Abstract
At present, glioma is the most common intracranial tumor and accounts for 40–60% of intracranial tumors. Glioma is highly anaplastic and demonstrates invasive growth. Although considerable progression has been achieved in the treatment of malignant glioma, the prognosis of this disease remains poor. Over the previous decade, several studies have confirmed that human cytomegalovirus (HCMV) enhances the growth or survival of tumors. This is likely to occur through mechanisms distinct from those of classic tumor viruses, which express transforming viral oncoproteins in the majority of tumor cells. The immediate-early 2 protein (IE86; 86 kDa) of HCMV is a key regulator for viral replication and host cell proliferation. The present study aimed to identify the association between the acetylation level and HCMV IE86 expression in various histological types of glioma. Tissue samples were obtained from 60 patients with glioma, consisting of 25 patients with glioblastoma multiforme (GBM), 16 patients with anaplastic glioma and 19 patients with low-grade glioma, in addition to 9 tissue samples obtained from the normal cortex, which were used as the control. The in situ protein expression of IE86, which is encoded by the IE2 gene, activating transcription factor 5 (ATF5), P300, acetyl-histone H3K9 and acetyl-histone H3K14 was detected by immunohistochemistry. The mRNA levels of ATF5, IE2 and P300 were measured by reverse transcription-quantitative polymerase chain reaction in GBM, anaplastic glioma, low-grade glioma and normal cortex tissue specimens. The protein levels of ATF5, IE86, P300, acetyl-histone H3K9 and acetyl-histone H3K14 were assessed by western blot analysis in high-grade glioma, low-grade glioma and normal cortex tissues. Analysis of the expression of the proteins revealed that the excessive expression of the HCMV IE86 protein is associated with the malignancy degree and acetylation level in glioma. IE86 expression is also associated with ATF5, which is an anti-apoptotic protein that is highly expressed in malignant glioma, but not in normal brain tissues. The expression level of IE86 may demonstrate considerable importance for the evaluation of the malignancy degree of human gliomas and extensive application in diagnostic and therapeutic medicine.
Keywords: human cytomegalovirus, glioma, acetylation, activating transcription factor 5, immunohistochemistry
Introduction
Human cytomegalovirus (HCMV) is a β-herpesvirus that causes persistent infection in humans and may cause severe disease in immunocompromized individuals. There is evidence that HCMV infection and expression may be associated with human malignancies, including malignant glioma, and colon and prostate cancer (1–5). HCMV may increase the malignancy of infected cells by disrupting cellular pathways involved in the cell cycle, apoptosis, angiogenesis, cell invasion and host immune response (6). Glioma is the most common primary brain tumor and arises from astrocytes and the precursors of astrocytes. Although the treatment of glioma has markedly advanced, the treatment efficacy is not so promising. It is of vital importance for HCMV infection to affect the central nervous system, and glioma is one of the most common primary diseases of the central nervous system (1,7–9).
In 2002, Cobbs et al initially reported the association between HCMV and malignant glioma (10). This study confirmed that HCMV nucleic acids and proteins were present in a high proportion of high- and low-grade gliomas, classified as World Health Organization (WHO) grades II–IV. The expression of early and delayed gene products occurred in these tumors (10). It is well-known that the HCMV genome encodes >200 proteins with expression that occurs in a program of three sequential stages. Immediate-early (IE) proteins are necessary for replication and activate early genes. Early proteins are essential for certain functional enzymes and, therefore, for the virus. Late proteins comprise the structural components of the virus. The HCMV immediate-early-1 (IE-1) protein was detected in >90% of gliomas. Almost 80% of newly diagnosed patients with glioblastoma multiforme (GBM) contain HCMV DNA in the peripheral blood, which indicates either systemic reactivation or viral shedding from tumor cells to the periphery. The immediate-early 2 (IE86) protein of human cytomegalovirus (HCMV) binds to the tumor suppressor p53 and inactivates the function of this protein through unknown mechanisms. IE86 is able to suppress the histone acetyltransferase (HAT) activity of the p53 transcriptional coactivators P300 and cyclic adenosine monophosphate response element-binding protein (CREB) binding protein (CBP). IE86 deletion mutants lacking the minimal N-terminal HAT inhibitory domain fail to repress the in vivo DNA binding of p53 and local histone acetylation.
The activating transcription factor 5 (ATF5) is a member of the ATF/CREB family of basic zipper proteins (11,12). It has previously been confirmed that ATF5 is highly expressed in malignant glioma and is essential for glioma cell survival (13). It has also been demonstrated that HCMV infection inhibits apoptosis by regulating the ATF5 signaling pathway in human malignant glioma cells (14,15). ATF5 expression is downregulated by trophic factors and this is required for the promotion of neuroprogenitor cell cycle exit and differentiation into other neurons, oligodendroglia or astrocytes. Examination of the role of ATF5 in glioblastoma cells indicates that interference with the expression or activity of ATF5 results in the cells undergoing apoptotic death. By contrast, normal astrocytes and neurons do not appear to require ATF5 for survival, indicating that ATF5 may be a selective target for the treatment of glioblastoma and other neural neoplasias (16,17).
Histone acetylation is closely associated with certain biological effects, including gene transcription, and is catalyzed by HATs. P300 is one of the numerous HATs that are able to change the structure of the chromosome and then trigger the process of transcription (18). In the meantime, the HAT P300 acetylates the various cellular proteins, and numerous studies indicate that the interaction between P300 and certain viral proteins promote the replication of the virus. It was revealed that IE86 binds to the HAT domain of the p53 coactivators P300 and CBP, and blocks the acetyltransferase activity of histones and p53 (19–22). HCMV IE86 downregulates p53-dependent gene activation by inhibiting P300/CBP-mediated local histone acetylation and that IE86 may have oncogenic activity.
