality of sample, time and type of preservation (preferable −80C), fixation method and efficiency of microdissection (less than an hour). FFPE tissue is the standard for preservation of tissue morphology and has been used by most pathology laboratories for decades. However, it creates cross-links between nucleic acids and proteins and between different proteins [37]. Although proteins are not extractable from FFPE samples, RNA can be isolated from FFPE tissue for downstream applications such as RT-PCR and cDNA microarray [48].
Fig. 2.
Schematic diagram of sample sources and applications for LCM
2.4. Advantages and Limitations of LCM
The most important advantages of LCM are its speed, precision, and versatility. Depending on the laser spot size, the architectural features of the tissue and the desired precision of the microdissection, thousands of cells can be collected within a fraction of time [43]. Morphology of both the captured cells as well as the residual tissue is well preserved and reduces the danger of tissue loss. LCM is very fast and does not destroy adjacent tissues; several tissue components can be sampled sequentially from the same slide, i.e. normal and neoplastic cells [15]. Because of easy handling LCM microscope can be easily calibrated and adjusted thereby shortening the learning curve, thus integrating into procedures for molecular genetic tissue diagnostics. LCM can be applied to a wide range of cell and tissue preparations. Even stained, archival sections can be microdissected successfully after removal of the coverslip [43]. In addition, tissues can be stained by conventional haematoxylin and eosin or by immunohistochemistry to identify particular cells of interest. The film and heat produced by the low power laser do not affect the integrity of DNA, RNA, or protein. In 1999 Banks et al compared tissue samples collected by LCM with tissue collected by more conventional methods and interestingly found no gross changes in protein profiles between LCM-collected and conventionally collected tissue when compared to electrophoresis [49]. Two tested proteins, HSP-60 and β2-microglobulin, retained their antigenicity on Western blot.
The few limitations of LCM mostly reflect the difficulties of microdissection in general. Among them, one significant limitation of LCM is that the microdissected tissue section is not cover slipped. Cover slipping would prevent physical access to the tissue surface, which is a requirement of any current microdissection method. Without a coverslip, and the index matching between the mounting media and the tissue, the dry tissue section has a refractile quality which might obscure cellular detail at high magnifications. Additionally, lack of cover slipping may create difficulties in the capture of particular cell types from certain complex tissues lacking architectural features, as noted in lymphoid tissues or diffusely infiltrating carcinomas [43]. This problem can be circumvented by specialized staining techniques, in particular immunohistochemistry, which help to differentiate the cell population to be isolated or avoided [50]. In contrast, most other microdissection techniques, with the exception of LMM with laser pressure catapulting, require the removal of the isolated cells with the help of a needle tip or a microcapillary—a precarious step requiring skill and practice [12]. The minimum laser spot size of 7.5 μm poses a limit to the precision of single cell or subcellular microdissection. Small cells may be difficult to isolate without contaminating fragments of adjacent cells. Compared to LCM from tissue sections, cytological preparations allow faster and more precise collection of pure cell populations than LCM because the cells are already physically separated [50]. The other issuue occasionally encountered in LCM is failure to remove the selected cells from the slide. This can result from a lack of adherence of the cells to the membrane, usually because of incomplete tissue dehydration or a laser setting that is too low for complete permeation of the melted polymer into the section. This is mainly encountered in frozen sections, if they are subjected to prolonged drying, whereas paraffin wax sections normally do not require special handling. Finally, the NIH software that accompanies the LCM has been designed so that a pathologist can take separate images with cover slipped slides and draw in landmarks of the areas of interest for subsequent LCM [16, 51]. Depending on the type of tissue and the disease state, a trained pathologist might be required to visually discriminate specific diseased cell populations, such as premalignant cancer. If cDNA libraries or diagnostic allele typing are performed on a patient’s tissue specimen, large amounts of time and resources would be wasted if the original diagnosis was not correct [16, 37]. Therefore, combination of an automatic imaging analyzer with LCM represents a future direction for expanding LCM applications [37]. Another problem is related to many dyes, such as eosin, used routinely to stain tissue during LCM can interfere with proteomic tools such as 2DE [52]. Fortunately, hematoxylin and methyl green seem to have no effect on protein migration; indicating they potentially could be used alone to stain tissue for LCM [52]. Overall high speed, easy handling, good control and documentation of dissected tissue make LCM an ideal tool for both the rapid collection a large amounts of tissue and the pooling of larger numbers of single cells.
3. Applications of LCM to ‘omics’ studies
The extensive advances during the past decade in genes and genomes knowledge (genomics) have yielded several new ‘omics’ technologies that are useful for the study of biological responses of organisms to several diseases and to understand the action underlying mechanisms [53]. ‘Omics’ differs from traditional hypothesis-driven research because it is a discovery-driven approach. Genomics deals with the analysis of the complete genome in order to understand the function of single genes. The majority of functional genomics is based on the analysis of gene expression (transcriptomics) and comprehensive proteins/metalloproteins analysis (proteomics/metallomics) [54]. In recent years, metabolomics (based on the complete study of metabolites involved in different metabolic processes of organisms) has become an emerging field in analytical biochemistry and can be regarded as the end point of ‘omics’ cascade [53]. Thus, while genomics or proteomics indicate the probability that a process may occur, metabolomics and ionomics provides more functional information. Because metabolomic and ionomic profiles of gene expression involve external factors (metal exposure, diet and others), they allow us to understand the consequences of complex biological mechanisms inside the organism [53]. In the future, it may be possible to perform genome-wide functional screening of gene function in humans [54].
Successful examination of molecular biological analysis methods depends on maximum precision and absolute freedom from contamination. Therefore, the contact-free isolation and separation offered by LCM is especially suited for isolation of single cells from tissue sections, cell components, chromosomes, living cells from cell cultures and native material. Once successfully removed by the LCM, the dissectates may be subjected to molecular biological and biochemical methods such as nucleic acid analysis (DNA and RNA) and protein investigations [55].It is thus possible to perform genomic analyses on samples derived from single cell, whereas for protein this may not be possible with the current generation of proteomic tests [56]. Recent studies involving the identification of prostate specific genes by the analysis of prostate expression sequence tags (ESTs) have shown the power of LCM in creating tissue specific expression libraries. In order to produce useful information, it is essential to have primary tissues of superior quality [57].
