Abstract
BACKGROUND. Activation of the NLRP3 inflammasome is associated with metabolic dysfunction, and intermittent fasting has been shown to improve clinical presentation of NLRP3 inflammasome–linked diseases. As mitochondrial perturbations, which function as a damage-associated molecular pattern, exacerbate NLRP3 inflammasome activation, we investigated whether fasting blunts inflammasome activation via sirtuin-mediated augmentation of mitochondrial integrity.
METHODS. We performed a clinical study of 19 healthy volunteers. Each subject underwent a 24-hour fast and then was fed a fixed-calorie meal. Blood was drawn during the fasted and fed states and analyzed for NRLP3 inflammasome activation. We enrolled an additional group of 8 healthy volunteers to assess the effects of the sirtuin activator, nicotinamide riboside, on NLRP3 inflammasome activation.
RESULTS. In the fasting/refeeding study, individuals showed less NLRP3 inflammasome activation in the fasted state compared with that in refed conditions. In a human macrophage line, depletion of the mitochondrial-enriched sirtuin deacetylase SIRT3 increased NLRP3 inflammasome activation in association with excessive mitochondrial ROS production. Furthermore, genetic and pharmacologic SIRT3 activation blunted NLRP3 activity in parallel with enhanced mitochondrial function in cultured cells and in leukocytes extracted from healthy volunteers and from refed individuals but not in those collected during fasting.
CONCLUSIONS. Together, our data indicate that nutrient levels regulate the NLRP3 inflammasome, in part through SIRT3-mediated mitochondrial homeostatic control. Moreover, these results suggest that deacetylase-dependent inflammasome attenuation may be amenable to targeting in human disease.
TRIAL REGISTRATION. ClinicalTrials.gov NCT02122575 and NCT00442195.
FUNDING. Division of Intramural Research, NHLBI of the NIH.
Introduction
Sterile inflammation linked to obesity is mediated in part by the NLRP3 (Nod-like receptor family protein 3) inflammasome (1). The activation of this program, as a component of the innate immune system, similarly exacerbates obesity-linked diseases, including insulin resistance, diabetes, and asthma (2, 3). The biological pathways driving this innate immune program are well defined (4). In the context of obesity, triggers that engage Toll-like receptors to initiate transcriptional priming of the NLRP3 inflammasome include adipose tissue hypertrophy with macrophage infiltration and cytokine secretion, elevated circulating saturated fatty acids, and/or obesity-linked endotoxemia (5–8). These, in turn activate NF-κB–dependent transcription to upregulate genes encoding NLRP3 and pro–IL-1β. Subsequent inflammasome activation/execution, as the cornerstone of intracellular surveillance, is initiated in response to additional pathogen-associated molecular patterns or host cell–derived damage-associated molecular patterns (DAMPs), which promote assembly and self-oligomerization of inflammasome components. The NLRP3 complex then promotes caspase-1 activation and cleavage of pro–IL-1β and pro–IL-18 into bioactive cytokines that amplify inflammation (9, 10).
Obesity and diabetes are associated with mitochondrial perturbations, and in these diseases, mitochondrial dysfunction itself may function as an inflammasome-activating DAMP (11, 12). The mitochondrial role as a DAMP linked to NLRP3 activation includes, but is not limited to, the leak of mitochondrial content into the cytoplasm (13, 14), into the circulation (15), and/or via disruption of mitochondrial quality control programs (16). The mechanisms whereby mitochondrial content extrusion into the cytoplasm functions as a DAMP include a role of the mitochondrial membrane cardiolipin via its direct interaction with NLRP3 (17), via the release of mitochondrial ROS (16), and/or due to the intrinsic composition of hypomethylated CpG motifs of mitochondrial DNA that resembles the immunogenic properties of bacterial CpG DNA motifs (18).
Interestingly, intermittent fasting and caloric restriction, which can counter the effects of obesity, also confer beneficial effects against canonical NLRP3 inflammation-linked pathologies, such as insulin resistance and asthma (19–21). As the nutrient-sensing NAD-dependent lysine deacetylase sirtuin proteins are activated by fasting/caloric restriction (22, 23), ameliorate nutrient excess–linked pathology (24, 25), and can enhance mitochondrial integrity (26, 27), we reasoned that intermittent fasting amelioration of inflammation may be mediated, in part, via sirtuin-regulated mitochondrial quality control, with subsequent blunting of the NLRP3 inflammasome.
We explored this hypothesis by comparing NLRP3 inflammasome activation in 19 healthy volunteers in the refed state and the fasted state. Activation of the NLRP3 inflammasome was assessed using peripheral blood mononuclear cell (PBMC) and monocyte extraction from fasted and postprandial samples. We demonstrated multiple levels of regulation of the NLRP3 inflammasome program, including diminished SIRT3-dependent NLRP3 inflammasome activity in the fasted state and an NF-κB–mediated priming effect of refeeding. To gain mechanistic insight into the role of SIRT3, genetic manipulation studies showed an essential role of SIRT3 maintenance of mitochondrial respiration and control of ROS production, underpinning its role in this nutrient-sensing inflammatory program. In parallel, a SIRT3 pharmacologic agonist, nicotinamide riboside (NR), enhanced mitochondrial function and blunted the NLRP3 inflammasome in leukocytes extracted from a group of healthy control subjects and from cells extracted in the refed state but not the fasted state from subjects enrolled in the index fasting inflammasome study.
Results
Clinical study.
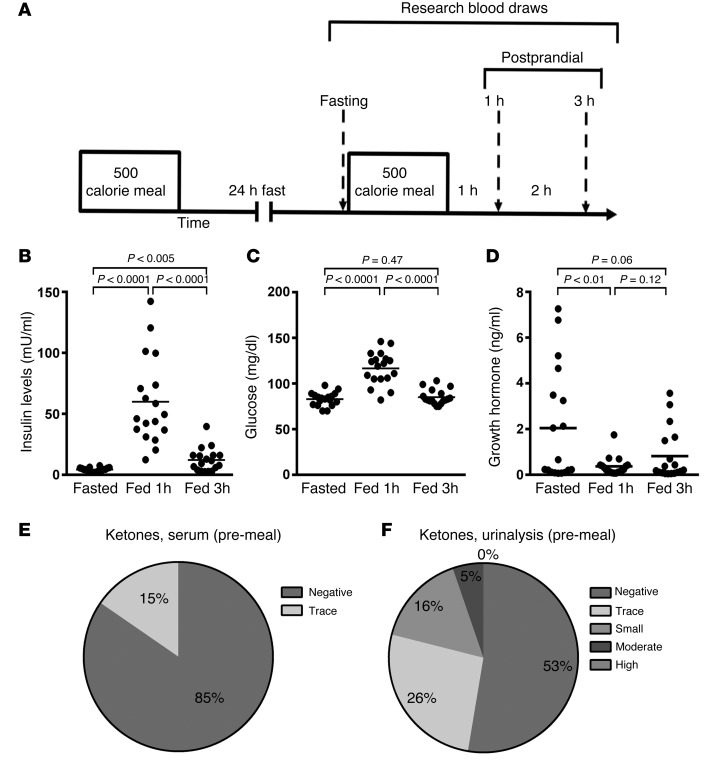
The participants in the pilot study to examine the fasting inflammasome tolerated the 24-hour fast without adverse effects. A schematic illustrating the temporal organization of the study is shown in Figure 1A. In brief, research blood draws were completed 24 hours after a fast and then at 1 and 3 hours following refeeding. One subject withdrew from the study at the time of the screening visit, but all subjects that initiated the 24-hour fast (n = 20) completed the study. One subject who completed all the blood draws was excluded from analysis due to a protocol violation. Laboratory studies consistent with fasting compliance included findings of low glucose and insulin levels at the time of fasting and induction of glucose and insulin and suppression of growth hormone levels in response to refeeding (Figure 1, B–D). Significantly lower insulin and glucose levels at baseline compared with levels 3 hours following the fixed caloric meal would be consistent with protocol compliance. As recent data show that ketone production can suppress inflammasome activity (28), we measured fasting serum and urine ketone levels at screening and following the 24-hour fast. No subjects had measurable ketones at the screening visit. Thirteen of nineteen subjects had fasting serum ketone levels measured and eighteen of nineteen subjects had fasting urine ketone levels determined. Eleven of thirteen subjects had undetectable serum ketone levels in the fasted state (Figure 1E), and 53% of subjects had no detectable urine ketone levels in the fasted state (Figure 1F).
