Abstract
Increasing evidence has suggested that microRNA-133b (miR-133b) is important in regulating the genesis of different types of cancer. However, the effects and the underlying mechanisms of miR-133b in the development of glioblastoma (GBM) remain largely unknown. The aim of the present study was to investigate the role of miR-133b in GBM and to determine the molecular mechanisms underlying its action. Reverse transcription-quantitative polymerase chain reaction was used to measure the expression levels of miR-133b in 21 human GBM samples and 9 normal brain tissue samples. A wound healing assay, and Transwell migration and invasion assays were used to evaluate the effects of miR-133b on cell migration and invasion. Western blotting and a luciferase reporter assay were used to identify the target genes of miR-133b. It was found that miR-133b suppressed GBM cell migration and invasion, and matrix metalloproteinase 14 (MMP14) was identified as a direct target gene. In conclusion, miR-133b may suppress GBM migration and invasion through directly targeting MMP14, highlighting its potential as a novel agent for the treatment of GBM invasion.
Keywords: glioblastoma, matrix metalloproteinase 14, migration, microRNA-133b, invasion
Introduction
Glioblastoma (GBM) is one of the most common and fatal tumors of the central nervous system (1–3). In spite of the therapeutic advances in strategies for treatment, the poor prognosis of patients diagnosed with GBM has not improved and the median survival is <1 year (2–4). One of the major reasons leading to the poor prognosis of patients is recurrence as a result of the diffuse infiltration of tumor cells into the surrounding brain tissue. Therefore, understanding the molecular mechanisms of astrocytic tumor cell invasion in the brain is essential for developing effective strategies for the control of GBM.
MicroRNAs (miRNAs/miRs) belong to a class of small non-coding single-stranded RNAs of 18–23 nucleotides, which take part in the control of gene expression by destabilizing target mRNA or by repressing protein translation. It is now clear that miRNAs are involved in regulating important cellular functions, including the stress response, death, differentiation, apoptosis, invasion, proliferation and development (5,6). Moreover, emerging evidence has shown that the dysfunction or deregulation of miRNAs is involved in the processes of cancer initiation, progression and metastasis (7,8). The aberrant expression of miR-133b has been reported to be downregulated in certain types of cancer, including cervical carcinoma, lung cancer, renal cell carcinoma (9), colon cancer (10), osteosarcoma (11) and prostate cancer (1,12). Furthermore, miR-133b has been suggested to function as a tumor suppressor in specific types of cancer by the direct targeting of certain oncogenes (13). However, the role of miR-133b in GBM is not clear.
The aim of the present study was to investigate the expression of miRNA-133b on the migration and invasion of GBM cells.
Materials and methods
Tumor and normal tissue samples
Normal brain tissue and human GBM samples were obtained from the Department of Neurosurgery at the Tumor Hospital (Harbin Medical University Harbin, Heilongjiang, China). None of the patients had received radiotherapy or chemotherapy prior to surgery. Samples were snap-frozen in liquid nitrogen and stored at −80°C until RNA extraction. Study approval was obtained from the Ethics Committee of The Tumor Hospital, Harbin Medical University and all patients involved provided written informed consent.
Cell culture and transfection
Two human glioma cell lines, U87 and U251, purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China), were included in this study. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone, Logan, UT, USA) containing 100 mg/ml streptomycin (Beyotime Institute of Biotechnology, Haimen, China), 100 U/ml penicillin ((Beyotime Institute of Biotechnology) and 10% fetal bovine serum (FBS; Hyclone) in a humidified atmosphere containing 5% CO2 at 37°C. On the day prior to transfection, the cells were seeded onto 6-well plates (5×105 cells/well) and inoculated in complete medium without antibiotics (2 ml/well).
Transfections with mature miR-133b mimics, negative control (NC) and luciferase reporter plasmid were carried out using 50 nm Lipofectamine 2000 (Shanghai GenePharma Co., Ltd., Shanghai, China) according to the manufacturer's instructions. The sequences were as follows: hsa-miR-133b mimic, 5′-UUUGGUCCCCUUCAACCAGCUA-3′; and NC, 5′-UUCUCCGAACGUGUCACGUTT-3.