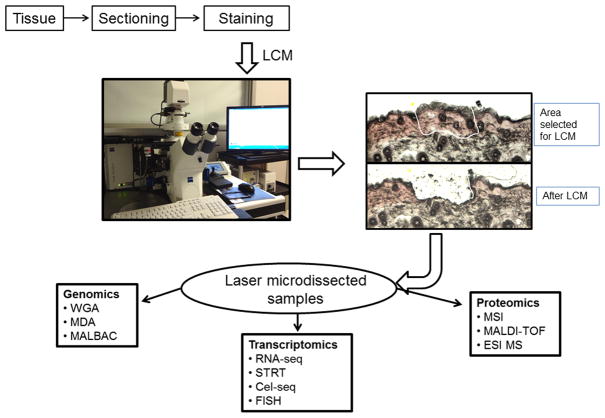
Fig. 3. Applications of LCM on ‘omics’ study.
LCM physically isolates the cells of interest, which then can be analyzed further. The remaining tissues on the slide are undisturbed, and other cell types can be isolated subsequently by LCM. (ESI MS, electrospray ionization mass spectrometry; FISH, fluorescence in situ hybridization; MALBAC, multiple annealing and looping-based amplification cycle; MALDI-TOF, matrix-assisted laser desorption ionization/time-of-flight; MDA, multiple displacement amplification; MSI, mass spectrometry imaging; STRT, single-cell tagged reverse transcription; WGA, whole-genome amplification.)
3.1. Proteomics
Proteins perform all the necessary functions of the cell. The existence of a DNA sequence does not guarantee the synthesis of a corresponding protein, nor is it sufficient to describe its function and cellular locations [56]. Detection of a DNA sequence also does not give information about context dependent post-translational processes such as glycosylation, phosphorylation or sulfation or how proteins link together into networks as functional machinery in the cell [58]. Proteomics is related with structure elucidation, quantitative analysis of protein expression and protein interactions, and provides information on components of metabolic pathways and regulatory circuits. It thus supplements and validates the data obtained in gene expression analysis [59]. It is a complementary approach to study gene expression and provide additional information regarding the effects of post-translational modification. A variety of techniques such as Western blotting, high resolution two dimensional polyacrylamide gel electrophoresis (2D PAGE), mass spectrometry and peptide sequencing may be used for analysis [47, 52]. With the advances in analytical technology, a variety of separation methods have been applied to facilitate the proteomic study of complex biological samples, including liquid chromatography (e.g., strong-cation-exchange, reversed phase, size exclusion), electrophoresis, solid phase extraction and immunoaffinity. Multidimensional separation can be applied to diagonally fractionate a complex sample at either the protein or peptide level to enhance the analytical dynamic range and detection sensitivity [60]. Mass spectrometry, such as surface enhanced laser desorption ionization (SELDI) mass spectrometry, has facilitated the study of gene expression at the protein level leading to the recent expansion of proteomics-based research [47, 52]. High-resolution instruments, such as Fourier transform–ion cyclotron resonance (FTICR), Orbitrap, quadrupole time-of-flight (TOF) and TOF/TOF are now available, greatly enhancing the quality of proteomic data. In addition to the widely used collision-induced dissociation method for ion fragmentation, soft collision techniques, such as electron transfer dissociation, have been introduced recently, which allows more sophisticated analysis of post-translational modifications (PTMs) including phosphorylation and glycosylation [60]. Briefly, a proteomics data analysis pipeline includes data conversion, database search and verification of peptide/protein identification.
3.1.1. LCM and Proteomics
LCM is powerful for selection and isolation of cells for the preparation of proteomic analysis. Early papers in the medical literature mostly concentrated on the feasibility of using LCM with 2DE and mass spectrometry. Recent papers have identified proteins that are differentially expressed in benign versus malignant cells and thus could potentially be used as new diagnostic biomarkers or targets for therapy [61]. In 2012, our lab describes a simple, highly efficient and robust proteomic workflow for routine liquid chromatography tandem mass spectrometry analysis of Laser Microdissection Pressure Catapulting (LMPC) isolates [26]. Highly efficient protein recovery was achieved by optimization of a “one-pot” protein extraction and digestion allowing for reproducible proteomic analysis on as few as 500 LMPC isolated cells. The method was combined with label-free spectral count quantitation to characterize proteomic differences from 3,000–10,000 LMPC isolated cells. To demonstrate the capability of this approach with human tissues, we analyzed punch biopsies of normal skin and chronic wound keratinocytes from a diabetic patient and glomeruli from needle biopsies of patients with diabetic, lupus and genetic kidney diseases. It was found that LC-MS/MS base peak chromatograms of peptides from LCM isolates demonstrates a high degree of sample complexity. LC-MS/MS base peak chromatograms from three separate LCM keratinocyte captures indicate excellent reproducibility. We also saw good reproducibility data across biological replicate biopsies for kidney samples from normal individuals and the patients. In 2000 Palmer-Toy et al reported on the feasibility of analyzing a lysate of captured cells with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) without intervening separation or purification steps such as 2DE [62]. They took frozen sections from one modified radical mastectomy specimen, stained with hematoxylin and eosin, and captured 2500 normal breast epithelial cells, normal stromal cells, in situ carcinoma cells, invasive carcinoma cells, and metastatic cells from a lymph node. Several high mass peaks were identified in the 45–60 kDa range that were different in malignant versus benign epithelial cells and between in situ carcinoma cells and benign epithelial cells but they did not go on to identify the proteins that produced the peaks because enough protein was not available for determination of protein composition by mass spectra alone. A similar study, using SELDI-TOF, found that a combination of mass spectra peaks might be used to discriminate between benign and malignant cells in the prostate gland [63]. Another interesting application of LCM was demonstrated by Simone et al. who utilized LCM to microdissect populations of normal prostate epithelial cells, cells from prostate intraepithelial neoplasia and invasive prostate adenocarcinoma cells, and analyzed using an automated sandwich chemiluminescent immunoassay to calculate the number of prostate specific antigen (PSA) molecules per cell [58, 64]. They confirmed that PSA expression is heterogeneous in prostate adenocarcinoma, showing that it varies considerably from carcinoma cells to carcinoma cells within the same tumor. Normal prostate epithelium, by contrast, had a more constant PSA content from cell to cell and suggested that cellular quantitation of HER-2/neu might be a future application for this technique [58, 64]. Using SELDI biochip, protein population was classified successfully into molecular weight