Figure 1. Clinical protocol and intervention characteristics.
(A) Schematic of the protocol, showing the intervals between the fixed caloric meals and temporal drawing of research blood. (B–D) Data points and means (horizontal bars) of subjects’ sera (B) insulin, (C) glucose, and (D) growth hormone levels at the end of the 24-hour fast and 1 and 3 hours following the fixed caloric meal (n = 19). The fasting insulin level was found to be less than 50% of the insulin level 3 hours after the 500 caloric meal in this young healthy cohort, supporting the integrity of the fast. (E and F) Percentage of subjects that displayed ketones in (E) sera (n = 13) or (F) urine (n = 18). Statistical testing for changes in metabolite and hormone levels were performed using randomized block design ANOVA.
The nutrient status modulates the NLRP3 inflammasome.
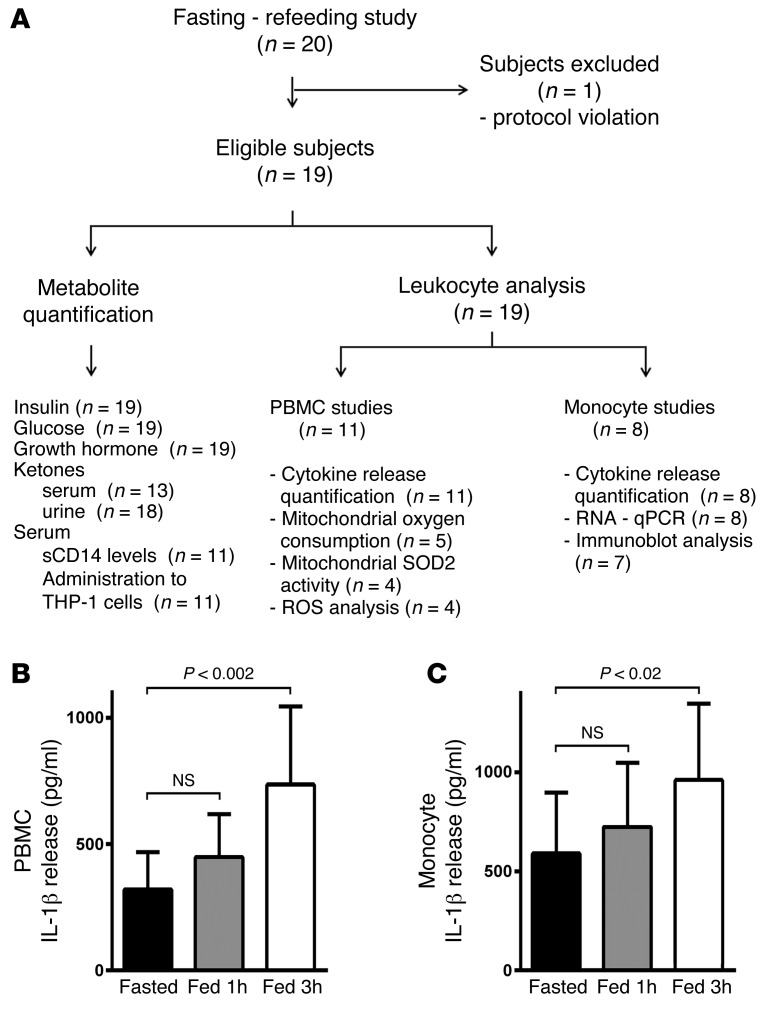
Due to the restricted volume of blood drawn at each time point, selected laboratory studies were performed in sequential subjects, as illustrated in the flow diagram (Figure 2A). As the measurement of the generation of IL-1β in response to NLRP3 triggers was the primary endpoint, PBMCs (initial 11 subjects) or monocytes (subsequent 8 subjects) from all study subjects were exposed to ATP as an NLRP3 trigger with or without LPS inflammasome priming. IL-1β secretion was substantially induced at 1 and 3 hours after feeding compared with fasting levels independent of inflammasome priming by LPS (Figure 2, B and C).
Figure 2. Nutrient status modulates the NLRP3 inflammasome.
(A) Fasting/refeeding study flow diagram. (B) ELISA assay measurement of IL-1β secretion from primary PBMCs exposed to 3 mM ATP (n = 11). (C) ELISA assay measurement of IL-1β secretion from primary monocytes exposed to 3 mM ATP (n = 8). Statistical testing for changes in IL-1β levels was performed using randomized block design ANOVA. As the refeeding response was more robust at 3 hours, the data at this time point were used as the index postprandial time for all subsequent analyses.