The acetylation histones affect the accessibility of chromatin for DNA replication, repair and transcription. Acetylation is the addition of an acetyl group from the co-factor acetyl coenzyme A to the amino group of lysine residues on histones, which is catalyzed by group enzymes termed HATs. Targeting of cellular HATs by viral proteins is important in the development of virus-associated diseases. Acetylation on histones, in general, demonstrates two distinct functions (20). First, histone acetylation may loosen the interactions between histones and DNA, as acetylation neutralizes the positive charge on a lysine residue, and thereby reduces electrostatic interactions between DNA and histones. Second, acetylation on a particular lysine residue may also recruit proteins to chromatin to perform specific functions. DNA is tightly bound around the nucleosome core, which consists of histone residues H2A, H2B, H3 and H4. It is known that histone modifications regulate diverse cellular processes that are critical for cancer prevention. In recent years, with the development of molecular biology and the progression of studies investigating glioma, it was found that the incidence and development of glioma were associated with acetyl-histone H3K9 and acetyl-histone H3K14, which were the most common histone acetylation forms. The identification of histone acetylation profiles in human neoplasia has been a major factor in constructing a novel paradigm, in which acetylation histones contribute markedly to human disease (23).
Materials and methods
Tissue specimens and clinical data
In total, 60 tissue samples were obtained from the Department of Pathology of the Affiliated Hospital of Qingdao University Medical College (Qingdao, Shandong, China) between March and December 2012. No patients underwent radiation or chemotherapy prior to surgical therapy. Ethical approval was obtained from the Institutional Review Board of the Affiliated Hospital of Qingdao University Medical College.
The pathological diagnosis and grading for each glioma was assessed by neuropathologists, according to the 2007 WHO Classification of Nervous System Tumors (24). In total, 60 patients with glioma were analyzed, consisting of 25 patients with GBM, 16 patients with anaplastic glioma and 19 patients with low-grade glioma. The anaplastic tumors consisted of 9 anaplastic astrocytomas, 3 anaplastic ependymomas, 3 anaplastic oligodendrogliomas and 1 anaplastic oligoastrocytoma. The low-grade tumors consisted of 3 astrocytomas, 2 pilocytic astrocytomas, 2 ependymomas, 1 ganglioma, 10 oligodendrogliomas and 1 subependymoma. In addition, 9 tissue specimens obtained from the normal cortex of patients without glioma were used as the negative control. All tissue samples were collected in accordance with Institutional Review Board-approved protocols. The mean age of the patients at the time of diagnosis was 55, 45, 43 and 42 years for GBM, anaplastic glioma, low-grade glioma and normal cortex, respectively. Overall, 36% of all patients were female. The formalin-fixed, paraffin-embedded tissue blocks were archived in the Section of Neuropathology of the Department of Pathology. The present study was approved by the Institutional Review Board of the Affiliated Hospital of Qingdao University Medical College and written informed consent was obtained from all patients.
Immunohistochemical analysis
The paraffin-embedded sections were heated overnight on slides in an oven at 60°C. The slides were removed from the oven and deparaffinized by placing in xylene (BioGenex, San Ramon, CA, USA). Subsequently, the slides were soaked in serial dilutions of ethanol (100, 95, 75 and 50% ethanol) to deparaffinize the slides, and then post-fixed in neutral-buffered saline. The sections were blocked for endogenous peroxidase in 3% H2O2 for 12 min. Antigen retrieval was performed for 20 min in a microwave oven on high power using citrate-buffered saline (pH 6.0). The slides were incubated in 5% bovine serum albumin to block non-specific binding. The sections were then incubated with the primary antibody overnight at 4°C, washed in phosphate-buffered saline (PBS) and incubated with secondary antibody for 1 h at 37°C. The sections were stained using 3,3-diaminobenzidine, which was stored at room temperature without light for 10 min, and the staining was finished using distilled water. The slides were then counterstained using hematoxylin. An additional step of hydrochloric acid alcohol differentiation was performed. Subsequently, dehydration, clearing and mounting with neutral gums were performed in turn.
Mouse anti-human HCMV IE2-86 monoclonal antibody (cat no. 0841) in the present experiment was purchased from Virostat (Westbrook, ME, USA) and used at a 1:30 dilution. Rabbit anti-human P300 polyclonal antibodies (cat no. sc-585) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA) and used at a dilution of 1:200. Rabbit anti-human acetyl-histone H3K9 polyclonal antibodies (cat no. YK0006) were obtained from Immunoway Biotechnology and used at a dilution of 1:200. Rabbit anti-human acetyl-histone H3K14 monoclonal antibodies (cat no. ab52946) were purchased from Abcam (Cambridge, UK) and used at a dilution of 1:200. Rabbit anti-human ATF5 polyclonal antibodies (cat no. ab6012) were also purchased from Abcam and used at a dilution of 1:250.
