Abstract
Cellular metabolism is increasingly recognized as a controller of immune cell fate and function. MicroRNA-33 (miR-33) regulates cellular lipid metabolism and represses genes involved in cholesterol efflux, HDL biogenesis, and fatty acid oxidation. Here, we determined that miR-33–mediated disruption of the balance of aerobic glycolysis and mitochondrial oxidative phosphorylation instructs macrophage inflammatory polarization and shapes innate and adaptive immune responses. Macrophage-specific Mir33 deletion increased oxidative respiration, enhanced spare respiratory capacity, and induced an M2 macrophage polarization–associated gene profile. Furthermore, miR-33–mediated M2 polarization required miR-33 targeting of the energy sensor AMP-activated protein kinase (AMPK), but not cholesterol efflux. Notably, miR-33 inhibition increased macrophage expression of the retinoic acid–producing enzyme aldehyde dehydrogenase family 1, subfamily A2 (ALDH1A2) and retinal dehydrogenase activity both in vitro and in a mouse model. Consistent with the ability of retinoic acid to foster inducible Tregs, miR-33–depleted macrophages had an enhanced capacity to induce forkhead box P3 (FOXP3) expression in naive CD4+ T cells. Finally, treatment of hypercholesterolemic mice with miR-33 inhibitors for 8 weeks resulted in accumulation of inflammation-suppressing M2 macrophages and FOXP3+ Tregs in plaques and reduced atherosclerosis progression. Collectively, these results reveal that miR-33 regulates macrophage inflammation and demonstrate that miR-33 antagonism is atheroprotective, in part, by reducing plaque inflammation by promoting M2 macrophage polarization and Treg induction.
Introduction
Macrophages are critical effectors of inflammation and innate immunity; they also regulate adaptive immunity by recruiting and/or activating other immune cells at inflammatory foci (1). In addition, macrophages play important roles in tissue homeostasis and resolution of inflammation (2). To fulfill these functions, macrophages can adopt a spectrum of activation programs depending on the context. M1, or classical activation, represents one end in response to products from bacterial infection such as LPS and IFN-γ, whereas M2, or alternative activation, occurs in response to the cytokines IL-4 and IL-13 (2). Although originally defined in the context of host defense against parasitic helminths (3), M2 macrophages are now appreciated as being important orchestrators of inflammation resolution and metabolic homeostasis through their secretion of antiinflammatory cytokines (e.g., IL-10 and TGF-β), mediators of tissue repair (eg. collagen), and catecholamines (4). Moreover, monocyte-derived M2 macrophages were recently shown to be an important source of retinoic acid (5), a hormone-like metabolite that promotes the differentiation of immunosuppressive Tregs (6, 7).
Recent evidence points to a pivotal role for cellular metabolism in macrophage activation. In particular, M1 and M2 macrophages use different metabolic programs to fuel their effector functions. M1 macrophages use aerobic glycolysis to rapidly provide energy needed for intense, short-lived bactericidal or proinflammatory responses, whereas M2 macrophages use a more efficient ATP-generating program of fatty acid oxidation (FAO) capable of being sustained for long periods of time (8, 9). Although these metabolic programs were originally thought to merely reflect the cell’s energy substrate utilization, recent findings indicate that disrupting cellular energy metabolism can directly alter macrophage M1/M2 fate and inflammatory functions. When the macrophage metabolism is shifted toward glycolysis, this drives a proinflammatory phenotype (10), whereas inhibition of glycolysis by 2-deoxyglucose (2-DG) decreases the production of inflammatory cytokines such as IL-1β (11). Similarly, M2 activation is hindered by inhibiting FAO (12), whereas overexpression of the nuclear receptor PPARγ coactivator peroxisome proliferator–activated receptor γ coactivator 1β (PGC1β) to drive mitochondrial oxidative phosphorylation primes macrophages for alternative activation and inhibits proinflammatory cytokine production (9). These studies suggest a paradigm in which cellular metabolism and the M1/M2 inflammatory axis are intimately linked, each influencing the other. There is thus considerable interest in identifying the molecular regulation of metabolic pathways controlling M1/M2 macrophage polarization and harnessing these to favorably intervene in chronic inflammatory conditions.
One clinically important chronic inflammatory condition is atherosclerotic cardiovascular disease, in which persistent macrophage accumulation in the artery wall underlies the pathogenesis. In the setting of hypercholesterolemia, monocyte-derived macrophages infiltrate the arterial intima to clear retained apolipoprotein B–containing (apoB-containing) lipoproteins (e.g., LDL) and are transformed into lipid-laden macrophage foam cells (13). For reasons that are poorly understood, these macrophage foam cells persist in the artery wall, setting off a maladaptive immune response that promotes the formation of plaques. These macrophages are a source of inflammatory mediators, including cytokines and chemokines that mediate the recruitment and/or activation of other immune cells, thereby chronically sustaining the inflammation that fuels plaque progression. While both M1 and M2 macrophages have been documented in human and mouse atherosclerotic plaques (14), the preponderance of evidence suggests that M1 macrophages promote plaque progression. Studies in mouse models of atherosclerosis have shown that conditions that increase macrophage polarization toward an M1 phenotype (15–17) or attenuate polarization toward M2 (18–20) accelerate atherosclerotic plaque formation, whereas administration of the M2-polarizing factor IL-13 inhibits disease progression (21). Moreover, aggressive lipid lowering or raising of HDL levels in mouse models induces atherosclerosis regression that is characterized by a switch from M1 to M2 macrophages in plaques (22, 23), suggesting that M2 polarization is central to resolving atherosclerotic inflammation. Nonetheless, the factors in the plaque microenvironment that regulate the polarization of these cells in vivo remain incompletely defined.
An appealing possibility that the regulatory factors include microRNAs (miRNAs) is suggested by their roles as important posttranscriptional fine tuners of many biological and metabolic programs. Recent studies from our lab and others identified microRNA-33 (miR-33a) and miR-33b as intronic miRNAs that are coexpressed with their host genes, SREBF2 and SREBF1, which code for transcription factors that regulate cholesterol and fatty acid synthesis/uptake (24–26). miR-33a/b repress the expression of genes involved in pathways that these oppose SREBP-driven functions, including cholesterol efflux (ABCA1, ABCG1, NPC1) (24–26) and FAO (HADHB, CROT, CPT1A, PRKAA1) (27–29). Studies in mice and nonhuman primates show that inhibitors of miR-33 may hold promise for the treatment of atherosclerosis, as they increase the expression of the cholesterol transporter ABCA1 in the liver and macrophages, thereby raising plasma levels of HDL and macrophage cholesterol efflux (24–26, 30–32). This is supported by studies of miR-33 inhibition or targeted deletion in atherosclerotic mouse models, which showed increased plasma HDL cholesterol (HDL-C) and reduced atherosclerotic plaque size (31, 33). However, recent studies in hyperlipidemic LDL receptor–knockout (Ldlr–/–) mice fed a Western diet concurrent with anti–miR-33 treatment showed atheroprotection even in the absence of changes in plasma levels of HDL-C (34), indicating that miR-33 inhibition has antiatherosclerotic properties that are distinct from its HDL-raising abilities. Given that miR-33 is a metabolic regulator, we investigated whether miR-33 regulation of cellular metabolism can control macrophage inflammatory phenotype and function to alter atherosclerotic plaque inflammation and disease progression.
We show herein that, indeed, miR-33 directly controls macrophage polarization and that this is independent of its effects on cholesterol efflux. Furthermore, we show that the effects are a result of targeting the AMP-activated protein kinase (AMPK), a key integrator of cellular energy homeostasis, thereby reducing FAO and promoting a proglycolytic state that fuels M1 activation. Inhibitors of miR-33 reverse this shift in metabolism by upregulating cellular FAO and driving macrophage polarization to the M2 state in vitro and in vivo, as evidenced by the enrichment of markers that characterize alternatively activated macrophages. Furthermore, anti–miR-33 increased macrophage expression and activity of the retinoic acid–producing enzyme aldehyde dehydrogenase (ALDH) family 1, subfamily A2 (ALDH1A2), endowing these cells with the capacity to promote the differentiation of inducible forkhead box P3–positive (FOXP3+) Tregs. Consistent with this, anti–miR-33 treatment of Western diet–fed Ldlr–/– mice increased the accumulation of atheroprotective M2 macrophages and FOXP3+ Tregs in plaques, while reducing atherosclerotic plaque size (–35%) independently of changes in plasma HDL-C. Collectively, these data indicate that miR-33 promotes a proinflammatory M1 state by altering macrophage FAO and retinoid metabolism, consequently affecting both innate and adaptive immune responses.
Results
miR-33 controls the macrophage inflammatory phenotype by altering the balance of cellular FAO and glycolysis.
