Abstract
MicroRNAs (miRs) are short, non-coding RNAs with post-transcriptional regulatory functions. Previous studies have demonstrated that miR-34c is involved in diverse biological processes, including carcinogenesis. The aim of the present study was to investigate the role of miR-34c and its target genes in hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Expression levels of miR-34c and its predicted target genes were measured. The target genes were validated by a luciferase assay. The effects of miR-34c restoration were evaluated by the detection of HBV antigens, cell proliferation and apoptosis in vitro, in addition to the tumor growth in vivo. The data demonstrated that miR-34c was downregulated in HBV-associated HCC clinical tissues and HCC cell lines compared with their corresponding controls. transforming growth factor-β-induced factor homeobox 2 (TGIF2), a transcription factor repressing transforming growth factor-β (TGFβ) signaling, was observed to be upregulated and was identified as a target gene of miR-34c. The restoration of miR-34c in HepG2.2.15 cells suppressed TGIF2 expression, HBV replication and viral antigen synthesis; inhibited cell proliferation; and induced apoptosis. miR-34c also inhibited tumor growth in a mouse model. The present study indicates that miR-34c may act as a tumor suppressor by targeting TGIF2 during HBV-associated hepatocellular carcinogenesis. miR-34c and TGIF2 may represent key regulatory factors, diagnostic markers and therapeutic targets for the prevention and treatment of HBV-associated HCC.
Keywords: microRNA, hepatitis B virus, hepatocellular carcinoma, microRNA-34c, TGFB-induced factor homeobox 2
Introduction
Previous epidemiological studies have demonstrated that hepatitis B virus (HBV) infection is a key risk factor for hepatocellular carcinoma (HCC). HBV is greatly detrimental to human health by inducing hepatitis, cirrhosis and HCC (1). The currently available diagnostic methods, including B-mode ultrasound and α-fetal protein (AFP) detection, lack the ability to detect liver cancer at an early stage (2,3). In addition, therapy is currently only partially effective against HBV. Therefore, investigation into the pathogenesis of HBV-related HCC is of great significance. Studies on microRNAs (miRs) may provide a valuable approach for devising novel strategies for the diagnosis, prevention and treatment of HCC.
miRs are non-coding, single-stranded RNA molecules containing 19–25 nucleotides. A previous study demonstrated that the majority of miRs regulate gene expression by imperfect base-pairing with the 3′-untranslated region (3′-UTR) of target mRNAs (4). miRs have been identified as key regulators in diverse biological processes, including cell cycle regulation, cell differentiation, proliferation and apoptosis (5,6). The significance of miRs in the tumorigenesis of different types of human cancer is highlighted in the present study.
miR-34c belongs to a family of evolutionarily conserved miRs (miR-34a, b and c) that are involved in apoptosis and negative feedback control of the cell cycle (7,8). The miR-34 family is downregulated in HCC and contributes to hepatocarcinogenesis by targeting genes that include NOTCH1, MET, E2F1-3, MYC, cyclin-dependent kinase (CDK)4, CDK6, cyclin D1, sirtuin 1 (SIRT1) and B-cell lymphoma 2 (BCL2) (9–11). In a preliminary study, it was revealed that miR-34c was significantly downregulated in HBV-infected transgenic mice and HBV-related HCC cell lines compared with their corresponding controls. Therefore, miR-34c was selected for further investigation. Transforming growth factor-β-induced factor homeobox 2 (TGIF2) is a transcriptional co-repressor that interacts with Smad and negatively regulates the transforming growth factor (TGF)-β/Smad response (12). Furthermore, it was previously reported that TGF-β signaling is involved in cell apoptosis and proliferation in HBV-associated HCC by upregulating Smad7 (13–15). Therefore, the present study investigated the roles and regulatory mechanisms of miR-34c and TGIF2 in the development of HBV-associated HCC.