classes and shown distinct protein expression patterns of normal, premalignant and malignant cancer cells procured by LCM from human tissues [58]. LCM was initially used to evaluate the genetic alterations in PCA. In 1999 Lutchman et al analyzed dermatin, a cytoskeleton protein encoded by a gene on chromosome 8p21 [65]. Rubin et al studied loss of heterozygosity at 10q23, a region that has been associated with many tumors including glioblastoma multiforme, melanoma, endometrial carcinoma, and PCA [66]. In 2004 Zheng et al. reported a putative marker for prostate cancer using microdissected benign and malignant epithelial cells, which they named PCa-24, by comparing the mass spectra from whole sections of benign and tumor tissue using SELDI TOF MS [67]. LCM can also aid in the diagnosis of many dermatological diseases. Routine diagnosis of cutaneous B- or T-cell lymphomas is challenging. In 2004, Yazdi et al have introduced a LCM-based clonality assay to overcome these diagnostic dilemmas [68]. Using this technique, the authors were able to determine whether clonal T-cell receptor (TCR) gene rearrangement, obtained by PCR, stems from lymphoma or some inflammatory skin diseases [68]. In 2013 Humerick et al., described a strategy to selectively dissect and collect specific neuronal phenotypes in the CNS for analysis using LCM and explained the role of oxytocin and vasopressin in magnocellular neurons in the hypothalamus [69]. In 2009, Johann et al., had used LCM techniques to examine solid tumor heterogeneity on a cellular basis using tissue proteomics that relies on a functional relationship between LCM and biological MS [70]. With the use of LCM, homogeneous regions of cells exhibiting uniform histology were isolated and captured from fresh frozen tissue specimens, which were obtained from a human lymph node containing breast carcinoma metastasis. Western blot analysis confirmed specific linkage of validated proteins to underlying pathology and their potential role in solid tumor heterogeneity. With continued research and optimization of this method, including analysis of additional clinical specimens, this approach may lead to an improved understanding of tumor heterogeneity and serve as a platform for solid tumor biomarker discovery [70]. The proteomes of glomeruli isolated by LCM from biopsies of normal kidneys (living-related donor kidneys) were compared to those patients with diabetic nephropathy, lupus nephritis, and fibronectin glomerulopathy. Glomerular proteins were extracted, trypsin digested and subjected to liquid chromatography-tandem mass spectrometry for identification and quantitation [71].
3.2. Genomics
Evaluation of the human genome has become quite efficient, using techniques such as loss of heterozygosity (LOH) screening [72–74] and comparative genomic hybridization (CGH)[75]. In combination with whole genome amplification [76], comparative genomic hybridization has been applied successfully to microdissected neoplastic and preneoplastic lesions of breast, cervix, and oral epithelium [77]. These techniques can also be used to identify and confirm both known and unknown alterations (deletions, mutations) in the genomes of a variety of tumors. For example, in 2002 Sobol et al. demonstrated the number and location of many genome alterations, by using polymorphic markers in a genome wide search for LOH in Burkitt lymphoma cell lines and their normal counterparts [74]. They were successful in distinguishing two types of altered allelic patterns; a bona fide LOH profile, indicative of a deletion and a profile associated with increased dosage (ID) [72]. Validation of these genomic changes at the transcriptional and translational levels is an important step in identifying biologically relevant candidates for further investigation as molecular targets in disease diagnosis and treatment [3]. High-throughput techniques to identify concomitant alterations in RNA and proteins have already been used to confirm data that is extracted from interrogation of the genome [3].
3.2.1. LCM and Genomics
For loss of heterozygosity (LOH) analysis, pure populations of tumor cells or preneoplastic foci are required because the contamination by even a few unwanted cells would mean the second allele “lost” in the cell population of interest will be amplified in the PCR reaction [56]. LOH analysis has been invaluable for the mapping of tumor suppressor genes (Tags), localization of putative chromosomal “hot spots” and the study of sequential genetic changes in preneoplastic lesions [47, 78, 79]. Before the widespread availability of laser capture microdissection techniques, many valuable samples had to be discarded because the desired purity could not be achieved. The use of microdissection has made a significant difference in the application of LOH analysis [78, 80].
Besides LOH analysis, other genome analyses can be performed from microdissected samples, such as analysis of patterns of X-chromosome inactivation to assess clonality, restriction fragment length polymorphism (RFLP) and single strand conformation polymorphism (SSCP) analysis for assessment of mutations in critical genes such as Ki-ras and P53, and most recently, the analysis of promoter hypermethylation [56]. The analysis of microdissected endocrine tumors played an instrumental role in identifying the multiple endocrine neoplasia type 1 (MEN1) gene by positional cloning[81]. Darling et al used LCM to analyze the partial re-expression of type XVII collagen in a patient with generalized atrophic benign epidermolysis bullosa, who was germ line homozygous for a 2-bp deletion in the COL17A1 gene [82]. LCM has also been used to demonstrate intratumoral hetrogeneity of p53 mutations in aflatoxin induced murine lung tumors [83]. Fend et al., analyzed cases of malignant non-Hodgkin’s lymphoma with two phenotypically and morphologically distinct cell populations in the same tumor site using LCM [84]. It has been used successfully to microdissect Reed-Sternberg (RS)-like cells from peripheral T cell lymphomas [85]. LCM combined with microarray analysis has also been used to identify genomic aberrations in hepatocellular carcinoma (HCC) [86]. In 2014, Pascarella et al., have identified the genome-wide collection of active promoters in the mouse Main Olfactory Epithelium (MOE) by coupling LCM to nano CAGE technology and next generation sequencing [9]. Recently, LCM has been also used to collect mouse embryonic day 16 (E16) meniscus, articular cartilage, and cruciate ligaments to perform genome-wide microarray analysis [87]. Because it can rapidly sample large numbers of purified cells from heterogeneous tissues, LCM is also a promising tool for other DNA based analyses, such as comparative genomic hybridization [43].