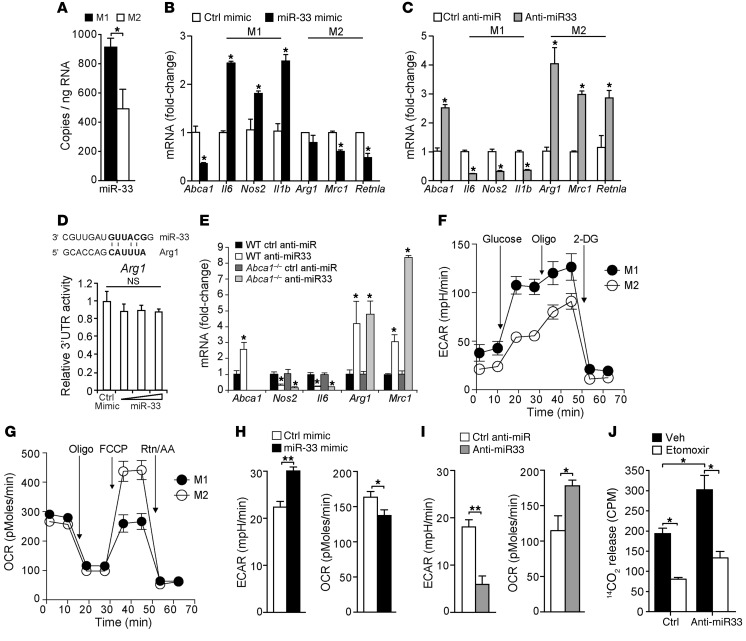
To understand the relationship between miR-33 and macrophage inflammation, we measured levels of miR-33 in BM-derived macrophages (BMDMs) classically (M1) activated or alternatively activated (M2) by treatment with IFN-γ and LPS or IL-4 for 24 hours, respectively. Expression of miR-33 was higher in M1 macrophages as compared with M2, pointing to a potential role for miR-33 in macrophage polarization (Figure 1A). To investigate this, we next profiled the expression of key genes that mark commitment to M1 or M2 activation in mouse peritoneal macrophages transfected with miR-33 mimic or control oligonucleotides. Overexpression of miR-33 increased macrophage mRNA expression of M1 markers such as Il6, Nos2, and Il1b, and reduced expression of the M2 macrophages Mrc1 (mannose receptor CD206) and Fizz1/Retnla (Figure 1B). Conversely, macrophages transfected with inhibitors of miR-33 showed a marked increase in expression of M2 markers (Arg1, Mrc1, Fizz1) and reduced expression of M1 markers (Il6, Nos2, Il1b) compared with macrophages treated with control oligonucleotides (Figure 1C), suggesting that miR-33 may alter macrophage inflammatory polarization. Furthermore, miR-33 inhibition or targeted deletion in BMDMs reduced the expression of M1 markers in macrophages classically activated with IFN-γ/LPS and increased expression of M2 markers in macrophages alternatively activated with IL-4 (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI81676DS1). Conversely, treatment with miR-33 mimics enhanced macrophage polarization to M1 with IFN-γ/LPS and reduced macrophage polarization to M2, as measured by the expression of M1 and M2 marker genes (Supplemental Figure 1). Notably, none of the altered genes contain potential miR-33–binding sites in their 3′ UTRs, with the exception of Arg1. However, overexpression of miR-33 failed to alter Arg1 3′ UTR luciferase activity, excluding Arg1 as a direct target of miR-33 (Figure 1D). Previous studies in macrophages lacking the cholesterol transporter ABCA1, a bona fide target of miR-33, suggested that enrichment of cellular membranes with free cholesterol can enhance macrophage proinflammatory responses (35, 36). However, the effects of miR-33 inhibition on macrophage M1/M2 gene programs were maintained in Abca1–/– macrophages (Figure 1E), indicating that miR-33 alters macrophage polarization by a mechanism independent of its regulation of ABCA1 expression.
Figure 1. miR-33 regulates macrophage polarization.
(A) Relative expression levels of miR-33 in BMDMs activated to M1 or M2 by treatment with LPS and IFN-γ, or IL-4, for 24 hours, respectively. Data are representative of 3 independent experiments. (B and C) Real-time PCR analysis of markers of classical M1 and alternative M2 activation in peritoneal macrophages transfected with (B) control (ctrl) or miR-33 mimics or (C) control or miR-33 inhibitors for 48 hours. Data are representative of 4 independent experiments. (D) 3′ UTR luciferase reporter activity of mouse Arg1 in HEK293 cells transfected with miR-33 or control mimics. Data are representative of 3 independent experiments. (E) Real-time PCR analysis of M1 and M2 markers in BMDMs isolated from WT or Abca1–/– mice and transfected with control anti-miR or anti–miR-33 for 48 hours. Data represent the mean ± SEM of 3 experiments. (F) ECAR of BMDMs cultured for 24 hours with IFN-γ and LPS (M1) or with IL-4 (M2), followed by sequential treatment (arrows) with glucose, oligomycin (Oligo), and 2-DG. (G) OCR of M1 and M2 macrophages prepared as in E and sequentially treated with oligomycin, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), and rotenone plus antimycin (Rtn/AA). (H and I) Maximal ECAR and OCR of macrophages treated with (H) control or miR-33 mimic or (I) control anti-miR or anti–miR-33 at 48 hours after transfection, measured in real time and presented as change in mpH per unit time (ECAR) and pMoles per unit time (OCR). Data are representative of 3 independent experiments (F–I). (J) 14C-oleic acid oxidation in macrophages treated with control or miR-33 inhibitors in the presence or absence of the FAO inhibitor etomoxir. Veh, vehicle. Data are representative of 2 independent experiments. Statistical comparisons were made using 2-tailed Student’s t test (A–C and H–J) or ANOVA (E). *P ≤ 0.05; **P ≤ 0.005, compared with controls. All values are mean ± SEM.
As recent studies indicate that the activation of macrophages to M1 and M2 phenotypes is marked by differences in core metabolic programs (4), we next investigated whether miR-33–induced changes in macrophage polarization gene profiles were accompanied by changes in cellular energy metabolism. As shown in Figure 1F and consistent with published reports (12), classical M1 activation of macrophages using IFN-γ and LPS treatment increased aerobic glycolysis and lactate production, which was reflected by a high extracellular acidification rate (ECAR). On the other hand, alternatively activated M2 macrophages induced by IL-4 preferentially rely on β-oxidation of fatty acids to fuel their functional activation, which was reflected by increased mitochondrial oxygen consumption rates (OCRs) in cells supplemented with carnitine and palmitate to support FAO (Figure 1G). Consistent with enhanced M1 activation, miR-33 overexpression shifted macrophage metabolism toward aerobic glycolysis and reduced FAO, as evidenced by increased cellular ECAR and reduced OCR (Figure 1H). Furthermore, this pattern was reversed upon inhibition of endogenous miR-33 in macrophages, which markedly increased the mitochondrial oxidation rate in the presence of palmitate and decreased glycolytic activity (Figure 1I). Increased FAO in macrophages treated with anti-miR33 compared with control anti-miR was confirmed by measuring 14-C-oleic acid oxidation, and this increase was abolished in the presence of the CPT1 inhibitor etomoxir, which blocks FAO (Figure 1J). Thus, modulation of miR-33 expression can promote distinct activation programs in macrophages, measurable by changes in both M1/M2 markers and the metabolic programs that fuel their bioenergetic needs.
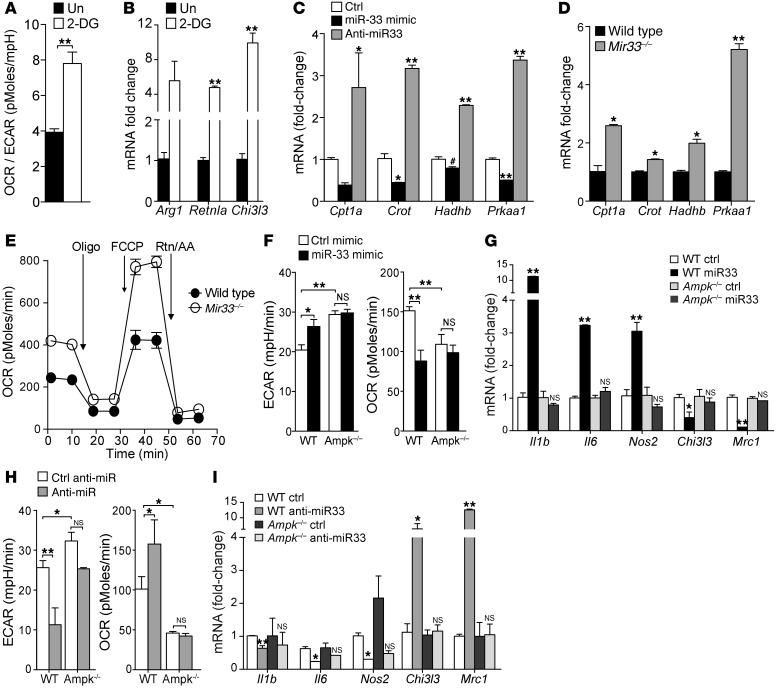
Recent studies suggest that manipulation of cellular metabolism can directly affect the inflammatory state of immune cells (e.g., refs. 9, 10). Consistent with these reports, we show that treatment of macrophages for 24 hours with 2-DG, a glucose analog that acts as a competitive inhibitor for glycolysis, increases the ratio of oxidative phosphorylation to aerobic glycolysis, and promotes the expression of genes that define M2 activation (Figure 2, A and B). Thus, we considered whether miR-33 targeting of genes involved in fatty acid or glucose metabolism could be responsible for reprogramming the macrophage inflammatory phenotype. We previously demonstrated that miR-33 targets genes involved in FAO in hepatocytes, including Cpt1a, Crot, Hadhb, and Prkaa1 (27), and show here that miR-33 overexpression and inhibition in macrophages reciprocally regulates the expression of Cpt1a, Crot, Hadhb, and Prkaa1 mRNA (Figure 2C). Consistent with the active repression of these genes in macrophages by miR-33, we show that BMDMs from miR-33–deficient mice have increased levels of Cpt1a, Crot, Hadhb, and Prkaa1 mRNA (Figure 2D) and this is associated with a marked upregulation of basal mitochondrial OCR and spare respiratory capacity (SRC) (the quantitative difference between maximal uncontrolled OCR and the initial basal OCR) (Figure 2E). Of these miR-33 target genes, Prkaa1 was the most highly derepressed in Mir33–/– macrophages and codes for AMPKα, an evolutionary conserved kinase that acts as a cellular energy sensor. Upon activation, AMPK switches off ATP-consuming pathways and promotes ATP-producing pathways, including FAO (37). We show that inhibition of miR-33 in BMDMs increases both AMPK protein and activity, as evidenced by elevated levels of phosphorylated AMPK (Supplemental Figure 2). To investigate whether miR-33 regulates macrophage activation by targeting AMPK, we overexpressed and silenced miR-33 in WT and Prkab1–/– (also known as Ampkβ1–/–, herein referred to as Ampk–/–) macrophages, which have approximately 90% reduced AMPK activity (38). Whereas WT macrophages transfected with miR-33 showed increased ECAR and reduced OCR, these effects were abolished in macrophages lacking AMPK activity (Figure 2F). Furthermore, miR-33 overexpression failed to increase markers of M1 macrophage activation (Il6, Nos2, Il1b) in Ampk–/– macrophages consistent with a loss of metabolic reprogramming by miR-33 in the absence of AMPK (Figure 2G). Similarly, miR-33 inhibition failed to reduce ECAR or increase OCR in Ampk–/– macrophages, and this was paralleled by a loss of macrophage polarization to the M2 phenotype (Figure 2, H and I). Together, these results indicate that miR-33 targeting of AMPK to alter macrophage metabolism is central to its immunomodulatory function.