The slides were examined independently by two investigators blinded to the clinical and pathological data of the patients. Protein expression was quantified using a visual grading system based on the extent of staining, determined by the percentage of positive tumor cells, which was graded on a scale of 0–4, as follows: 0, none; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. The staining intensity was graded on a scale of 0–3, as follows: 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. For additional analysis, the product of the staining extent and intensity grades was used to define the cut-off value for high expression of the proteins, and the protein expression was classified into two categories, high (grades 12–4) and low (grades 0–3).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) to detect ATF5, P300 and IE2 mRNA expression in glioma
The PCR primers were as follows: ATF5 forward, 5′-AAGTCGGCGGCTCTGAGGTA-3′ and reverse, 5′-TCC GAACTCAAAGAAGGCCA-3′; P300 forward, 5′-GACCCT CAGCTTTTAGGAATCC-3′ and reverse, 5′-TGCCGTAGC AACACAGTGTCT-3′; IE2 forward, 5′-GCGCAATATCAT GAAAGATAAGAACA-3′ and reverse, 5′-GATTGGTGTTGC GGAACATG-3′; and β-actin forward, 5′-TGGAACGGTGAA GGTGACAG-3′ and reverse, 5′-GGCTTTTAGGATGGC AAGGG-3′. β-actin was used as an internal control. The primers were synthetized by Shanghai Sangon Biological Co., Ltd. (Shanghai, China). PCR was performed at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 55°C for 1 min. Each reaction was performed in triplicate and analyzed individually relative to β-actin, which was calculated using the 2−∆∆Ct method.
RNA extraction and RT-qPCR
RNA was extracted using TRIzol reagent (Takara Bio, Inc., Otsu, Shiga, Japan), according to the manufacturer's instructions. To obtain cDNA, 1 µg RNA was reverse-transcribed using the PrimeScript RT reagent kit with gDNA Eraser (Takara Bio, Inc.), according to the manufacturer's instructions. RT-qPCR was performed using the GoTaq qPCR Master Mix (Promega, Madison, WI, USA). The primers used were as follows: ATF5 forward, 5′-AGTGGGCTGGGATGGCTCGTAGAC-3′ and reverse, 5′-CTCGGGTGGTGGCAGGATGTGG-3′; IE2 forward, 5′-GCGCAATATCATGAAAGATAAGAACA-3′ and reverse, 5′-GATTGGTGTTGCGGAACATG-3′; and β-actin forward, 5′-TGGAACGGTGAAGGTGACAG-3′ and reverse, 5′-GGC TTTTAGGATGGCAAGGG-3′.
Western blot analysis
For each tissue specimen, 50 mg of tissue was transferred into a 1.5 ml microcentrifuge tube. A total of 500 µl cell lysis buffer was added to the tube. The tissue was homogenized on ice by 10–15 strokes (3–4 sec/stroke) of a mini-homogenizer and a plastic pestle. The sample was centrifuged at 12,000 × g for 15 min at 4°C and the supernatant was then transferred to a fresh tube. A total of 50 µg protein and an equal volume of 2X sample buffer were heated at 94°C for 5 min. The proteins were separated on an 8% sodium dodecyl sulfate-polyacrylamide gel and then transblotted onto polyvinylidene difluoride membranes. The membranes were blocked in PBS with Tween 20 (Tianjin Guangfu Fine Chemical Research Institute, Tianjin, China) and 5% skimmed dried milk at 37°C for 1 h. The membranes were incubated with anti-ATF5 (dilution, 1:2,000), anti-IE86 (dilution, 1:40), anti-P300 (dilution, 1:1,000), anti-acetyl-histone H3K9 (dilution, 1:1,000) and anti-acetyl-histone H3K14 (dilution, 1:2,000) primary antibodies at 4°C overnight. The membranes were then treated with goat anti-rabbit (dilution, 1:2,000; cat. no. BA1054-1) and goat anti-mouse (dilution, 1:2,000; cat. no. BA1050-0.5) IgG secondary antibodies (BosterBio, Pleasanton, CA, USA). The western blot membranes were developed using enhanced chemiluminescence reagents and visualized using the BIO-PRINT ST4 gel imaging system (Vilber Lourmat, Marne-la-Vallée, France).
Statistical analysis
Statistical analyses were performed using SPSS 19.0 (IBM, Armonk, NY, USA). One-way analysis of variance and least significant difference test were used to analyze the present data. P<0.05 was considered to indicate a statistically significant difference. The values are expressed as the mean ± standard deviation.
