Abstract
Background
The incidence of tuberculosis (TB) remains high among Chinese Uygurs (a long-dwelling ethnic minority in Xinjiang) in China and the variants in IL-23R likely contribute to individual’s diversity in host response during infection.
Methods
A hospital based one to one matched case–control study was performed to assess the role of single nucleotide polymorphisms (SNPs) and copy number variation (CNV) of IL-23R in susceptibility and clinical features of pulmonary TB in Chinese Uygurs. Thirteen SNPs in IL-23R were genotyped by multiplex SNaPshot and a CNV was analyzed using Taqman real-time PCR in 250 pairs of pulmonary TB patients and controls.
Results
The SNP rs7518660 (OR = 4.78, 95 % CI 3.14–8.52) and the CNV in IL23R (OR = 2.75, 95 % CI 1.51–4.98) were significantly associated with susceptibility to pulmonary TB. The SNP rs11465802 (OR = 3.23, 95 % CI 1.85–5.62) was significantly associated with drug-resistance and the SNP rs1884444 (OR = 3.61, 95 % CI 1.90–6.85) was significantly related to cavitary lesion in Chinese Uygurs.
Conclusions
Our study shows for the first time that SNP and CNV in IL23R were associated with susceptibility, drug resistance and cavity formation of pulmonary TB. Our findings indicate that these IL-23R polymorphisms may be considered as risk factors for active pulmonary TB and its severe clinical forms.
Electronic supplementary material
The online version of this article (doi:10.1186/s12879-015-1284-2) contains supplementary material, which is available to authorized users.
Keywords: IL23 receptor, Tuberculosis, Susceptibility, SNP, Copy number variation, Drug-resistant, Cavitary lesion
Background
China has the world’s second largest tuberculosis (TB) burden, accounting for 12 % of the global total cases in 2012 [1], and the Xinjiang Uygur Autonous Region (Xinjiang) in northwestern China, has one of the highest rates of incidence and mortality of TB in China [2–4]. The Chinese Uygur, one of the minority ethnic groups, has higher prevalence of TB than the Chinese Han and other Chinese minorities in Xinjiang, China [3], indicating that this group of people may be susceptible to TB.
The essential role of human genetic factors in susceptibility to TB has been confirmed by twin studies [5], linkage analysis [6], candidate gene association analysis (reviewed in [7]) and genome-wide association studies (GWAS) [8]. However, cumulative evidence suggests that there is an ethnic difference in the polymorphisms of candidate genes associated with TB susceptibility [9, 10]. Thus, identification of variants of the genes linked with TB susceptibility is still crucial for understanding both the mechanism underlying TB susceptibility and gene function.
IL-23, a member of the IL-12 cytokine family, is a key proinflammatory cytokine in innate and adaptive immune systems. This cytokine plays a pivotal role in the differentiation of the native CD4+ T cells into Th17 [11]. It has been shown that IL-17, mostly produced by Th17 cells, plays a crucial role in control of M. tuberculosis infection [12]. Furthermore, IL-23 is required for long-term control of M. tuberculosis [13]. The activity of IL-23 is mediated by its binding to IL23R complex, which is composed of IL-12Rβ1 and IL-23R [14]. Recent studies have shown that IL23R might also affect other dependent cytokines, such as IL-1, IL-6 and tumor necrosis factor (TNF) [15]. These cytokines are also critical for the progression of tuberculosis [7, 16]. However, there has been only one study showing one single nucleotide polymorphism (SNP) in IL-23R was associated with susceptibility to TB [17]. In addition, IL23R exhibits a copy number variation (CNV) [18, 19], which could contribute to individual’s diversity in host response during infection and inflammation, and is possibly associated with TB.
A hospital based one to one matched case–control study was conducted to determine whether the SNPs and CNV of IL23R are associated with susceptibility and clinical forms of pulmonary TB in Chinese Uygurs.
Methods
Study population
All participants were from Kashgar Prefecture, Xinjiang, China. The area was selected due to the high incidence of TB as well as the uniform ethnicity (with common genetic background of Chinese Uygur [20]) and socioeconomic status. The ethnicity recording of all the participants as Chinese Uygur were derived from their self-declaration and checked by their identity (ID) card. 250 unrelated Uygur patients diagnosed with active pulmonary TB at Kashgar Prefecture Chest Hospital were enrolled from October 2013 to April 2014. Criteria for inclusion as pulmonary TB cases were: 1) positive sputum culture; 2) age range 18-75 years; 3) excluding those with comorbidities such as lung carcinoma, asthma, bronchiectasis, diabetes mellitus and other immunosuppressive conditions. Patients meeting all the criteria were recruited. An equal number of age- (within 3 years) and sex-matched Uygur healthy controls from the Health Checkup Center of Kashgar Prefecture People’s Hospital were enrolled during the same period. Those controls were negative both for history of TB and T. SPOT. TB assay. All participants had BCG vaccination and were HIV seronegative.
The protocol was approved by the Ethics Committee of The First Affiliated Hospital of Xinjiang Medical University, Urumqi, China. Written informed consents were obtained from all participants.
Clinical features of pulmonary TB including drug resistance of the strain, elapsed time to sputum culture conversion, pulmonary infiltration and cavitary lesion were reviewed as well as age, gender, body measure index (BMI), smoking status and education background possibly to be risk factors for developing pulmonary TB. The culture, identification and drug sensitivity test of M. tuberculosis strains were performed according to the national criteria in China [21]. Four first-line anti-TB drugs (isoniazid, rifampicin, ethambutol, and streptomycin) and seven second-line anti-TB drugs (Paminosalicylicacid, Ofloxacin, Capreomycin, Amikacin, Kanamycin, Ethionamide, and Cycloserine) were adopted to detect drug resistance, as described elsewhere [22]. Drug sensitivity was defined as sensitivity to all the tested drugs and drug resistance as resistance to at least one tested drug, as described elsewhere [23]. Pulmonary infiltration and cavity were evaluated on the basis of chest CT scanning obtained at the beginning of the treatment, which was read by independent physician not affiliated with the study. Pulmonary infiltration on chest CT scanning was divided into two categories, one involving one third or less of one lung, and the other involving more than that. The commercially available T. SPOT. TB assay (Oxford Immunotec, Abingdon, UK) was performed to exclude latent M. tuberculosis infection (LTBI) in controls, due to some candidate genes being related to TB as well as LTBI [24].
Genomic DNA extraction and marker selection
Genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) of the samples with the TIANamp blood Genomic DNA kit (Tiangen, Beijing, China).