Figure 2. miR-33 regulates the macrophage metabolic and inflammatory phenotype via targeting of AMPK.
(A) Ratio of OCR to ECAR in BMDM in the presence or absence of 2-DG to decrease glycolysis. Data represent mean ± SEM of 4 experiments. Un, untreated. (B) Real-time PCR analysis of markers of M2 activation in macrophages in A treated with vehicle or 2-DG for 24 hours. Data represent mean ± SEM of 3 experiments. (C and D) Real-time PCR analysis of (C) BMDMs transfected with control or miR-33 mimics and inhibitors for 48 hours and (D) BMDMs from WT or Mir33–/– mice. (E) ECAR of WT or Mir33–/– BMDMs sequentially treated (arrows) with oligomycin, FCCP, and rotenone plus antimycin. Data are representative of 3 independent experiments (C–E). (F) Maximal ECAR and OCR of WT or Ampk–/– BMDMs transfected with miR-33 or control mimics and (G) real-time PCR analysis of markers of classical M1 and alternative M2 activation in those cells. (H) Maximal ECAR and OCR in WT or Ampk–/– BMDMs treated with anti–miR-33 or control anti-miR and (I) real-time PCR analysis of markers of classical M1 and alternative M2 activation in those cells. Data are representative of 2 independent experiments (F–I). Statistical comparisons were made using 2-tailed Student’s t test (A, B, D, and F–H) or ANOVA (C, G, and I). *P ≤ 0.05; **P ≤ 0.005, compared with controls. All values are mean ± SEM.
Anti–miR-33 treatment inhibits the progression of atherosclerosis independently of HDL levels.
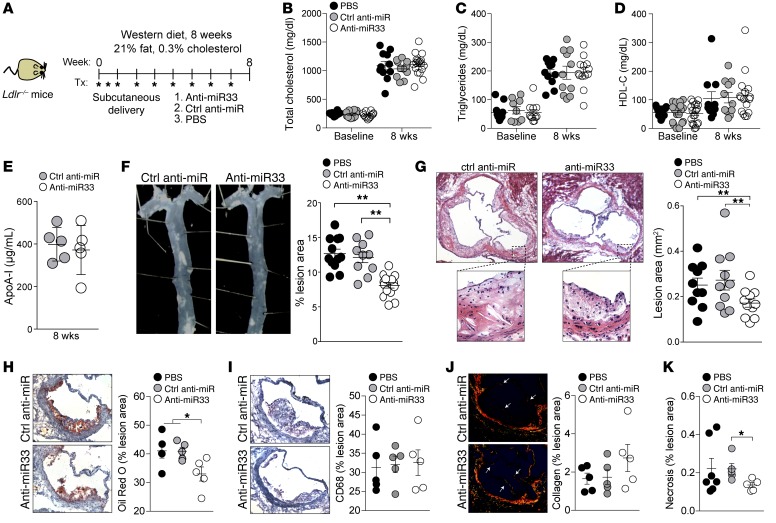
A recent study of atherosclerosis progression in Ldlr–/– mice fed a Western diet concurrent with anti–miR-33 treatment for 8 weeks reported reduced plaque size in the absence of changes in plasma levels of HDL-C (34); however, the mechanisms by which miR-33 inhibition reduces atherosclerosis under these conditions were unclear. Furthermore, this finding remains controversial, as a second study found no difference in atherosclerosis progression in control and miR-33 locked nucleic acid (LNA) inhibitor–treated Ldlr–/– mice fed a high cholesterol–containing diet for 12 weeks (39). To investigate whether miR-33 inhibitors could protect against atherosclerosis by directly altering the plaque macrophage phenotype and associated inflammatory responses, we performed a similar study in Ldlr–/– mice treated with PBS, a control anti-miR, or anti–miR-33 for 8 weeks concurrent with Western diet feeding (Figure 3A). We observed no differences among the groups in weight (data not shown) or plasma total cholesterol and triglyceride levels (Figure 3, B and C). In agreement with previous reports (34, 39), anti–miR-33 treatment of Ldlr–/– mice during Western diet feeding failed to increase plasma HDL-C (Figure 3D), and consistent with this, we observed no difference in plasma levels of the HDL-associated apolipoprotein ApoA1 between the treatment groups (Figure 3E). Despite this, anti–miR-33 treatment reduced plaque burden and size compared with control anti-miR or PBS treatment, as assessed by morphometric analyses of lesion area in the aorta en face (Figure 3F; 37% reduction) and cross-sections of the aortic root (Figure 3G; 33% reduction). Characterization of the aortic root atherosclerotic plaques of mice treated with anti–miR-33 showed that they contained less neutral lipid by oil red O staining, but had a content of macrophages similar to that found in the plaques of control anti-miR– or PBS-treated mice (Figure 3, H and I). Furthermore, there was a trend toward increased plaque collagen content measured by picrosirius red staining (Figure 3J) and significant reduction in necrotic area (Figure 3K) in plaques of anti–miR-33–treated mice compared with those of control anti-miR or PBS-treated mice, which are markers of increased plaque stability in humans.
Figure 3. Anti–miR-33 reduces atherosclerosis without altering plasma levels of HDL in Western diet–fed Ldlr–/– mice.
(A) Schematic diagram showing the treatment regimen of male Ldlr–/– mice on a Western diet. (B–E) Baseline (week 0) and terminal (week 8) plasma levels of (B) total cholesterol, (C) triglycerides, (D) HDL-C, and (E) HDL-associated apoA-I. n = 11–19 mice/group (B–D); n = 5 mice/group (E). (F and G) Quantification of atherosclerotic lesion area in (F) the aorta en face and (G) the aortic root of mice receiving the indicated treatment (n = 10–12). Representative images of the (F) aorta and (G) aortic root lesions stained with H&E are presented at left of graphs. Original magnification, ×40 (F); ×50 (top images, G); ×200 (bottom images, G). (H and I) Characterization of aortic root plaques by (H) oil red O staining of neutral lipids, (I) immunohistochemical staining for the macrophage marker CD68, (J) picrosirius red staining of collagen, and (K) quantification of necrotic area. Original magnification, ×200 (H–J). Quantification of images from n = 5 mice/group is shown at right for H–J and n = 5–7 mice/group for K. All values are mean ± SEM. Statistical comparisons were made by ANOVA. *P ≤ 0.05; **P ≤ 0.005.
Anti–miR-33 treatment alters the plaque macrophage phenotype in situ.
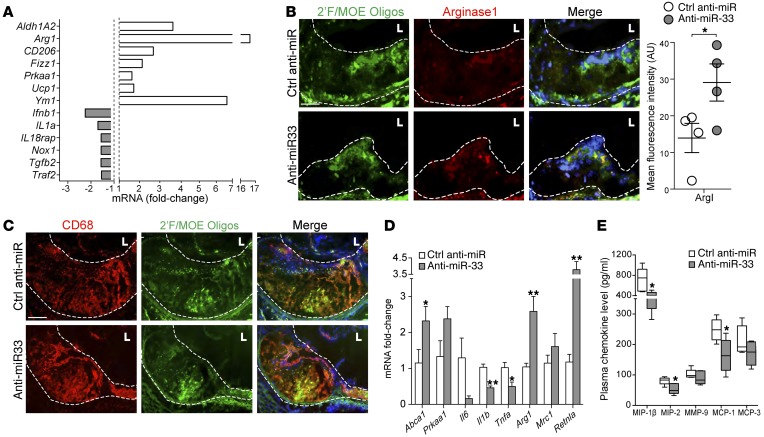
To interrogate the gene expression profiles of plaque macrophages from anti–miR-33– and control anti-miR–treated mice, we isolated lesional macrophages by laser-capture microdissection (LCM) and performed NanoString profiling of mRNA using a custom panel of 230 inflammatory genes. Strikingly, this analysis revealed an enrichment in markers of alternatively activated M2 macrophages including Arg1, Mrc1, Retnla, and Ym1 and a parallel decrease in inflammatory gene expression (Ifnb1, Il1a, Nox1) in anti–miR-33–treated mice (Figure 4A). Using fluorescence microscopy, we showed increased expression of the prototypic M2 marker arginase 1 in plaques of anti–miR-33– versus control anti-miR–treated mice, which colocalized to cells that stained positive for the anti–miR-33 2′F/MOE (Figure 4B). Costaining for the macrophage marker CD68 showed that control and anti–miR-33 oligonucleotides both localize to macrophages within the plaque (Figure 4C). Similar polarization to M2 was observed in peritoneal macrophages isolated from these mice (Figure 4D). Furthermore, measurement of plasma chemokine levels revealed lower levels of monocyte chemotactic protein 1 (MCP-1), macrophage inflammatory protein-1β (MIP-1β), and MIP-2 in mice treated with anti–miR-33 compared with control anti-miR (Figure 4E). Together, these data indicate that anti–miR-33 can reduce local and systemic inflammation by altering the plaque macrophage inflammatory phenotype in the face of continued hyperlipidemia.
Figure 4. miR-33 inhibitors target plaque macrophages to alter macrophage polarization and reduce systemic inflammation.
(A) Gene expression profile of macrophages isolated from aortic root plaques of control anti-miR or anti–miR-33–treated Ldlr–/– mice using LCM. Data represent the mean of n = 4 mice/group. (B and C) Immunofluorescent staining of aortic root plaques for the 2′F/MOE moiety of the control anti-miR or anti–miR-33 oligonucleotides (green) and (B) arginase 1 (red) or (C) the macrophage marker CD68. Colocalization of staining is seen as yellow in the merged image, with DAPI-stained nuclei in blue. Quantification of arginase 1 staining is shown at right. Data represent the mean ± SEM of n = 4 mice/group. Scale bars: 100 μM. (D) Real-time PCR analysis of peritoneal macrophages isolated from control anti-miR and anti–miR-33–treated Ldlr–/– mice. Data represent the mean ± SEM of n = 4–5 mice per group. (E) Plasma chemokine levels in control anti-miR– or anti–miR-33–treated Ldlr–/– mice. Data represent the mean ± SEM of 5 mice per group. Statistical comparisons were made using 2-tailed Student’s t test (B, D, and E). *P ≤ 0.05; **P ≤ 0.005, compared with control.