Results
Expression of ATF5, IE86, P 300, acetyl-histone H3K9 and acetyl-histone H3K14 in GBM, anaplastic glioma, low-grade glioma and normal cortex tissues
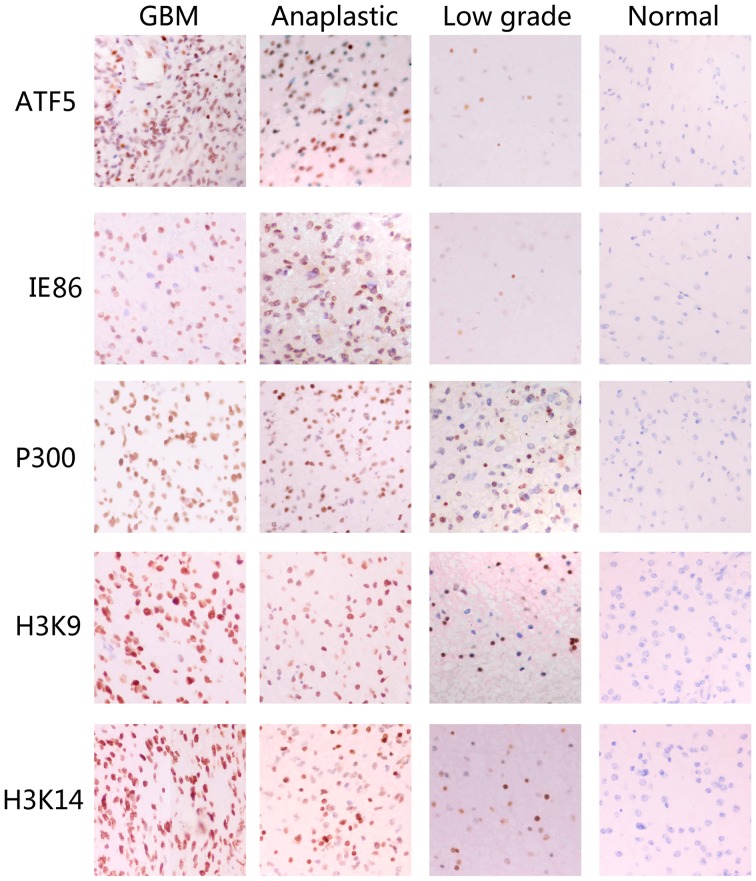
Immunohistochemical analysis revealed that the expression of the ATF5, IE86, P300, acetyl-histone H3K9 and acetyl-histone H3K14 proteins in GBM and anaplastic glioma tissues was significantly increased compared with the expression in the normal cortex tissues. However, there was little difference between the expression levels of the ATF5, IE86, P300, acetyl-histone H3K9 and acetyl-histone H3K14 proteins in the low-grade glioma tissues and normal cortex tissues (Fig. 1). The results indicated that the protein expression levels of ATF5, IE86, P300, acetyl-histone H3K9 and acetyl-histone H3K14 may be associated with the clinical development and classification of gliomas.
Figure 1.
IE86, ATF5, P300, acetyl-histone H3K9 and acetyl-histone H3K14 overexpression is associated with the histological level of human glioma tissue specimens. Immunohistochemical staining with the indicated specific antibodies was performed on 25 glioblastoma, 16 anaplastic astrocytoma, 19 low-grade gloma and 9 normal cortex tissues samples. The representative images of three tumor samples indicate the pattern of IE86, ATF5, P300, acetyl-histone H3K9 and acetyl-histone H3K14 expression in different sections. IE86, immediate-early 2 protein; ATF5, activating transcription factor 5; GBM, glioblastoma multiforme.
Analysis of protein expression in the tissue specimens revealed that the expression level of ATF5 in GBM tissues was evidently increased compared with the expression in anaplastic glioma, low-grade glioma and normal cortex tissues (P<0.05). The expression level of the ATF5 protein in anaplastic glioma tissues was significantly increased compared with the expression in low-grade glioma and normal cortex tissues (P<0.05). However, the expression level of the ATF5 protein in low-grade glioma tissues was slightly increased compared with the expression in the normal cortex (P>0.05). It was concluded that patient age and gender are not associated with the expression levels of ATF5 in glioma tissues, but ATF5 expression is associated with the classification of gliomas.
Immunohistochemical staining also revealed that IE expression was markedly increased in GBM tissues compared with the expression in low-grade glioma and normal cortex tissues (P<0.05). However, the expression level of IE86 was not evidently increased in GBM tissues compared with the expression in anaplastic glioma tissues (P>0.05). In addition, the expression level of IE86 in low-grade glioma tissues was not significantly different from the expression in the normal cortex tissues (P>0.05). However, the expression level of IE86 in anaplastic glioma tissues was significantly increased compared with the expression in low-grade glioma and normal cortex tissues (P<0.05). The expression level of IE86 in low-grade glioma tissues was also significantly increased (P<0.05) compared with the expression in the normal cortex tissues. Therefore, the expression level of IE86 in glioma was associated with the classification of gliomas, which was independent of patient gender and age.
The expression level of P300 in GBM tissues was significantly increased (P<0.05) compared with the expression in anaplastic glioma, low-grade glioma and normal cortex tissues. The expression level of P300 in GBM tissues was significantly increased (P<0.05) compared with the expression in low-grade glioma and normal cortex tissues. However, there was no difference in the expression levels of P300 between the low-grade glioma and normal cortex (P>0.05). It was concluded that the expression level of P300 is associated with the classification of glioma, but is not associated with patient gender or age.
There are marked variations in the expression levels of acetyl-histone H3K9 in GBM tissues compared with the expression levels in anaplastic glioma, low-grade glioma and normal cortex tissues (P<0.05). The expression level of acetyl-histone H3K9 in anaplastic glioma tissues is extremely different from the expression level in low-grade glioma and normal cortex tissues (P<0.05). However, there is little difference between the expression levels of acetyl-histone H3K9 in low-grade glioma tissues and normal cortex tissues (P>0.05). The expression level of acetyl-histone H3K9 was associated with the classification of gliomas, but was not associated with age or gender.
The expression level of acetyl-histone H3K14 in GBM tissues was markedly different from the expression in anaplastic glioma, low-grade glioma and normal cortex tissues (P<0.05). The expression level of acetyl-histone H3K14 in anaplastic glioma was significantly different from the expression levels in low-grade glioma and normal cortex tissues (P<0.05). There was no difference between the expression levels of acetyl-histone H3K14 in low-grade glioma and that in the normal cortex (P>0.05). The expression levels of acetyl-histone H3K14 demonstrated no association with age or gender.