3.3. Transcriptomics
Transcriptomics is the study of transcriptome-the complete set of RNA transcripts produced by genome under specific circumstances or in a specific cell using high throughput techniques. Comparison of transcriptomes allows the identification of genes that are differentially expressed in distinct cell populations, or in response to different treatments. This provides both quantitative and qualitative information on genetic activity. Microarray technology has become increasingly widespread in genome analysis, diagnostics and gene expression analysis. By means of these so-called gene chips it is possible to examine the expression of many genes at the same time.
Tissue heterogeneity confounds assigning expressed genes to specific cell populations when gross tissue extracts are used as an mRNA source. Confirmation by in situ techniques, such as mRNA in situ hybridization or immunohistochemistry, might not always be possible and is laborious and time consuming when large numbers of messages need to be examined [43]. Furthermore, mRNA in situ hybridization lacks sensitivity for the detection of low abundance mRNA. Therefore, many groups have tried to develop microdissection protocols that yield mRNA of sufficient quality for the subsequent analysis of gene expression. In contrast to DNA, mRNA is more sensitive to fixation, is quickly degraded by ubiquitous RNases, and requires stringent RNase free conditions during specimen handling and preparation. Despite these limitations, several groups have recovered good quality mRNA from microdissected samples by reverse transcription PCR (RT–PCR), down to the single cell level [11, 43, 88].
In response to growing requirements to carry out global gene expression profiling on limited sample material, further advances in RNA amplification have arisen, providing numerous technologies claiming the ability to process RNA amounts within the nanogram range [89–91]. Using these methods, several groups have reported global transcriptomics data from LCM and flow cytometry collected cells by in vitro transcription-based methods using 100 and 200 ng of input RNA, respectively, followed by hybridization to Affymetrix GeneChip.
3.3.1. LCM and Transcriptomics
Transcriptomic analysis in normally developing and diseased tissue progression requires the microdissection and extraction of a microscopic homogeneous cellular subpopulation from its complex tissue milieu. This subpopulation can then be compared with adjacent interacting, but distinct, subpopulations of cells in the same tissue. The method of procurement of pure cell populations from heterogeneous tissue should fully preserve the state of the cell molecules if it is to allow quantitative analysis, particularly in sensitive amplification methods based on polymerase chain reaction (PCR), reverse transcriptase- PCR, or enzymatic function. LCM has been developed to provide a fast and dependable method of capturing and preserving specific cells from tissue, under direct microscopic visualization [16]. It is quite possible that the identification of gene expression patterns related to neoplastic transformation, inflammation or tissue repair will have far reaching consequences in the prognostic and diagnostic field, preventive medicine and for novel treatments tailored for specific genetic alterations [43, 56]. The growing potential of sequencing to carry out transcriptomics has been recently demonstrated by reports of this technology being used at the single cell level [92].
LCM offers several advantages for mRNA analysis; its superior speed allows sampling of large numbers of cells without appreciable RNA degradation. Rapid sampling of large quantity of cells should help to reduce artifacts caused by a high number of amplification cycles or a lack of reproducibility as a result of variability of gene expression in small sample size. In addition, the dehydration of the tissue section during LCM likely inhibits the activity of tissue RNases, in contrast to some techniques where microdissection is performed on buffer covered sections. Several groups have tried to assess the optimal conditions for RNA recovery from tissue subjected to LCM [38, 50, 93]. Because LCM helps to collect an exact determinable number of purified cells under controlled conditions, its combination with methods such as real time quantitative RT–PCR will allow for a more precise determination of cell specific gene expression on a microscopic scale [11, 94].
In 2007 our laboratory presented evidence demonstrating that that selective microdissection of blood vessels, high-density microarray analysis, quantitative PCR-based validation of microarray data, and immunohistochemistry can all be performed by using no more than one 3-mm punch biopsy from the affected tissue[27]. This provides a powerful tool to interrogate blood vessels isolated from patients of different disease settings with the goal to understand the molecular aspects of vascular biology in actual clinical setting. The approach described herein is applicable to a broad range of clinical research and therefore represents a powerful tool to enable sophisticated translational research. Comparison of results from blood vessels at the edge of chronic wound tissue with that of vessels in intact human skin demonstrated a revealing contrast between the transcriptome of the two vessels. Of the 18,400 transcripts and variants screened, a focused set of 53 up-regulated and 24 down-regulated genes were noted in wound derived blood vessels compared with blood vessels from intact human skin. The mean abundance of periostin in wound-site blood vessels was 96-fold higher. Forty-fold higher expression of heparan sulfate 6-Oendosulfatase1(Sulf1)n and lower expression of CD 24 was noted in wound-site vessels. The outcomes of this work provide a unique opportunity to appreciate the outstanding contrast in the transcriptome composition in blood vessels collected from the intact skin and from the wound-edge tissue using LCM.
The power in combining LCM and cDNA microarray hybridization was demonstrated in 20th century by Luo et al., where they reported reproducible differences in gene expression between large and small neurons isolated from rat dorsal root ganglia [95]. For each experiment, 1000 cells of one population were captured, and the RNA was amplified with T7 RNA polymerase to obtain sufficient material to generate the fluorescent probe for microarray hybridization. Similar combination of LCM, cDNA with real time quantitative PCR was used to show altered gene expression patterns at various stages of breast cancer progression [96]. The power of LCM for creating tissue specific expression libraries had been demonstrated by studies aimed at the identification of prostate specific genes by the analysis of expressed sequence tags (ESTs). A highly expressed T cell receptor gamma transcript found in prostate libraries generated from microdissected tissue was initially believed to stem from contaminating T cells in the prostatic interstitium. However, subsequent studies showed that the transcript did originate from prostate epithelial cells [97]. In 2002, Sluka et al., have used RT-PCR in LCM-procured seminiferous tubules to study transition protein-1 (TP-1) gene expression in the various stage groupings; this gene is involved in the compaction of the spermatid nucleus during elongation [98]. LCM has also been used to isolate foam cells from atherosclerotic arteries; RNA was extracted from isolated cells and used for molecular analysis by real time quantitative polymerase chain reaction [99]. Recently, we have also shown the use of LCM in capturing biofilms from porcine wounds, allowing for transcriptomic analysis of biofilms being performed [100]. The combination of LCM and next-generation sequencing is a powerful tool which may be used to resolve the entire transcriptome of specific cell types and tissues. Cañas et al., recently developed a protocol for transcriptomic analyses of conifer tissue types using LCM and 454 pyrosequencing; this technical approach will facilitate global gene expression analysis in individual tissues of conifers, and may also be applied to other plant species [101].