Anti–miR-33 treatment restores impaired ALDH1A2 expression and activity in macrophages and macrophage foam cells.
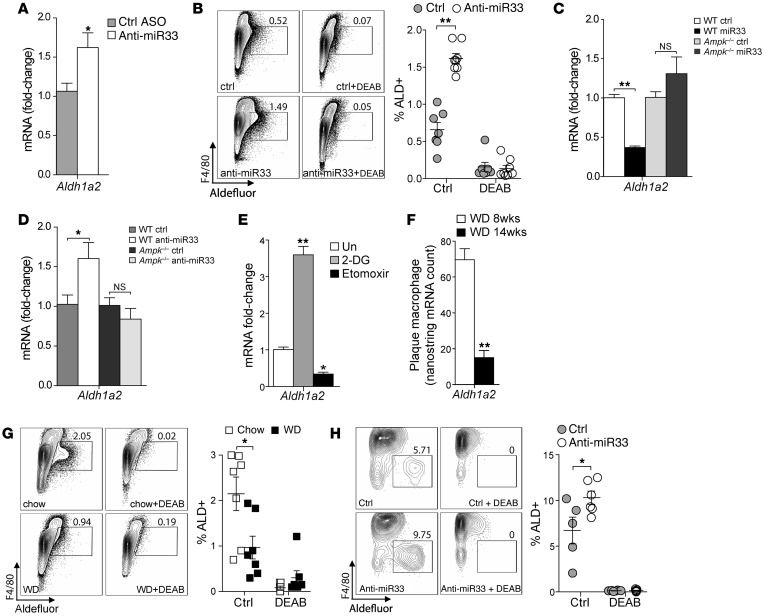
Among the genes induced by anti–miR-33 in plaque macrophages, we noted an increase in Aldh1a2 (Figure 4A), a gene recently reported to be expressed in alternatively activated macrophages (6). Aldh1a2 codes for the enzyme retin ALDH type 2 (RALDH2), which regulates the synthesis of retinoic acid, a metabolite with immunoregulatory function (7). Although not a predicted target of miR-33, we found that peritoneal macrophages elicited from mice treated with anti–miR-33 also had increased Aldh1a2 mRNA expression 3 days after injection, compared with macrophages from control anti-miR–treated mice (Figure 5A). Correspondingly, F4/80+ peritoneal macrophages from these mice treated with anti–miR-33 showed increased ALDH activity, as measured by flow cytometry, which could be blocked by the ALDH-specific enzyme inhibitor diethylaminobenzaldehyde (DEAB) (Figure 5B). Consistent with a key role for miR-33 targeting of AMPK in regulating macrophage polarization, we found that modulation of Aldh1a2 mRNA by miR-33 overexpression or inhibition was abolished in Ampk–/– macrophages (Figure 5, C and D). These data suggest that miR-33 modulation of Aldh1a2 expression is downstream of AMPK and metabolic pathways that dictate macrophage polarization. Indeed, acute inhibition of glycolysis in macrophages using 2-DG increased the expression of Aldh1a2, whereas inhibition of FAO using etomoxir reduced Aldh1a2 mRNA (Figure 5E), showing that expression of Aldh1a2 in macrophages is controlled by metabolic programs that determine M1/M2 polarization.
Figure 5. miR-33 regulates Aldh1a2 expression and activity in vitro and in vivo.
(A) Real-time PCR analysis of Aldh1a2 mRNA expression in thioglycollate-elicited peritoneal macrophages from C57BL/6 mice isolated 72 hours after i.p. injection of control or anti–miR-33 antisense oligonucleotides. Data are representative of 3 independent experiments. (B) Flow cytometric analysis of ALDH activity of macrophages prepared as indicated in A in the presence and absence of the ALDH inhibitor DEAB to show specificity. Data represent the mean ± SEM of n = 7–8 mice/group. (C and D) Real-time PCR analysis of Aldh1a2 mRNA expression in BMDMs from WT or Ampk–/– mice transfected with (C) control or miR-33 mimics or (D) control anti-miR or anti–miR-33 at 48 hours after transfection. Data are representative of 2 independent experiments. (E) Real-time PCR analysis of Aldh1a2 mRNA in BMDMs cultured for 24 hours with 2-DG to inhibit glycolysis or etomoxir to inhibit FAO. Data represent mean ± SEM of 3 experiments. (F) Nanostring measurement of Aldh1A2 mRNA expression in macrophages isolated from aortic root plaques of Ldlr–/– mice fed a Western diet for 8 or 14 weeks (n = 5 mice/group). (G and H) Flow cytometric analysis of ALDH activity in (G) thioglycollate-elicited peritoneal macrophages from Ldlr–/– mice fed a chow or Western diet (n = 6 mice/group) and (H) resident peritoneal macrophages isolated from Western diet–fed Ldlr–/– mice treated with control anti-miR or anti–miR-33 for 8 weeks. ALDH activity was measured in the presence and absence of DEAB to show specificity. Statistical comparisons were made using 2-tailed Student’s t test (A, B, and F–H) or ANOVA (C–E). *P ≤ 0.05; **P ≤ 0.005, compared with controls. All values are mean ± SEM.
Recent studies indicate that in progressing atherosclerotic plaques, M1 macrophages predominate over M2 macrophages (40). Consistent with this idea, the expression of Aldh1a2 in plaque macrophages declined with plaque progression in Western diet–fed Ldlr–/– mice (Figure 5F). Interestingly, we also observed reduced ALDH activity in peritoneal macrophages isolated from Western diet–fed Ldlr–/– mice compared with chow-fed WT mice (Figure 5G), suggesting that Aldh1a2 function may become impaired in macrophage foam cells. Notably, anti–miR-33 treatment enhanced ALDH activity in peritoneal macrophages isolated from Western diet–fed Ldlr–/– mice compared with those treated with control oligo (Figure 5H), suggesting that miR-33 inhibition can restore defective ALDH1A2 function in cholesterol-loaded macrophages.
Inhibition of miR-33 in macrophages promotes Treg differentiation in vitro and in vivo.
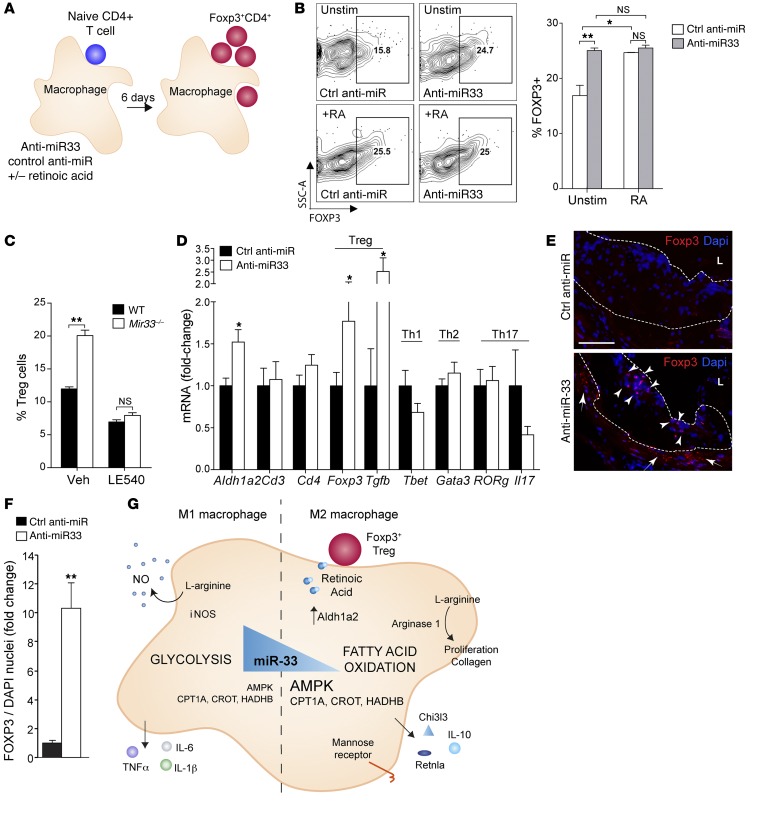
The metabolic product of ALDH activity, retinoic acid, can promote the development of extrathymic inducible FOXP3+ Tregs (5), which exert a protective role in atherosclerosis (reviewed in ref. 41). To determine whether inhibition of miR-33 in macrophages could promote Treg differentiation, we cocultured naive T cells with macrophages that were transfected with anti–miR-33 or control oligonucleotides (Figure 6A). After 6 days of coculture, a significant increase in FOXP3+CD4+ cells was detected in the T cells coincubated with anti–miR-33–treated macrophages as compared with control anti-miR–treated macrophages (Figure 6B). Notably, this effect was of a magnitude similar to the addition of exogenous retinoic acid to the naive T cells. Importantly, in macrophage cocultures, the addition of retinoic acid to T cells cocultured with anti–miR-33–treated macrophages showed no further enhancement in the extent of Foxp3+ induction (Figure 6B). Consistent with these findings, we observed a significant increase in Treg expansion when naive CD4+ T cells were coincubated with Mir33–/– macrophages compared with WT macrophages, and this was inhibited when RA signaling was blocked using the retinoic acid receptor pan-antagonist LE540 (Figure 6C). Collectively, these results indicate that anti–miR-33–treated macrophages have an enhanced capacity to induce Treg differentiation from a naive T cell population in vitro via increased retinoic acid production. To assess whether miR-33 inhibition increased Treg populations in vivo, we measured the expression of T cell markers and their relevant cytokines in the aortic arches of mice treated with control or anti–miR-33 oligonucleotides. Notably, we observed significant increases in Aldh1a2, Foxp3, and Tgfb mRNA in anti–miR-33–treated mice compared with control oligonucleotide–treated mice (Figure 6D). Consistent with this finding, immunofluorescence staining of the aortic root of atherosclerotic lesions revealed greater numbers of FOXP3+ cells in plaques and surrounding adventitia of anti–miR-33–treated mice compared with the control group (Figure 6, E and F). We observed no evidence for the expansion of FOXP3+ Tregs in secondary lymphoid organs such as the spleen and lymph nodes of anti–miR-33–treated mice as compared with control-treated mice (data not shown), consistent with our hypothesis that enhanced ALDH1A2 activity of anti–miR-33–treated plaque macrophages induces a local increase in inducible FOXP3+ Tregs.