Overall, the present results indicated that as the malignancy degree of glioma increased, the increased expression of ATF5, IE86 and P300 demonstrated a notable significance in evaluating the malignancy degree of human glioma. The level of histone acetylation, indicated by H3K9 and H3K14, was positively associated with the malignant grade of brain glioma (Tables I–V).
Table I.
Expression of ATF5 in GBM, anaplastic glioma, low-grade glioma and the normal cortex.
| Expression of ATF5 | |||
|---|---|---|---|
| Clinical characteristics | High, n | Low, n | P-value |
| Age, years | |||
| Median | 61 | 58 | 0.261 |
| Range | 42–80 | 41–75 | |
| Gender | |||
| Male | 18 | 14 | 0.171 |
| Female | 21 | 16 | |
| Grade | |||
| GBM | 24 | 1 | |
| Anaplastic glioma | 12 | 4 | 0.045a |
| Low-grade glioma | 2 | 17 | 0.045a |
| 0.000b | |||
| Normal cortex | 1 | 8 | 0.000a |
| 0.002b | |||
| 0.963c | |||
Compared with GBM;
Compared with anaplastic glioma;
Compared with low-grade glioma. ATF5, activating transcription factor 5; GBM, glioblastoma multiforme.
Table V.
Expression of Acetyl-Histone H3K14 in GBM, anaplastic glioma, low-grade glioma and the normal cortex.
| Expression of acetyl-histone H3K14 | |||
|---|---|---|---|
| Clinical characteristics | High, n | Low, n | P-value |
| Age, year | |||
| Median | 60 | 58 | 0.277 |
| Range | 50–70 | 49–67 | |
| Gender | |||
| Male | 18 | 15 | 0.168 |
| Female | 22 | 14 | |
| Grade | |||
| GBM | 25 | 0 | |
| Anaplastic glioma | 12 | 4 | 0.008a |
| Low-grade glioma | 3 | 16 | 0.000a |
| 0.000b | |||
| Normal cortex | 0 | 9 | 0.000a |
| 0.000b | |||
| 0.207c | |||
Compared with GBM;
Compared with anaplastic glioma;
Compared with low-grade glioma. GBM, glioblastoma multiforme.
mRNA expression of ATF5 and P300 increased with IE2 as the malignancy of glioma increased
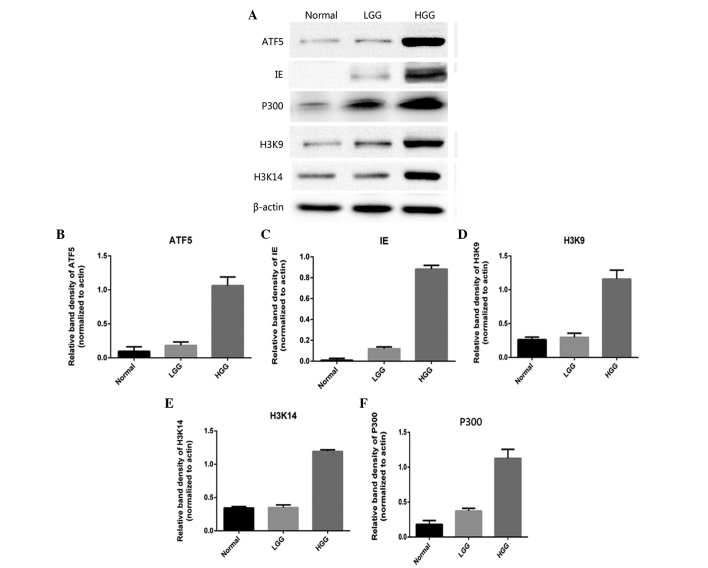
RT-qPCR and western blot analysis were performed to investigate the expression of ATF5, IE2, P300, H3K9 and H3K14 mRNA in human glioma and to elucidate the clinical significance of this mRNA expression. As shown in Fig. 2, the expression levels of ATF5, IE2 and P300 mRNA demonstrated a sustainable increase between the normal cortex tissues and GBM tissues. The highest levels of ATF5, IE2 and P300 mRNA expression were identified in the samples obtained from patients with GBM. The expression levels of ATF5, IE2 and P300 mRNA were by far the lowest percentage in the LGG and normal cortex groups. GBM and anaplastic gliomas were considered to be high-grade gliomas. As shown in Fig. 3, western blot analysis revealed that the expression of ATF5, IE2, P300, acetyl-histone H3K9 and acetyl-histone H3K14 mRNA was significantly increased in the high-grade glioma tissue specimens compared with the normal cortex tissues. However, this increase in expression was not evident in low-grade glioma tissues.
Figure 2.
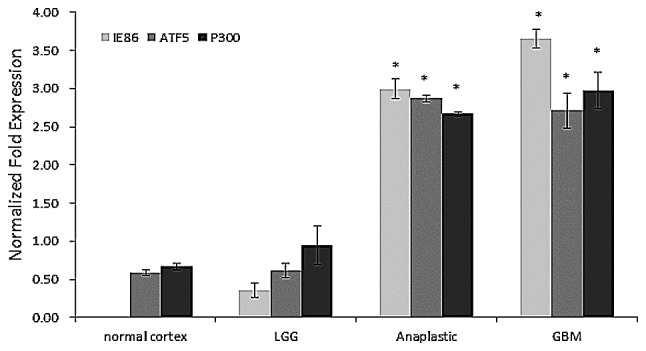
Reverse transcription-quantitative polymerase chain reaction analysis of the expression level of IE2, ATF5 and P300 mRNA in various glioma types. The mRNA levels of ATF5, IE2 and P300 demonstrated an increasing trend between the normal cortex and GBM tissues. *P<0.05 vs. LGG. IE2, immediate-early 2; ATF5, activating transcription factor 5; GBM, glioblastoma multiforme; LGG, low-grade glioma.