To confirm SNPs in Chinese Uygurs, we initially sequenced all 11 exons of IL-23R using ABI 3730 DNA Analyzer (Additional file 1: Figure S1A) in 10 pairs of samples, with each exon flanked with partial intronic sequences. The primers were designed by Primer 5.0 software (Additional file 2: Table S1). Thirteen common SNPs were identified in those regions and all were selected for further genotyping (Additional file 3: Table S2). The CNV in exon 11 of IL23R demonstrated previously [19] was selected for further analysis.
SNP genotyping and copy number measurement
SNP genotyping was performed by the multiplex SNaPshot technique (Additional file 1: Figure S1B). In brief, the primers and probes were designed by Primer 5.0 software (Additional file 4: Table S3). Multiplex PCR products amplified by HotStarTaq (Qiagen) were purified with shrimp alkaline phosphatase (SAP) (Promega, Madison, USA) and Exonuclease I (ExoI) (Epicentre, Madison, USA). The purified Multiplex PCR products were then used as templates for a minisequencing extension reaction using the SNaPshot Multiplex kit (Applied Biosystems, Carlsbad, CA, United States). The SNaPshot PCR products were also purified by SAP, and then detected by capillary electrophoresis on an ABI 3130XL DNA Analyzer (Applied Biosystems). The call rate per SNP exceeded 98 %. The CNV in IL-23R of all participants was determined by Taqman real-time PCR, as described elsewhere [19]. The call rate for copy number measurement reached 100 %. Furthermore, Accucopy assay [25] and droplet digital PCR (ddPCR) [26] were used to validate the copy number of 25 pairs randomly selected from those tested samples (Additional file 5: Figure S2 and Additional file 6: Figure S3), which account for 10 % of the total samples.
Statistical analysis
Continuous variables were compared using the Student’s t test. Hardy–Weinberg equilibrium (HWE) was assessed for all SNPs by χ2 goodness of fit test or Fisher’s exact test. SNPs that deviated from the HWE (P < 0.05) or had frequencies of variants of < 5 % in the control group were excluded. Linkage disequilibrium blocks were structured and haplotype frequencies were calculated using Haploview version 4.2 (Daly Lab at the Broad Institute Cambridge, MA, United States). The Linkage Disequilibrium (LD) between SNPs and CNV was determined both at the genotype and haplotype levels in the control group, also using Haploview version 4.2, as described elsewhere [27]. The associations between pulmonary TB and Il-23R polymorphisms were calculated by conditional logistic regression analysis, for three different genetic models (dominant, recessive and additive), then adjusted by covariants for all analyses. The association of each polymorphism with pulmonary TB clinical forms was confirmed by unconditional logistic regression. Bonferroni correction was applied for multiple testing. All P values reported were two-tailed, and a level of 0.05 was considered statistically significant. Statistical analysis was performed using SAS version 9.2 (SAS institute, Cary, NC, United States).
Results
This study consisted of 250 pulmonary TB cases and 250 controls, matched with age and sex. Males accounted for 55.6 % and females 44.4 % in each group. The age was 44.34 ± 14.17 years for cases and 47.34 ± 16.20 years for controls. Among those 250 pulmonary TB patients, there were 172 drug-sensitive patients and 78 drug-resistant ones. In those 78 drug-resistant pulmonary TB patients, there were 10 multidrug-resistant tuberculosis (MDR). There was no significant difference in age, gender, smoking, alcohol intake, presence of TB history of houshold or education background between pulmonary TB patients and controls. As expected, BMI showed significant difference between these two groups, demonstrating that thin individuals (BMI < 18.5) (OR = 3.93, 95 % CI 2.71–5.71) are more likely to suffer from pulmonary TB in Chinese Uygurs (Table 1).
Table 1.
Basic characteristics of the participants in the study
| Characteristics | Controls (N = 250) | PTB Cases (N = 250) | OR (95 % CI) |
|---|---|---|---|
| Age, year | |||
| Mean ± SD | 47.34 ± 16.20 | 44.34 ± 14.17 | |
| Gender | |||
| Male : Female | 139:111 | 139:111 | |
| BMI | |||
| <18.5 : ≥18.5 | 92:158 | 174:76 | 3.93 (2.71–5.71) |
| Smoking status | |||
| Smoker : No-smoker | 9:241 | 5:245 | 0.55 (0.18–1.65) |
| Alcohol intake status | |||
| Drinker : No-drinker | 12:238 | 8:242 | 0.66 (0.26–1.63) |
| Presence of TB history of houshold | |||
| Presence : Absence | 24:226 | 36:224 | 1.51 (0.87–2.62) |
| Education background | |||
| Junior high school or below : Senior high school or above | 116:134 | 124:126 | 1.14 (0.80–1.62) |
PTB pulmonary tuberculosis, OR odds ratio, CI confidence interval, SD standard deviation, BMI body measure index
The ethnicity recording of the study population as Chinese Uygur were derived from their self-declaration and checked by their identity (ID) card
Association of the IL-23R genotype and allele frequencies with pulmonary tuberculosis
Genotype frequencies for 9 SNPs (rs1884444, rs11465770, rs10889664, rs11465788, rs7518660, rs11465802, rs11465804, rs11209016 and rs10889677) included were in HWE in both case and control groups (P > 0.05), whereas other 4 SNPs (rs6687620, rs2863212, rs7530511 and rs10889671) were excluded, due to their deviation from HWE (P < 0.05) in the control group. Of the 9 SNPs included initially, 3 SNPs (rs11465770, rs11465804 and rs11209026) were also excluded, because of their frequencies of variants of < 5 % in the controls. Finally, 6 SNPs (rs1884444, rs10889664, rs11465788, rs7518660, rs11465802 and rs10889677) were included for further analysis.
The SNP rs7518660 (AA vs GG, OR = 12.73, 95 % CI 7.16–22.6; AG vs GG, OR = 2.99, 95 % CI 1.87–4.79; AA + AG vs GG, OR = 4.78, 95 % CI 3.14–8.52) showed significant association with pulmonary TB susceptibility (Table 2, Table 3). The genotypes distribution frequencies of rs10889677 (CC vs AA OR = 1.85, 95 % CI 1.13–3.05) showed significant difference between the PTB cases and controls (Table 2). However, after adjusting for the covariant of BMI, there was no association between rs10889677 (CC + AC vs AA OR = 0.88, 95 % CI 0.42–1.23) and pulmonary TB (Table 3). There was moderate to high LD in the control group between these 2 SNPs (r2 = 0.59, D' = 0.96). There was also moderate to high LD in controls between rs7518660 and rs11465802 (r2 = 0.59, D' = 0.97), and high LD between rs10889677 and rs11465802 (D' = 0.97) (Fig. 1). Unexpectedly, there was no association between rs11465802 and susceptibility to pulmonary TB in Chinese Uygurs. The alleles of rs7518660 (OR = 3.56, 95 % CI 2.74–4.62) and rs10889677 (OR = 1.37, 95 % CI 1.07–1.76) also showed significant difference between the cases and the controls (Table 2).