Figure 6. Anti–miR-33–treated macrophages induce differentiation of Tregs.
(A) Naive CD4+ T cells were cultured in the presence or absence of retinoic acid for 6 days with macrophages transfected with control anti-miR or anti-miR33 and (B) subsequently analyzed by flow cytometry for nuclear FoxP3 expression to determine Treg differentiation. Data are representative of 2 independent experiments. (C) Percentage of naive CD4+ T cells differentiated into Tregs in coculture with WT and Mir33–/– macrophages in the presence or absence of LE540 to block RA signaling. Data are representative of 2 independent experiments. (D) Real-time PCR analysis of mRNA from aortic arches of Western diet–fed Ldlr–/– mice treated with control anti-miR or anti–miR-33 (8 weeks) for markers of T cell subsets and associated cytokines. Data represent the mean ± SEM of n = 6 mice/group. (E) Immunofluorescent staining for Foxp3 in plaques of Western diet–fed Ldlr–/– mice treated with control anti-miR or anti–miR-33 (8 weeks). Dotted line indicates the outline of the intima. L, lumen. FoxP3+ cells accumulate in both the intima (arrowheads) and adventitia (arrows) of plaques of anti–miR-33–treated mice. Data represent the mean ± SEM of n = 6 mice/group. Scale bar: 100 μM. (F) Quantification of FoxP3 staining is shown in D. n = 5 mice/group. (B–D and F) All values are mean ± SEM. Statistical comparisons were made using 2-tailed Student’s t test. *P ≤ 0.05; **P ≤ 0.005, compared with controls. (G) Model of molecular mechanisms involved in miR-33 regulation of macrophage polarization and Treg differentiation. When miR-33 levels are high, AMPK, CPT1A, CROT, and HADHB are repressed, leading to reduced FAO and increased glycolysis, which polarizes macrophages to the inflammatory M1 phenotype. Conversely, when miR-33 levels are low, AMPK, CPT1A, CROT, and HADHB are no longer repressed, leading to increased FAO and polarization of macrophages to the M2 phenotype. M2 macrophages suppress inflammation through their production of antiinflammatory cytokines (e.g., IL-10) and retinoic acid, which can foster the differentiation of iTregs.
Discussion
Our study shows that miR-33 regulates macrophage cellular metabolism in a way that is intimately linked to the cell’s inflammatory phenotype, with important consequences on innate and adaptive immune responses. We demonstrate that by reducing FAO and related SRC in macrophages, miR-33 promotes an inflammatory M1-like macrophage phenotype that is associated with metabolic diseases such as atherosclerosis. Notably, inhibition of miR-33 metabolically reprograms macrophages to the M2 phenotype by upregulating FAO (via the master FAO switch AMPK as well as multiple enzymes that execute the process), which fuels alternatively activated macrophage responses involved in resolving inflammation and tissue repair (Figure 6G). In atherosclerosis, where macrophages are key integrators of inflammatory and metabolic signals that drive plaque progression, metabolic reprogramming by anti–miR-33 promoted the accumulation of M2 macrophages and FOXP3+ Tregs in plaques, decreased markers of systemic inflammation, and reduced plaque size. Importantly, we found that miR-33 exerts its effects on macrophage polarization and inflammation independently of its previously described roles in regulating macrophage cholesterol efflux and plasma HDL levels. Together, these data define what we believe to be a novel role for miR-33 in regulating the cellular pathways that underpin macrophage polarization and suggest that metabolic reprogramming of plaque macrophages to alternatively activated M2 cells by miR-33 inhibition promotes the resolution of atherosclerosis.
Recent studies indicate that metabolic changes in cells that participate in inflammation are critical to their function. Classically activated macrophages and dendritic cells as well as Th17 cells exhibit a shift toward aerobic glycolysis that accompanies their secretion of proinflammatory mediators and reactive oxygen species (8, 42, 43). Conversely, cells that limit inflammation, such as alternatively activated macrophages and Tregs, exhibit oxidative metabolism and the secretion of antiinflammatory cytokines (8, 43). Studies manipulating glycolytic and oxidative pathways have shown that these metabolic programs can control cellular inflammatory responses, revealing a close relationship between metabolism and immunity. Our data indicate that, while miR-33 is not required for macrophage polarization, it tempers macrophage M1/M2 polarization programs by altering the balance of cellular glycolysis/FAO. Consistent with this, we identify a major role for miR-33 targeting of AMPK, a master metabolic switch, in mediating its effects on macrophage inflammation. AMPK increases FAO via a 2-pronged mechanism: (a) inhibitory phosphorylation and inhibition of acetyl-CoA carboxylase, lowering malonyl-CoA–mediated inhibition of CPT1 and stimulating uptake of activated fatty acids into the mitochondria (44, 45), and (b) enhancement of expression of SIRT1 and PGC-1α, resulting in transcriptional activation of genes involved in mitochondrial FAO and mitochondrial biogenesis (46, 47). Mechanistically, we show that miR-33 overexpression reduces cellular oxygen consumption and increases extracellular acidification indicative of increased glycolysis, which is paralleled by an increase in markers of M1 macrophages (e.g., Il1b, Il6, Nos2) in WT, but not AMPK-deficient, macrophages. Furthermore, miR-33 silencing decreases ECARs and promotes FAO and the expression of M2 markers (e.g., Ym1, Cd206) only in AMPK-expressing macrophages, effects that could be reproduced by blocking glycolysis with 2-DG. Our findings are in agreement with studies showing that expression of a constitutively activated form of AMPK in macrophages can counterregulate inflammatory signaling, whereas siRNA targeting or genetic deletion of AMPK increases the expression of inflammatory cytokines in macrophages and obesity-induced insulin resistance (38, 48). However, it is possible that miR-33 targeting of other genes involved in fatty acid β-oxidation, including Crot, Cpt1a, and Hadhb (27–29), may also contribute to its effects on macrophage polarization by reinforcing the effects of AMPK. It is interesting to note that AMPK-dependent mechanisms have also been shown to contribute to differentiation of CD4+ T lymphocytes into inducible Tregs (iTregs), where AMPK acts to simultaneously reduce levels of the glucose transporter Glut1 and increase FAO (49). Finally, although previous studies have shown that free cholesterol enrichment of cellular membranes can enhance inflammatory signaling from lipid rafts, particularly TLR signaling and activation of NF-κB (35, 50, 51), we did not observe a role for miR-33 targeting of the cholesterol transporter ABCA1 in macrophage inflammatory polarization.
Our findings that miR-33 inhibition can promote M2 polarization independently of effects on ABCA1 provide a framework for understanding the HDL-independent mechanisms by which miR-33 inhibitors protect from atherosclerosis. miR-33 was one of the earliest identified miRNAs to regulate cholesterol homeostasis, and much of the research involving this miRNA has focused on its role in repressing cholesterol efflux and plasma levels of HDL-C by targeting ABCA1 (52). Studies in mice and nonhuman primates showed that miR-33 inhibitors increase plasma levels of HDL (24–26, 30–32), a lipoprotein whose plasma level in observational studies is inversely associated with cardiovascular risk, suggesting that it may hold value as a therapeutic target for atherosclerosis. Indeed, a previous study of Ldlr–/– mice with established atherosclerotic plaques that were treated with anti–miR-33 in conjunction with chow diet showed increased plasma levels of HDL-C and regression of atherosclerosis (31). Notably, that study was designed to simulate a clinical scenario in which plasma LDL-C levels were lowered in conjunction with anti–miR-33 treatment. However, 2 studies subsequently reported that the HDL-raising effects of anti–miR-33 in mice are blunted during Western diet feeding, presumably due to decreased hepatic expression of SREBF2/miR-33 (34, 39). Despite this, in a study by Rotllan et al. there were significant reductions in atherosclerotic lesion area in the aortas of Western diet–fed (0.3% cholesterol) Ldlr–/– mice treated with anti–miR-33 for 8 weeks (34), suggesting that anti–miR-33’s atheroprotective effects might not be entirely explained by its ability to increase HDL. Yet this conclusion remained controversial, as Marquart et al. found no differences in either plasma HDL-C or atherosclerosis progression in anti–miR-33 LNA-treated Ldlr–/– mice fed a high cholesterol–containing diet (1.25%) for 12 weeks (39). In agreement with Rotllan et al, we observed a 37% reduction in atherosclerosis in the aorta en face in Ldlr–/– mice treated with anti–miR-33 for 8 weeks on a Western diet (0.3% cholesterol) in the absence of changes in plasma levels of total or HDL cholesterol. Furthermore, we also observed similar reductions (33%) in atherosclerotic plaque size in cross-sections of the aortic root of anti–miR-33–treated mice, and gene expression profiling of plaque macrophages in this site revealed increased expression of genes indicating M2 polarization (Arg1, Mrc1, Fizz1/Retnla, YM1/Chi3l3). Immunofluorescent staining of plaques showed that macrophages that had taken up anti–miR-33, but not control oligonucleotides, had increased expression of the canonical M2 marker arginase 1, supporting our in vitro findings that anti–miR-33 directly polarizes macrophages to the M2 phenotype. In addition, anti–miR-33 may alter macrophage M2 polarization and decrease Western diet–induced inflammation in other tissues, such as the liver and adipose tissue, which may contribute to the observed reductions in plasma chemokine levels and systemic inflammation. That such an enrichment of M2 macrophages is sufficient to reduce atherosclerosis is supported by studies in which atherosclerotic mice treated systemically with M2-polarizing factors had attenuated atherosclerosis progression in the face of continued hyperlipidemia (21, 53).