Wound healing assay
The U87 and U251 cells were plated onto 96-well plates and treated with miR-133b mimics and NC for 24 h. A 100 μl plastic pipette tip was then dragged across the plate to create a wound line that is a cell-free area. Images (magnification, ×40) of wound lines for each treatment were taken at 0 and 24 h using a digital camera (Nikon, Tokyo, Japan). The distance of wound closure (compared with the control at 0 h) was measured. These experiments were repeated three times.
Transwell migration and invasion assays
A Transwell invasion assay was performed using a 24-well Transwell chamber with polycarbonate membranes with 8-μm pores (Corning-Costar Inc., Corning, NY, USA) coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Transfected cells (miR-133 mimics or NC) were loaded onto the upper filter (0.2 ml) at a density of 5×105 cells/ml of serum-free medium. DMEM/F12 medium containing 15% FBS (0.5 ml) was added to the lower chamber. The cells were incubated for 24 h at 37°C in a humidified incubator containing 5% CO2. A cotton swab was used to scrape off cells and Matrigel on the upper side of the Transwell insert filter. Invading cells were fixed with 4% paraformaldehyde (Beyotime Institute of Biotechnology, Haimen, China) for 10 min, stained with hematoxylin for 2 min, rinsed in running tap water for 10 min, differentiated with 0.3% acid alcohol for 1 min, rinsed in running tap water for 10 min, stained with eosin for 2 min, and counted by capturing images in three independent fields for each Transwell insert filter (magnification, ×200). The numbers of invading cells were averaged. A migration assay was performed following the same procedure, except that the cells were incubated for 12 h and the polycarbonate filters were not coated with Matrigel. These experiments were repeated three times.
Western blotting
The cells were washed twice with phosphate-buffered saline and lysed directly in lysis buffer (1 mmol/l PMSF, 1 mmol/l EDTA, 40 mmol/l Tris-HCl, 100 mmol/l NaVO3, 150 mmol/l KCl and 1% Triton X-100) on ice for 15 min. Subsequent to centrifugation (12,000 × g for 20 min at 4°C), the Bradford protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to measure the protein concentrations of the lysates. Equal amounts of proteins were separated on 10% SDS-PAGE gels and then electrotransferred onto a polyvinylidene difluoride membrane. The membranes were blocked in 5% skimmed milk-Tris-buffered saline plus Tween-20 solution and incubated with rabbit MMP14 monoclonal antibody (1:1,000; Abcam, Cambridge, UK) or rabbit polyclonal β-actin antibody (1:1,000; Bioss, Beijing, China). Following incubation with peroxidase-conjugated AffiniPure goat anti-rabbit IgG (1:2,000; ZSGB-BIO, Beijing, China), protein bands were detected with Fujifilm Las-4000 (Fujifilm, Tokyo, Japan).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). RNA (~1 μg) was reverse transcribed using the miR-133b qPCR Quantitation kit (Shanghai GenePharma Co., Ltd.). The primers of miR-133b and U6 were synthesized by Shanghai Genepharma Co., Ltd., and RT-qPCR was performed on a 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR Master Mix (Applied Biosystems). The reaction conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec and 60°C for 1 min. The mRNA expression levels of miR-133b were normalized to U6 using the standard ΔΔCt method. Each assay was performed in triplicate and all experiments were repeated at least three times.
miRNA target prediction and luciferase reporter assay
The following target prediction software was used to identify miRNA-133b that could potentially bind MMP14: Pictar (http://www.pictar.mdc-berlin.de/), TargetScan (http://www.targetscan.org/), miRanda (http://www.microrna.org/) and miRWalk (http://www.umm.uni-heidelberg.de/). The U87 and U251 cells were seeded in 24-well plates. One day later, the cells were co-transfected with luciferase vectors, either the wild-type MMP14 3′-untranslated region (3′UTR) reporter plasmid or mutated MMP14 3′UTR reporter plasmid together with miR-133b mimics or miRNA-NC, and the Renilla control. After 48 h, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA) and normalized to Renilla luciferase activity.
Statistical analysis
SPSS statistical software, version 19.0 (IBM, Armonk, NY, USA) was used for statistical analysis. The expression of miR-133b in the tissue samples was analyzed using the Mann-Whitney U test for comparing the results of two groups. Other data were expressed as the mean ± standard deviation, and a two-sided Student's t-test was used for the statistical analysis. P<0.05 was used to indicate a statistically significant difference.