Table 2.
Genotype and allele frequencies distribution of IL-23R polymorphisms between controls and cases
| dbSNP ID/CNV | Genotype/Allele | Controls n (%) | PTB cases n (%) | OR (95 % CI) |
|---|---|---|---|---|
| rs1884444 | TT | 82 (33.1) | 87 (34.8) | 1 |
| GT | 122 (49.2) | 114 (45.6) | 0.88 (0.59–1.31) | |
| GG | 44 (17.7) | 49 (19.6) | 1.05 (0.63–1.74) | |
| T | 286 (57.7) | 288 (57.6) | 1 | |
| G | 210 (42.3) | 212 (42.4) | 1.00 (0.78–1.29) | |
| rs10889664 | CC | 93 (37.5) | 103 (41.2) | 1 |
| CT | 117 (47.2) | 109 (43.6) | 0.84 (0.57–1.23) | |
| TT | 38 (15.3) | 38 (15.2) | 0.90 (0.53–1.53) | |
| C | 303 (61.1) | 315 (63.0) | 1 | |
| T | 193 (38.9) | 185 (37.0) | 0.92 (0.71–1.19) | |
| rs11465788 | TT | 85 (34.7) | 87 (34.9) | 1 |
| CT | 120 (49.0) | 114 (45.8) | 0.93 (0.63–1.38) | |
| CC | 40 (16.3) | 48 (19.3) | 1.17 (0.70–1.96) | |
| T | 290 (59.2) | 288 (57.8) | 1 | |
| C | 200 (40.8) | 210 (42.2) | 1.06 (0.82–1.36) | |
| rs7518660 | GG | 105 (42.3) | 33 (13.2) | 1 |
| AG | 116 (46.8) | 109 (43.6) | 2.99 (1.87–4.79) | |
| AA | 27 (10.9) | 108 (43.2) | 12.73 (7.16–22.6) | |
| G | 326 (65.7) | 175 (35.0) | 1 | |
| A | 170 (34.3) | 325 (65.0) | 3.56 (2.74–4.62) | |
| rs11465802 | CC | 74 (30.0) | 70 (28.0) | 1 |
| AC | 121 (49.0) | 121 (48.4) | 1.06 (0.70–1.60) | |
| AA | 52 (21.0) | 59 (23.6) | 1.20 (0.73–1.97) | |
| C | 269 (54.5) | 261 (52.2) | 1 | |
| A | 225 (45.5) | 239 (47.8) | 1.10 (0.85–1.41) | |
| rs10889677 | AA | 76 (30.9) | 57 (22.9) | 1 |
| AC | 119 (48.4) | 121 (48.6) | 1.36 (0.89–2.08) | |
| CC | 51 (20.7) | 71 (28.5) | 1.85 (1.13–3.05) | |
| A | 271 (55.1) | 235 (47.2) | 1 | |
| C | 221 (44.9) | 263 (52.8) | 1.37 (1.07–1.76) | |
| CNV | CN < 2 | 3 (1.2) | 7 (2.8) | 2.66 (0.68–10.41) |
| CN = 2 | 230 (92.0) | 202 (80.8) | 1 | |
| CN > 2 | 17 (6.8) | 41 (16.4) | 2.75 (1.51–4.98) |
PTB pulmonary tuberculosis, OR odds ratio, CI confidence interval, CNV copy number variation, CN copy number
Table 3.
Association of IL-23R SNPs genotypes with pulmonary TB under different genotype models
| dbSNP ID | Allele | Genetic Model | OR (95 % CI) | ORc (95 % CI) | ORa (95 % CI) |
|---|---|---|---|---|---|
| rs1884444 | T > G | Dominant | 1.08 (0.75–1.56) | 1.12 (0.83–1.77) | 1.22 (0.89–1.83) |
| Recessive | 1.17 (0.72–1.79) | 1.21 (0.77–1.84) | 1.33 (0.79–1.94) | ||
| Additive | 0.86 (0.61–1.23) | 0.91 (0.81–1.34) | 0.93 (0.85–1.46) | ||
| rs10889664 | C > T | Dominant | 1.16 (0.81–1.67) | 1.22 (0.90–1.83) | 1.31 (0.94–1.92) |
| Recessive | 0.86 (0.62–1.65) | 0.89 (0.72–1.88) | 0.95 (0.76–1.97) | ||
| Additive | 1.16 (0.81–1.64) | 1.34 (0.93–1.77) | 1.54 (0.95–1.83) | ||
| rs11465788 | T > C | Dominant | 0.99 (0.68–1.43) | 1.02 (0.74–1.65) | 1.23 (0.82–1.75) |
| Recessive | 1.22 (0.77–1.30) | 1.24 (0.82–1.43) | 1.37 (0.85–1.55) | ||
| Additive | 0.88 (0.62–1.25) | 0.93 (0.81–1.38) | 0.95 (0.86–1.43) | ||
| rs7518660 | G > A | Dominant | 4.83 (3.10–7.53) | 4.91 (3.25–8.17) | 4.78 (3.14–8.52) |
| Recessive | 6.25 (3.85–10.00) | 6.54 (3.62–10.34) | 6.38 (3.51–10.46) | ||
| Additive | 0.88 (0.62–1.25) | 0.91 (0.58–1.36) | 0.93 (0.65–1.47) | ||
| rs11465802 | C > A | Dominant | 0.91 (0.62–1.33) | 0.95 (0.57–1.46) | 1.01 (0.48–1.52) |
| Recessive | 1.16 (0.60–1.75) | 1.23 (0.54–1.86) | 1.45 (0.76–1.92) | ||
| Additive | 1.02 (0.72–1.46) | 1.14 (0.65–1.57) | 1.26 (0.63–172)) | ||
| rs10889677 | A > C | Dominant | 0.66 (0.44–0.99) | 0.72 (0.39–1.12) | 0.88 (0.42–1.23) |
| Recessive | 1.53 (1.01–2.31) | 1.61 (0.99–1.98) | 1.75 (1.05–2.03) | ||
| Additive | 1.01 (0.71–1.43) | 1.15 (0.68–1.74) | 1.24 (0.73–1.82) |
OR odds ratio, CI confidence interval
OR for comparison of genetic models by conditioned logistic regression analysis
ORc = OR value after Bonferroni correction
ORa = OR value adjusted for BMI
Fig. 1.