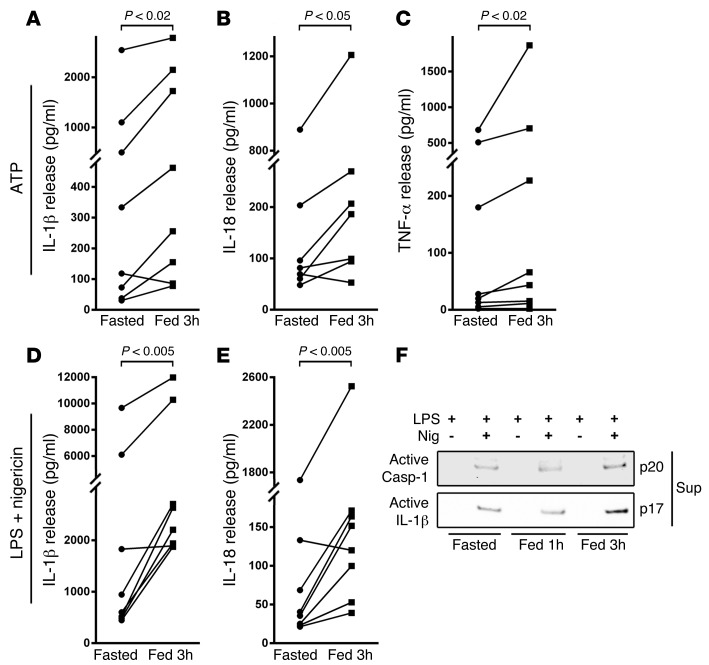
As we saw the greater effect 3 hours after feeding compared with that 1 hour after feeding and with fasting, as prespecified in the protocol, this later time point was evaluated in all subsequent studies. The IL-1β response of monocytes from individual subjects, comparing fasting to 3 hours following refeeding in response to ATP stimulation without prior priming, is shown in Figure 3A. In parallel with the IL-1β effect and independent of exogenous priming, IL-18 secretion was similarly induced 3 hours after refeeding (Figure 3B). In addition, the broader inflammatory milieu in the refed state was evident with induction of TNF-α release (Figure 3C). These findings of increased secretion of IL-1β and TNF-α were replicated in the PBMCs from subjects (Supplemental Figure 1, A and B). The priming effect of refeeding was supported by only a modest further induction of cytokine release when monocytes and PBMCs were preincubated with LPS (Supplemental Figure 1, C and D). In contrast, in response to nigericin, a less robust NLRP3 activator, LPS priming was necessary, and here again IL-1β and IL-18 secretion from monocytes was significantly elevated following refeeding (Figure 3, D and E). To confirm the activation of the NLRP3 program by nutrient status, we assayed the cleavage products of caspase-1 and IL-1β in the supernatant of monocytes exposed to LPS and nigericin. Here too, we found that, at the 3-hour postprandial time point, the steady-state levels of cleaved caspase-1 and IL-1β were markedly induced (Figure 3F).
Figure 3. Nutrient status modulates the NLRP3 inflammasome.
(A) ELISA assay measurement of IL-1β secretion from primary monocytes exposed to 3 mM ATP (n = 8). (B) IL-18 release in the same monocyte studies, showing that both NLRP3 cytokine levels are more robustly secreted in the refed state versus fasted state. (C) TNF-α release in the same monocytes. (D) IL-1β and (E) IL-18 secretion from monocytes primed with 1 ng/ml LPS followed by 5 μM nigericin administration as a second inflammasome trigger (n = 8). The fasting to 3-hour–refed cytokine levels were analyzed using paired 2-tailed t tests. (F) Representative immunoblot analysis showing secretion of mature caspase-1 and mature IL-1β in the supernatants of monocytes treated with LPS and with and without nigericin.
Refeeding initiates NLRP3 inflammasome priming.
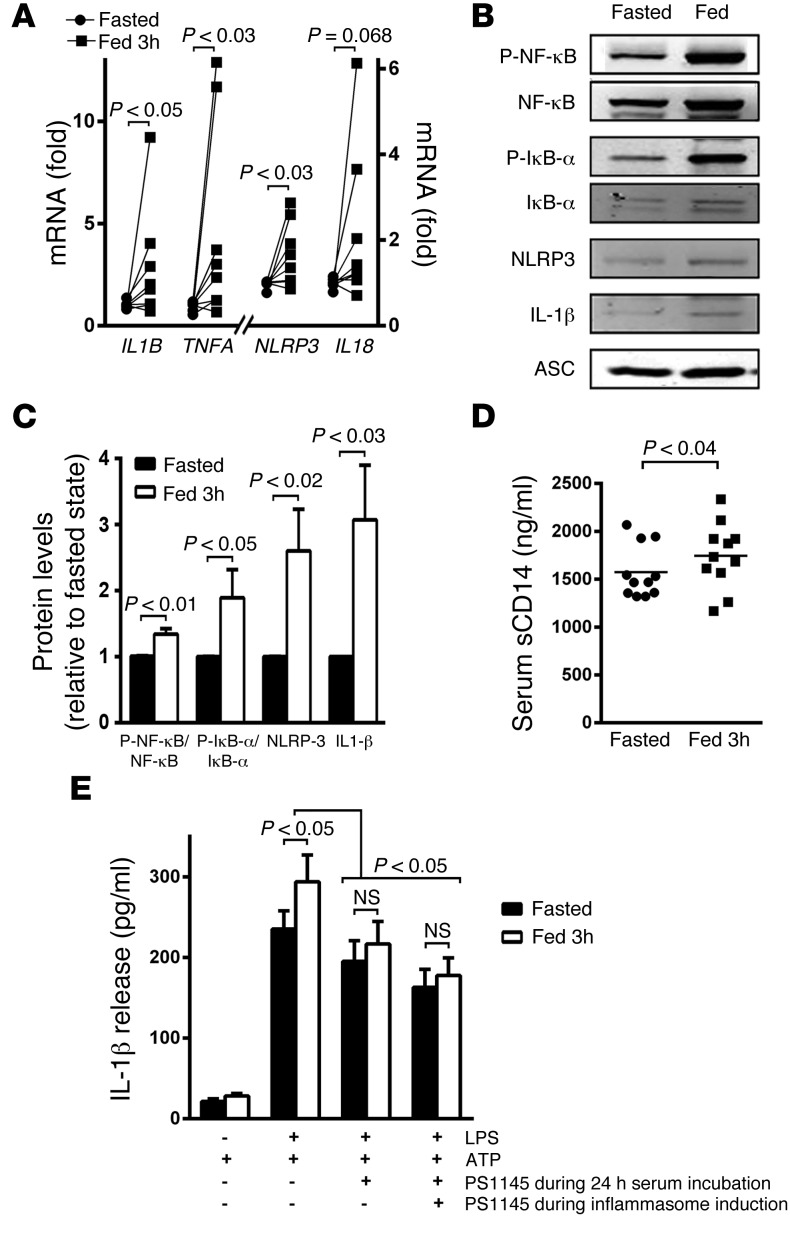
The increase in cytokine release in response to ATP in PBMCs and monocytes independent of priming suggested that a component of the differential effects of nutrient status on inflammasome activation might include refeeding-evoked transcriptional activation of this program. To explore this, we measured the transcript levels of IL1B, TNFA, NLRP3, and IL18 in unstimulated monocytes extracted from PBMCs drawn following the 24-hour fast compared with the draw 3 hours postprandial. All of the canonical transcript targets of NLRP3 priming and the transcript encoding TNF were substantially increased following refeeding (Figure 4A). As inflammasome priming is activated by NF-κB signaling, we then explored the activation of this pathway by measuring phosphorylation levels of NF-κB and of IκBα and by quantifying the steady-state protein levels of the downstream inflammasome targets NLRP3 and pro–IL-1β. The intrinsic induction of postprandial phosphorylation and inflammasome protein levels similarly supports that refeeding primes this pathway in monocytes (Figure 4, B and C, and Supplemental Figure 2, A–F). Although we did not directly measure serum LPS levels, we found a modest but significant induction of levels of soluble CD14, a marker of monocytic activation in response to LPS (29), in subject sera at the 3-hour postprandial time point compared with fasting levels (Figure 4D). In light of a potential serologic role in refeeding-induced priming, we evaluated whether sera from the fasted and 3-hour–refed time points could recapitulate inflammasome effects in human THP-1–derived macrophages. To test this, THP-1–derived macrophages were incubated in either the fasted or refed sera for 24 hours prior to priming and inflammasome activation. Following inflammasome priming and activation, the refed sera–supplemented THP-1 cells significantly increased IL-1β secretion by approximately 25% more than fasted sera–supplemented cells (Figure 4E). The establishment of this ex vivo system using fasted and refed serum supplementation of THP-1 cells then enabled us to examine the effects of NF-κB signaling inhibition. The administration of PS-1145 (30) either for the 24-hour period of serum supplementation prior to inflammasome priming and activation or for the duration of the experiments resulted in blunting and equalization of IL-1β secretion in cells supplemented with fasted or refed sera (Figure 4E).