Materials and methods
Tissue samples and cell lines
Thirty pairs of tumor and adjacent non-cancerous tissues were obtained from patients with HBV-associated HCC at the Provincial Hospital affiliated to Shandong University (Jinan, China) between 2008 and 2011. The tissue samples were snap-frozen in liquid nitrogen immediately following resection and stored at −80°C. Informed consent was obtained from all subjects (patient or the patient's family) and the study was approved by the Provincial Hospital Affiliated to Shandong University Ethics Committee. The human HCC cell lines HepG2, HepG2.2.15 and BEL7402 (American Type Culture Collection, Manassas, VA, USA) were cultured at 37°C in an atmosphere of 5% CO2. HepG2 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL, Grand Island, NY, USA) at 37°C in a 5% CO2 incubator. HepG2.2.15 cells were cultured in minimal essential medium (MEM; HyClone, Beijing, China) supplemented with 10% new bovine serum (NBS; Gibco-BRL) and 380 µg/ml G418 (Promega, Madison, WI, USA). BEL7402 cells were cultured in RPMI-1640 (Gibco-BRL) supplemented with 10% FBS. BEL7402-HBV cells are BEL7402 cells transfected with pcDNA3-1.1HBV.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted by TRIzol® reagent (Invitrogen Life Technologies) according to the manufacturer's instructions. The mRNA was reverse transcribed into cDNA with the First Strand cDNA Synthesis kit ReverTra Ace-α (Toyobo, Osaka, Japan). The cDNA was then amplified using 2X Taq PCR Master Mix (Tiangen Biotech, Beijing, China). The sequences of the primers for the amplification of TGIF2 were as follows: F 5′-TCTCTGTGTTGCCTCCCTCT-3′ and R 5′-CCACCTCAGCCCAATACACT-3′. The human β-actin gene was used as an internal control and amplified with the following primers: F 5′-ACACTGTGCCCATCTACGAGGGG-3′ and R 5′-ATGATGGAGTTGAAGGTAGTTTCGTGGAT-3′. The PCR products were analyzed on 1% agarose gels by electrophoresis.
Quantitative (q)PCR
qPCR was performed to detect the expression levels of miR-34c and TGIF2. The NCode VILO miRNA cDNA Synthesis kit (Invitrogen Life Technologies) and the all-in-one miR qPCR primer set (GeneCopoeia, Rockville, MD, USA) were used for miR-34c amplification. The PCR reaction was performed using the Express SYBR® GreenER™ miRNA qRT-PCR kit (Invitrogen Life Technologies) on a preheated qPCR instrument (ABI 7000; Applied Biosystems, Foster City, CA, USA). All reactions were performed in triplicate. The relative expression level of miR-34c compared with small nuclear RNA U6 was determined using the 2−ΔΔCt method (16).
The ReverTra Ace qPCR RT kit (Toyobo) and SYBR® Premix Ex Taq Perfect Real-Time reagent (Takara Bio, Inc., Shiga, Japan) were used for TGIF2 amplification. The primers used for the qPCR and RT-PCR were the same. Human β-actin RNA served as an internal control. All qPCR reactions were performed in triplicate.
Target prediction
The miR-34c target genes were predicted using bioinformatics analysis, and the miR-34c target list was obtained from a comparison of three established miR target prediction programs, including TargetScan (http://www.targetscan.org/), PicTar (http://pictar.bio.nyu.edu/) and MiRanda (http://microrna.sanger.ac.uk/). The duplex structure between miR-34c and the 3′-UTR of the target genes was predicted using RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/).
Construction and transfection of recombinant plasmids
Primer Premier 5.0 (Premier Biosoft, Palo Alto, CA, USA) was used to design the miR-34c primers according to the sequence of precursor miR-34c cDNA provided by GenBank (National Center for Biotechnology Information, Bethesda, MD, USA). The primer sequences were as follows: F 5′-CGCGGATCCTCTATTTGCCATCGTCTA-3′ and R 5′-CTGAAGCTTCAGGCAGCTCATTTGGAC-3′. The restriction enzyme cutting sites of BamHI and HindIII are included in the primer sequence. The recombinant plasmid was named pS-miR-34c and confirmed by PCR, restriction enzyme digestion and DNA sequencing. Additionally, a control vector containing an antisense sequence was constructed and denoted as pS-control.
HepG2.2.15 cells were plated 24 h prior to transfection at a density of 2×105 cells per well in 6-well plates. The cell medium (Invitrogen Life Technologies) was replaced with serum-free Opti-MEM® (Invitrogen Life Technologies) prior to transfection. A total of 2 µg of the constructed plasmids were transfected into HepG2.2.15 cells using FuGENE® 6 transfection reagent (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's instructions. Following incubation for 6 h, the medium was replaced with fresh medium supplemented with 10% FBS. The supernatant and cells were collected 48 h following transfection for further studies.
Luciferase assay
The 3′-UTRs of human TGIF2 mRNA sense and antisense strands were amplified using the following primers: Sense, F 5′-TAGGTACCCAGAAGCAGCAGGACCCA-3′; sense, R 5′-GCAGATCTTCATCACTGAGCGGAGGC-3′; antisense, F 5′-AGATCTCAGAAGCAGCAGGACCCA-3′; antisense, R 5′-GGTACCTCATCACTGAGCGGAGGC-3′. The segments were digested with the restriction enzymes KpnI and BglII (Takara) and cloned into the corresponding restriction sites of the pGL3-REPORT vector (Promega Corporation) to produce the pGL3-TGIF2 and pGL3-anti-TGIF2 plasmids. HepG2 cells were seeded in 24-well culture plates at a density of 2×105 cells per well and cultured to 90% confluence. The cells were transfected with pGL3-TGIF2 or pGL3-anti-TGIF2 (100 ng) and co-transfected with pRL-TK plasmid (40 ng) by FuGENE® HD transfection reagent (Roche). The cells were harvested in 100 µl of cell lysis buffer (NP40; Invitrogen Life Technologies) 24 h post-transfection. Firefly and Renilla luciferase activity levels were detected using the Dual-Luciferase Reporter assay (Promega Corporation) according to the manufacturer's instructions. Experiments were performed in triplicate.