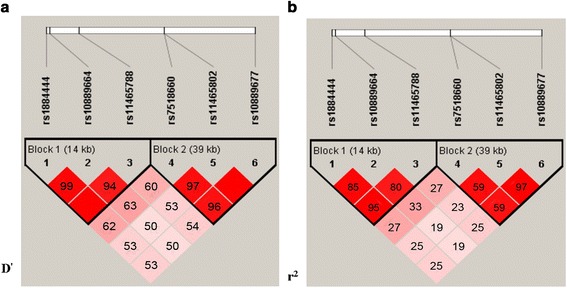
Linkage Disequilibrium Plot of SNPs of IL-23R in the control group. Linkage disequilibrium (LD) blocks in the IL23R polymorphisms black squares indicate significant allelic association between groups of SNPs from the control subjects measured by the D' (left panels) and r2 (right panels) statistics. A D' or r2 cutoff of 0.8 is used for LD. High D' values are dark, low D' values are light
Association of IL-23R CNV with pulmonary TB
We initially sequenced 11 exons of IL23R from 10 pair of samples. However, we did not find the CNV reported previously [19] in those subjects. Given the much lower frequency distribution of CNV compared to SNPs [28] and the small sample size we selected, the negative result was reasonable. Taking this into consideration, we still further identified the CNV using Taqman real-time PRC in 250 pairs of samples.
Delta Ct value of IL23R for each sample was calculated by normalizing with the Ct value of RnaseP (Additional file 1: Figure S1C). Copy number was then transferred from the Delta Ct value. The copy number frequency distribution of IL-23R showed a significant difference (P = 0.001) between controls and cases (Fig. 2). The further analysis showed that the frequencies of copy number increase (OR = 2.75, 95 % CI 1.51–4.98) in patients were significantly higher than those in controls (Table 2).
Fig. 2.

Copy number frequencies distribution of IL-23R between controls and pulmonary TB patients
It is important to determine the degree of LD between the CNV of IL-23R and rs7518660 and rs10889677, given that these variants were all associated with susceptibility to pulmonary TB in Chinese Uygurs. LD between the CNV and these two SNPs was determined in the control group at the haplotype level, using estimated phased as well as genotype level. There was weak LD between the CNV and rs7518660 and rs10889677 in the control group, as measured by r2 (r2 < 0.01), which was not affected by the MAF of the haplotypes (Additional file 7: Table S4).
We then used AccuCopy assay and ddPCR to validate the copy number of IL-23R calculated by Taqman real-time PCR with 25 pairs randomly selected from those 500 tested samples (Additional file 5: Figure S2 and Additional file 6: Figure S3). The results both showed 98 % concordant. Due to the relatively small sample size, we then calculated the power of the test. It is over 0.8, meaning that the result is reliable.
Association of IL-23R haplotypes with pulmonary TB
Pairwise LD between the 6 SNPs of IL-23R was calculated by D' and r2 from controls (Fig. 1). Based on the LD, 2 block haplotypes were constructed using Haploview version 4.2. The common haplotypes (frequency > 3 %) in each block accounted for 97.3 % and 96.4 % for the cases and 100 % and 97.5 % for the controls. We found 5 haplotypes (GTC and GCC in Block1; GAA, ACA and GAC in Block2) were significantly associated (OR = 6.10, 95 % CI 4.99–7.47; OR = 6.50, 95 % CI 4.36–9.71; OR = 1.97, 95 % CI 1.57–2.47; OR = 0.61, 95 % CI 0.44–0.85; OR = 6.76, 95 % CI 4.76–9.60, respectively) with pulmonary TB (Table 4). Among these 5 haplotypes, ACA in Block2 was a protective factor for Pulmonary TB, while other 4 haplotypes were risk factors.
Table 4.
Association between haplotypes frequencies of IL-23R and pulmonary TB susceptibility
| Block | Haplotypes | Controls (%) | PTB Cases (%) | OR (95 % CI) |
|---|---|---|---|---|
| Block1 | ||||
| rs1884444 rs10889664 rs11465788 | TCT | 70.8 | 27.5 | 1 |
| GTC | 25.4 | 60.2 | 6.10 (4.99–7.47) | |
| GCC | 3.8 | 9.6 | 6.50 (4.36–9.71) | |
| Block2 | ||||
| rs7518660 rs11465802 rs10889677 | ACC | 44.6 | 30.4 | 1 |
| GAA | 22.8 | 30.6 | 1.97 (1.57–2.47) | |
| ACA | 14.2 | 5.9 | 0.61 (0.44–0.85) | |
| AAA | 11.3 | 8.3 | 1.08 (0.78–1.48) | |
| GAC | 4.6 | 21.2 | 6.76 (4.76–9.60) | |
PTB pulmonary tuberculosis, OR odds ratio, CI confidence interval
Association of IL-23R polymorphisms with clinical forms of pulmonary TB
The SNP rs11465802 (AA + AC vs CC, OR = 3.23, 95 % CI 1.85–5.62) showed significant association with drug resistance of the strain and the SNP rs1884444 (GG + GT vs TT, OR = 3.61, 95 % CI 1.90–6.85) demonstrated significant association with cavitary formation on chest CT scanning in Pulmonary TB patients. There were no significant association between the polymorphisms of IL-23R and conversion of sputum culture and pulmonary infiltration in the pulmonary TB patients (Table 5, Table 6).
Table 5.
Association of IL-23R polymorphisms and drug-sensitivity and conversion of sputum culture in pulmonary TB patients
| dbSNP ID/ | Genotype | Drug-sensitivity (N = 250) | OR (95 % CI) | Months of treatment for Negative culture (N =250) | OR (95 % CI) | ||
|---|---|---|---|---|---|---|---|
| CNV | Drug-sensitive | Drug-resistant | ≤2 months | > 2 months | |||
| N = 172 n (%) | N = 78 n (%) | N = 195 n (%) | N = 55 n (%) | ||||
| rs1884444 | TT | 72 (41.9) | 31 (39.7) | 1 | 79 (40.5) | 24 (43.6) | 1 |
| GG + GT | 100 (58.1) | 47 (60.3) | 1.09 (0.63–1.88) | 116 (59.5) | 31 (56.4) | 0.88 (0.48–1.61) | |
| rs10889664 | CC | 85 (49.4) | 38 (48.7) | 1 | 96 (49.2) | 27 (49.1) | 1 |
| TT + CT | 87 (50.6) | 40 (51.3) | 1.03 (0.60–1.76) | 99 (50.8) | 28 (50.9) | 1.01 (0.55–1.83) | |
| rs11465788 | TT | 73 (42.1) | 32 (41.0) | 1 | 83 (42.6) | 22 (40.0) | 1 |
| CC + CT | 99 (57.9) | 46 (59.0) | 1.06 (0.62–1.83) | 112 (57.4) | 33 (60.0) | 1.11 (0.60–2.05) | |
| rs7518660 | GG | 8 (4.7) | 4 (5.1) | 1 | 9 (4.6) | 3 (5.5) | 1 |
| AA + AG | 164 (95.3) | 74 (94.9) | 0.90 (0.29–3.09) | 186 (95.4) | 52 (94.5) | 0.84 (0.22–3.21) | |
| rs11465802 | CC | 117 (32.0) | 31 (60.3) | 1 | 116 (59.5) | 32 (58.2) | 1 |
| AA + AC | 55 (68.0) | 47 (39.7) | 3.23 (1.85–5.62) | 79 (40.5) | 23 (41.8) | 1.06 (0.58–1.94) | |
| rs10889677 | AA | 37 (21.5) | 16 (20.5) | 1 | 41 (21.0) | 12 (21.8) | 1 |
| CC + AC | 135 (78.5) | 62 (79.5) | 1.06 (0.55–2.05) | 154 (79.0) | 43 (78.2) | 0.95 (0.46–1.97) | |
| CNV | CN = 2 | 138 (80.2) | 62 (79.5) | 1 | 157 (80.5) | 43 (78.2) | 1 |
| CN = 1, 3, 4 | 34 (19.8) | 16 (20.5) | 1.05 (0.54–2.04) | 38 (19.5) | 12 (21.8) | 1.15 (0.56–2.40) | |
OR odds ratio, CI confidence interval
Table 6.
