EGFR-mutant lung cancers represent a paradigm for the use of tyrosine kinase inhibitors (TKIs) to treat molecular subsets of cancer, with randomized trials demonstrating the efficacy of first-generation, reversible epidermal growth factor receptor (EGFR) TKIs. However, acquired resistance invariably develops, and rebiopsy of patients with clinical progression has elucidated EGFR T790M mutation as the major resistance mechanism.1
To address this clinical challenge, third-generation pyrimidine-based EGFR TKIs have been designed to have selectivity for EGFR T790M over wild-type EGFR.2 They form a covalent bond with cysteine 797 in the adenosine triphosphate–binding cleft. Early-phase clinical trials have demonstrated their efficacy in patients with double-mutant tumors (EGFR L858R/T790M and ex19del/T790M) and acquired resistance to first-generation EGFR inhibitors.3 Specificity for EGFR T790M may be the result of hydrophobic interactions between the bulky methionine moiety of EGFR T790M and these pyrimidine-based drugs.
Mechanisms of resistance to these third-generation EGFR TKIs have been identified in preclinical in vitro models, but none have so far been identified in patients. Mutations at the EGFR C797 codon, located within the kinase-binding site, are a predicted resistance mechanism to irreversible EGFR inhibitors also confirmed in vitro.4 Loss of the potential for covalent bond formation at position 797 by the missense mutation C797S results in a markedly reduced cellular potency of this class of TKIs.5 Interestingly, an analogous cysteine-to-serine mutation is a mechanism of resistance to the irreversible Bruton tyrosine kinase inhibitor ibrutinib in patients with chronic lymphocytic leukemia.6
Report of a Case
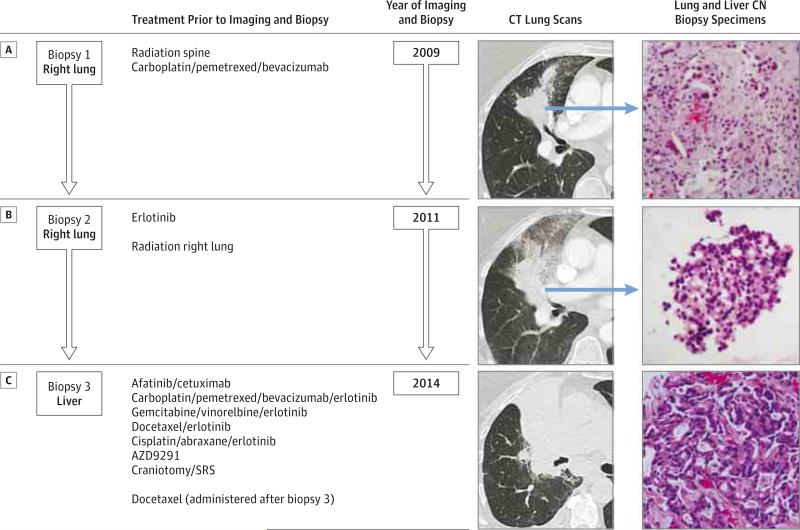
A former smoker in her 60s presented with stage IV lung adenocarcinoma metastatic to the lung, lymph nodes, bone, and brain. After progression during first-line chemotherapy, a biopsy of the right lung tumor (biopsy 1, Figure 1) identified a 15–base pair deletion in EGFR exon 19. She experienced a partial response with erlotinib treatment for 15 months. After further progression, the enlarging right lung mass was rebiopsied (biopsy 2, Figure 1), and in addition to the previous EGFR exon 19 deletion, Sanger sequencing identified EGFR T790M.
Figure 1. Patient Clinical Course Including Treatment History and Relevant Imaging Studies and Tumor Biopsy Specimens Demonstrating Adenocarcinoma.
A, Right lung tumor before treatment with erlotinib (lung biopsy specimen positive for EGFR exon 19 deletion). B, Enlarging right lung tumor after acquired resistance to erlotinib (lung biopsy specimen positive for EGFR exon 19 deletion and EGFR T790M mutation). C, Further enlarged lung tumor CT after acquired resistance to AZD9291. In 2014, the pictured biopsy specimen is from a metastatic liver lesion and was positive for EGFR exon 19 deletion, EGFR T790M mutation, and EGFR C797S mutation. All biopsy specimens were stained with hematoxylin-eosin, original magnification ×200; CN indicates core needle; CT, computed tomographic; SRS, stereotactic radiosurgery.
The patient was treated with a sequence of EGFR TKIs and chemotherapies (Figure 1). She also enrolled in a phase 1 study of AZD9291 (NCT01802632) and received AZD9291 for 9 months until further disease progression. She then underwent a biopsy of a hepatic metastasis (biopsy 3, Figure 1) in which Sanger sequencing identified a third mutation, EGFR C797S, in addition to the exon 19 deletion and T790M.