Alternatively activated M2 macrophages have numerous inflammation-suppressive and tissue reparative functions that may contribute to reducing the chronic inflammation that promotes atherosclerotic plaque progression (13). As a result of high arginase 1 expression, M2 macrophages metabolize l-arginine to urea and l-ornithine, which sustain collagen synthesis and cell proliferation needed for tissue repair (54). In addition, M2 macrophages have high expression of MERTK, endowing these cells with an enhanced capacity for the clearance of apoptotic cells (55), a process that becomes defective in atherosclerotic plaques and contributes to necrotic core formation (56). M2 macrophages are also a source of inhibitory cytokines such as IL-10 and TGF-β, which help to resolve inflammation and have been shown to protect from atherosclerosis (57, 58). Moreover, of particular relevance to the present report, myeloid cells treated in vitro with M2-polarizing factors (e.g., IL-4, PPARγ agonists) and in vivo monocyte-derived M2 macrophages have recently been shown to express high levels of retinal dehydrogenase (ALDH1A2/RALDH), the rate-limiting enzyme for the production of retinoic acid, which, together with TGF-β, can promote the differentiation and expansion of a population of FoxP3+ Tregs known as iTregs (5–7, 59).
iTregs develop extrathymically in response to several soluble mediators, including TGF-β, IL-2, and retinoic acid, and in the gut, retinoic acid–mediated trafficking of T cells has been shown to be required for expansion of iTreg numbers (60). Indeed, we show that macrophages treated with anti–miR-33 have increased Aldh1a2 expression in vitro and in vivo and have an enhanced capacity to induce FOXP3 expression by naive CD4+ cells. Tregs exert immunosuppressive functions through their expression of IL-10 and TGF-β and have been shown to protect from atherosclerosis by suppressing effector Th1 and Th17 T cells, dendritic cell activation and migration, endothelial cell activation, and macrophage foam cell formation (reviewed in ref. 41). Impaired Treg numbers and function are found in patients with cardiovascular disease compared with healthy controls, and studies in mice indicate that Treg numbers in plaques decline with time of Western diet feeding, resulting in an accumulation of effector T cells that promote atherosclerosis (41). We observed a similar decline in plaque macrophage expression of Aldh1a2 with atherosclerosis progression, suggesting that the loss of this iTreg differentiation signal may contribute to the decline. While the mechanisms regulating Aldh1a2 expression in macrophages are poorly understood, we show that modulating cellular glycolysis and FAO programs alters Aldh1a2 mRNA levels, suggesting that this gene is under metabolic control. Notably, addition of exogenous retinoic acid has been shown to sustain iTreg differentiation under conditions that would normally oppose it (e.g., in the presence of IL-6 or in high costimulatory environments), and consistent with this, we observed an increase in aortic Foxp3 mRNA and FOXP3+ cells in plaques of anti–miR-33–treated mice under continued Western diet feeding. As Tregs also reduce macrophage inflammation and promote differentiation to the M2 phenotype, their increase in plaques of anti–miR-33–treated mice may initiate a positive feedback loop that further restores the imbalance of M1>M2 macrophages and Th1>Tregs to break the cycle of chronic inflammation.
In conclusion, we have shown that the macrophage phenotypic state can be controlled by miR-33 through the regulation of metabolic cues that determine M1/M2 polarization and that this in turn can increase numbers of Tregs, which would be expected to influence immunopathology. Although the source of iTregs in atherosclerotic plaques is unclear, we observed that anti–miR-33 treatment promoted the accumulation of FOXP3+ cells within both the adventitia and the plaque intima, suggesting that these cells may originate from adventitial tertiary lymphoid organs, which have been described in atherosclerotic mice (61). While we cannot exclude the possibility that anti–miR-33 might also have direct effects on T cell metabolism and polarization in vivo, on balance, our findings that anti–miR-33 colocalizes with M2 macrophage markers in plaques and is associated with increased plaque macrophage expression of Aldh1a2 (which promotes the expansion of FoxP3+ Tregs) suggest a macrophage-intrinsic effect. Given the protective effect of anti–miR-33 in atherosclerosis, silencing miR-33 may be an effective therapeutic approach in the treatment of other chronic inflammatory diseases in which M1 macrophages predominate.
Methods
Oligonucleotides.
The 2′-fluoro-methoxyethyl–modified control (TTATCGCCATGTCCAATGAGGCT) or anti–miR-33 (TGCAATGCAACTACAATGCAC) oligonucleotides were provided by Regulus Therapeutics and were characterized as described (30, 31).
Mice.
Ampkβ1–/– mice (Prkab1–/–, referred to as Ampk–/–), which lack more than 95% of all AMPK activity, were generated as previously described (38). Mir33–/– mice were generated as previously described (62). Abca1–/– mice were provided by Yves Marcel (University of Ottawa, Ottawa, Ontario, Canada). C57BL/6 and Ldlr–/– mice on a C57BL/6 background were from Jackson Laboratories. All mice were housed in a pathogen-free facility at an ambient temperature of 22–25°C. For studies of atherosclerosis, 8-week-old Ldlr–/– mice were randomized into 3 groups (n = 10) and treated with PBS (no treatment) or 2′-fluoro-methoxyethyl–modified control (TTATCGCCATGTCCAATGAGGCT) or anti–miR-33 (TGCAATGCAACTACAATGCAC) oligonucleotides concurrent with Western diet (21% [wt/wt] fat, 0.3% cholesterol; Research Diets) feeding for 8 weeks. Mice received 2 subcutaneous injections of control or anti–miR-33 oligonucleotides (10 mg/kg in PBS; provided by Regulus Therapeutics) or an equivalent volume of PBS the first week, spaced 2 days apart, and weekly injections of the treatment thereafter. At sacrifice, mice were anesthetized with isoflurane, exsanguinated by cardiac puncture, and perfused with PBS, followed by 10% sucrose in PBS. The heart was separated from the aorta at the root and embedded in OCT medium and snap-frozen. The aorta was stored in paraformaldehyde until analysis. Additional tissue samples were snap-frozen in liquid nitrogen and stored at –80°C for analysis of RNA.
Plasma analysis.
Plasma was collected at baseline by tail vein bleeding and at sacrifice by cardiac puncture. Total cholesterol was assayed using the Cholesterol-E Kit (Wako Chemicals) as described (25). For HDL measurements, apoB-containing lipoproteins were precipitated by the phosphotungstate-magnesium method, and HDL-C was measured using the HDL Cholesterol Kit (Wako) (25). Triglycerides were measured using the L-type enzymatic triglyceride assay (Wako). Analysis of plasma chemokines and apoA-I was performed by Myriad RBM using the rodent Multi-Analyte Profile.
Atherosclerosis analysis.
Hearts were sectioned through the aortic root (6 μm) and stained with H&E for lesion quantification or used for immunohistochemical analysis as previously described (63). For morphometric analysis of lesions, 6 sections per mouse were imaged (Nikon Eclipse) with Image-Pro Plus, spanning the entire aortic root, and lesions and necrotic area were quantified using iVision software. For collagen analysis, 6 sections per mouse were stained with picrosirius red and imaged under polarized light using a Zeiss Axioplan microscope. For detection of neutral lipid, oil red O staining was performed as previously described (63, 64). For macrophage analysis, 6 sections per mouse were incubated with an anti-CD68 antibody (AbD Serotec) and antibody reactivity was visualized using the Vectastain ABC Alkaline Phosphatase Kit (Vector Laboratories) and the Vector Red Substrate Kit (Vector Laboratories). For costaining of 2′F/MOE oligonucleotides with macrophage markers, frozen sections were fixed in acetone and blocked with 5% BSA (MP Biomedicals LLC), followed by incubation with primary antibodies raised against the phosphorothioate backbone of the 2′F/MOE oligonucleotides (Regulus Therapeutics) and arginase I (Santa Cruz Biotechnology Inc.) or CD68 antibodies. Fluorophore-conjugated secondary antibodies (Invitrogen) were used along with DAPI (Sigma Aldrich) and visualized under a fluorescence microscope (Zeiss Axiovert 200 M with Axio Vision 4.8).
Gene profiling of plaque macrophages.
LCM was performed as previously described using the Leica DM-6000B instrument and Leica LMD CC7000 camera (Leica Microsystems) (65). A guide slide for each animal was prepared by staining CD68+ macrophages, and the CD68+ areas in serial sections were collected by LCM. RNA was extracted from CD68+ plaque macrophages using the Arcturus Picopure RNA Isolation Kit (Life Technologies). Total RNA was amplified using the Ovation WT Pico Amp Kit (NuGen), purified using the QIAquick PCR Purification Kit (QIAGEN), and used for quantitative PCR as described below; alternatively, 2 ng of unamplified total RNA was hybridized to the Nanostring nCounter GX Mouse Inflammation gene code set (Nanostring Technologies) using the single cell method.
Cell culture.
Peritoneal macrophages were isolated from mice by peritoneal lavage 4 days after i.p. injection of 1 ml of 3% thioglycollate, as previously described (66). Cells were cultured in DMEM with 10% FBS and 20% L929-conditioned medium. Alternatively, mice were injected i.p. with 1 ml of 3% thioglycollate and 2 days later were injected with 10 mg/kg of antisense miR-33 or mismatch control (Regulus Therapeutics) 3 days prior to macrophage isolation. BMDMs were prepared by flushing the marrow from the tibiae and femorae of 6- to 8-week-old mice. Cells were differentiated into macrophages in DMEM media supplemented with 10% FBS, 1% penicillin/streptomycin (P/S) (Life Technologies), and 15% L929-conditioned media for 7 days. For experiments with miRNA mimics or inhibitors, macrophages were transfected overnight with 80 to 120 nM miRIDIAN miRNA mimics (miR-33 mimic) or inhibitors (anti–miR-33; Dharmacon) or with antisense oligonucleotides against miR-33 (Regulus Therapeutics) using Lipofectamine RNAiMax Transfection Reagent (Invitrogen) and analyzed 48 hours after transfection. All experimental control samples were treated with an equal concentration of a nontargeting control mimic or inhibitor sequence for use as controls for nonsequence-specific effects in the miRNA overexpression or inhibition experiments. For macrophage activation experiments, BMDMs were stimulated for 24 hours with ultrapure lipopolysaccharide (1 μg/ml; Invivogen) plus IFN-γ (50 ng/ml; R&D Systems) for M1 activation, with IL-4 (20 ng/ml; PeproTech) for M2 activation. In some experiments, BMDMs were treated with etomoxir (100 μM, Sigma-Aldrich) to inhibit FAO and promote M1 activation or with 2-DG (1 mM, Seahorse Bioscience) to inhibit glycolysis and promote M2 activation.