Results
Expression of miR-133b is lower in GBM specimens than normal brain tissues
The expression levels of miR-133b were analyzed by RT-qPCR in 21 GBM specimens and 9 normal brain tissue samples. As shown in Fig. 1, the expression of miR-133b was significantly lower in the GBM specimens than in the normal brain tissues (P<0.05).
Figure 1.
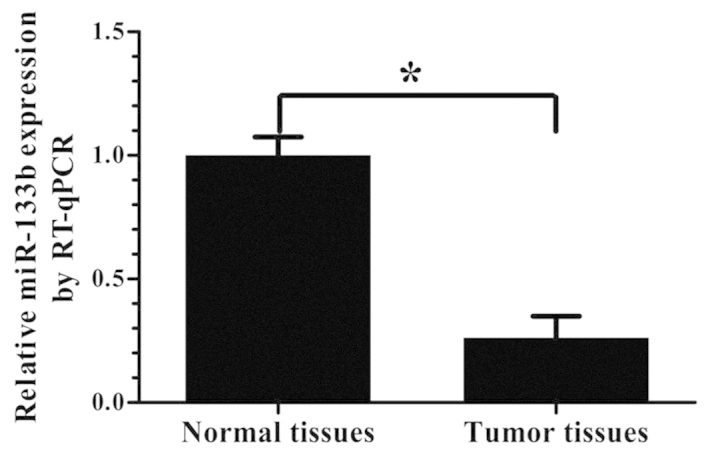
Expression of miR-133b in tissue samples. The relative expression of miR-133b in 21 GBM tissues and 9 normal brain tissues was measured by RT-qPCR, and U6 small nuclear RNA was used as an internal control (means ± standard deviation; n=3; *P<0.05). miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; GBM, glioblastoma.
Expression of miR-133b following transfection of miR-133b mimics in U87 and U251 cells
miR-133b or NC mimics were transfected into the U87 and U251 cells. After 24 h, the ectopic expression of miR-133b in the cells was analyzed by RT-qPCR. Compared with the NC group, the miR-133b mimic group showed significantly upregulated miR-133b expression in the two cell lines (P<0.05; Fig. 2).
Figure 2.
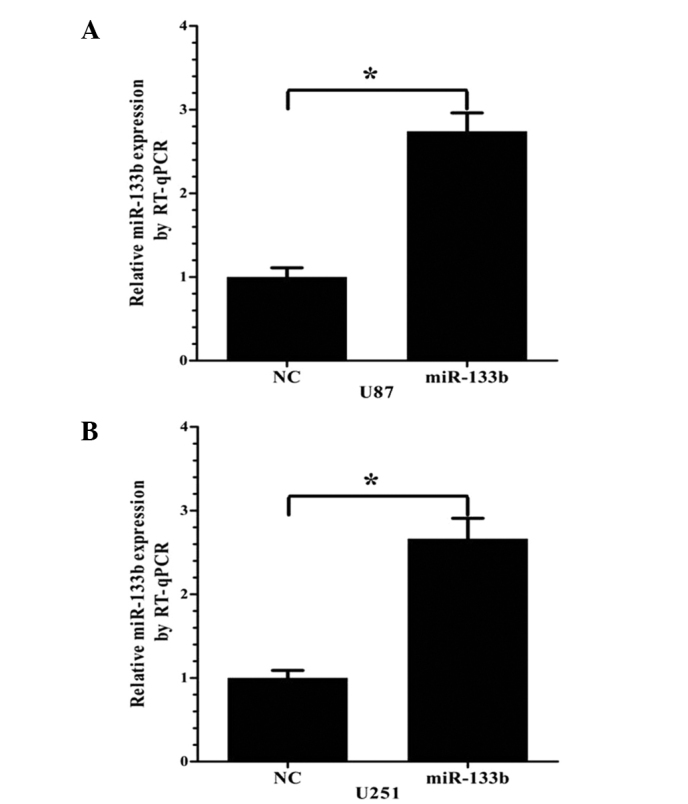
Relative expression of miR-133b in GBM cell lines at 24 h post-transfection, as measured by RT-qPCR. Following miR-133b mimic transfection, miR-133b expression was markedly increased in the (A) U87 and (B) U251 cells (mean ± standard deviation; n=3; *P<0.05). miR, microRNA; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; GBM, glioblastoma.