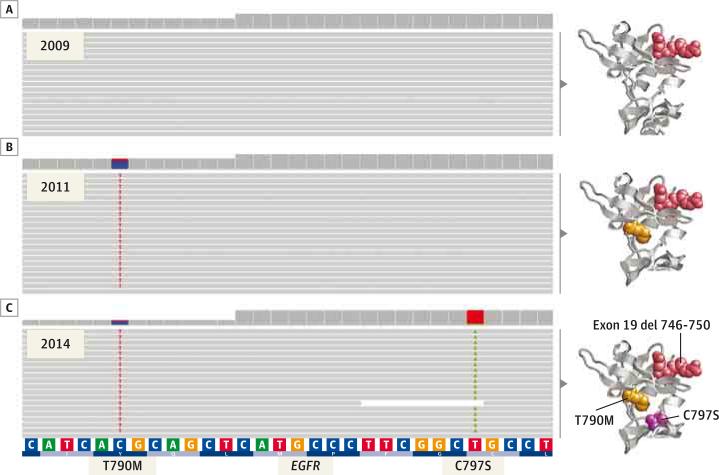
The patient signed an institutional biospecimen protocol, and next-generation sequencing (NGS) was performed on all 3 biopsy specimens: before treatment with erlotinib (biopsy 1, Figure 1), after acquired resistance to erlotinib (biopsy 2, Figure 1), and after acquired resistance to AZD9291 (biopsy 3, Figure 1). The EGFR mutations seen on Sanger sequencing were confirmed by NGS (Figure 2), which also showed that the C797S mutation was acting in cis with the T790M; ie, all alleles with C797S also had T790M, and conversely all alleles with T790M had C797S. In addition, PTEN Y27C and CTNNB1 S37F mutations were present in all 3 samples, and a TSC2 N486I was seen in the third biopsy sample only.
Figure 2. Mutation Analysis of Exon 20 of EGFR in the 2009, 2011, and 2014 Tumor Samples.
Integrative genomic viewer screenshots of next-generation sequencing readings from sequential tumor biopsy specimens. A, The 2009 pre–erlotinib treatment sample shows absence of both T790M and C797S EGFR mutations. B, The 2011 post–erlotinib treatment sample shows the T790M EGFR mutation only (23% of readings). C, The 2014 post–AZ9291 treatment sample shows both T790M and C797S mutations acting in cis; the T790M mutation is present at an allele frequency of 25%, with all mutant readings also harboring the C797S mutation; conversely, all C797S mutant readings had the concurrent T790M mutation. The corresponding in silico 3-dimensional models for the 3 sequential states of the EGFR (epidermal growth factor receptor) kinase domain are shown on the right side of each panel.
Discussion
We describe herein a patient whose tumor acquired an EGFR C797S mutation after treatment with a third-generation EGFR TKI. The acquired EGFR C797S should confer resistance to all third-generation EGFR TKIs, similar to the emergence of EGFR T790M and its cross-resistance to all first-generation EGFR TKIs. The original sensitizing EGFR mutation was present in all obtained tumor samples, demonstrating the continued dependence of the tumor on EGFR signaling for growth and survival. Under the strong selective pressure of EGFR TKIs, the tumor developed secondary and tertiary mutations in EGFR (T790M and C797S, respectively) to maintain EGFR signaling. To our knowledge, this is the first report of a tertiary acquired mutation identified in a clinical lung cancer sample.
Acknowledgments
Funding/Support: This work was supported in part by National Institutes of Health (NIH) grant P01 CA129243 to Memorial Sloan Kettering Cancer Center (Dr Ladanyi).
Role of the Funder/Sponsor: The NIH had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Dr Ladanyi had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Yu, Drilon, Riely, Arcila.
Acquisition, analysis, or interpretation of data: Yu, Tian, Drilon, Borsu, Arcila, Ladanyi.
Drafting of the manuscript: Yu, Tian, Drilon, Borsu, Arcila, Ladanyi.
Critical revision of the manuscript for important intellectual content: Yu, Tian, Drilon, Riely, Arcila, Ladanyi.
Interpretation of molecular results: Arcila.
Obtained funding: Ladanyi.
Administrative, technical, or material support: Yu, Tian, Drilon, Borsu.
Study supervision: Drilon, Riely, Arcila, Ladanyi.
Conflict of Interest Disclosures: Dr Yu has consulted for Clovis Oncology and has received research support from Clovis Oncology, AstraZeneca, Pfizer, Astellas, and Incyte. Dr Drilon has consulted for Ignyta and Genentech and has received research funding from Foundation Medicine. Dr Riely has consulted for Ariad, Celgene, Mersana, and Novartis and has received research support from Novartis, Roche/Genentech, Millennium, GlaxoSmithKline, Pfizer, and Infinity. No other disclosures are reported.
Additional Contributions: Justyna Sadowska, MBA, Raghu Chandramohan, MS, and Donavan Cheng, PhD, Department of Pathology, Memorial Sloan Kettering Cancer Center, contributed to the bioinformatic data analysis of the tumor samples. Boris Reva, PhD, Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, created the structural images in Figure 2. Andrew Plodkowski, MD, Department of Radiology, Memorial Sloan Kettering Cancer Center, edited the radiographic images. Charles Rudin, MD, PhD, and Mark Kris, MD, Department of Medicine, Memorial Sloan Kettering Cancer Center, provided editorial comments. No compensation was received by any of the acknowledged persons for their contributions.
References
- 1.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462(7276):1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janne PA, Ramalingam SS, Yang JC- H, et al. Clinical activity of the mutant-selective EGFR inhibitor AZD9291 in patients (pts) with EGFR inhibitor–resistant non-small cell lung cancer (NSCLC) [suppl; abstr 8009]. J Clin Oncol. 2014;32:5s. doi: 10.3978/j.issn.2218-6751.2014.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Z, Boggon TJ, Kobayashi S, et al. Resistance to an irreversible epidermal growth factor receptor (EGFR) inhibitor in EGFR-mutant lung cancer reveals novel treatment strategies. Cancer Res. 2007;67(21):10417–10427. doi: 10.1158/0008-5472.CAN-07-1248. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz PA, Kuzmic P, Solowiej J, et al. Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance. Proc Natl Acad Sci U S A. 2014;111(1):173–178. doi: 10.1073/pnas.1313733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woyach JA, Furman RR, Liu TM, et al. Resistance mechanisms for the Bruton's tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370(24):2286–2294. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]