Association of IL-23R polymorphisms and pulmonary infiltration and cavitary formation in pulmonary TB patients
| dbSNP ID/ | Genotype | Cavitary formation (N = 250) | OR (95 % CI) | Pulmonary infiltration (N = 250) | OR (95 % CI) | ||
|---|---|---|---|---|---|---|---|
| No Cavity lesion | Cavity lesion | ≤ 1/3 of one lung | >1/3 of one lung | ||||
| N = 179 n (%) | N = 71 n (%) | N = 138 n (%) | N = 112 n (%) | ||||
| rs1884444 | TT | 88 (49.2) | 15 (21.1) | 1 | 58 (42.0) | 45 (40.2) | 1 |
| GG + GT | 91 (50.8) | 56 (78.9) | 3.61 (1.90–6.85) | 80 (58.0) | 67 (59.8) | 1.08 (0.65–1.79) | |
| rs10889664 | CC | 87 (48.6) | 36 (50.7) | 1 | 68 (49.3) | 55 (49.1) | 1 |
| TT + CT | 92 (51.4) | 35 (49.3) | 0.92 (0.53–1.59) | 70 (50.7) | 57 (50.9) | 1.01 (0.61–1.66) | |
| rs11465788 | TT | 74 (41.3) | 31 (43.7) | 1 | 57 (41.3) | 48 (42.9) | 1 |
| CC + CT | 105 (58.7) | 40 (56.3) | 0.91 (0.52–1.59) | 81 (58.7) | 64 (57.1) | 0.94 (0.57–1.55) | |
| rs7518660 | GG | 7 (4.1) | 5 (7.0) | 1 | 7 (5.1) | 5 (4.5) | 1 |
| AA + AG | 172 (95.9) | 66 (93.0) | 0.54 (0.17–1.75) | 131 (94.9) | 107 (95.5) | 1.14 (0.35–3.71) | |
| rs11465802 | CC | 72 (40.2) | 30 (42.3) | 1 | 56 (40.6) | 46 (41.1) | 1 |
| AA + AC | 107 (59.8) | 41 (57.7) | 0.92 (0.53–1.61) | 82 (59.4) | 66 (58.9) | 0.98 (0.59–1.63) | |
| rs10889677 | AA | 39 (21.8) | 14 (19.7) | 1 | 31 (22.5) | 22 (19.6) | 1 |
| CC + AC | 140 (78.2) | 57 (80.3) | 1.13 (0.57–2.25) | 107 (77.5) | 90 (80.4) | 1.19 (0.64–2.19) | |
| CNV | CN = 2 | 144 (80.4) | 56 (78.9) | 1 | 109 (79.0) | 91 (81.3) | 1 |
| CN = 1, 3, 4 | 35 (19.6) | 15 (21.1) | 1.10 (0.56–2.17) | 29 (21.0) | 21 (18.7) | 0.87 (0.46–1.62) | |
OR odds ratio, CI confidence interval
Discussion
In the present study, we found that the SNP rs7518660 and the CNV in exon 11 of IL-23R were significantly associated with susceptibility to active pulmonary TB, meanwhile, we observed that rs11465802 was associated with drug-resistance and rs1884444 was related to cavity formation in Chinese Uygurs in China. To our knowledge, this is the first report showing that the CNV of IL-23R are associated with active pulmonary TB and the first association of polymorphisms in IL-23R with drug-sensitivity and cavitary lesion in pulmonary TB patients.
To date, the G1186A SNP in MRC1 [29] has been reported to be associated with TB in Chinese Uygurs. There was only one study reporting the only SNP (rs11209026) of IL-23R associated with the development of a severe form (extensive lung infiltration) of active pulmonary TB in Tunisians [17]. Unfortunately, we couldn’t analyze the association between this polymorphism with TB, because it was excluded from further analysis due to its low minor allele frequency in the control group. The results indicate that there was ethnic difference of the SNP rs11209026 among different racial groups.
One function of IL-23 mediated by IL-23R complex is driving T cell differentiation toward Th17, which leads to an increased release of other cytokines causing inflammation, such as IL-17 and TNF [12]. The exact mechanisms how these polymorphisms regulate the function of IL-23R is currently unclear. Both the SNPs rs7518660 and rs11465802 are located in intron 7, and they may influence the expression of IL-23R by regulating differential splicing [30].
The signals in gene expression from SNP and CNV had little overlap, thus it is important to realize the linkage of these two polymorphisms with gene expression for understanding the mechanisms of susceptibility to TB [28]. Furthermore, both M. tuberculosis and M. leprae are intracellular bacteria, so the patients with TB and leprosy may have a similar immune response. An increase in the copy number of exon 11 in the IL23R gene has been reported to be associated with the paucibacillary form of leprosy [19]. Therefore, we propose a hypothesis that the CNV of IL-23R, if present in Chinese Uygurs, may be associated with sputum positive pulmonary TB. The result supports our hypothesis that the increase in the copy number of IL23R is associated with pulmonary TB in the Chinese Uygur population. Interestingly, there was weak LD between the CNV in exon 11 and rs7518660, which was significantly associated with susceptibility to pulmonary TB in Chinese Uygurs. It suggested that the CNV was an independent risk factor for pulmonary TB in contrast with SNPs among Chinese Uygurs. However, a recent study found that there was an association of CCL3L1 copy number with TB in a northwestern Colombian population [31], but not in African and Peruvian populations [32]. All these data indicate that CNVs in some special genes may also be associated with susceptibility to TB, although the ethnic difference exists. CNV can have influence on gene expression levels by altering gene dosage, disrupting coding sequences or perturbing long-range gene regulation [33, 34]. An increase in the copy number of IL-23R coincided with a relatively higher fraction of memory T cells [19], but the influence of the CNV in the exon 11 of IL23R on its expression level needs to be further elucidated.