4. Conclusion and future directions
The interest to culture animal cells, was driven by need to understand how different cell types respond differently to same biological stimuli. Conditions for cell culture were properly developed during 20th century in standardize culture medium and an approach that relies on the culture of isolated cells in synthetic culture plates. While this approach has many strains, the isolation of cells from natural tissue habitat and culturing them under artificial conditions are confounding factors that distance cell culture experiments from in vivo biology. LCM is a technology that enables sophisticated cell biology studies on cells that were grown in their natural tissue habitat. The development of advance technologies enabling molecular analysis of small samples, ranging from subcellular to single cell structures, substantially strengthen the overall capabilities of LCM. On one hand, big data can be collected from small clinical samples making LCM a powerful translational tool. On the other hand the use of advanced tweezers and techniques to study simpler life forms in vivo make LCM a versatile tool to advance basic science.
The development of LCM allows investigators to determine specific gene-expression patterns from tissues of individual patients. Pure populations of cells can be obtained, RNA extracted, copied to cDNA and hybridized to thousands of genes on a cDNA microchip microarray. In this manner, an individualized molecular profile can be obtained for each histologically identified pathology. Using this multiplex analysis, investigators will be able to correlate the pattern of expressed genes with the etiology and response to treatment. A patient’s risk for disease and appropriate choice of treatment could, in the future, be personalized based on the profile. A growing clinical database of such results could be used to develop a minimal subset of key markers that will lead to a unique manner for the early detection and accurate diagnosis of disease. In future LCM can also be used for capturing pathogen associated macrophages and leading edge cells from wound tissue. LCM can also assist in the examination and separation of single cells. One of the greatest problems in oncology is the selective isolation of DNA from cancer cells in growing tumors. In the early stage, the affected area of tissue is often only small, and little material is available. The solution is to pre-select the relevant cells using the laser microdissection technique, isolate them and compare them with healthy surrounding tissue to look for signs of mutation.
Depending on the focus of research, it is also possible, to use LCM to isolate selected areas or distinct clones from living cell cultures for further cultivation or additional analysis such as PCR. The advantage is the relevant region can be examined without the surroundings falsifying the result. Such techniques are also suitable for cell surgery and similar manipulations on living cells. Even sensitive stem cells can be selected with LCM without losing their division potential. In future, these cells can be used for stem cell therapy, regenerative medicine and drug screening.
Acknowledgments
This study was partly supported by The Ohio State University and the National Institutes of Health awards GM 077185, GM 069589, NR013898 to CKS, and DK076566 to SR.
6. Abbreviations
- cDNA
Complementary deoxyribonucleic acid
- CGH
Comparative genomic hybridization
- DNA
Deoxyribonucleic acid
- ESI MS
Electrospray ionization mass spectrometry
- ESTs
Expression sequence tags
- FFPE
Formalin fixed paraffin embedded
- FISH
Fluorescence in situ hybridization
- FTICR
Fourier transform–ion cyclotron resonance
- HCC
Hepatocellular carcinoma
- IR LCM
Infrared Laser Capture Microdissection
- LCM
Laser Capture Microdissection
- LMM
Laser Microbeam Microdissection
- LOH
Loss of heterozygosity
- MALBAC
Multiple annealing and looping-based amplification cycle
- MALDI
Matrix-assisted laser desorption/ionization
- MDA
Multiple displacement amplification
- MEN 1
Multiple endocrine neoplasia type 1
- MOE
Main olfactory epithelium
- MS
Mass spectrometry
- NIH
National Institute of Health
- OCT
Optimal cutting temperature
- PAGE
Polyacrylamide gel electrophoresis
- PCR
Polymerase chain reaction
- PSA
Prostate specific antigen
- PTM
Post transcriptional modification
- RFLP
Restriction fragment length polymorphism
- RNA
Ribonucleic acid
- SELDI
Surface enhanced laser desorption ionization
- SSCP
Single strand conformation polymorphism
- STRT
Single-cell tagged reverse transcription
- TCR
T cell receptor
- TOF
Time-of-flight
- UV LCM
Ultraviolet laser capture microdissection
- WGA
Whole-genome amplification
References
- 1.Wiener MC, et al. Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem. 2004;76(20):6085–96. doi: 10.1021/ac0493875. [DOI] [PubMed] [Google Scholar]
- 2.Brioschi M, et al. A mass spectrometry-based workflow for the proteomic analysis of in vitro cultured cell subsets isolated by means of laser capture microdissection. Anal Bioanal Chem. 2014;406(12):2817–25. doi: 10.1007/s00216-014-7724-9. [DOI] [PubMed] [Google Scholar]
- 3.Ardekani AM, Akhondi MM, Sadeghi MR. Application of genomic and proteomic technologies to early detection of cancer. Arch Iran Med. 2008;11(4):427–34. [PubMed] [Google Scholar]
- 4.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 6.Carninci P, Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Curr Opin Genet Dev. 2007;17(2):139–44. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Forrest AR, Carninci P. Whole genome transcriptome analysis. RNA Biol. 2009;6(2):107–12. doi: 10.4161/rna.6.2.7931. [DOI] [PubMed] [Google Scholar]
- 8.Washietl S, et al. Computational analysis of noncoding RNAs. Wiley Interdiscip Rev RNA. 2012;3(6):759–78. doi: 10.1002/wrna.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascarella G, et al. NanoCAGE analysis of the mouse olfactory epithelium identifies the expression of vomeronasal receptors and of proximal LINE elements. Front Cell Neurosci. 2014;8:41. doi: 10.3389/fncel.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernsen MR, et al. Identification of multiple mRNA and DNA sequences from small tissue samples isolated by laser-assisted microdissection. Lab Invest. 1998;78(10):1267–73. [PubMed] [Google Scholar]