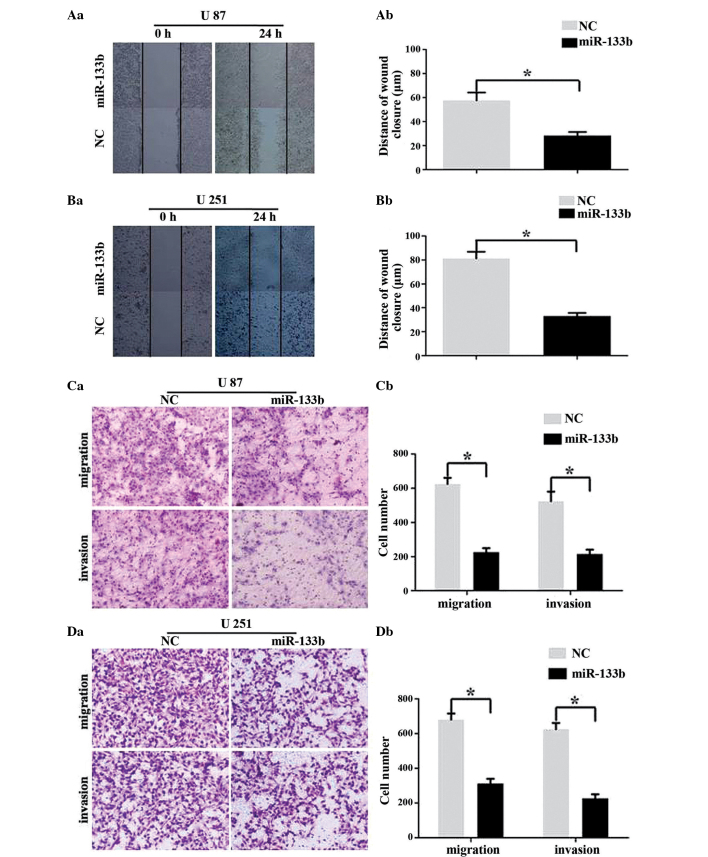
miR-133b inhibits GBM cell migration and invasion
To determine whether the overexpression of miR-133b has an effect on the migration and invasion of GBM cells, wound healing and Transwell assays were performed. As demonstrated in Fig. 3A and B (wound healing assays), the overexpression of miR-133b using mimics significantly inhibited the migratory ability of the U87 and U251 cells (P<0.05). As shown in Fig. 3C and D, the ability of the U87 and U251 cells to migrate through a Transwell insert filter was clearly inhibited by miR-133b (P<0.05). The Transwell invasion assay also showed that the number of invasive cells transfected with miR-133b mimics was significantly lower compared with the NC transfected cells (P<0.05). Therefore, these findings indicated that miR-133b acts as an invasive suppressor in GBM cells.
Figure 3.
miR-133b inhibits the migration and invasion of human GBM cell lines. (A and B) Wound healing assays using U87 and U251 cells transfected with miR-133b mimics or NC. (Aa and Ba) Representative images and (Ab and Bb) quantification of 3 randomly selected fields (mean ± standard deviation; n=3; *P<0.05). (C and D) Transwell migration and invasion assays using U87 and U251 cells transfected with miR-133b mimics or NC. (Ca and Da) Representative images and (Cb and Db) quantification of 3 randomly selected fields (mean ± standard deviation; n=3; *P<0.05). miR, microRNA; NC, negative control; GBM, glioblastoma.