Among 250 clinical isolates of Mycobacterium tuberculosis, 172 isolates (68.8 %) were drug-sensitive and 78 (31.2 %) were resistant to at least one drug and 10 (4 %) were multidrug-resistant (MDR), consistent with the previous epidemiological results [22]. Interestingly, we observed that rs11465802 was significantly associated with drug-resistance, whereas it was not related to susceptibility to pulmonary TB. To date, variants in the HLA-DRB1, -DQB1 [23] and SLC11A1 [35] have shown association with M. tuberculosis drug sensitivity. The results demonstrate that human genetic factors are associated with the development of drug resistance of the strain in pulmonary TB patients. These data suggest that patients carrying particular gene polymorphisms may be incapable of acquiring adequate immune response to M. tuberculosis, and thus the infected bacteria is prone to develop drug resistance.
In addition, in this study, it was revealed that rs1884444 was associated with cavity formation, while it was unassociated with susceptibility to pulmonary TB or drug-resistance. It was reported that SLC11A1 polymorphinisms were associated with the presence of cavitary lesions among pulmonary TB patients [35, 36]. These results suggest that human gene polymorphisms may affect their inflammatory response after the infection with M. tuberculosis, and thus result in the severe pathological lesion.
The IL-23/IL-17 pathway is mainly composed of IL-23, its functional receptor complex (a combination of IL-23R and IL-12Rβ1), the downstream cytokines (for example, IL-17) and other key signal molecules such as STAT4. This immune pathway and its constituent molecular members play crucial roles in autoimmune inflammatory disease pathogenesis [15]. In this study, we found the SNP rs7518660 in IL-23R were associated with pulmonary TB in the Chinese Uygurs. It has been reported that polymorphisms in the IL-12 receptorβ1 gene were associated with TB in Moroccan [37] and Japanese populations [38], but not in Koreans [39]. The SNPs in IL-17 F were associated with pulmonary TB in Chinese Han [40] and northern Spain patients [41], but not in north Indians [42]. In addition, the polymorphisms of STAT4 promoter-region were reported to be associated with pulmonary TB in a Moroccan population, which might impact STAT4 expression [43]. All these findings indicate that the IL-23/IL-17 pathway may be a key pathway in charging M. tuberculosis infection [13], although ethnic difference exists. Thus, further research on the function of the IL-23/IL-17 pathway in pulmonary TB is desired. The IL-23/IL-17 pathway was thought to be an attractive target for experimental tuberculosis therapies [44, 45]. Therefore, IL-23R, a key member in the IL-23/IL-17 pathway, also may be a potential target molecule for the therapeutic management of tuberculosis.
In our study, it was revealed that individuals with BMI < 18.5 kg/m2 are more likely to suffer from pulmonary TB in Chinese Uygurs. It was reported that TB incidence increased exponentially as BMI (range 18.5–30 kg/m2) decreased across a variety of settings with different levels of TB burden, however, the dose–response relationship was uncertain at BMI levels <18.5 kg/m2. There were only a few studies reporting data on TB incidence for people with BMI <18.5 kg/m2, but the results were inconsistent. Lonnroth K et al. [46] and Cegielski JP et al. [47] reported that people with BMI <18.5 kg/m2 increased TB incidence compared with those with BMI ≥18.5 kg/m2(including normal BMI, overweight and obesity). However, Hanrahan CF et al. [48] revealed that people with BMI <18.5 kg/m2 didn’t increase TB incidence compared with those with normal BMI (range 18.6-25 kg/m2) in HIV-infected adults.
Our study had several inherent limitations. Firstly, selection bias arose in the observation, for it was conducted using a hospital based case–control study. Secondly, the sample size was relative small, although the power of the test was over 0.8, the results were still needed to be validated in a large sample size. Additionally, because the patients were only limited to sputum positive pulmonary TB individuals, we didn’t know the association between those polymorphisms and sputum negative ones. Finally, the controls were only defined as those without LTBI. This study were conducted in TB-endemic settings, therefore we assumed that all individuals were exposed to TB. Given that the controls were uninfected, it’s unclear whether an association implies susceptibility for developing active pulmonary TB or just acquisition of LTBI.
In fact, human genetic effect on TB susceptibility is more complicated than many other diseases because of the serious confounding effects of behavioral and environmental factors [49]. However, we did not find those behavioral factors were associated with pulmonary TB, as evidenced by smoking or drinking, possibly because few participants smoke or drink due to the Muslim custom. Therefore, further study of confounding effects between IL-23/IL-17 pathway genes polymorphisms and behavioral and environmental risk factors in other ethnic groups is needed.
Conclusions
Our study showed for the first time that IL23R SNPs and CNV are associated with susceptibility to pulmonary TB. Our results also revealed a significant association of IL23R SNPs with the severe clinical forms of pulmonary TB in the Chinese Uygur population.
Acknowledgements
The authors thank Reyangguli N (Department of Laboratory, Kashgar Chest Hospital, Xinjiang, China) and Maimaitiaili T (Department of Pulmonology, Kashgar Prefecture People’s Hospital, Xinjiang, China) for their assistance in recruiting participants. They thank Maosen Liao and his colleagues (Ou Yi Biotechnology Co. Ltd, Urumqi, china) for their technical assistance. They thank Jianghong D (Public Health College, Xinjiang Medical University, Xinjiang, China) for her assistance in study design and statistics. They thank Leonard Paul Arthur Daly (New Zealand) for his assistance in language editing of the manuscript. The genomic DNA extraction in this research was performed at Center for Disease Prevention and Control, Xinjiang Uygur Autonous Region. This work was supported by a grant from the Open-up Program of Xinjiang Key Medical Laboratory for Major Disease (SKLIB-XJMDR-2012-2) and National Natural Science Foundation of China (81160004).