Western blot analysis
Transfected cells were lysed with 1% radioimmunoprecipitation assay lysis buffer (Beyotime Biotechnology, Haimen, China) 72 h after transfection. The supernatants were reserved and the concentration of protein was determined using the Pierce BCA protein assay reagent kit (Pierce, Rockford, IL, USA). Subsequently, equal amounts (30 µg) of protein were separated by 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% milk and incubated with a mouse monoclonal primary antibody against human TGIF2 (dilution, 1:1,000; cat no. ab57527; Abcam, Cambridge, MA, USA). Following incubation with goat anti-mouse horseradish peroxidase secondary antibody, the signals were detected using enhanced chemiluminescence (ECL Plus™; GE Healthcare, Little Chalfont, UK).
ELISA assay for the detection of hepatitis B virus surface antigen (HBsAg) and hepatitis B virus e antigen (HBeAg)
The concentrations of HBsAg and HBeAg in culture supernatants were measured by an ELISA assay using HBsAg and HBeAg enzyme diagnostic kits (Autobio, Zhengzhou, China) according to the manufacturer's instructions. The levels of HBsAg and HBeAg were determined from the optical density at 450 nm using an absorbance microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The assays were performed in triplicate. Inhibitory rates were calculated according to the following formula: Inhibitory rate (%) = (Ccontrol-Ctested)/Ccontrol × 100%, in which C is the optical density of the HBV antigen.
qPCR of HBV DNA
HBV DNA in cell culture supernatants collected post-transfection were extracted and purified by standard methods. qPCR was performed using the HBV PCR Fluorescence Quantitative Detection kit (Bioer Technology Co., Ltd., Hangzhou, China) following the manufacturer's instructions. Experiments were performed in triplicate, and the average cycle threshold (Ct) values were used to determine the concentration of HBV DNA.
Cell proliferation and apoptosis assays
HepG2.2.15 cells were seeded in 96-well culture plates (Costar®; Costar, Corning, NY, USA). As indicated, 10 µl of Cell counting kit-8 (CCK-8) reagents were added into each well and incubated for 1 h at 0, 24, 48 or 72 h post-transfection. The cell proliferation rates were calculated by measuring the optical density at 450 nm using an absorbance microplate reader (Bio-Rad 680; Bio-Rad Laboratories, Inc.).
Flow cytometry (FCM) was performed 48 h post-transfection in 6-well plates. The cells were resuspended in binding buffer containing Annexin V-fluorescein isothiocyanate and propidium iodide (PI) according to the manufacturer's instructions (KeyGen Biotech Co., Ltd., Nanjing, China). The samples were analyzed using a Beckman Coulter Flow Cytometer (Beckman Coulter, Fullerton, CA, USA) and experiments were performed in triplicate.
Mouse model
Male Balb/c nude mice (n=15) were purchased from the Animal Center of Shandong University. Mice were maintained in laminar flow rooms, with exposure to 12 h light and 12 h dark cycles, at a temperature of 21°C and 50% humidity, with free access to food and water. Animals were fed according to protocols approved by the Shandong University Animal Care Committee. A total of 2×106 BEL7402-1.1-HBV cells were injected subcutaneously into the flanks of each BALB/c nude mouse (6–8 weeks of age). The mice were randomly assigned to 3 groups (5 mice per group). When the tumor grew to 90 mm3 in volume, plasmids of pS-miR-34c or pS-control (20 µg each) was injected into tumor of each mouse every two days five times. Tumor volume was measured by caliper and calculated as length × width × height (mm3).
Statistical analysis
All experiments were performed in triplicate. The results were expressed as the mean ± standard deviation and analyzed using GraphPad Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Differences between experimental groups were evaluated by analysis of variance or the Student's t-test, and P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-34c and TGIF2 in HBV-associated HCC is inversely correlated
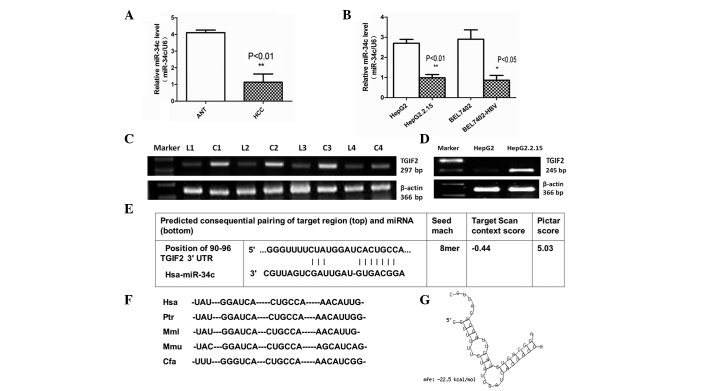
In the present study, qPCR revealed that miR-34c was downregulated in HBV-related HCC tissues compared with adjacent non-cancerous liver tissues (Fig. 1A). The levels of miR-34c expression were also detected by qPCR in two cell lines of HBV-associated HCC (Fig. 1B). Analysis of the microarray data demonstrated that TGIF2 was upregulated in HBV-infected transgenic mice compared with syngeneic BALB/c mice (unpublished data). As presented in Fig. 1C and D, TGIF2 was also significantly upregulated in HepG2.2.15 cells compared with HepG2 cells, and in HBV-associated HCC tissues (C1-C4) compared with adjacent non-cancerous tissues (L1-L4).