- 11.Fink L, et al. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4(11):1329–33. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- 12.Schutze K, Lahr G. Identification of expressed genes by laser-mediated manipulation of single cells. Nat Biotechnol. 1998;16(8):737–42. doi: 10.1038/nbt0898-737. [DOI] [PubMed] [Google Scholar]
- 13.Gutstein HB, et al. Microproteomics: analysis of protein diversity in small samples. Mass Spectrom Rev. 2008;27(4):316–30. doi: 10.1002/mas.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohm M, et al. Microbeam MOMeNT: non-contact laser microdissection of membrane-mounted native tissue. Am J Pathol. 1997;151(1):63–7. [PMC free article] [PubMed] [Google Scholar]
- 15.Bonner RF, et al. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278(5342):1481, 1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 16.Simone NL, et al. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14(7):272–6. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- 17.Suarez-Quian CA, et al. Laser capture microdissection of single cells from complex tissues. Biotechniques. 1999;26(2):328–35. doi: 10.2144/99262rr03. [DOI] [PubMed] [Google Scholar]
- 18.Espina V, et al. Laser-capture microdissection. Nat Protoc. 2006;1(2):586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 19.Mustafa D, Kros JM, Luider T. Combining laser capture microdissection and proteomics techniques. Methods Mol Biol. 2008;428:159–78. doi: 10.1007/978-1-59745-117-8_9. [DOI] [PubMed] [Google Scholar]
- 20.Okuducu AF, et al. Laser-assisted microdissection, techniques and applications in pathology (review) Int J Mol Med. 2005;15(5):763–9. [PubMed] [Google Scholar]
- 21.Clement-Sengewald A, Buchholz T, Schutze K. Laser microdissection as a new approach to prefertilization genetic diagnosis. Pathobiology. 2000;68(4–5):232–6. doi: 10.1159/000055929. [DOI] [PubMed] [Google Scholar]
- 22.Kleeberger W, et al. High frequency of epithelial chimerism in liver transplants demonstrated by microdissection and STR-analysis. Hepatology. 2002;35(1):110–6. doi: 10.1053/jhep.2002.30275. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann U, Versmold A, Kreipe H. Combined laser-assisted microdissection and short tandem repeat analysis for detection of in situ microchimerism after solid organ transplantation. Methods Mol Biol. 2005;293:113–23. [PubMed] [Google Scholar]
- 24.Burnet PW, Eastwood SL, Harrison PJ. Laser-assisted microdissection: methods for the molecular analysis of psychiatric disorders at a cellular resolution. Biol Psychiatry. 2004;55(2):107–11. doi: 10.1016/s0006-3223(03)00642-5. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura N, et al. Laser capture microdissection for analysis of single cells. Methods Mol Med. 2007;132:11–8. doi: 10.1007/978-1-59745-298-4_2. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro JP, et al. A quantitative proteomic workflow for characterization of frozen clinical biopsies: laser capture microdissection coupled with label-free mass spectrometry. J Proteomics. 2012;77:433–40. doi: 10.1016/j.jprot.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy S, et al. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci U S A. 2007;104(36):14472–7. doi: 10.1073/pnas.0706793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esposito G. Complementary techniques: laser capture microdissection--increasing specificity of gene expression profiling of cancer specimens. Adv Exp Med Biol. 2007;593:54–65. doi: 10.1007/978-0-387-39978-2_6. [DOI] [PubMed] [Google Scholar]
- 29.Kase M, et al. Laser microdissection combined with immunohistochemistry on serial thin tissue sections: a method allowing efficient mRNA analysis. Histochem Cell Biol. 2007;127(2):215–9. doi: 10.1007/s00418-006-0241-y. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, et al. Detection of CCND1 amplification using laser capture microdissection coupled with real-time polymerase chain reaction in human esophageal squamous cell carcinoma. Cancer Genet Cytogenet. 2007;175(1):19–25. doi: 10.1016/j.cancergencyto.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Jensen LH, et al. Laser microdissection and microsatellite analysis of colorectal adenocarcinomas. Anticancer Res. 2006;26(3A):2069–74. [PubMed] [Google Scholar]
- 32.Geho DH, Petricoin EF, Liotta LA. Blasting into the microworld of tissue proteomics: a new window on cancer. Clin Cancer Res. 2004;10(3):825–7. doi: 10.1158/1078-0432.ccr-1223-3. [DOI] [PubMed] [Google Scholar]
- 33.Liotta LA, Petricoin EF. Beyond the genome to tissue proteomics. Breast Cancer Res. 2000;2(1):13–4. doi: 10.1186/bcr23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders ME, et al. Differentiating proteomic biomarkers in breast cancer by laser capture microdissection and MALDI MS. J Proteome Res. 2008;7(4):1500–7. doi: 10.1021/pr7008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung CH, et al. Genomics and proteomics: emerging technologies in clinical cancer research. Crit Rev Oncol Hematol. 2007;61(1):1–25. doi: 10.1016/j.critrevonc.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Jares P, Campo E. Genomic platforms for cancer research: potential diagnostic and prognostic applications in clinical oncology. Clin Transl Oncol. 2006;8(3):161–72. doi: 10.1007/s12094-006-0006-z. [DOI] [PubMed] [Google Scholar]
- 37.Liu A. Laser capture microdissection in the tissue biorepository. J Biomol Tech. 2010;21(3):120–5. [PMC free article] [PubMed] [Google Scholar]
- 38.Emmert-Buck MR, et al. Laser capture microdissection. Science. 1996;274(5289):998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 39.Gjerdrum LM, et al. Laser-assisted microdissection of membrane-mounted paraffin sections for polymerase chain reaction analysis: identification of cell populations using immunohistochemistry and in situ hybridization. J Mol Diagn. 2001;3(3):105–10. doi: 10.1016/S1525-1578(10)60659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunt JL, Finkelstein SD. Microdissection techniques for molecular testing in surgical pathology. Arch Pathol Lab Med. 2004;128(12):1372–8. doi: 10.5858/2004-128-1372-MTFMTI. [DOI] [PubMed] [Google Scholar]