Figure 4. Refeeding initiates NLRP3 inflammasome priming via NF-κB signaling.
(A) Transcript levels encoding TNFA and inflammasome components IL1B, IL18, and NLRP3. Refeeding increases the expression of all the mRNAs (n = 8). (B) Representative protein levels of signaling molecules activating the NLRP3 inflammasome. Increased phosphorylation of NF-κB and IκBα shows their activation in the fed state. The NLRP3 complex component — NLRP3 and IL-1β — levels are also increased in the fed state. The relative quantitative changes (n = 5–7) are shown in C. Note, the refeeding effects on gene transcript, phosphoprotein, and protein levels are evident at baseline and do not require exogenous inflammasome priming. The fasting to 3-hour–refed transcript and steady-state protein levels were analyzed using paired 2-tailed t tests. (D) Serum levels of soluble CD14, as a marker of endotoxin activity (n = 11). (E) Histogram showing changes in IL-1β release from THP-1–derived macrophages supplemented with fasted and refed sera in the presence and absence of the NF-κB pathway inhibitor PS-1145 (n = 11). The inhibition of NF-κB signaling abolished the excess IL-1β secretion using the refed sera, whether it was administered during the serum incubation alone and/or when maintained through the priming and activation of the inflammasome. Statistical testing for changes in IL-1β levels was performed using 2-way ANOVA.
SIRT3 levels modify NLRP3 activation and mitochondrial function.
To investigate a potential role of mitochondrial functioning in the regulatory control of the NLRP3 inflammasome, we then measured mitochondrial respiration and ROS levels during the fasted and fed states. Interestingly, basal and maximal oxygen consumption was significantly higher in parallel with lower mitochondrial ROS levels in PBMCs isolated in the fasted state compared with that in the refed state (Supplemental Figure 3, A and B).
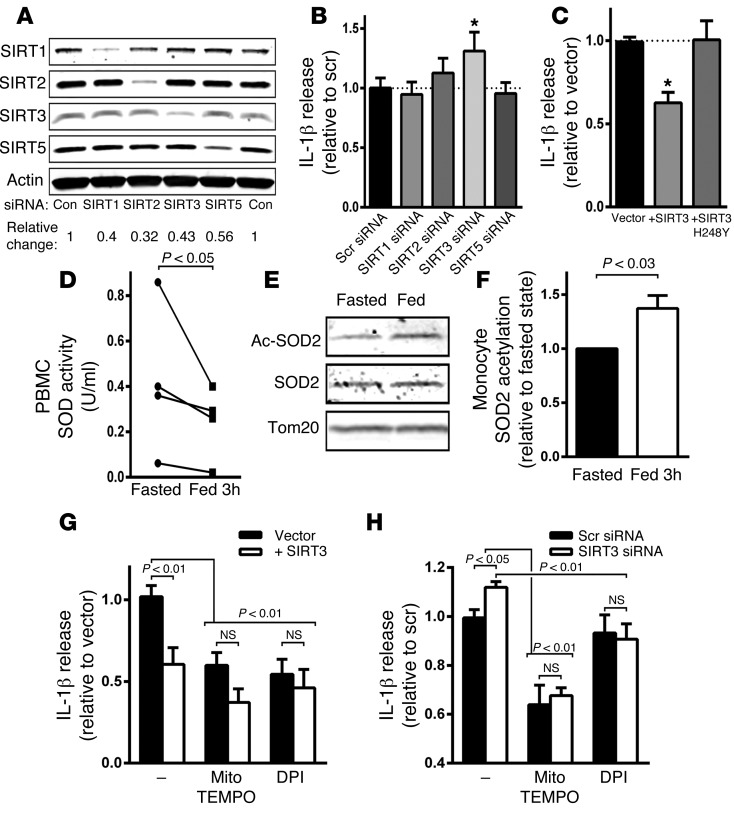
As sirtuins are activated by fasting and can modulate mitochondrial function and ROS production (31), we then evaluated whether select sirtuins may be operational in the modulation of this program. We used siRNAs targeted to individually deplete SIRT1, SIRT2, SIRT3, and SIRT5 levels in human THP-1–derived macrophages. The knockdown of the sirtuin enzyme was confirmed by immunoblot analysis (Figure 5A). Subsequent priming and activation of the NLRP3 inflammasome in the sirtuin-depleted cells by LPS and ATP showed that SIRT3 knockdown significantly augmented IL-1β secretion (Figure 5B). In parallel, overexpression of wild-type SIRT3 but not the deacetylase mutant SIRT3 (22) suppressed IL-1β release (Figure 5C).
Figure 5. SIRT3 levels modify NLRP3 activation and acetylation of SOD2.
(A) Representative immunoblot of steady-state levels of SIRT1, SIRT2, SIRT3, and SIRT5 following the siRNA knockdown of each isoform in THP-1 cells. The relative changes represent the values from 3 separate experiments. (B) IL-1β release in response to inflammasome activation in the sirtuin knockdown THP-1 cells (n = 11). (C) Inflammasome activation with wild-type and deacetylase mutant SIRT3 overexpression in THP-1 cells (n = 4). *P < 0.05. (D) Mitochondrial superoxide dismutase activity in PBMC mitochondria isolated in the fasted and fed states (n = 4). (E) Representative immunoblot analysis of relative SOD2 protein acetylation on K68 in the fed and fasted states. The relative quantitative change in SOD2 acetylation is shown in F (n = 4). (G) Histogram showing relative release of IL-1β in control and SIRT3-overexpressing cells in response to inflammasome activation in the absence of or presence of ROS inhibition with mitoTEMPO and DPI (n = 5). (H) Histogram showing the same studies as performed in F, with the exception of testing the effect of SIRT3 knockdown rather than overexpression (n = 5). Statistical analysis of changes in IL-1β secretion levels was performed using 2-way ANOVA and analysis of changes in SOD2 activity and acetylation was performed using a paired 2-tailed t test.