Figure 1.
miR-34c expression was downregulated in HBV-associated HCC, and TGIF2 is a putative target of miR-34c. (A) Expression levels of miR-34c were significantly reduced in HBV-associated HCC clinical specimens compared with ANT (**P<0.01). (B) Quantitative PCR results showed that the expression levels of miR-34c were reduced in HepG2.2.15 compared with HepG2 cells (**P<0.01), as well as in BEL7402-HBV compared with BEL7402 cells (*P<0.05). (C) mRNA expression levels of TGIF2 were significantly upregulated in HBV-associated HCC tissues compared with ANT. (D) Reverse transcription-PCR results showed that TGIF2 was upregulated in HepG2.2.15 compared with HepG2 cells. All of the above data were obtained from three independent experiments. (E) TGIF2 was identified as a potential target gene of miR-34c using TargetScan and PicTar software. (F) The TargetScan prediction showed a common binding sequence on the 3′-UTR of TGIF2 that was conserved across vertebrate species, including human, chimpanzee, rhesus macaque, mouse and dog. (G) The duplex structure of miR-34c and the 3′-UTR of TGIF2 is shown. The free energy of miR-34c bound to the target site of TGIF2 was calculated by RNAhybrid (minimum free energy, −22.5 kcal/mol). miR-34c, microRNA-34c; ANT, adjacent non-cancerous tissue; PCR, polymerase chain reaction; HCC, hepatocellular carcinoma; HBV, hepatitis B virus; 3′-UTR, 3′-untranslated region; TGIF2, TGFB-induced factor homeobox 2.
TGIF2 is a candidate target gene of miR-34c
TGIF2 was identified as a candidate target gene of miR-34c by querying MiRanda and TargetScan. The analysis of potential miR-34c binding sites within the published 3′-UTR of TGIF2 using the miR-target prediction programs and RNAhybrid revealed one putative miR-34c target site (Fig. 1E). TargetScan also demonstrated that the 3′-UTR of TGIF2 targeted by miR-34c was conserved across vertebrate species, including human, chimpanzee, rhesus macaque, mouse and dog (Fig. 1F). These findings indicate that the targeting of TGIF2 by miR-34c may be evolutionarily conserved. The free energy of miR-34c binding to the TGIF2 targeting site was calculated (minimum free energy, −22.5 kcal/mol), and the duplex structure between miR-34c and the 3′-UTR of TGIF2 was identified by RNAhybrid (Fig. 1G).
miR-34c downregulated TGIF2 expression in HBV-associated HCC
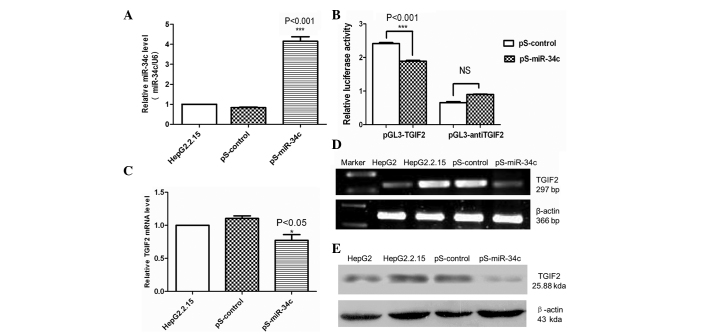
qPCR was used to confirm that HepG2.2.15 cells transfected with pS-miR-34c stably expressed miR-34c. The results demonstrated that miR-34c was significantly upregulated in HepG2.2.15 cells transfected with pS-miR-34c (P<0.001) (Fig. 2A). No difference was identified between HepG2.2.15 cells transfected with pS-control or untreated cells.
Figure 2.