- 41.Vandewoestyne M, et al. Laser capture microdissection: should an ultraviolet or infrared laser be used? Anal Biochem. 2013;439(2):88–98. doi: 10.1016/j.ab.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 42.Zanni KL, Chan GK. Laser capture microdissection: understanding the techniques and implications for molecular biology in nursing research through analysis of breast cancer tumor samples. Biol Res Nurs. 2011;13(3):297–305. doi: 10.1177/1099800411402054. [DOI] [PubMed] [Google Scholar]
- 43.Fend F, Raffeld M. Laser capture microdissection in pathology. J Clin Pathol. 2000;53(9):666–72. doi: 10.1136/jcp.53.9.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldstein SR, McQueen PG, Bonner RF. Thermal modeling of laser capture microdissection. Appl Opt. 1998;37(31):7378–91. doi: 10.1364/ao.37.007378. [DOI] [PubMed] [Google Scholar]
- 45.von Eggeling F, Melle C, Ernst G. Microdissecting the proteome. Proteomics. 2007;7(16):2729–37. doi: 10.1002/pmic.200700079. [DOI] [PubMed] [Google Scholar]
- 46.Curran S, et al. Laser capture microscopy. Mol Pathol. 2000;53(2):64–8. doi: 10.1136/mp.53.2.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maitra A, Gazdar AF. Tissue microdissection and processing. Cancer Treat Res. 2001;106:6–84. doi: 10.1007/978-1-4615-1657-6_3. [DOI] [PubMed] [Google Scholar]
- 48.Coudry RA, et al. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2007;9(1):70–9. doi: 10.2353/jmoldx.2007.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banks RE, et al. The potential use of laser capture microdissection to selectively obtain distinct populations of cells for proteomic analysis--preliminary findings. Electrophoresis. 1999;20(4–5):689–700. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<689::AID-ELPS689>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 50.Fend F, et al. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am J Pathol. 1999;154(1):61–6. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mouledous L, et al. Navigated laser capture microdissection as an alternative to direct histological staining for proteomic analysis of brain samples. Proteomics. 2003;3(5):610–5. doi: 10.1002/pmic.200300398. [DOI] [PubMed] [Google Scholar]
- 52.Craven RA, et al. Laser capture microdissection and two-dimensional polyacrylamide gel electrophoresis: evaluation of tissue preparation and sample limitations. Am J Pathol. 2002;160(3):815–22. doi: 10.1016/S0002-9440(10)64904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Sevillano MA, et al. Omics technologies and their applications to evaluate metal toxicity in mice M. spretus as a bioindicator. J Proteomics. 2014;104:4–23. doi: 10.1016/j.jprot.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 54.Kiechle FL, Zhang X, Holland-Staley CA. The -omics era and its impact. Arch Pathol Lab Med. 2004;128(12):1337–45. doi: 10.5858/2004-128-1337-TOEAII. [DOI] [PubMed] [Google Scholar]
- 55.Standaert DG. Applications of laser capture microdissection in the study of neurodegenerative disease. Arch Neurol. 2005;62(2):203–5. doi: 10.1001/archneur.62.2.203. [DOI] [PubMed] [Google Scholar]
- 56.Domazet B, et al. Laser capture microdissection in the genomic and proteomic era: targeting the genetic basis of cancer. Int J Clin Exp Pathol. 2008;1(6):475–88. [PMC free article] [PubMed] [Google Scholar]
- 57.Suarez-Quian CA, Goldstein SR, Bonner RF. Laser capture microdissection: a new tool for the study of spermatogenesis. J Androl. 2000;21(5):601–8. [PubMed] [Google Scholar]
- 58.Simone NL, et al. Laser capture microdissection: beyond functional genomics to proteomics. Mol Diagn. 2000;5(4):301–7. doi: 10.1007/BF03262091. [DOI] [PubMed] [Google Scholar]
- 59.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 60.Caudle WM, et al. Using ‘omics’ to define pathogenesis and biomarkers of Parkinson’s disease. Expert Rev Neurother. 2010;10(6):925–42. doi: 10.1586/ern.10.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kunz GM, Jr, Chan DW. The use of laser capture microscopy in proteomics research--a review. Dis Markers. 2004;20(3):155–60. doi: 10.1155/2004/913436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palmer-Toy DE, et al. Direct acquisition of matrix-assisted laser Desorption/Ionization time-of-flight mass spectra from laser capture microdissected tissues. Clin Chem. 2000;46(9):1513–6. [PubMed] [Google Scholar]
- 63.Cazares LH, et al. Normal, benign, preneoplastic, and malignant prostate cells have distinct protein expression profiles resolved by surface enhanced laser desorption/ionization mass spectrometry. Clin Cancer Res. 2002;8(8):2541–52. [PubMed] [Google Scholar]
- 64.Simone NL, et al. Sensitive immunoassay of tissue cell proteins procured by laser capture microdissection. Am J Pathol. 2000;156(2):445–52. doi: 10.1016/S0002-9440(10)64749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lutchman M, et al. Loss of heterozygosity on 8p in prostate cancer implicates a role for dematin in tumor progression. Cancer Genet Cytogenet. 1999;115(1):65–9. doi: 10.1016/s0165-4608(99)00081-3. [DOI] [PubMed] [Google Scholar]
- 66.Rubin MA. Use of laser capture microdissection, cDNA microarrays, and tissue microarrays in advancing our understanding of prostate cancer. J Pathol. 2001;195(1):80–6. doi: 10.1002/path.892. [DOI] [PubMed] [Google Scholar]
- 67.Zheng Y, et al. Prostate carcinoma tissue proteomics for biomarker discovery. Cancer. 2003;98(12):2576–82. doi: 10.1002/cncr.11849. [DOI] [PubMed] [Google Scholar]
- 68.Yazdi AS, et al. Laser-capture microdissection: applications in routine molecular dermatopathology. J Cutan Pathol. 2004;31(7):465–70. doi: 10.1111/j.0303-6987.2004.00221.x. [DOI] [PubMed] [Google Scholar]
- 69.Humerick M, et al. Analysis of transcription factor mRNAs in identified oxytocin and vasopressin magnocellular neurons isolated by laser capture microdissection. PLoS One. 2013;8(7):e69407. doi: 10.1371/journal.pone.0069407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johann DJ, et al. Approaching solid tumor heterogeneity on a cellular basis by tissue proteomics using laser capture microdissection and biological mass spectrometry. J Proteome Res. 2009;8(5):2310–8. doi: 10.1021/pr8009403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Satoskar AA, et al. Characterization of glomerular diseases using proteomic analysis of laser capture microdissected glomeruli. Mod Pathol. 2012;25(5):709–21. doi: 10.1038/modpathol.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bielas JH, Venkatesan RN, Loeb LA. LOH-proficient embryonic stem cells: a model of cancer progenitor cells? Trends Genet. 2007;23(4):154–7. doi: 10.1016/j.tig.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 73.Radford DM, et al. Allelic loss on a chromosome 17 in ductal carcinoma in situ of the breast. Cancer Res. 1993;53(13):2947–9. [PubMed] [Google Scholar]