As SIRT3 is known to deacetylate and activate mitochondrial superoxide dismutase 2 (SOD2) (32), we explored the activity and acetylation of this protein in leukocytes from subjects in the fasting and refeeding study. We found that SOD2 activity was significantly lower in PBMCs extracted in the fed state (Figure 5D). Furthermore and consistent with the concept that fasting increased SIRT3 and mitochondrial SOD activity, we found that SOD2 was deacetylated to a greater extent on the cognate SIRT3-regulated lysine residues (K68) in monocytes from subjects in the fasted state compared with those from subjects in the refed state (Figure 5, E and F, and Supplemental Figure 3C).
To directly interrogate the effect of the modulation of mitochondrial ROS by SIRT3 in inflammasome activation, we explored the effect of mitoTEMPO, the mitochondria-targeted superoxide dismutase mimetic with superoxide scavenging properties, and diphenylene iodonium (DPI), the flavin-containing enzyme inhibitor, which blocks mitochondrial ROS production (33). MitoTEMPO and DPI reduced IL-1β release in control and SIRT3-expressing THP-1–derived macrophages, with a blunting of the difference in cytokine release between control and SIRT3-overexpressing cells (Figure 5G). In parallel, the administration of these mitochondrial ROS–reducing agents decreased IL-1β release to the same levels in scrambled control and SIRT3-depleted THP-1 cells (Figure 5H).
A SIRT3 agonist blunts inflammasome activation and improves mitochondrial function.
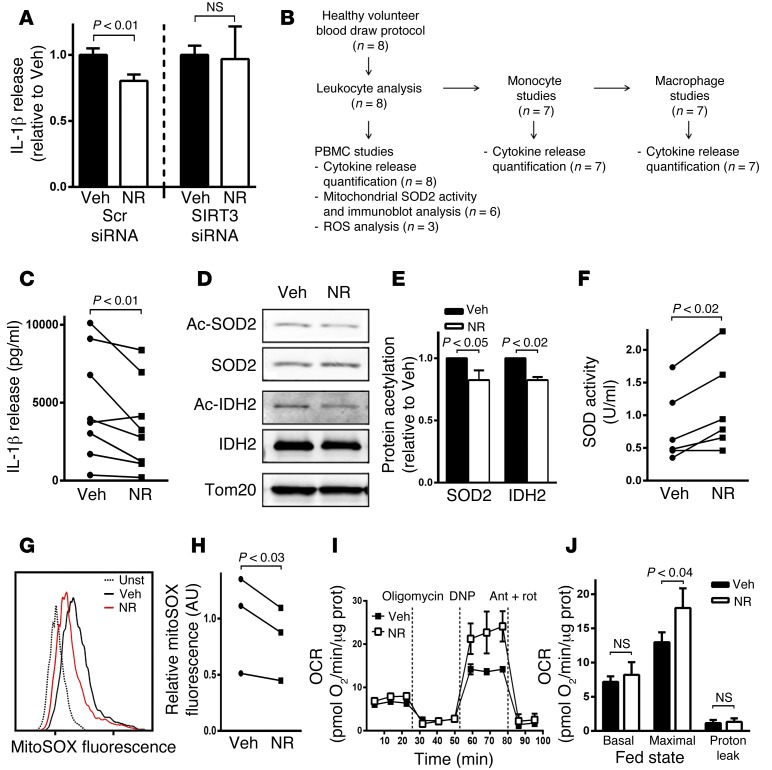
We then assessed whether NR, as an NAD+ precursor that functions as a SIRT3 agonist (34), could replicate the fasting phenotype. We first confirmed NR SIRT3 specificity, showing that this compound blunted the inflammasome in control siRNA THP-1–derived macrophages but not in the SIRT3 siRNA THP-1–derived macrophages (Figure 6A).
Figure 6. A SIRT3 agonist blunts inflammasome activation and improves mitochondrial function.
(A) Evaluation of IL-1β release in response to inflammasome activation in control and SIRT3 siRNA–treated THP-1–derived macrophages to assess the effect of NR on IL-1β secretion in the presence of SIRT3 or its depletion (n = 4). (B) Flow diagram of studies performed on blood draw from the normal volunteer blood draw protocol. (C–H) NR administration to PBMCs from healthy volunteers to evaluate the effect on (C) inflammasome induction–mediated IL-1β secretion (n = 8), (D and E) SIRT3 substrate acetylation levels (n = 6), (F) mitochondrial superoxide dismutase activity (n = 6), and (G and H) ATP-induced mitochondrial ROS levels (n = 3). (I and J) NR effect on PBMCs extracted in the refed state, showing an augmentation of maximal respiratory capacity. A representative oxygen consumption rate (OCR) tracing and a histogram are shown, including the data from all the subject cells studied (n = 5). Maximal respiration was assessed in response to dinitrophenol (DNP) and proton leak as the difference in oxygen consumption between inhibition of ATP synthase with oligomycin and the inhibition of mitochondrial respiration with antimycin A (Ant) and rotenone (Rot). Statistical analyses of experiments in this figure were performed using paired 2-tailed t tests.
To explore the potential role of NR in primary cells, an additional group of healthy control subjects donated blood through the NHLBI Healthy Volunteer Blood Collection Protocol. Blood from these volunteers was used to extract PBMCs and monocytes and for the culturing of primary macrophages to assess the effect of the SIRT3 agonist NR on the NLRP3 inflammasome. No dietary restrictions were placed on these subjects to enroll in the study, and no adverse effects were reported during these blood draws. The flow diagram illustrating the investigation of these blood samples is shown in Figure 6B. The 24-hour administration of NR to PBMCs from healthy volunteers mimicked the fasting effect with blunting of inflammasome activation (Figure 6C). Consistent with NR’s known role in SIRT3 activation, we found that NR reduced SOD2 and isocitrate dehydrogenase 2 (IDH2) acetylation (Figure 6, D and E), augmented mitochondrial superoxide dismutase activity (Figure 6F), and diminished mitochondrial ROS levels (Figure 6, G and H). NR similarly decreased cytokine release in PBMCs and monocytes and in primary human macrophages from the healthy control population (Supplemental Figure 4, A–C). The maintenance of fed and fasted monocytes in culture for 24 hours annulled the dietary inflammasome effect, which precluded our assessment of the efficacy of NR in reversing the feeding effect in this context. Nevertheless, we found that 4 hours of administration of NR enhanced maximal oxygen consumption in PBMCs extracted from refed subjects (Figure 6, I and J), whereas there was no additive effect on respiration in PBMCs extracted in the fasted state (Supplemental Figure 4D). Together, these data support that SIRT3 is constitutively activated in the fasted state and show that NR can activate SIRT3 to recapitulate the blunted inflammasome effects in leukocytes extracted in the refed state.