TGIF2 expression was downregulated by the transfection of miR-34c. (A) Expression of miR-34c was significantly upregulated by ~4-fold in HepG2.2.15 cells transfected with pS-miR-34c compared with untreated cells and pS-control-transfected cells (***P<0.001). (B) The luciferase reporter constructs containing the sense or antisense 3′-UTR of TGIF2 were co-transfected with pS-miR-34c or pS-control into HepG2.2.15 cells. Luciferase activity was measured 48 h after transfection. The results showed that TGIF2 was a direct target of miR-34c. Three different transfections were carried out and the luciferase activity was measured twice for each reaction. (C) The mRNA expression level of TGIF2 was shown to be significantly reduced in cells transfected with pS-miR-34c by quantitative PCR (*P<0.05). (D) The mRNA expression level of TGIF2 in HepG2.2.15 cells transfected with pS-miR-34c was shown to be significantly reduced compared with pS-control-transfected cells and untreated cells using reverse transcription-PCR. β-actin was used as the endogenous control for RNA normalization. (E) Protein expression of TGIF2 was reduced following pS-miR-34c transfection compared with pS-control-transfected cells and untreated cells, which was consistent with the mRNA expression levels. All data were obtained from three independent experiments. PCR, polymerase chain reaction; miR-34c, microRNA-34c; 3′-UTR, 3′-untranslated region; pS, plasmid; TGIF2, TGFB-induced factor homeobox 2.
To test whether miR-34c directly binds to the 3′-UTR of TGIF2, a dual luciferase reporter assay was performed. As shown in Fig. 2B, the luciferase activity of the reporter containing the 3′-UTR of TGIF2 reduced in cells expressing miR-34c compared with cells expressing pS-control (P<0.001). This result indicated that TGIF2 was a direct target of miR-34c.
To determine whether miR-34c overexpression affects TGIF2, the levels of TGIF2 mRNA and protein were measured 48 and 72 h after transfection. qPCR and RT-PCR demonstrated that TGIF2 mRNA was significantly reduced following pS-miR-34c transfection compared with pS-control-transfected and untreated cells (P<0.05) (Fig. 2C and D). Western blot analysis revealed that the protein levels of TGIF2 were reduced in HepG2.2.15 cells transfected with pS-miR-34c compared with pS-control-transfected or untreated cells (Fig. 2E). Therefore, these results were consistent with the changes in TGIF2 mRNA expression.
miR-34c inhibited HBV DNA replication and viral antigen synthesis
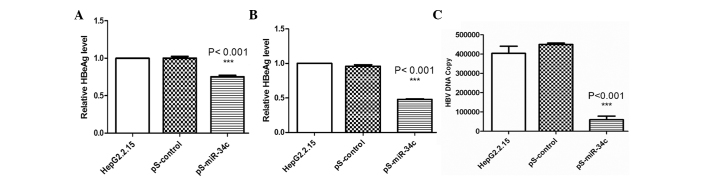
To determine the effect of miR-34c on HBV gene expression, HBsAg and HBeAg levels were measured 48 h after transfection. As shown in Fig. 3A and B, the inhibitory rates of HBsAg and HBeAg were 24.39 and 48.67%, respectively (P<0.001). No significant reduction was observed in untreated or pS-control-transfected cells.
Figure 3.
HBsAg and HBeAg secretion and HBV DNA replication were inhibited by transfection with pS-miR-34c, as shown by ELISA assay. (A) Transfection with pS-miR-34c had a significant inhibitory effect on HBsAg levels compared with pS-control-transfected and untreated cells, and the inhibitory ratio was 24.39% (***P<0.001). (B) Transfection with pS-miR-34c inhibited the expression of HBeAg, and the inhibitory ratio was 48.67% (***P<0.001). (C) DNA copy number of HBV in cells transfected with pS-miR-34c was reduced by 83.27% 48 h after transfection compared with untreated cells and cells transfected with the pS-control vector (***P<0.001). All data were obtained from three independent experiments. miR-34c, microRNA-34c; HBV, hepatitis B virus; pS, plasmid; HBsAg, hepatitis B virus surface antigen; HBeAg, hepatitis B virus e antigen.
qPCR was performed to determine whether transfection with pS-miR-34c would result in a reduction in HBV DNA replication. Quantitative assays revealed that the copy number of HBV DNA was reduced by 83.27% in cells transfected with pS-miR-34c 48 h after transfection (P<0.001) (Fig. 3C). No evident changes were detected in the cells transfected with pS-control, or untreated cells.