Figure 3.
(A) Western blot analysis of the expression level of IE86, ATF5, P300, acetyl-histone H3K9 and acetyl-histone H3K14 acetylation in various types of glioma. Western blot analysis performed on various grades of human glioma specimens confirmed the immunohistochemical data. The data indicated that the expression of (B) ATF5, (C) IE86, (D) acetyl-histone H3K9, (E) acetyl-histone H3K14 and (F) P300 was highest in HGG, consisting of GBM and anaplastic glioma, followed by LGG and then the normal cortex tissues. IE86, immediate-early 2 protein; ATF5, activating transcription factor 5; GBM, glioblastoma multiforme; LGG, low-grade glioma; HGG, high-grade glioma.
Discussion
Based on the findings and results of previous studies that have investigated the activity of HCMV proteins in glioma tissues and glioma cell lines, there is considered to be sufficient evidence to conclude that HCMV sequences and viral gene expression exist in the majority of malignant gliomas (3–5,8). However, previous studies have not focused on determining the association between the host cell proteins and high expression of HCMV, or on the role of HCMV as a glioma-initiating event. In the present study, the expression of ATF5, a basic leucine zipper transcription factor, and the cell acetylation level were revealed to be positively associated with the expression of the HCMV immediate-early regulator protein IE86.
In general, there is considered to be sufficient evidence to support the hypothesis that HCMV can alter the malignant phenotype in glioma via modulation of the proliferation, apoptosis and differentiation of glioma cells . It is becoming clear that the post-translational modification of histone and non-histone proteins by acetylation is a component of an important cellular signaling process that controls tumor progression. Previous studies have identified this signaling pathway as one of the primary targets of viral proteins subsequent to HCMV infection. IE proteins are the proteins that are initially expressed following HCMV infection. There is also an association between high levels of IE86 expression and the malignant progression of glioma (4,10,25). A previous study has reported that IE86 interacts directly with chromatin acetylation factor P300/CBP-associated factor during infection. The interaction of the IE86 protein with P300 has been revealed to lead to the dissociation of the myoblast determination protein (MyoD)-P300 complex and to the downregulation of MyoD transcriptional activity (20,26). The acetylation of histone H3 at K9 (H3K9) or K14 (H3K14) is increased during the initial phases of infection at the immediate-early promoters in human MRC-5 embryonic lung cells (27). Overall, these results indicate that high levels of IE86 expression are associated with the malignant progression of glioma and may affect the acetylation of glioma cells. In the present study, the expression of acetyl transferase P300 and the acetylation level of H3K9 and H3K14 were detected. The high expression of P300, H3K9 and H3K14 in high-grade glioma tissues and the association with the malignant progression of glioma suggested that the presence of HCMV IE86 in glioma cells may regulate the cell signaling pathway by acetylating histone and non-histone proteins.
IE86 has been revealed to bind to a number of cellular factors, indicating that IE86 may also act as a bridge between transcription factors and increase the recruitment of general transcription factors to the pre-initiation complex (21,22,28–30). Previous studies have reported that IE2-86 forms complexes with P300 and CBP, which in turn bind to CREB and may act as adaptor proteins for the function of CREB (22). As a member of the ATF/CREB family, ATF5 contains associated basic leucine zipper domains. The basic region is enriched with lysine and arginine residues and is involved in DNA binding, whereas the leucine zipper motif mediates protein-protein interactions (12,31). Notably, it has previously been indicated that ATF5 may also form a complex with P300 and enhance ATF5 acetylation (32). Therefore, it was hypothesized that ATF5 is positively associated with the increased expression of IE86. In the present study, immunohistochemical analysis revealed that IE86 expression is associated with ATF5 expression and the clinical development and classification of gliomas, but the expression of IE86 is independent of patient gender and age. GBM and anaplastic glioma were classified as high-grade gliomas, and the present study identified no significant difference between the expression levels of ATF5, IE86 and histone acetylation protein in these two tissue types. RT-qPCR and western blot analysis were then performed to investigate the expression of ATF5, IE86, P300, acetyl-histone H3K9 and acetyl-histone H3K14 in the high-grade and low-grade glioma tissues. The expression of all proteins was significantly increased in the high-grade glioma tissues compared with low-grade glioma tissues and tissues from the normal cortex.
The exact role of HCMV in glioma remains under investigation. There is no conclusive evidence of the transformation of normal human cells subsequent to HCMV infection (33). Therefore, HCMV may modulate the malignant properties of the tumor cells through mechanisms that affect the cell cycle, survival, invasive potential, chromosomal stability, immunodetection and angiogenic properties of the cells. Glioma cells provide a genetic environment, including the high level of ATF5 and histone acetylation, that is characterized by disturbances in intracellular signaling pathways, transcriptional control and tumor suppressor proteins, which enables HCMV to exert oncomodulatory effects on tumor cells, but not in normal cells (12). Thus, the excessive expression of the HCMV IE86 and ATF5 proteins is a predictor of the malignancy level in glioma patients and may be a novel target of molecular therapies for glioma. However, the exact role of HCMV in regulating glioma malignancy through ATF5 should be investigated in future studies.
Table II.