Discussion
Obesity is linked to NLRP3-linked sterile inflammation, and intermittent nutrient and/or chronic caloric restriction suppress inflammation (20, 21). Whether caloric restriction signaling directly modulates the inflammasome or whether the nutrient depletion effect on inflammation is due to concomitant weight reduction from these dietary interventions has not been assessed. To begin to explore this, we assessed the control of the NLRP3 inflammasome at the level of priming and execution in response to a 24-hour fast and refeeding protocol. We showed that fasting blunted the NLRP3 inflammasome by sirtuin-dependent enhancement of mitochondrial quality control and that refeeding activated NLRP3 priming. Together, these data show that “nutrient sensing” orchestrates control of the NLRP3 inflammasome at multiple regulatory levels.
The metabolic, neuroendocrine, and molecular control mechanisms governing the biological effects of fasting, refeeding, and caloric restriction are extensive (19, 35) and will ultimately require the integration of reductionist studies and broad systems biological modeling to advance our understanding of all the changes and their consequences. One component of these nutrient-deprivation interventions includes the modulation of the immune system (21, 36). In this context, the objective of this study was to interrogate a component of innate immunity by exploring the role of nutrient-sensing sirtuin-dependent control of mitochondrial integrity in the modulation of the NLRP3 inflammasome. In addition to this, we found that refeeding following the 24-hour fast had a significant effect on NLRP3 inflammasome priming. This regulation appears to be operational by the modulation of transcript levels and at the level of protein expression and posttranslational control of protein function in the canonical NLRP3 inflammasome pathway. Although the mechanisms orchestrating the refeeding priming effect have not been extensively characterized, we found that an inflammatory signature was linked to LPS signaling (29), as shown by soluble CD14 levels, which were significantly, albeit modestly, elevated in the refed sera. Additionally, we show that the refed sera increases inflammasome-induced IL-1β secretion from THP-1 cells compared with fasted sera and that inhibition of NF-κΒ signaling uniformly blunts IL-1β secretion levels and abrogates excessive IL-1β secretion in cells supplemented with the sera from refed subjects. Together, these data suggest that refeeding, which is known to evoke postprandial endotoxemia (37), may, at least in part, induce inflammasome priming via the activation of NF-κB signaling. Further studies would be required to evaluate whether the content of the diet and what additional factors modulate this refeeding effect.
The functioning of mitochondria as a DAMP in the activation of the NLRP3 inflammasome has been well established and appears to be orchestrated via the extrusion of numerous components of the mitochondria, including cardiolipin and mitochondrial genomic DNA. An emerging concept concomitantly suggests that the enhancement of mitochondrial integrity and/or quality control should ameliorate this innate immune response. This has been shown in cases in which the augmentation of mitochondrial integrity via enhanced autophagy has an ameliorative effect (16) and in cases in which impaired autophagy, which causes the accumulation of damaged mitochondrial content in the cytosol, exacerbates NLRP3 inflammasome activation (38). The questions we posed here were whether this biological program was operational in humans and whether the sirtuin-mediated mitochondrial housekeeping program plays a role in the modulation of this activation. The finding in this study shows that, via a SIRT3-dependent mechanism, the maintenance of mitochondrial function and the attenuation in ROS levels decrease NLRP3 inflammasome activation. Additionally and in keeping with the nutrient level–dependent activation of SIRT3 (23), 24 hours of fasting in human subjects similarly blunts inflammasome activation. Interestingly, the disruption of mitochondrial homeostasis via alternate regulatory perturbations has also been found to evoke innate immunological reactions (39, 40), which, together with our data, support the emerging understanding of the intimate interaction between mitochondrial integrity and the innate immune system (41). Strategies to enhance mitochondrial integrity to ameliorate diseases have been long sought after, and the findings in this study, showing that NR can recapitulate inflammasome modulatory effects of fasting on the NLRP3 inflammasome, highlight the potential to target SIRT3 activation in NLRP3 inflammasome–linked diseases.
The limitations of this study include the lack of the contribution of myeloid cell autonomous and serum-derived factors in contributing to increased susceptibility to NLRP3 inflammasome activation in the refed state and the more comprehensive analysis of the numerous potential SIRT3 effects that could account for the inflammasome-blunting effects seen in prolonged fasting human subjects. At the same time, we cannot exclude effects of the sirtuins that we did not study or the effects of other nutrient-sensing programs, such as the AMP kinase pathway (42), in response to the fasting intervention. Finally, there was a lack of samples for comparison at a time point prior to the 24-hour fast, and the assessment of the minimum fasting time sufficient to blunt inflammasome activation was not performed.
In conclusion, this study shows that refeeding after a 24-hour fast increases susceptibility to inflammasome activation, in part, via refeeding-mediated activation of NF-κB signaling. Moreover, a 24-hour fast in human subjects activates SIRT3 biology, and this, in turn, appears to confer resistance to NLRP3 inflammasome activation via blunting of mitochondrial ROS levels. This inflammasome-provoking effect of refeeding after a 24-hour fast can be inhibited by SIRT3 activation using NR. These data have identified a potentially drug-modifiable SIRT3-dependent program to attenuate the NLRP3 inflammasome and suggest that this pathway may be modulated as a strategy to alleviate/attenuate NLRP3-linked inflammation.
Methods
Study participants and protocol.
Twenty consenting healthy volunteers were studied in the index fasting inflammasome study. This study was composed of 8 female and 12 male subjects, with ages ranging from 21 to 36 years. Subjects initiated the study with an early morning fixed caloric meal followed by fasting, with the exception of unrestricted water intake for 24 hours prior to undergoing the fasting blood draw. This was followed by a fixed calorie meal and postprandial blood draws 1 and 3 hours later. One subject was excluded for a protocol violation, and all other subjects’ samples were assayed for cytokine release, using either PBMCs or monocytes. Due to limitations in sample acquisition per study subject, leukocytes from sequential subjects were used for the additional experiments in this study. An additional group of 8 healthy volunteers enrolled in a blood collection protocol to enable the collection of blood cells to test the effects of NR. This group consisted of 6 women and 2 men, with an age range of 23 to 48 years. These subjects had no history of acute or chronic disease.
Cell culture and transfection.
PBMCs were isolated from human blood by density centrifugation using LSM Lymphocyte Separation Medium (MP Biomedicals), with PBMCs collected at the interface. Monocytes were obtained from PBMCs using the Monocyte Isolation Kit II (Miltenyi Biotec). PBMCs were preferentially used for experiments that required a large number of cells, such as flow cytometry and measurement of oxygen consumption and superoxide dismutase activity. To generate human macrophages, human monocytes were incubated with 15% human serum and 20 ng/ml M-CSF for 7 days.
THP-1 monocyte cells obtained from ATCC were cultured in RPMI 1640 plus 25 mM HEPES and 10% heat-inactivated FBS. They were differentiated into macrophages by incubation with 5 ng/ml PMA for 48 hours (43). THP-1 cells were transiently transfected 18 hours before differentiation with the Cell Line Nucleofector Kit V (Lonza), program V-001 (THP-1 high efficiency), and 0.5 μg DNA or 400 pmol siRNA (SMARTpool siRNA, Dharmacon). The number of cells was adjusted to 2.5 × 106 cells per transfection cuvette to increase cell viability after transfection (44). To evaluate the effect of the sera from subjects on the activation of the inflammasome, THP-1 cells were incubated with 5 ng/ml PMA in RPMI 1640 with 25 mM HEPES and 10% heat-inactivated FBS for 24 hours, washed, and then incubated with PMA in RPMI and HEPES, with 10% of the fasted or refed sera of subjects for further 24 hours to allow complete differentiation into macrophages.