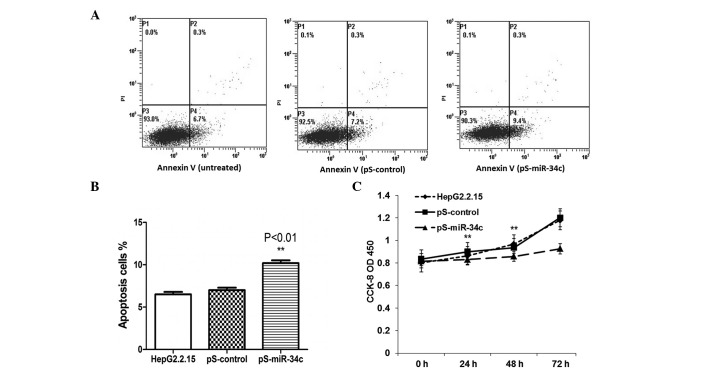
miR-34c induces apoptosis and suppresses cell proliferation in HepG2.2.15 cells
Subsequently, the effects of miR-34c transfection on the induction of apoptosis were examined. The data indicated that miR-34c expression had a positive effect on apoptosis in HepG2.2.15 cells at 48 h post-transfection compared with untreated and pS-control-transfected cells (P<0.01) (Fig. 4A and B). A CCK-8 assay was used to observe the biological effects of miR-34c on tumor cell growth. As shown in Fig. 4C, the overexpression of miR-34c suppressed cell proliferation at 48 h and 72 h post-transfection (P<0.05).
Figure 4.
Restoration of miR-34c induced apoptosis and inhibited cell proliferation. (A and B) Flow cytometric analysis showed that enhanced miR-34c expression had a significant effect on apoptosis induction in HepG2.2.15 cells at 48 h post-transfection compared with untreated cells and pS-control-transfected cells (***P<0.001). (C) Cell proliferation was analyzed using CCK-8 assays. Each value represents the mean of three replicates. The proliferation of HepG2.2.15 cells transfected with pS-miR-34c was significantly suppressed at 48 and 72 h post-transfection compared with untreated cells and cells transfected with the control vector (P<0.05). All data were obtained from three independent experiments. miR-34c, microRNA-34c; pS, plasmid; CCK-8, Cell Counting kit-8; OD, optical density.
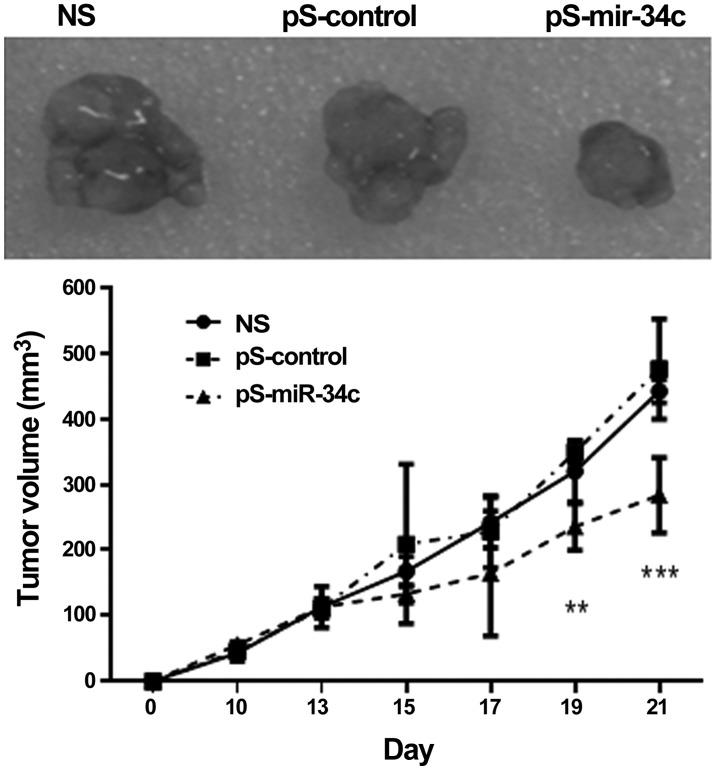
miR-34c inhibits tumor growth in vivo
BALB/c nude mice were transplanted with BEL7402-1.1-HBV cells and the tumors were formed in 2 weeks. As shown in Fig. 5, tumor growth in mice injected with miR-34c was significantly inhibited compared with the control mice (P<0.001 at day 21).
Figure 5.
miR-34c inhibits tumor growth in vivo. A total of 2×106 BEL7402-HBV cells were injected subcutaneously into the flanks of each BALB/c nude mouse. Fifteen mice were used and randomly assigned to 3 groups (5 mice per group). When the tumor grew to a volume of 90 mm3 at 2 weeks, pS-miR-34c or pS-control (20 µg each) was injected into the tumor of each mouse five times every two days. Tumor volume was measured by a caliper and calculated as: Length × width × height (mm3). Tumor growth in mice injected with miR-34c was significantly inhibited compared with the the NS or pS-control mice (n=5; day 19, **P<0.01; day 21, ***P<0.001). miR-34c, microRNA-34c; NS, normal saline; pS, plasmid Silencer.