RNA isolation and quantitative PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen) and Direct-zol RNA MiniPrep columns (Zymo Research) and quantified using Nanodrop 2000 software (Nanodrop Products). For mRNA quantification, cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative real-time PCR was performed in triplicate using KAPA SYBR FAST Universal 2× qPCR Master Mix (Kapa Biosystems) on the iCycler Real-Time Detection System (Bio-Rad). The mRNA levels were normalized to Gapdh as a housekeeping gene. The following primer sequences were used: Abca1, 5′-AAAACCGCAGACATCCTTCAG-3′ and 5′-CATACCGAAACTCGTTCACCC-3′; Gapdh, 5′-TGTGAGGGAGATGCTCAGTG-3′ and 5′TGTTCCTACCCCCAATGTGT-3′; Prkaa1, 5′-GTCAAAGCCGACCCAATGATA-3′ and 5′-CGTACACGCAAATAATAGGGGTT-3′; Nos2, 5′-GTTCTCAGCCCAACAATACAAGA-3′ and 5′-GTGGACGGGTCGATGTCAC-3′; Il1b, 5′-GCAACTGTTCCTGAACTCAACT-3′ and 5′-ATCTTTTGGGGTCCGTCAACT-3′; Il6, 5′-TAGTCCTTCCTACCCCAATTTCC-3′ and 5′-TTGGTCCTTAGCCACTCCTTC-3′; Arg1, 5′-CTCCAAGCCAAAGTCCTTAGAG-3′ and 5′-AGGAGCTGTCATTAGGGACATC-3′; Retnla, 5′-CTGGGTTCTCCACCTCTTCA-3′ and 5′-TGCTGGGATGACTGCTACTG-3′; Mrc1, 5′-CTCTGTTCAGCTATTGGACGC-3′ and 5′-CGGAATTTCTGGGATTCAGCTTC-3′; Chi3l3, 5′-GTCTTGCTCATGTGTGTAAGTGA-3′ and 5′-CAGGTCTGGCAATTCTTCTGAA-3′
miRNA quantification.
For mature miRNA quantification, the miScript II RT Kit (QIAGEN) was used for reverse-transcription reactions, using HiSpec Buffer, according to the manufacturer’s protocol. miR-33 and miR-33* miScript miRNA mimics were used to generate a standard curve of Ct values (y axis) against log copy number (x axis), according to the instructions in QIAGEN’s miScript PCR System Handbook, to ensure equivalent miR-33 and miR-33* primer efficiencies and for absolute quantification. miScript Primer Assays and miScript SYBR Green PCR were used on the iCycler Real-Time Detection System (Bio-Rad). For relative quantification, miRNA levels were normalized to RNU6-2 miScript PCR control (QIAGEN).
3′ UTR luciferase reporter assays.
The mouse Arg1 (MmiT026580) 3′ UTR target clone was purchased from GeneCopoeia. Point mutations in the predicted miR-33 site within the 3′ UTR of mouse Arg1 were generated using Multisite-Quickchange (Stratagene) according to the manufacturer’s protocol. HEK293T cells (ATCC) were plated in antibiotic-free media in 96-well plates and cotransfected with 0.2 μg of 3′ UTR luciferase reporter vectors and the miR-33 mimic or negative control mimic (Dharmacon) utilizing Lipofectamine 2000 (Invitrogen). Luciferase activity was measured using the Secrete-Pair Dual Luminescence Assay Kit (GeneCopoeia). Renilla luciferase activity was normalized to the corresponding firefly luciferase activity and plotted as fold-change relative to control. Experiments were performed in quadruplicate wells of a 96-well plate and repeated 3 times.
Seahorse extracellular flux analysis.
BMDMs were seeded into XF24 cell culture microplates (Seahorse Bioscience) and transfected with anti–miR-33 or control anti-miR as described above, and 48 hours later, OCRs and ECARs were quantified using the XFe24 instrument (Seahorse Bioscience) according to the manufacturer’s protocol. For glycolysis assessments, the XF Glycolysis Stress Test Kit was used according to the manufacturer’s protocol. For cellular FAO quantification, the OCR was measured following exposure to BSA-conjugated palmitate (200 μM) in Krebs-Henseleit buffer (KHB; 111 mM NaCl, 4.7 mM KCl, 2 mM MgSo4, 1.2 mM Na2HPO4, 2.5 mM glucose, 0.5 mM carnitine) using the XFe24 instrument and the XF Cell Mito Stress Test kit.
FAO analysis.
1 × 106 BMDMs were cultured in 12-well plates and transfected with control or miR-33 miRNA inhibitors. Cells were incubated for 3 hours with 0.3% BSA/100 μM oleic acid/0.4 μCi/ml 1-14C-oleic acid (PerkinElmer) in DMEM, in the presence or absence of etomoxir (25 μM, Sigma-Aldrich), as previously described (67). Oxidation was stopped by the addition of 80 μl of 50% TCA per well, and 14CO2 was trapped on Whatman paper disks soaked with 20 μl of 1M NaOH for 1 hour in a fume hood, after which trapped 14CO2 was measured by liquid scintillation counting.
Flow cytometry.
Cells were stained with LIVE/DEAD Blue (Invitrogen), blocked with 4 μg/ml aCD16/32 (2.4G2; Bioxcell), and stained with the cell surface markers CD11B eFluor450 (eBioscience), F4/80 PE-Cy7 (eBioscience), Siglec-F BV421 (BD Biosciences), CD4 BV650 (BioLegend), and CD25 PB (BioLegend). For intracellular nuclear staining of FOXP3, cells were washed with PhosFlow permeabilization buffer (BD Biosciences) and blocked with anti-mouse CD16/32 following surface staining with antibodies against CD4 and CD25, and stained with a FOXP3 antibody (PE-CF594, BD Biosciences) for 1 hour at 4°C in Foxp3/Transcription Factor Fixation/Permeabilization Concentrate and Diluent (eBioscience) as previously described (5). ALDH activity was measured using the ALDEFLUOR staining kit (StemCell Technologies) in the presence of the ALDH inhibitor DEAB (15 μM), as previously described (5, 6). In this assay, BODIPY aminoacetaldehyde, the nontoxic substrate for ALDH that is able to diffuse freely into cells, is converted by ALDH into BODIPY aminoacetate, a fluorescent product that is retained in cells and can be detected using flow cytometry. Cells were acquired using an LSR II (BD Biosciences) and analyzed using FlowJo software (Treestar). FOXP3+ Tregs were quantified by using the following gating strategy: singlets, live, CD4+, and FOXP3+ cells were selected. To quantify macrophage ALDH activity, macrophages were gated as Siglec-F negative, CD11B+, and F4/80+, and the ALDHhi macrophage population was considered as ALDH+.
Treg differentiation assay.
Naive T cells were isolated from the spleens and lymph nodes of C57BL/6 mice using the Naive CD4+ T Cell Isolation Kit II (Miltenyi Biotec) and were cultured together with macrophages as previously described (6). 4 × 105 naive T cells were cocultured with 4 × 105 peritoneal macrophages that were transfected with control anti-miR or anti–miR-33 at 48 hours prior to the coculture. Cells were cultured in complete RPMI containing soluble anti-CD3 (1 μg/ml), soluble anti-CD28 (1 μg/ml), and recombinant human IL-2 (5 ng/ml) for 6 days. The cocultures were supplemented with fresh medium and IL-2 on day 3. Retinoic acid (100 nM) was added to some culture wells. Cells were harvested on day 6 and stained for FOXP3 by intracellular nuclear staining as described above.
Western blots.
Protein was extracted in RIPA buffer with protease and phosphatase inhibitors, and samples (60 μg/well) were electrophoresed on 8% SDS-polyacrylamide gels and transferred to nitrocellulose membranes at 125 V for 2 hours. Membranes were incubated overnight with specified antibodies (AMPKα [catalog 2603] and phospho-AMPα Thr172 [catalog 2535] were purchased from Cell Signaling Technology, and tubulin [catalog T6074] was obtained from Sigma-Aldrich), and proteins were visualized using appropriate secondary antibodies conjugated to IR dyes (Rockland) and scanned using the Odyssey Imaging System (Licor).
Statistics.
For atherosclerosis and immunohistochemical analyses, all comparisons were made using 1-way ANOVA (P ≤ 0.05), and data are expressed as mean ± SEM, unless otherwise noted. Other comparisons were made using 2-tailed Student’s t test.
Study approval.
All studies in animals were approved by the NYU School of Medicine Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We would like to acknowledge Regulus Therapeutics for providing the control and anti–miR-33 oligonucleotides and the NYU Langone Medical Center Immune Monitoring Core for the use of the XFe24 Extracellular Flux Analyzer (supported by NYU-HHC CTSI grant UL1 TR000038 and the NYU Cancer Institute’s Cancer Center support grant P30CA016087). This work was supported by the NIH (R01HL108182 to K.J. Moore; R01HL119047 to K.J. Moore and K.J. Rayner; R01AI093811 and R01AI094166 to P. Loke; HL098055 and HL084312 to E.A. Fisher), the Canadian Institutes of Health Research (postdoctoral fellowship to M. Ouimet), and the American Heart Association (15POST25090199 to M. Ouimet; 13PRE14190001 to H.N. Ediriweera; 14POST20180018 to C. van Solingen). G.R. Steinberg is a Canada Research Chair and the J. Bruce Duncan Chair in Metabolic Diseases.
Footnotes
Conflict of interest: Gregory R. Steinberg has received more than $10,000 and research support from Esperion Therapeutics and is an inventor on patent application WO 2014124523 A1, A method of treating obesity. Edward A. Fisher and Katey J. Moore receive research support from Merck Research Laboratories for work on chronic kidney disease.