Additional files
A. A fragment of exon11 sequenced directly by ABI 3730XL Genetic Analyzer (rs10889677, genotype AA). B. Three genotypes of rs 7518660 performed by the multiplex SNaPshot technique. C. Representative amplification plots of twofold copy number difference for IL23R in pulmonary TB samples compared to controls. (PDF 296 kb)
Primer sets used for the initial exons sequencing of IL23R. (PDF 111 kb)
Thirteen SNPs identified in IL-23R by exons resequencing from ten pairs of pulmonary TB cases and controls. (PDF 106 kb)
Primer sets used for the multiplex SNaPshot of IL23R. (PDF 106 kb)
IL-23R copy number genotyped by AccuCopy assay (Genesky Bio-Tech Co., Ltd, Shanghai, China). (PDF 161 kb)
IL-23R copy number genotyped by ddPCR. A. 2-copy genotype; B. 3-copy genotype. (PDF 228 kb)
LD between the CNV in IL-23R and rs7518660 and rs10889677 in the control group. (PDF 96 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
QW conceived and designed the study. WZ participated in its design and helped to draft the manuscript. DJ participated in its design, carried out the molecular genetic studies and drafted the manuscript. AK performed the statistical analysis and helped to draft the manuscript. XH participated in its design and the sequence alignment. SI participated in its design and the molecular biological study. AJ participated in its design and the molecular genetic studies. All authors read and approved the final manuscript.
Contributor Information
Daobin Jiang, Email: fangxueer1027@163.com.
Atikaimu Wubuli, Email: 492242282@qq.com.
Xin Hu, Email: 147951422@qq.com.
Syed Ikramullah, Email: dr.ikram96@yahoo.com.
Abudoujilili Maimaiti, Email: 276576643@qq.com.
Wenbao Zhang, Email: wenbaozhang2013@163.com.
Qimanguli Wushouer, Email: qimenw@hotmail.com.
References
- 1.World Health Organization . Global tuberculosis report 2014. Geneva: World Health Organization; 2014. pp. 23–28. [Google Scholar]
- 2.National Technical Steering Group of the Epidemiological Sampling Survey for Tuberculosis, Implementing Office of the Sampling Survey for Tuberculosis The prevalence of pulmonary tuberculosis in a national survey across China in 2010. Chin J Tuberc Respir Dis. 2012;35:665–668. [PubMed] [Google Scholar]
- 3.Yang JM, Simahule JES, Tai XR, Li YH, Zhao Z. Analysis of tuberculosis epidemiological survey conducted in 2010-2011 in Xinjiang Uygur Autonomous Region. Chin J Antituberc. 2013;35:960–964. [Google Scholar]
- 4.Wang L, Zhang H, Ruan Y, Chin DP, Xia Y, Cheng S, et al. Tuberculosis prevalence in China, 1990-2010; a longitudinal analysis of national survey data. Lancet. 2014;383(9934):2057–2064. doi: 10.1016/S0140-6736(13)62639-2. [DOI] [PubMed] [Google Scholar]
- 5.Comstock GW. Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis. 1978;117(4):621–624. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- 6.Stein CM, Zalwango S, Chiunda AB, Millard C, Leontiev DV, Horvath AL, et al. Linkage and association analysis of candidate genes for TB and TNFalpha cytokine expression: evidence for association with IFNGR1, IL-10, and TNF receptor 1 genes. Hum Genet. 2007;121(6):663–673. doi: 10.1007/s00439-007-0357-8. [DOI] [PubMed] [Google Scholar]
- 7.Yim JJ, Selvaraj P. Genetic susceptibility in tuberculosis. Respirology. 2010;15(2):241–256. doi: 10.1111/j.1440-1843.2009.01690.x. [DOI] [PubMed] [Google Scholar]
- 8.Thye T, Owusu-Dabo E, Vannberg FO, van Crevel R, Curtis J, Sahiratmadja E, et al. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat Genet. 2012;44(3):257–259. doi: 10.1038/ng.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stead WW, Senner JW, Reddick WT, Lofgren JP. Racial differences in susceptibility to infection by Mycobacterium tuberculosis. N Engl J Med. 1990;322(7):422–427. doi: 10.1056/NEJM199002153220702. [DOI] [PubMed] [Google Scholar]
- 10.Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis. 2002;186(10):1463–1468. doi: 10.1086/344891. [DOI] [PubMed] [Google Scholar]
- 11.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10(3):314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khader SA, Cooper AM. IL-23 and IL-17 in tuberculosis. Cytokine. 2008;41(2):79–83. doi: 10.1016/j.cyto.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khader SA, Guglani L, Rangel-Moreno J, Gopal R, Junecko BA, Fountain JJ, et al. IL-23 is required for long-term control of Mycobacterium tuberculosis and B cell follicle formation in the infected lung. J Immunol. 2011;187(10):5402–5407. doi: 10.4049/jimmunol.1101377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168(11):5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 15.Kikly K, Liu L, Na S, Sedgwick JD. The IL-23/Th(17) axis: therapeutic targets for autoimmune inflammation. Curr Opin Immunol. 2006;18(6):670–675. doi: 10.1016/j.coi.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Stein CM, Guwatudde D, Nakakeeto M, Peters P, Elston RC, Tiwari HK, et al. Heritability analysis of cytokines as intermediate phenotypes of tuberculosis. J Infect Dis. 2003;187(11):1679–1685. doi: 10.1086/375249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Selma W, Boukadida J. IL23R(Arg381Gln) functional polymorphism is associated with active pulmonary tuberculosis severity. Clin Vaccine Immunol. 2012;19(8):1188–1192. doi: 10.1128/CVI.00135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 19.Ali S, Srivastava AK, Chopra R, Aggarwal S, Garg VK, Bhattacharya SN, et al. IL12B SNPs and copy number variation in IL23R gene associated with susceptibility to leprosy. J Med Genet. 2013;50(1):34–42. doi: 10.1136/jmedgenet-2012-101214. [DOI] [PubMed] [Google Scholar]
- 20.Ablimit A, Qin W, Shan W, Wu W, Ling F, Ling KH, et al. Genetic diversities of cytochrome B in Xinjiang Uyghur unveiled its origin and migration history. BMC Genet. 2013;14:100. doi: 10.1186/1471-2156-14-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.China Antituberculosis Association The laboratory science procedure of diagnostic bacteriology in tuberculosis. Bulletin of the Chinese Antituberculosis Association. 1996;18:28–31. [Google Scholar]