Discussion
miRs have been shown to serve important roles in viral infection and carcinogenesis. Chronic infection of HBV is a key factor in the development of HCC (17). Previously, certain mammalian miRs have been identified that exhibit antiviral effects (9). miR-34c is involved in negative feedback control of the cell cycle, including cell cycle arrest, senescence and apoptosis. Previous studies have shown that miR-34c is downregulated in different tumors and cell lines, including laryngeal and nasopharyngeal carcinoma, and prostate, colorectal, bronchial squamous, breast, gastric and lung cancer (18–25). The present study extended these findings by demonstrating that miR-34c was downregulated in HBV-associated HCC tissues as well as in HepG2.2.15 cells that constitutively replicate HBV, compared with HepG2 cells that do not. Previous studies have indicated that miRs influence cell proliferation, apoptosis, migration and invasion (26–28). The present study also demonstrated that miR-34c inhibited cell proliferation in HepG2.2.15 cells, which was accompanied by the induction of cell apoptosis.
Viral replication and gene expression may be inhibited by virus-specific small interfering RNAs. For example, previous studies have shown that human miR-125a-5p interfered with HBsAg expression (29), miR-199a-3p and miR-210 suppressed HBV replication (30), and miR-122 suppressed HBV replication through the downregulation of heme oxygenase-1 and N-myc downstream-regulated gene 3 (31,32). The present study demonstrated that HBV DNA replication and viral antigen secretion were significantly inhibited in HepG2.2.15 cells by miR-34c transfection. Although further studies are required to elucidate the mechanisms behind the antiviral effect, the present data are important for understanding HBV-host interactions.
miR-34c is downregulated in HCC and contributes to hepatocarcinogenesis by targeting genes, including MET, E2F1-3, MYC, SIRT1 and BCL2. The present study combined data from bioinformatics, gene chip analysis and a dual luciferase assay to identify the target gene of miR-34c. TGIF2 is a transcriptional co-repressor that interacts with Smad and represses the TGF-β signaling pathway (12). TGF-β is a multifunctional cell factor that inhibits cell proliferation and induces apoptosis (33). TGIF2 may function as an oncogene by regulating the Ras/MAPK pathway (34). The present study identified that TGIF2 was directly targeted and repressed by miR-34c in HBV-associated HCC cell lines, indicating that TGIF2 participates in HBV-associated hepatocarcinogenesis. However, TGIF2 is not the only target gene of miR-34c that is relevant to tumorigenesis and metastasis in HBV-associated HCC. The prediction and identification of other target genes may be important to improve understanding of the role of miR-34c in HBV-related HCC.
Previous studies highlight miR-34a, b and c as direct, conserved p53 target genes that presumably mediate cell cycle arrest, induction of apoptosis and senescence by p53 (8,9,35,36). Since miRs may regulate the expression of hundreds of target genes, these findings add new challenges and opportunities for studies focused on the p53 network. As reported previously, HBV DNA may bind to the p53 protein and form DNA-protein complexes in human hepatoma cell lines (37). Cotransfection with p53 and HBV DNA increased the replication of HBV, chloramphenicol acetyltransferase activity, tumor cell apoptosis and cytoplasmic p53 accumulation in hepatoma cells (37). To further substantiate the connection between p53 and the miR-34c target genes, additional studies are required to determine the relevance of miR-34c-mediated target gene downregulation for p53 tumor suppression. Studies in gene knockout mouse models of HBV transfection-induced hepatitis and HCC may aid in determining the function of miR-34c genes and their relevance for tumor suppression in vivo.
In conclusion, miR-34c may serve a significant role as a tumor suppressor miR by targeting TGIF2 during HBV-related HCC. The precise mechanisms that result in miR-34c downregulation following HBV infection are not well understood, but the present results indicate that miR-34c represses HBV replication and viral antigen expression. HepG2.2.15 cells were influenced by miR-34c through the inhibition of cell proliferation and induction of cell apoptosis. Thus, miR-34c and TGIF2 represent key regulatory factors, diagnostic markers and therapeutic targets for the prevention and treatment of HBV-associated HCC.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant nos. 30772031 and 30801036) and the Doctorate Fund for New Teachers from the National Education Ministry of China (grant no. 200804221065).