Reference information:J Clin Invest. 2015;125(12):4334–4348. doi:10.1172/JCI81676.
References
- 1.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11(11):762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496(7446):445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gause WC, Wynn TA, Allen JE. Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat Rev Immunol. 2013;13(8):607–614. doi: 10.1038/nri3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annu Rev Immunol. 2014;32:609–634. doi: 10.1146/annurev-immunol-032713-120236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broadhurst MJ, et al. Upregulation of retinal dehydrogenase 2 in alternatively activated macrophages during retinoid-dependent type-2 immunity to helminth infection in mice. PLoS Pathog. 2012;8(8): doi: 10.1371/journal.ppat.1002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gundra UM, et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood. 2014;123(20):e110–e122. doi: 10.1182/blood-2013-08-520619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall JA, Grainger JR, Spencer SP, Belkaid Y. The role of retinoic acid in tolerance and immunity. Immunity. 2011;35(1):13–22. doi: 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Prados JC, et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol. 2010;185(1):605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 9.Vats D, et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 2006;4(1):13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freemerman AJ, et al. Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J Biol Chem. 2014;289(11):7884–7896. doi: 10.1074/jbc.M113.522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496(7444):238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang SC, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15(9):846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13(10):709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chinetti-Gbaguidi G, et al. Human atherosclerotic plaque alternative macrophages display low cholesterol handling but high phagocytosis because of distinct activities of the PPARγ and LXRα pathways. Circ Res. 2011;108(8):985–995. doi: 10.1161/CIRCRESAHA.110.233775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buono C, Come CE, Stavrakis G, Maguire GF, Connelly PW, Lichtman AH. Influence of interferon-γ on the extent and phenotype of diet-induced atherosclerosis in the LDLR-deficient mouse. Arterioscler Thromb Vasc Biol. 2003;23(3):454–460. doi: 10.1161/01.ATV.0000059419.11002.6E. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S, Pablo AM, Jiang X, Wang N, Tall AR, Schindler C. IFN-γ potentiates atherosclerosis in ApoE knock-out mice. J Clin Invest. 1997;99(11):2752–2761. doi: 10.1172/JCI119465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitman SC, Ravisankar P, Elam H, Daugherty A. Exogenous interferon-γ enhances atherosclerosis in apolipoprotein E–/– mice. Am J Pathol. 2000;157(6):1819–1824. doi: 10.1016/S0002-9440(10)64820-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanna RN, et al. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ Res. 2012;110(3):416–427. doi: 10.1161/CIRCRESAHA.111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamers AA, et al. Bone marrow-specific deficiency of nuclear receptor Nur77 enhances atherosclerosis. Circ Res. 2012;110(3):428–438. doi: 10.1161/CIRCRESAHA.111.260760. [DOI] [PubMed] [Google Scholar]
- 20.Liao X, et al. Kruppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121(7):2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardilo-Reis L, et al. Interleukin-13 protects from atherosclerosis and modulates plaque composition by skewing the macrophage phenotype. EMBO Mol Med. 2012;4(10):1072–1086. doi: 10.1002/emmm.201201374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feig JE, et al. Reversal of hyperlipidemia with a genetic switch favorably affects the content and inflammatory state of macrophages in atherosclerotic plaques. Circulation. 2011;123(9):989–998. doi: 10.1161/CIRCULATIONAHA.110.984146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feig JE, et al. HDL promotes rapid atherosclerosis regression in mice and alters inflammatory properties of plaque monocyte-derived cells. Proc Natl Acad Sci U S A. 2011;108(17):7166–7171. doi: 10.1073/pnas.1016086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107(27):12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328(5985):1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davalos A, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108(22):9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerin I, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285(44):33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rottiers V, et al. MicroRNAs in metabolism and metabolic diseases. Cold Spring Harb Symp Quant Biol. 2011;76:225–233. doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rayner K, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rayner KJ, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121(7):2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rottiers V, et al. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med. 2013;5(212): doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horie T, et al. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE–/– mice. J Am Heart Assoc. 2012;1(6): doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr–/– mice — brief report. Arterioscler Thromb Vasc Biol. 2013;33(8):1973–1977. doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yvan-Charvet L, et al. Increased inflammatory gene expression in ABC transporter-deficient macrophages: free cholesterol accumulation, increased signaling via toll-like receptors, and neutrophil infiltration of atherosclerotic lesions. Circulation. 2008;118(18):1837–1847. doi: 10.1161/CIRCULATIONAHA.108.793869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu X, et al. Increased cellular free cholesterol in macrophage-specific Abca1 knock-out mice enhances pro-inflammatory response of macrophages. J Biol Chem. 2008;283(34):22930–22941. doi: 10.1074/jbc.M801408200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galic S, et al. Hematopoietic AMPK beta1 reduces mouse adipose tissue macrophage inflammation and insulin resistance in obesity. J Clin Invest. 2011;121(12):4903–4915. doi: 10.1172/JCI58577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marquart TJ, Wu J, Lusis AJ, Baldan A. Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2013;33(3):455–458. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peled M, Fisher EA. Dynamic aspects of macrophage polarization during atherosclerosis progression and regression. Front Immunol. 2014;5(doi): doi: 10.3389/fimmu.2014.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foks AC, Lichtman AH, Kuiper J. Treating atherosclerosis with regulatory T cells. Arterioscler Thromb Vasc Biol. 2015;35(2):280–287. doi: 10.1161/ATVBAHA.114.303568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krawczyk CM, et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115(23):4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi LZ, et al. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fullerton MD, et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med. 2013;19(12):1649–1654. doi: 10.1038/nm.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGarry JD, Leatherman GF, Foster DW. Carnitine palmitoyltransferase I. The site of inhibition of hepatic fatty acid oxidation by malonyl-CoA. J Biol Chem. 1978;253(12):4128–4136. [PubMed] [Google Scholar]
- 46.Lin JD, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 47.O’Neill HM, Holloway GP, Steinberg GR. AMPK regulation of fatty acid metabolism and mitochondrial biogenesis: implications for obesity. Mol Cell Endocrinol. 2013;366(2):135–151. doi: 10.1016/j.mce.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 48.Sag D, Carling D, Stout RD, Suttles J. Adenosine 5′-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J Immunol. 2008;181(12):8633–8641. doi: 10.4049/jimmunol.181.12.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michalek RD, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu X, et al. Macrophage ABCA1 reduces MyD88-dependent Toll-like receptor trafficking to lipid rafts by reduction of lipid raft cholesterol. J Lipid Res. 2010;51(11):3196–3206. doi: 10.1194/jlr.M006486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mogilenko DA, et al. Endogenous apolipoprotein A-I stabilizes ATP-binding cassette transporter A1 and modulates Toll-like receptor 4 signaling in human macrophages. FASEB J. 2012;26(5):2019–2030. doi: 10.1096/fj.11-193946. [DOI] [PubMed] [Google Scholar]
- 52.Rayner KJ, Moore KJ. MicroRNA control of high-density lipoprotein metabolism and function. Circ Res. 2014;114(1):183–192. doi: 10.1161/CIRCRESAHA.114.300645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfs IM, et al. Reprogramming macrophages to an anti-inflammatory phenotype by helminth antigens reduces murine atherosclerosis. FASEB J. 2014;28(1):288–299. doi: 10.1096/fj.13-235911. [DOI] [PubMed] [Google Scholar]
- 54.Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annu Rev Pathol. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zizzo G, Hilliard BA, Monestier M, Cohen PL. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J Immunol. 2012;189(7):3508–3520. doi: 10.4049/jimmunol.1200662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145(3):341–355. doi: 10.1016/j.cell.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Von Der Thusen JH, Kuiper J, Fekkes ML, De Vos P, Van Berkel TJ, Biessen EA. Attenuation of atherogenesis by systemic and local adenovirus-mediated gene transfer of interleukin-10 in LDLr–/– mice. FASEB J. 2001;15(14):2730–2732. doi: 10.1096/fj.01-0483fje. [DOI] [PubMed] [Google Scholar]
- 58.Robertson AK, Rudling M, Zhou X, Gorelik L, Flavell RA, Hansson GK. Disruption of TGF-β signaling in T cells accelerates atherosclerosis. J Clin Invest. 2003;112(9):1342–1350. doi: 10.1172/JCI18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szatmari I, et al. PPARγ controls CD1d expression by turning on retinoic acid synthesis in developing human dendritic cells. J Exp Med. 2006;203(10):2351–2362. doi: 10.1084/jem.20060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hadis U, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34(2):237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 61.Grabner R, et al. Lymphotoxin β receptor signaling promotes tertiary lymphoid organogenesis in the aorta adventitia of aged ApoE–/– mice. J Exp Med. 2009;206(1):233–248. doi: 10.1084/jem.20080752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horie T, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107(40):17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore KJ, et al. Loss of receptor-mediated lipid uptake via scavenger receptor A or CD36 pathways does not ameliorate atherosclerosis in hyperlipidemic mice. J Clin Invest. 2005;115(8):2192–2201. doi: 10.1172/JCI24061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manning-Tobin JJ, et al. Loss of SR-A and CD36 activity reduces atherosclerotic lesion complexity without abrogating foam cell formation in hyperlipidemic mice. Arterioscler Thromb Vasc Biol. 2009;29(1):19–26. doi: 10.1161/ATVBAHA.108.176644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trogan E, Choudhury RP, Dansky HM, Rong JX, Breslow JL, Fisher EA. Laser capture microdissection analysis of gene expression in macrophages from atherosclerotic lesions of apolipoprotein E-deficient mice. Proc Natl Acad Sci U S A. 2002;99(4):2234–2239. doi: 10.1073/pnas.042683999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallily R, Feldman M. The role of macrophages in the induction of antibody in x-irradiated animals. Immunology. 1967;12(2):197–206. [PMC free article] [PubMed] [Google Scholar]
- 67.Huynh FK, Green MF, Koves TR, Hirschey MD. Measurement of fatty acid oxidation rates in animal tissues and cell lines. Methods Enzymol. 2014;542:391–405. doi: 10.1016/B978-0-12-416618-9.00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.