Expression of IE86 in GBM, anaplastic glioma, low-grade glioma and the normal cortex.
| Expression of IE86 | |||
|---|---|---|---|
| Clinical characteristics | High, n | Low, n | P-value |
| Age, year | |||
| Median | 59 | 57 | 0.243 |
| Range | 40–68 | 43–71 | |
| Gender | |||
| Male | 23 | 15 | 0.147 |
| Female | 20 | 11 | |
| Grade | |||
| GBM | 25 | 0 | |
| Anaplastic glioma | 15 | 1 | 0.206a |
| Low-grade glioma | 3 | 16 | 0.000a |
| 0.000b | |||
| Normal cortex | 0 | 9 | 0.000a |
| 0.000b | |||
| 0.207c | |||
Compared with GBM;
Compared with anaplastic glioma;
Compared with low-grade glioma. IE86, immediate-early-2; GBM, glioblastoma multiforme.
Table III.
Expression of P300 in GBM, anaplastic glioma, low-grade glioma and the normal cortex.
| Expression of P300 | |||
|---|---|---|---|
| Clinical characteristics | High, n | Low, n | P-value |
| Age, year | |||
| Median | 58 | 56 | 0.198 |
| Range | 45–71 | 44–68 | |
| Gender | |||
| Male | 19 | 17 | 0.145 |
| Female | 18 | 15 | |
| Grade | |||
| GBM | 25 | 0 | |
| Anaplastic glioma | 11 | 5 | 0.003a |
| Low-grade glioma | 1 | 18 | 0.000a |
| 0.000b | |||
| Normal cortex | 0 | 9 | 0.000a |
| 0.001b | |||
| 0.483c | |||
Compared with GBM;
Compared with anaplastic glioma;
Compared with low-grade glioma. GBM, glioblastoma multiforme.
Table IV.
Expression of Acetyl-Histone H3K9 in GBM, anaplastic glioma, low-grade glioma and the normal cortex.
| Expression of acetyl-histone H3K9 | |||
|---|---|---|---|
| Clinical characteristics | High, n | Low, n | P-value |
| Age, year | |||
| Median | 58 | 61 | 0.254 |
| Range | 40–76 | 42–80 | |
| Gender | |||
| Male | 18 | 18 | 0.183 |
| Female | 16 | 17 | |
| Grade | |||
| GBM | 25 | 0 | |
| Anaplastic glioma | 7 | 9 | 0.000a |
| Low-grade glioma | 2 | 17 | 0.000a |
| 0.025b | |||
| Normal cortex | 0 | 9 | 0.000a |
| 0.019b | |||
| 0.312c | |||
Compared with GBM;
Compared with anaplastic glioma;
Compared with low-grade glioma. GBM, glioblastoma multiforme.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant no. 81471958) and the Natural Science Foundation of Shandong Province (grant no. J 122211).
Glossary
Abbreviations
- ATF5
activating transcription factor 5
- HCMV
human cytomegalovirus
- IE
immediate early genes
- WHO
World Health Organization
- CREB
cyclic adenosine monophosphate response element-binding protein
- CBP
CREB-binding protein
- GBM
glioblastoma
- HGG
high-grade glioma, consisting of GBM and anaplastic glioma
- HAT
histone acetyltransferase
References
- 1.Ranganathan P, Clark PA, Kuo JS, Salamat MS, Kalejta RF. Significant association of multiple human cytomegalovirus genomic loci with glioblastoma multiforme samples. J Virol. 2012;86:854–864. doi: 10.1128/JVI.06097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dziurzynski K, Wei J, Qiao W, Hatiboglu MA, Kong LY, Wu A, Wang Y, Cahill D, Levine N, Prabhu S, et al. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin Cancer Res. 2011;17:4642–4649. doi: 10.1158/1078-0432.CCR-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2011;157:193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barami K. Oncomodulatory mechanisms of human cytomegalovirus in gliomas. J Clin Neurosci. 2010;17:819–823. doi: 10.1016/j.jocn.2009.10.040. [DOI] [PubMed] [Google Scholar]
- 5.Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: Increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cinatl J, Scholz M, Kotchetkov R, Vogel JU, Doerr HW. Molecular mechanisms of the modulatory effects of HCMV infection in tumor cell biology. Trends Mol Med. 2004;10:19–23. doi: 10.1016/j.molmed.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Duan YL, Ye HQ, Zavala AG, Yang CQ, Miao LF, Fu BS, Seo KS, Davrinche C, Luo MH, Fortunato EA. Maintenance of large numbers of virus genomes in human cytomegalovirus-infected T98 G glioblastoma cells. J Virol. 2014;88:3861–3873. doi: 10.1128/JVI.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dziurzynski K, Chang SM, Heimberger AB, Kalejta RF, Dallas SR McGregor, Smit M, Soroceanu L, Cobbs CS. HCMV and Gliomas Symposium: Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol. 2012;14:246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo MH, Fortunato EA. Long-term infection and shedding of human cytomegalovirus in T98 G glioblastoma cells. J Virol. 2007;81:10424–10436. doi: 10.1128/JVI.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, Nabors LB, Cobbs CG, Britt WJ. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–3350. [PubMed] [Google Scholar]
- 11.Liu X, Liu D, Qian D, Dai J, An Y, Jiang S, Stanley B, Yang J, Wang B, Liu X, Liu DX. Nucleophosmin (NPM1/B23) interacts with activating transcription factor 5 (ATF5) protein and promotes proteasome- and caspase-dependent ATF5 degradation in hepatocellular carcinoma cells. J Biol Chem. 2012;287:19599–19609. doi: 10.1074/jbc.M112.363622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng Z, Evans SK, Green MR. An activating transcription factor 5-mediated survival pathway as a target for cancer therapy? Oncotarget. 2010;1:457–460. doi: 10.18632/oncotarget.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angelastro JM, Canoll PD, Kuo J, Weicker M, Costa A, Bruce JN, Greene LA. Selective destruction of glioblastoma cells by interference with the activity or expression of ATF5. Oncogene. 2006;25:907–916. doi: 10.1038/sj.onc.1209116. [DOI] [PubMed] [Google Scholar]