- 22.Zhang J, Mi L, Wang Y, Liu P, Liang H, Huang Y, et al. Genotypes and drug susceptibility of Mycobacterium tuberculosis Isolates in Shihezi, Xinjiang Province, China. BMC Res Notes. 2012;5:309. doi: 10.1186/1756-0500-5-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HS, Park MH, Song EY, Park H, Kwon SY, Han SK, et al. Association of HLA-DR and HLA-DQ genes with susceptibility to pulmonary tuberculosis in Koreans: preliminary evidence of associations with drug resistance, disease severity, and disease recurrence. Hum Immunol. 2005;66(10):1074–1081. doi: 10.1016/j.humimm.2005.08.242. [DOI] [PubMed] [Google Scholar]
- 24.Thuong NT, Dunstan SJ, Chau TT, Thorsson V, Simmons CP, Quyen NT, et al. Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS Pathog. 2008;4(12):e1000229. doi: 10.1371/journal.ppat.1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You GL, Ding QL, Lu YL, Dai J, Xi XD, Wang XF, et al. Characterization of large deletions in the F8 gene using multiple competitive amplification and the genome walking technique. J Thromb Haemost. 2013;11(6):1103–1110. doi: 10.1111/jth.12205. [DOI] [PubMed] [Google Scholar]
- 26.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84(2):1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niederer HA, Willcocks LC, Rayner TF, Yang W, Lau YL, Williams TN, et al. Copy number, linkage disequilibrium and disease association in the FCGR locus. Hum Mol Genet. 2010;19(16):3282–3294. doi: 10.1093/hmg/ddq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Li X, Zhang W, Wei L, Jiang T, Chen Z, et al. The novel human MRC1 gene polymorphisms are associated with susceptibility to pulmonary tuberculosis in Chinese Uygur and Kazak populations. Mol Biol Rep. 2013;40(8):5073–5083. doi: 10.1007/s11033-013-2610-7. [DOI] [PubMed] [Google Scholar]
- 30.Zwiers A, Kraal L, van de Pouw Kraan TC, Wurdinger T, Bouma G, Kraal G. Cutting edge: a variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. J Immunol. 2012;188(4):1573–1577. doi: 10.4049/jimmunol.1101494. [DOI] [PubMed] [Google Scholar]
- 31.Mamtani M, Mummidi S, Ramsuran V, Pham MH, Maldonado R, Begum K, et al. Influence of variations in CCL3L1 and CCR5 on tuberculosis in a northwestern Colombian population. J Infect Dis. 2011;203(11):1590–1594. doi: 10.1093/infdis/jir145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpenter D, Taype C, Goulding J, Levin M, Eley B, Anderson S, et al. CCL3L1 copy number, CCR5 genotype and susceptibility to tuberculosis. BMC Med Genet. 2014;15:5. doi: 10.1186/1471-2350-15-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76(1):8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38(1):86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi K, Hasegawa Y, Abe T, Yamamoto T, Nakashima K, Imaizumi K, et al. SLC11A1 (formerly NRAMP1) polymorphisms associated with multidrug-resistant tuberculosis. Tuberculosis (Edinb) 2008;88(1):52–57. doi: 10.1016/j.tube.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Abe T, Iinuma Y, Ando M, Yokoyama T, Yamamoto T, Nakashima K, et al. NRAMP1 polymorphisms, susceptibility and clinical features of tuberculosis. J Infect. 2003;46(4):215–220. doi: 10.1053/jinf.2002.1064. [DOI] [PubMed] [Google Scholar]
- 37.Remus N, El Baghdadi J, Fieschi C, Feinberg J, Quintin T, Chentoufi M, et al. Association of IL12RB1 polymorphisms with pulmonary tuberculosis in adults in Morocco. J Infect Dis. 2004;190(3):580–587. doi: 10.1086/422534. [DOI] [PubMed] [Google Scholar]
- 38.Kusuhara K, Yamamoto K, Okada K, Mizuno Y, Hara T. Association of IL12RB1 polymorphisms with susceptibility to and severity of tuberculosis in Japanese: a gene-based association analysis of 21 candidate genes. Int J Immunogenet. 2007;34(1):35–44. doi: 10.1111/j.1744-313X.2007.00653.x. [DOI] [PubMed] [Google Scholar]
- 39.Lee HW, Lee HS, Kim DK, Ko DS, Han SK, Shim YS, et al. Lack of an association between interleukin-12 receptor beta1 polymorphisms and tuberculosis in Koreans. Respiration. 2005;72(4):365–368. doi: 10.1159/000086249. [DOI] [PubMed] [Google Scholar]
- 40.Peng R, Yue J, Han M, Zhao Y, Liu L, Liang L. The IL-17 F sequence variant is associated with susceptibility to tuberculosis. Gene. 2013;515(1):229–232. doi: 10.1016/j.gene.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 41.Ocejo-Vinyals JG, de Mateo EP, Hoz MA, Arroyo JL, Aguero R, Ausin F, et al. The IL-17 G-152A single nucleotide polymorphism is associated with pulmonary tuberculosis in northern Spain. Cytokine. 2013;64(1):58–61. doi: 10.1016/j.cyto.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 42.Abhimanyu, Bose M, Komal, Varma-Basil M. Lack of association between IL17A and IL17F polymorphisms and related serum levels in north Indians with tuberculosis. Gene. 2013;529(1):195–198. doi: 10.1016/j.gene.2013.06.090. [DOI] [PubMed] [Google Scholar]
- 43.Sabri A, Grant AV, Cosker K, El Azbaoui S, Abid A, Abderrahmani Rhorfi I, et al. Association Study of Genes Controlling IL-12-dependent IFN-gamma Immunity: STAT4 Alleles Increase Risk of Pulmonary Tuberculosis in Morocco. J Infect Dis. 2014;210(4):611–618. doi: 10.1093/infdis/jiu140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuniga J, Torres-Garcia D, Santos-Mendoza T, Rodriguez-Reyna TS, Granados J, Yunis EJ. Cellular and humoral mechanisms involved in the control of tuberculosis. Clin Dev Immunol. 2012;2012:193923. doi: 10.1155/2012/193923. Epub 2012 May 17. [DOI] [PMC free article] [PubMed]
- 45.Matucci A, Maggi E, Vultaggio A. Cellular and humoral immune responses during tuberculosis infection: useful knowledge in the era of biological agents. J Rheumatol Suppl. 2014;91:17–23. doi: 10.3899/jrheum.140098. [DOI] [PubMed] [Google Scholar]
- 46.Lonnroth K, Williams BG, Cegielski P, Dye C. A consistent log-linear relationship between tuberculosis incidence and body mass index. Int J Epidemiol. 2010;39(1):149–155. doi: 10.1093/ije/dyp308. [DOI] [PubMed] [Google Scholar]
- 47.Cegielski JP, McMurray DN. The relationship between malnutrition and tuberculosis: evidence from studies in humans and experimental animals. Int J Tuberc Lung Dis. 2004;8(3):286–298. [PubMed] [Google Scholar]
- 48.Hanrahan CF, Golub JE, Mohapi L, Tshabangu N, Modisenyane T, Chaisson RE, et al. Body mass index and risk of tuberculosis and death. AIDS. 2010;24(10):1501–1508. doi: 10.1097/QAD.0b013e32833a2a4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein CM. Genetic epidemiology of tuberculosis susceptibility: impact of study design. PLoS Pathog. 2011;7(1):e1001189. doi: 10.1371/journal.ppat.1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]