Cell stimulation and cytokine assays.
PBMCs were incubated at 2 × 106 cells per ml in 96-well plates in RPMI medium plus 25 mM HEPES and 10% heat-inactivated FBS with or without 1 ng/ml LPS (Ultrapure Salmonella minnesota R595; Enzo Life Sciences) for 4 hours. Human monocytes were incubated at 1.5 × 106 cells per ml in a 96-well plate in RPMI medium plus 25 mM HEPES and 10% heat-inactivated FBS with or without 1 ng/ml LPS for 4 hours. THP-1–derived macrophages were incubated at 1.5 × 106 cells per ml in 96-well plates with or without 10 ng/ml LPS for 4 hours. To stimulate the release of IL-1β, 3–5 mM ATP (Sigma-Aldrich) or 5 μM nigericin was added for the last 30 minutes of incubation. Supernatants were collected, centrifuged to remove cells and debris, and stored at –80°C for later analysis. IL-1β, IL-18, and TNF-α cytokine analysis was performed by ELISA (R&D Systems). Results were normalized to cell number, as determined by the CyQuant cell proliferation assay (Invitrogen). ROS inhibitors (5 μM DPI, 500 μM MitoTEMPO) were added to THP-1 cells in parallel with LPS. The NF-κB pathway reversible inhibitor PS-1145 (20 μM) or DSMO as vehicle was added to THP-1 cells during the 24-hour incubation with subject sera. The cells were then washed twice with RPMI and incubated with LPS in the presence or absence of both subject sera and PS-1145. Soluble CD14 in human sera was determined by ELISA (R&D Systems).
Immunoblot analysis.
For Western blots, cell lysates were prepared using RIPA buffer, separated by SDS-PAGE, and transferred to nitrocellulose membranes. Images were captured using the Odyssey system (Li-Cor). The following antibodies were used: caspase-1 (2225; Cell Signaling), IL-1β (ab9722; Abcam), NLRP3 (AG-20B-0014-C100; Adipogen), ASC (sc-22514; Santa Cruz), SIRT1 (sc-15404; Santa Cruz), SIRT2 (sc-20966; Santa Cruz), SIRT3 (5490S; Cell Signaling), SIRT5 (HPA022002; Sigma-Aldrich), phospho-IκBα (2859S; Cell Signaling), IκBα (9242S; Cell Signaling), phospho–NF-κB P65 (3033; Cell Signaling), NF-κB P65 (8242; Cell Signaling), and Tom20 (sc-11415; Santa Cruz). We also used acetyl-IDH2 and acetyl-SOD2 (Lys68) antibodies, which were provided by David Gius from Northwestern University, Chicago, Illinois, USA.
To measure caspase-1 and IL-1β release into the supernatant, cells were seeded in 24-well plates to a cell density of 1 × 106 cells per well. Cells were primed with LPS for 4 hours at 37°C, followed by washing once with PBS and transfer to serum-free media with or without nigericin for 1 hour (45). Supernatants were collected and centrifuged at 10,000 g for 10 seconds to remove any detached cells, followed by transfer to a fresh tube. The supernatant was concentrated by TCA precipitation. Then, the precipitated pellets were washed with acetone, dissolved in 10 μl 0.2 M NaOH, diluted with 40 μl H2O, supplemented with 10 μl 6× SDS-PAGE sample buffer, and boiled for 5 minutes.
Quantitative PCR analysis.
mRNA was isolated using Tripure (Roche), and cDNA was produced using a First-Strand Synthesis Kit (Invitrogen). Transcript levels were measured using validated gene-specific primers (Qiagen).
Measurement of cellular oxygen consumption.
The oxygen consumption rate was measured using a Seahorse XF24 Extracellular Flux Analyzer (Seahorse Bioscience). Briefly, 1 × 106 PBMCs per well were resuspended in unbuffered DMEM (DMEM supplemented with 5.5 mM glucose, 1 mM sodium pyruvate, 31 mM NaCl, 2 mM GlutaMax, pH 7.4) and plated on CellTak-coated (BD Biosciences) XF24-well plates, as previously described (46). Cells were maintained at 37°C in a non-CO2 incubator for at least 1 hour before the assay. The measurement of basal and maximal respiration was performed and the proton leak was calculated as previously described (47). For normalization, protein concentration in each well was determined using a BCA assay (Thermo Fisher Scientific).
Mitochondrial ROS detection.
For detection of mitochondrial ROS production, human PBMCs were incubated with 5 μM MitoSOX Red (Life Technologies) for 30 minutes in Hank’s Balanced Salt Solution (Life Technologies), washed, and incubated with 3 mM ATP for 15 minutes before cells were analyzed by flow cytometry on a BD FACSCanto (BD Biosciences). The monocyte subset was identified on the basis of size and complexity.
Superoxide dismutase activity.
Mitochondrial fractions were obtained from 1.5 × 107 LPS-treated PBMCs using the Q-proteome Mitochondria Isolation Kit (Qiagen). SOD activity was tested in 0.1 μg PBMC mitochondria using the SOD Assay Kit (Cayman Chemical), and to ensure the detection of only Mn-SOD (SOD2) activity, 5 mM potassium cyanide was added to the assay to inhibit cytosolic Cu/Zn-SOD (SOD1) and extracellular Cu/Zn-SOD (SOD3) activities.
Statistics.
The primary outcome was defined as the change in IL-1β secretion in response to inflammasome stimulation in PBMCs, comparing the fasted response to the refed response. As there were two postprandial blood draws, it was predetermined that higher mean IL-1β levels between the two fed states would be defined as the index fed response for comparison to fasting levels. Statistical tests for the primary outcome analysis were performed using randomized block design ANOVA. After a global difference was found (P < 0.005 for PBMCs) in the first 11 subjects, we then evaluated whether this was also evident in monocytes. The same comparison was performed in monocytes in the subsequent 8 subjects in the index study. Here, too the ANOVA test showed a global difference of P < 0.05. For all other studies, as predetermined, we compared the fasted state to the 3-hour–refed state by paired 2-tailed t tests at α = 0.05. Error bars in all figures represent SEM.
Study approval.
Human subjects provided written informed consent. The IRB of NHLBI approved the study protocols (Fasting Inflammasome Study — NCT02122575; Healthy Volunteer Blood Collection Protocol — NCT00442195).
Author contributions
JT, MAW, MP, RMS, and MNS designed research studies. JT, MK-S, TCO, RDH, AB, and KH conducted experiments and acquired data. AAS generated and provided reagents. JT, MAW, KH, MP, and MNS wrote and edited the manuscript. JL conducted experiments and acquired data.