References
- 1.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J, Sherman M. American Association for the Study of Liver Diseases: Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki K, Okuda Y, Ota M, Kojima F, Horimoto M. Diagnosis of hepatocellular carcinoma nodules in patients with chronic liver disease using contrast-enhanced sonography: Usefulness of the combination of arterial- and kupffer-phase enhancement patterns. J Ultrasound Med. 2015;34:423–433. doi: 10.7863/ultra.34.3.423. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 6.Skaftnesmo KO, Prestegarden L, Micklem DR, Lorens JB. MicroRNAs in tumorigenesis. Curr Pharm Biotechnol. 2007;8:320–325. doi: 10.2174/138920107783018390. [DOI] [PubMed] [Google Scholar]
- 7.Paris R, Henry RE, Stephens SJ, et al. Multiple p53-independent gene silencing mechanisms define the cellular response to p53 activation. Cell Cycle. 2008;7:2427–2433. doi: 10.4161/cc.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 9.He L, He X, Lim LP, et al. A microRNA component of the p53 tumor suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong YW, Cannell IG, de Moor CH, et al. The mechanism of microRNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci USA. 2008;105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Migliore C, Petrelli A, Ghiso E, et al. MicroRNAs impair MET mediated invasive growth. Cancer Res. 2008;68:10128–10136. doi: 10.1158/0008-5472.CAN-08-2148. [DOI] [PubMed] [Google Scholar]
- 12.Wotton D, Lo RS, Lee S, Massagué J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/S0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 13.Yang P, Li Q-J, Feng Y, Zhang Y, Markowitz GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karimi-Googheri M, Daneshvar H, Nosratabadi R, Zare-Bidaki M, Hassanshahi G, Ebrahim M, Arababadi MK, Kennedy D. Important roles played by TGF-β in hepatitis B infection. J Med Virol. 2014;86:102–108. doi: 10.1002/jmv.23727. [DOI] [PubMed] [Google Scholar]
- 15.Liu N, Jiao T, Huang Y, Liu W, Li Z, Ye X. Hepatitis B virus regulates apoptosis and tumorigenesis through the microRNA-15a-Smad7-transforming growth factor beta pathway. J Virol. 2015;89:2739–2749. doi: 10.1128/JVI.02784-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.McGlynn KA, London WT. Epidemiology and natural history of hepatocellular carcinoma. Best Pract Res Clin Gastroenterol. 2005;19:3–23. doi: 10.1016/j.bpg.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Cai KM, Bao XL, Kong XH, et al. Hsa-miR-34c suppresses growth and invasion of human laryngeal carcinoma cells via targeting c-Met. Int J Mol Med. 2010;25:565–571. doi: 10.3892/ijmm_00000378. [DOI] [PubMed] [Google Scholar]
- 19.Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Chen JX, Fu XP, Yang S, Zhang Z, Chen KH, Li Y. microRNA expression profiling of nasopharyngeal carcinoma. Oncol Rep. 2011;25:1353–1363. doi: 10.3892/or.2011.1204. [DOI] [PubMed] [Google Scholar]
- 21.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 22.Mascaux C, Laes JF, Anthoine G, Haller A, Ninane V, Burny A, Sculier JP. Evolution of microRNA expression during human bronchial squamous carcinogenesis. Eur Respir J. 2009;33:352–359. doi: 10.1183/09031936.00084108. [DOI] [PubMed] [Google Scholar]
- 23.Yu F, Jiao Y, Zhu Y, Wang Y, Zhu J, Cui X, Liu Y, He Y, Park EY, Zhang H, Lv X, et al. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J Biol Chem. 2012;87:465–473. doi: 10.1074/jbc.M111.280768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki H, Yamamoto E, Nojima M, et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 2012;31:2066–2073. doi: 10.1093/carcin/bgq203. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Chen Z, Gao Y, Li N, Li B, Tan F, Tan X, Lu N, Sun Y, Sun J, Sun N, He J. DNA hypermethylation of microRNA-34b/c has prognostic value for stage I non-small cell lung cancer. Cancer Biol Ther. 2011;11:490–496. doi: 10.4161/cbt.11.5.14550. [DOI] [PubMed] [Google Scholar]
- 26.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenouda SK, Alahari SK. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucl Acids Res. 2011;39:5157–5163. doi: 10.1093/nar/gkr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Qiu L, Fan H, Jin W, Zhao B, Wang Y, Ju Y, Chen L, Chen Y, Duan Z, Meng S. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem Biophys Res Commun. 2010;398:771–777. doi: 10.1016/j.bbrc.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Fan CG, Wang CM, Tian C, Wang Y, Li L, Sun WS, Li RF, Liu YG. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol Rep. 2011;26:1281–1286. doi: 10.3892/or.2011.1375. [DOI] [PubMed] [Google Scholar]
- 33.Yamada SD, Baldwin RL, Karlan BY. Ovarian carcinoma cell cultures are resistant to TGF-beta1-mediated growth inhibition despite expression of functional receptors. Gynecol Oncol. 1999;75:72–77. doi: 10.1006/gyno.1999.5535. [DOI] [PubMed] [Google Scholar]
- 34.Lo RS, Wotton D, Massagué J. Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. EMBO J. 2001;20:128–136. doi: 10.1093/emboj/20.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 36.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu J, Lin J, Zhang S, Zhu Z, Ni C, Zhang S, Gao H, Zhu M. HBV DNA can bind to P53 protein and influence p53 transactivation in hepatoma cells. Biochem Biophys Res Commun. 2009;386:504–509. doi: 10.1016/j.bbrc.2009.06.081. [DOI] [PubMed] [Google Scholar]