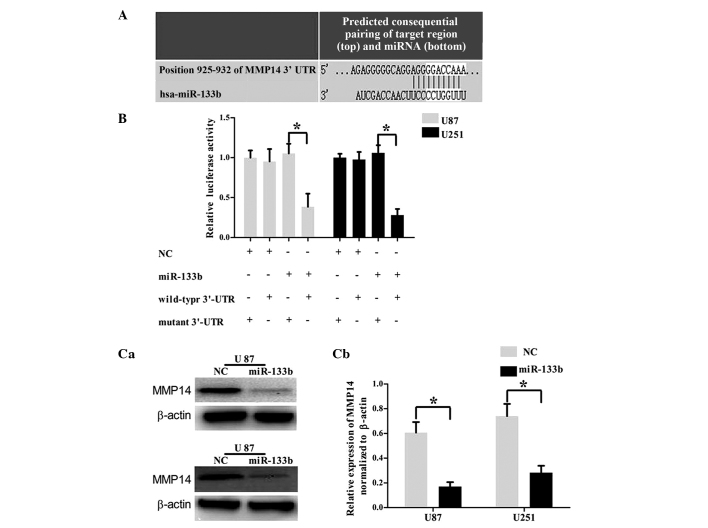
miR-133b targets the 3′UTR of MMP14
To investigate the mechanism by which miR-133b regulates cell invasion, an miRNA target search was performed with four target prediction databases (miRanda, TargetScan, PicTar and miRWalk), and all four databases showed that the MMP14 mRNA contained miR-133b binding sites (Fig. 4A). To examine the hypothesis that miR-133b targeted MMP14, a luciferase reporter assay was performed. The wild-type MMP-9 3′UTR reporter plasmid or mutated MMP-9 3′UTR reporter plasmid was cotransfected into the U87 and U251 cells with miR-377 mimic or NC. As shown in Fig. 4B, the U87 and U251 cell lines cotransfected with miR-133b mimic and wild-type MMP14 3′UTR reporter plasmid showed a significant decrease in reporter activity (P<0.05). To confirm that the expression of MMP14 is regulated by miR-133b, western blot analysis was performed. As shown in Fig. 4C, MMP14 was significantly downregulated in the U87 and U251 GBM cell lines following the overexpression of miR-131b (P<0.05). These findings demonstrated that the 3′UTR of MMP14 was the directed target of miR-133b.
Figure 4.
miR-133b directly targets MMP14. (A) Bioinformatics predicted interactions of miR-133b and their binding sites at the 3′UTR of MMP14 (TargetScan 6.0). (B) Luciferase activity assay demonstrated a direct targeting of the 3′UTR of MMP14 by miR-133b. Cells (U87 and U251) were co-transfected with luciferase vectors, either the wild-type MMP14 3′UTR reporter plasmid or mutated MMP14 3′UTR reporter plasmid, together with miR-133b mimics or NC. After 48 h, the luciferase activities were measured (mean ± standard deviation; n=3; *P<0.05). (C) Western blotting of MMP14 in U87 and U251 cells transfected with miR-133b mimics or NC. (a) Representative images of western blotting and (b) quantification of the bands (mean ± standard deviation; n=3; *P<0.05). miR, microRNA; NC, negative control; GBM, glioblastoma; MMP14, matrix metalloproteinase 14; UTR, untranslated region.
Discussion
The present study showed for the first time a novel molecular mechanism of miR-133b and MMP14 in glioma cell invasion. It was found that miR-133b was frequently downregulated in the tumor tissues of GBM patients. In addition, MMP14 was identified as target of miR-133b, and miR-133b significantly induced the protein expression of MMP14 in glioma cells. More significantly, it was also concluded that miR-133b suppressed glioma cell invasion via its target MMP14.
miRNAs are key regulators of gene expression at the post-transcriptional level and mediate a large variety of physiological and pathological processes (8,14). Mounting evidence has indicated that alterations in the expression of miRNAs correlate with the pathogenesis of different types of human malignancy, where they serve as either oncogenes or tumor suppressors. Accumulating studies have demonstrated that miR-133b functions as a tumor suppressive miRNA in various cancers (15). For instance, it was reported that miR-133b inhibited the cell viability and colony formation ability of non-small cell lung cancer (16). Wu et al showed that miR-133b suppressed the cell proliferation, migration and invasion of renal cell carcinoma cells (9). However, the role of miR-133b in GBM remains unknown. The present study first analyzed the expression of miR-133b in GBM and found that this expression was lower in GBM compared with normal brain tissues. The inhibition of migration and invasion caused by the overexpression of miR-133b in the human GBM U87 and U251 cell lines was also observed. This result demonstrates that miR-133b acts as a tumor suppressor and inhibits GBM cell migration and invasion.