- 74.Sobol H, et al. Genome-wide search for loss of heterozygosity in Burkitt lymphoma cell lines. Genes Chromosomes Cancer. 2002;33(2):217–24. doi: 10.1002/gcc.10022. [DOI] [PubMed] [Google Scholar]
- 75.Choi YW, et al. Gene expression profiles in squamous cell cervical carcinoma using array-based comparative genomic hybridization analysis. Int J Gynecol Cancer. 2007;17(3):687–96. doi: 10.1111/j.1525-1438.2007.00834.x. [DOI] [PubMed] [Google Scholar]
- 76.Dietmaier W, et al. Multiple mutation analyses in single tumor cells with improved whole genome amplification. Am J Pathol. 1999;154(1):83–95. doi: 10.1016/S0002-9440(10)65254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aubele M, et al. Distinct cytogenetic alterations in squamous intraepithelial lesions of the cervix revealed by laser-assisted microdissection and comparative genomic hybridization. Cancer. 1998;84(6):375–9. doi: 10.1002/(sici)1097-0142(19981225)84:6<375::aid-cncr10>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 78.Katona TM, et al. Molecular evidence for independent origin of multifocal neuroendocrine tumors of the enteropancreatic axis. Cancer Res. 2006;66(9):4936–42. doi: 10.1158/0008-5472.CAN-05-4184. [DOI] [PubMed] [Google Scholar]
- 79.McCarthy RP, et al. Molecular genetic evidence for different clonal origins of epithelial and stromal components of phyllodes tumor of the prostate. Am J Pathol. 2004;165(4):1395–400. doi: 10.1016/S0002-9440(10)63397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng L, et al. Allelic loss of the active X chromosome during bladder carcinogenesis. Arch Pathol Lab Med. 2004;128(2):187–90. doi: 10.5858/2004-128-187-ALOTAX. [DOI] [PubMed] [Google Scholar]
- 81.Chandrasekharappa SC, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276(5311):404–7. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 82.Darling TN, et al. Revertant mosaicism: partial correction of a germ-line mutation in COL17A1 by a frame-restoring mutation. J Clin Invest. 1999;103(10):1371–7. doi: 10.1172/JCI4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tam AS, et al. High frequency and heterogeneous distribution of p53 mutations in aflatoxin B1-induced mouse lung tumors. Cancer Res. 1999;59(15):3634–40. [PubMed] [Google Scholar]
- 84.Fend F, et al. Composite low grade B-cell lymphomas with two immunophenotypically distinct cell populations are true biclonal lymphomas. A molecular analysis using laser capture microdissection. Am J Pathol. 1999;154(6):1857–66. doi: 10.1016/S0002-9440(10)65443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quintanilla-Martinez L, et al. Peripheral T-cell lymphoma with Reed-Sternberg-like cells of B-cell phenotype and genotype associated with Epstein-Barr virus infection. Am J Surg Pathol. 1999;23(10):1233–40. doi: 10.1097/00000478-199910000-00008. [DOI] [PubMed] [Google Scholar]
- 86.Wei YM, et al. Aberrant gene expression profiles in hepatocellular carcinoma detected by microdissection. Genet Mol Res. 2013;12(4):5527–36. doi: 10.4238/2013.November.18.3. [DOI] [PubMed] [Google Scholar]
- 87.Pazin DE, et al. Gene signature of the embryonic meniscus. J Orthop Res. 2014;32(1):46–53. doi: 10.1002/jor.22490. [DOI] [PubMed] [Google Scholar]
- 88.Hiller T, Snell L, Watson PH. Microdissection RT-PCR analysis of gene expression in pathologically defined frozen tissue sections. Biotechniques. 1996;21(1):38–40. 42–44. doi: 10.2144/96211bm07. [DOI] [PubMed] [Google Scholar]
- 89.Bak M, et al. Evaluation of two methods for generating cRNA for microarray experiments from nanogram amounts of total RNA. Anal Biochem. 2006;358(1):111–9. doi: 10.1016/j.ab.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 90.Kennedy L, et al. Global array-based transcriptomics from minimal input RNA utilising an optimal RNA isolation process combined with SPIA cDNA probes. PLoS One. 2011;6(3):e17625. doi: 10.1371/journal.pone.0017625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh R, et al. Microarray-based comparison of three amplification methods for nanogram amounts of total RNA. Am J Physiol Cell Physiol. 2005;288(5):C1179–89. doi: 10.1152/ajpcell.00258.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 93.Maitra A, et al. Enrichment of epithelial cells for molecular studies. Nat Med. 1999;5(4):459–63. doi: 10.1038/7458. [DOI] [PubMed] [Google Scholar]
- 94.Goldsworthy SM, et al. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog. 1999;25(2):86–91. [PubMed] [Google Scholar]
- 95.Luo L, et al. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5(1):117–22. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 96.Sgroi DC, et al. In vivo gene expression profile analysis of human breast cancer progression. Cancer Res. 1999;59(22):5656–61. [PubMed] [Google Scholar]
- 97.Essand M, et al. High expression of a specific T-cell receptor gamma transcript in epithelial cells of the prostate. Proc Natl Acad Sci U S A. 1999;96(16):9287–92. doi: 10.1073/pnas.96.16.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sluka P, O’Donnell L, Stanton PG. Stage-specific expression of genes associated with rat spermatogenesis: characterization by laser-capture microdissection and real-time polymerase chain reaction. Biol Reprod. 2002;67(3):820–8. doi: 10.1095/biolreprod.102.004879. [DOI] [PubMed] [Google Scholar]
- 99.Trogan E, Fisher EA. Laser capture microdissection for analysis of macrophage gene expression from atherosclerotic lesions. Methods Mol Biol. 2005;293:221–31. doi: 10.1385/1-59259-853-6:221. [DOI] [PubMed] [Google Scholar]
- 100.Roy S, et al. Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J Pathol. 2014;233(4):331–43. doi: 10.1002/path.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Canas RA, et al. Transcriptome analysis in maritime pine using laser capture microdissection and 454 pyrosequencing. Tree Physiol. 2014 doi: 10.1093/treephys/tpt113. [DOI] [PubMed] [Google Scholar]