Supplementary Material
Acknowledgments
This study was supported by the Division of Intramural Research, NHLBI, NIH (to M.N. Sack —HL005102-11). M. Kwarteng-Siaw was funded through the NIH Biomedical Research Training Program of Underrepresented Minority Groups. A.A. Sauve has intellectual property related to methods of production of NR and receives royalties from a commercial license to ChromaDex Inc. of this intellectual property. Furthermore, A.A. Sauve is a consultant and cofounder of Metro Mid-Atlantic Biotech LLC. We thank Shahin Hassanzadeh and Kinneret Broder for coordinating blood collections, Jamie Grimes for clinical protocol administration, and Neal Young of the NHLBI for including M.N. Sack as an Associate Investigator on his blood collection protocol. We thank David Gius from Northwestern University for supplying antibodies to measure SOD2 and IDH2 protein acetylation.
Footnotes
Conflict of interest: Anthony A. Sauve has intellectual property related to the methods of production of NR and receives royalties from a commercial license to ChromaDex Inc. Furthermore, A.A. Sauve is a consultant for and cofounder of Metro Midatlantic Biotech LLC.
Reference information:J Clin Invest. 2015;125(12):4592–4600. doi:10.1172/JCI83260.
References
- 1.Vandanmagsar B, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011;17(2):179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee HM, Kim JJ, Kim HJ, Shong M, Ku BJ, Jo EK. Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes. Diabetes. 2013;62(1):194–204. doi: 10.2337/db12-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim HY, et al. Interleukin-17-producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat Med. 2014;20(1):54–61. doi: 10.1038/nm.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutterwala FS, Haasken S, Cassel SL. Mechanism of NLRP3 inflammasome activation. Ann N Y Acad Sci. 2014;1319:82–95. doi: 10.1111/nyas.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18(3):363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- 6.Kheirandish-Gozal L, et al. Lipopolysaccharide-binding protein plasma levels in children: effects of obstructive sleep apnea and obesity. J Clin Endocrinol Metab. 2014;99(2):656–663. doi: 10.1210/jc.2013-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goossens GH, et al. Expression of NLRP3 inflammasome and T cell population markers in adipose tissue are associated with insulin resistance and impaired glucose metabolism in humans. Mol Immunol. 2012;50(3):142–149. doi: 10.1016/j.molimm.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Stienstra R, et al. Inflammasome is a central player in the induction of obesity and insulin resistance. Proc Natl Acad Sci U S A. 2011;108(37):15324–15329. doi: 10.1073/pnas.1100255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henao-Mejia J, Elinav E, Strowig T, Flavell RA. Inflammasomes: far beyond inflammation. Nat Immunol. 2012;13(4):321–324. doi: 10.1038/ni.2257. [DOI] [PubMed] [Google Scholar]
- 10.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481(7381):278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 11.Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest. 2011;121(6):2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagel-Langenickel I, Bao J, Pang L, Sack MN. The role of mitochondria in the pathophysiology of skeletal muscle insulin resistance. Endocr Rev. 2010;31(1):25–51. doi: 10.1210/er.2009-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 14.Shimada K, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer SS, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39(2):311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- 19.Longo VD, Mattson MP. Fasting: molecular mechanisms and clinical applications. Cell Metab. 2014;19(2):181–192. doi: 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JB, et al. Alternate day calorie restriction improves clinical findings and reduces markers of oxidative stress and inflammation in overweight adults with moderate asthma. Free Radic Biol Med. 2007;42(5):665–674. doi: 10.1016/j.freeradbiomed.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontana L, Meyer TE, Klein S, Holloszy JO. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci U S A. 2004;101(17):6659–6663. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Z, et al. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 2011;12(8):840–846. doi: 10.1038/embor.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bao J, et al. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic Biol Med. 2010;49(7):1230–1237. doi: 10.1016/j.freeradbiomed.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kendrick AA, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433(3):505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sack MN, Finkel T. Mitochondrial metabolism, sirtuins, and aging. Cold Spring Harb Perspect Biol. 2012;4(12): doi: 10.1101/cshperspect.a013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster BR, Scott I, Traba J, Han K, Sack MN. Regulation of autophagy and mitophagy by nutrient availability and acetylation. Biochim Biophys Acta. 2014;1841(4):525–534. doi: 10.1016/j.bbalip.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Youm YH, et al. The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat Med. 2015;21(3):263–269. doi: 10.1038/nm.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandler NG, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203(6):780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vodanovic-Jankovic S, Hari P, Jacobs P, Komorowski R, Drobyski WR. NF-κB as a target for the prevention of graft-versus-host disease: comparative efficacy of bortezomib and PS-1145. Blood. 2006;107(2):827–834. doi: 10.1182/blood-2005-05-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webster BR, Lu Z, Sack MN, Scott I. The role of sirtuins in modulating redox stressors. Free Radic Biol Med. 2012;52(2):281–290. doi: 10.1016/j.freeradbiomed.2011.10.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozden O, et al. Acetylation of MnSOD directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging (Albany NY) 2011;3(2):102–107. doi: 10.18632/aging.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulua AC, et al. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS) J Exp Med. 2011;208(3):519–533. doi: 10.1084/jem.20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown KD, et al. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 2014;20(6):1059–1068. doi: 10.1016/j.cmet.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontana L, Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell. 2015;161(1):106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandhorst S, et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab. 2015;22(1):86–99. doi: 10.1016/j.cmet.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amar J, et al. Energy intake is associated with endotoxemia in apparently healthy men. Am J Clin Nutr. 2008;87(5):1219–1223. doi: 10.1093/ajcn/87.5.1219. [DOI] [PubMed] [Google Scholar]
- 38.van der Burgh R, et al. Defects in mitochondrial clearance predispose human monocytes to interleukin-1β hypersecretion. J Biol Chem. 2014;289(8):5000–5012. doi: 10.1074/jbc.M113.536920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, et al. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc Natl Acad Sci U S A. 2014;111(2):E245–E254. doi: 10.1073/pnas.1316941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West AP, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3):406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinberg GR, Schertzer JD. AMPK promotes macrophage fatty acid oxidative metabolism to mitigate inflammation: implications for diabetes and cardiovascular disease. Immunol Cell Biol. 2014;92(4):340–345. doi: 10.1038/icb.2014.11. [DOI] [PubMed] [Google Scholar]
- 43.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res. 2007;56(1):45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 44.Schnoor M, et al. Efficient non-viral transfection of THP-1 cells. J Immunol Methods. 2009;344(2):109–115. doi: 10.1016/j.jim.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 45.Qu Y, et al. Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J Immunol. 2011;186(11):6553–6561. doi: 10.4049/jimmunol.1100478. [DOI] [PubMed] [Google Scholar]
- 46.Avila C, et al. Platelet mitochondrial dysfunction is evident in type 2 diabetes in association with modifications of mitochondrial anti-oxidant stress proteins. Exp Clin Endocrinol Diabetes. 2012;120(4):248–251. doi: 10.1055/s-0031-1285833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott I, Webster BR, Chan CK, Okonkwo JU, Han K, Sack MN. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289(5):2864–2872. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.