To date, certain targets of miR-133b have been reported, including FGFR1 in gastric cancer (16), FSCN-1 in esophageal squamous cell carcinoma (17) and CXCR4 in colorectal cancer (18). In the present study, it was shown that miR-133b targets MMP14, revealing a potential mechanism associated with the development of GBM.
The MMPs, a family of proteolytic enzymes, have been indicated to be important in the tissue remodeling associated with various physiological or pathological processes including arthritis, cirrhosis, tissue repair, angiogenesis, morphogenesis, tumor invasion and neoplastic metastasis (19). There are 24 soluble and membrane-anchored members of the MMP family in humans, and MMP14 is one of them. MMP14 acts either as a pericellular collagenase or as a proMMP-2 activator via formation of a TIMP-2/roMMP-2/MMP14 complex. Multiple studies have demonstrated that MMP14 is expressed in various malignant tumors, and its overexpression in tumor cells promotes tumorigenesis and metastasis. For example, in previous studies, high MMP14 mRNA expression was found to be an independent factor of lymph node metastasis and tumor invasion in carcinoma of the cervix, lung and stomach (20,21). In addition, it is proposed that a ternary protein complex composed of αVβ3 integrin, ADAM12 and MMP14 at the tumor cell surface regulates the function of MMP14 to affect tumor growth (22). More and more studies have confirmed that MMP14 also plays significant role in glioma progression. Increases in MMP14 were reported to correlate with poor overall survival, and may represent a diagnostic and prognostic marker of glioma (23). Recently, certain studies have further investigated the mechanisms that regulate MMP14 expression. A study by Xie et al also found that the expression of MMP14 was high in human brain gliomas of different pathological grades and that this may be regulated by activation of the extracellular signal-regulated kinase 1/2 signaling pathway (24). Another study found that MMP14 could contribute to the invasion of glioma via HOXD10, which is a target of miR-10b (10). However, the results of the present study indicated that MMP14 was regulated by miR-133b and suggested that miR-133b may suppresses GBM cell migration and invasion via the downregulation of MMP14.
In summary, the present study showed that miR-133b was downregulated in GBM. The decreased expression of miR-133b promoted GBM cell invasion by modulating MMP14, which was identified as a novel target of miR-133b. Based on these observations, the study indicates that miR-133b may represent a potential target for an effective treatment strategy to suppress GBM invasion and metastasis.
Acknowledgements
This study was supported by the Scientific and Technological Project of Heilongjiang Province of China (grant no. GC12C303-1) and the Foundation of Heilongjiang Educational Committee (grant no. 12541467).
References
- 1.Nava F, Tramacere I, Fittipaldo A, Bruzzone MG, Dimeco F, Fariselli L, Finocchiaro G, Pollo B, Salmaggi A, Silvani A, et al. Survival effect of first- and second-line treatments for patients with primary glioblastoma: A cohort study from a prospective registry, 1997–2010. Neuro Oncol. 2014;16:719–727. doi: 10.1093/neuonc/not316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang JB, Dong DF, Gao K, Wang MD. Mechanisms underlying the biological changes induced by isocitrate dehydrogenase-1 mutation in glioma cells. Oncol Lett. 2014;7:651–657. doi: 10.3892/ol.2014.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Y, Xu Y, Wang H, Niu J, Hou H, Jiang Y. Histone deacetylase inhibitor, valproic acid, radiosensitizes the C6 glioma cell line in vitro. Oncol Lett. 2014;7:203–208. doi: 10.3892/ol.2013.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen SH, Kwan AL, Chen YY, Wang ZX. Effect of silencing HIF-1α on proliferation, invasion and migration of glioblastoma U87 cells. Neurol Sci. 2013;34:365–371. doi: 10.1007/s10072-012-1010-4. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu JH, Yao YL, Gu T, Wang ZY, Pu XY, Sun WW, Zhang X, Jiang YB, Wang JJ. MiR-421 regulates apoptosis of BGC-823 gastric cancer cells by targeting caspase-3. Asian Pac J Cancer Prev. 2014;15:5463–5468. doi: 10.7314/APJCP.2014.15.13.5463. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013;13:68. doi: 10.1186/1475-2867-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D, Pan H, Zhou Y, Zhou J, Fan Y, Qu P. microRNA-133b downregulation and inhibition of cell proliferation, migration and invasion by targeting matrix metallopeptidase-9 in renal cell carcinoma. Mol Med Rep. 2014;9:2491–2498. doi: 10.3892/mmr.2014.2116. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang J, Wang X, You Y, Yang Z, Liu N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Zhao H, Li M, Li L, Yang X, Lan G, Zhang Y. MiR-133b is down-regulated in human osteosarcoma and inhibits osteosarcoma cells proliferation, migration and invasion and promotes apoptosis. PLoS One. 8:e83571, 2013. doi: 10.1371/journal.pone.0083571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen D, Li S, Ji F, Cao H, Jiang W, Zhu J, Fang X. MiR-133b acts as a tumor suppressor and negatively regulates FGFR1 in gastric cancer. Tumour Biol. 2013;34:793–803. doi: 10.1007/s13277-012-0609-7. [DOI] [PubMed] [Google Scholar]
- 13.Nohata N, Hanazawa T, Enokida H, Seki N. MicroRNA-1/133a and microRNA-206/133b clusters: Dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karatas OF, Guzel E, Suer I, Ekici ID, Caskurlu T, Creighton CJ, Ittmann M, Ozen M. MiR-1 and miR-133b are differentially expressed in patients with recurrent prostate cancer. PLoS One. 2014;9:e98675. doi: 10.1371/journal.pone.0098675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Shao X, Gao W, Zhang Z, Liu P, Wang R, Huang P, Yin Y, Shu Y. MicroRNA-133b inhibits the growth of non-small-cell lung cancer by targeting the epidermal growth factor receptor. FEBS J. 2012;279:3800–3812. doi: 10.1111/j.1742-4658.2012.08741.x. [DOI] [PubMed] [Google Scholar]
- 17.Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. MiR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 18.Duan FT, Qian F, Fang K, Lin KY, Wang WT, Chen YQ. MiR-133b, a muscle-specific microRNA, is a novel prognostic marker that participates in the progression of human colorectal cancer via regulation of CXCR4 expression. Mol Cancer. 2013;12:164. doi: 10.1186/1476-4598-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Raawi D, Abu-El-Zahab H, El-Shinawi M, Mohamed MM. Membrane type-1 matrix metalloproteinase (MT1-MMP) correlates with the expression and activation of matrix metalloproteinase-2 (MMP-2) in inflammatory breast cancer. Int J Clin Exp Med. 2011;4:265–275. [PMC free article] [PubMed] [Google Scholar]
- 20.Arroyo AG, Genís L, Gonzalo P, Matí-as-Román S, Pollán A, Gálvez BG. Matrix metalloproteinases: New routes to the use of MT1-MMP as a therapeutic target in angiogenesis-related disease. Curr Pharm Des. 2007;13:1787–1802. doi: 10.2174/138161207780831284. [DOI] [PubMed] [Google Scholar]
- 21.Basile JR, Holmbeck K, Bugge TH, Gutkind JS. MT1-MMP controls tumor-induced angiogenesis through the release of semaphorin 4D. J Biol Chem. 2007;282:6899–6905. doi: 10.1074/jbc.M609570200. [DOI] [PubMed] [Google Scholar]
- 22.Albrechtsen R, Kveiborg M, Stautz D, Vikeså J, Noer JB, Kotzsh A, Nielsen FC, Wewer UM, Fröhlich C. ADAM12 redistributes and activates MMP-14, resulting in gelatin degradation, reduced apoptosis and increased tumor growth. J Cell Sci. 2013;126:4707–4720. doi: 10.1242/jcs.129510. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Yuan J, Tu Y, Mao X, He S, Fu G, Zong J, Zhang Y. Co-expression of MMP-14 and MMP-19 predicts poor survival in human glioma. Clin Transl Oncol. 2013;15:139–145. doi: 10.1007/s12094-012-0900-5. [DOI] [PubMed] [Google Scholar]
- 24.Xie H, Xue YX, Liu LB, Wang P, Liu YH, Ying HQ. Expressions of matrix metalloproteinase-7 and matrix metalloproteinase-14 associated with the activation of extracellular signal-regulated kinase1/2 in human brain gliomas of different pathological grades. Med Oncol. 2011;(28 Suppl 1):S433–S438. doi: 10.1007/s12032-010-9660-7. [DOI] [PubMed] [Google Scholar]