- 14.Liu DX, Qian D, Wang B, Yang JM, Lu Z. P300-Dependent ATF5 acetylation is essential for Egr-1 gene activation and cell proliferation and survival. Mol Cell Biol. 2011;31:3906–3916. doi: 10.1128/MCB.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang T, Qian D, Hu M, Li L, Zhang L, Chen H, Yang R, Wang B. Human cytomegalovirus inhibits apoptosis by regulating the activating transcription factor 5 signaling pathway in human malignant glioma cells. Oncol Lett. 2014;8:1051–1057. doi: 10.3892/ol.2014.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Zhang YD, Zhang YY, Qian SW, Zhang ZC, Li SF, Guo L, Liu Y, Wen B, Lei QY, et al. P300-dependent acetylation of activating transcription factor 5 enhances C/EBPβ transactivation of C/EBPα During 3T3-L1 differentiation. Mol Cell Biol. 2013;34:315–324. doi: 10.1128/MCB.00956-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu L, Zhang X, Che Y, Zhang Y, Tang S, Liao Y, Na R, Xiong X, Liu L, Li Q. A cellular response protein induced during HSV-1 infection inhibits viral replication by interacting with ATF5. Sci China Life Sci. 2013;56:1124–1133. doi: 10.1007/s11427-013-4569-y. [DOI] [PubMed] [Google Scholar]
- 18.Nevels M, Paulus C, Shenk T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci USA. 2004;101:17234–17239. doi: 10.1073/pnas.0407933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang ES, Zhang Z, Cai H, Huang DY, Huong SM, Cha CY, Huang ES. Human cytomegalovirus IE1-72 protein interacts with p53 and inhibits p53-dependent transactivation by a mechanism different from that of IE2-86 protein. J Virol. 2009;83:12388–12398. doi: 10.1128/JVI.00304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryant LA, Mixon P, Davidson M, Bannister AJ, Kouzarides T, Sinclair JH. The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF. J Virol. 2000;74:7230–7237. doi: 10.1128/JVI.74.16.7230-7237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodems SM, Clark CL, Spector DH. Separate DNA elements containing ATF/CREB and IE86 binding sites differentially regulate the human cytomegalovirus UL112-113 promoter at early and late times in the infection. J Virol. 1998;72:2697–2707. doi: 10.1128/jvi.72.4.2697-2707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz R, Helmich B, Spector DH. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J Virol. 1996;70:6955–6966. doi: 10.1128/jvi.70.10.6955-6966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Xu Y, Wang H, Niu J, Hou H, Jiang Y. Histone deacetylase inhibitor, valproic acid, radiosensitizes the C6 glioma cell line in vitro. Oncol Lett. 2014;7:203–208. doi: 10.3892/ol.2013.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lois DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. World Organization Classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheurer ME, Bondy ML, Aldape KD, Albrecht T, El-Zein R. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 2008;116:79–86. doi: 10.1007/s00401-008-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caron C, Col E, Khochbin S. The viral control of cellular acetylation signaling. Bioessays. 2003;25:58–65. doi: 10.1002/bies.10202. [DOI] [PubMed] [Google Scholar]
- 27.Cuevas-Bennett C, Shenk T. Dynamic Histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J Virol. 2008;82:9525–9536. doi: 10.1128/JVI.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stinski MF, Petrik DT. Functional roles of the human cytomegalovirus essential IE86 protein. Curr Top Microbiol Immunol. 2008;325:133–152. doi: 10.1007/978-3-540-77349-8_8. [DOI] [PubMed] [Google Scholar]
- 29.Taylor RT, Bresnahan WA. Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFkappaB-dependent gene expression. J Virol. 2006;80:10763–10771. doi: 10.1128/JVI.01195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo YD, Chiou CJ, Choi KS, Yi Y, Michelson S, Kim S, Hayward GS, Kim SJ. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor beta1 gene through an Egr-1 binding site. J Virol. 1996;70:7062–7070. doi: 10.1128/jvi.70.10.7062-7070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheng Z, Li L, Zhu LJ, Smith TW, Demers A, Ross AH, Moser RP, Green MR. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med. 2010;16:671–677. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu DX, Qian D, Wang B, Yang JM, Lu Z. p300-Dependent ATF5 acetylation is essential for Egr-1 gene activation and cell proliferation and survival. Mol Cell Biol. 2010;31:3906–3916. doi: 10.1128/MCB.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao XH, Dong X, Wu C, Wang T, Liu F, Zhou J, Zhang TC. Human cytomegalovirus immediate early protein 2 enhances myocardin-mediated survival of rat aortic smooth muscle cells. Virus Res. 2014;192:85–91. doi: 10.1016/j.virusres.2014.08.007. [DOI] [PubMed] [Google Scholar]