Abstract
Osteosarcoma (OS) is the most commonly diagnosed bone tumor in young adults under the age of 20. Metastasis is considered an important factor underlying cancer-associated morbidity and mortality, and, as a result, the survival rate of patients with metastatic OS is low. In spite of this, the mechanisms underlying metastasis in OS are currently not well understood. The present study compared gene expression levels between five non-metastatic and four metastatic OS tumor samples, using an Affymetrix microarray. A total of 282 genes were differentially expressed in the metastatic samples, as compared with the non-metastatic samples. Of these differentially expressed genes (DEGs), 212 were upregulated and 70 were downregulated. The following DEGs were associated with metastasis: Homeobox only protein; lysosomal-associated membrane protein-3; chemokine (C-C motif) ligand-18; carcinoembryonic antigen-related cell adhesion molecule-6; keratin-19; prostaglandin-endoperoxide synthase-2; clusterin; and nucleoside diphosphate kinase-1. Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathway analyses were conducted, which identified 529 biological processes (P<0.01) and 10 KEGG pathways (P<0.05) that were significantly over-represented in the metastatic samples, as compared with the non-metastatic samples. Interaction networks for the DEGs were constructed using the corresponding GO terms and KEGG pathways, and these identified numerous genes that may contribute to OS metastasis. Among the enriched biological processes, four DEGs were consistently over-represented: Jun proto-oncogene, caveolin-1, nuclear factor-κB-inhibitor-α and integrin alpha-4; thus suggesting that they may have key roles in OS metastasis, and may be considered potential therapeutic targets in the treatment of patients with OS.
Keywords: osteosarcoma, bone tumor, osteosarcoma lung metastases
Introduction
Osteosarcoma (OS) is the most common malignant bone tumor, which typically develops in children and adolescents. The estimated annual incidence worldwide is 10 patients per million people, with the majority of cases occurring in patients under 20 years of age (1,2). OS has a high propensity for metastasis, and ~80% of all metastases occur in the lungs (3). The 5-year survival rate for patients with primary OS tumors has greatly improved from ~20 to 70% since the introduction of chemotherapy in the 1970s; however, the survival rate for patients with metastatic OS is estimated to be <30%, based on the current therapeutic strategies (4). Conventional chemotherapy remains the standard method for treating patients with OS; however, patients with metastatic OS have previously demonstrated poor responses to chemotherapeutic drugs (5). Therefore, developing novel strategies to improve the treatment of patients with metastatic OS will require elucidation of the mechanisms underlying metastasis in patients with OS, and its corresponding biological processes.
Whole genome expression and proteomic analyses are able to detect the abnormal expression of genes in OS samples, and thus permit the identification of various metastasis-associated targets (6). It has previously been reported that thatezrin, a member of the ezrin-radixin-moesin family, is highly expressed in metastatic OS cell lines, and is required for OS metastasis, due to its functional connection with the actin cytoskeleton (7). In addition, the expression of Fas has previously been associated with OS metastasis, and an inhibitor of the Fas pathway (c-FLIP) has been developed as a potential treatment for patients with lung metastasis (8). Nagao-Kitamoto et al (9) demonstrated that knockdown of GLI family zinc finger 2 (GLI2) using RNA interference was able to significantly attenuate the migration and invasion of OS cells; thus suggesting that inhibition of GLI2 may be a potential strategy for the treatment of patients with metastatic OS. Furthermore, numerous microRNAs (miRNAs) have been implicated in the OS metastatic process, including miRNA-20a, miRNA-143, miRNA-202 and miRNA-9 (10–12).
In the present study, a high-throughput method was used to identify factors associated with the OS metastatic process, and potential novel targets that may be considered as biomarkers for the treatment of patients with metastatic OS. The aims of the present study were to identify metastasis-associated genes for OS tumor and to extend our mechanistic understanding of metastatic processes in OS cells. The results may provide new insight into therapeutic strategy for OS patients.
Materials and methods
Data collection
The Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) was searched, and microarray expression data (GSE14359) from two groups was obtained, which included five non-metastatic OS samples and four OS lung metastases tumor samples. Each sample had two replicates, and the data were analyzed using the Affymetrix Human Genome U133A Array (Affymetrix, Inc., Santa Clara, CA, USA). Unprocessed data sets (.cel files) were collected for further analysis. The probe annotation files were downloaded for further research.
Data processing and filtering
Numerous algorithms have been developed in order to quantify microarray signals, and the present study applied Guanine Cytosine Robust Multi-Array Analysis (13). The normalization process consisted of three steps: i) Model-based background correction; ii) quantile normalization; and iii) summarizing.
In order to filter out uninformative data, including control probe sets and other internal controls, as well as genes whose expression levels were uniformly close to the background detection levels, the nsFilter function from the genefilter package in R programming language was used (14). However, the filter was unable to remove probe-sets without Entrez Gene identifiers or with identical Entrez Gene identifiers.
Analysis of differentially expressed genes (DEGs)
Statistical comparisons between the two groups were conducted. Limma in the nsFilter function from the genefilter package in R programming language version 3.1.1 was used. to identify genes that were significantly differentially expressed between the two groups (15). For probes with identical Entrez Gene identifiers, only the probes occupying the biggest variance were retained for further DEG analysis. In addition, only DEGs with a log2 (fold change) >1.5 and an adjusted P<0.01 were recognized as statistically significant. The adjusted P-value was obtained by applying Benjamini and Hochberg's false discovery rate correction on the original P-value (16). The fold change threshold was selected based on the requirement for focusing on only genes that were significantly differentially expressed.
Hierarchical clustering
Hierarchical clustering was conducted using the DEGs in order to classify the samples according to their gene expression profiles and observe global alterations in gene expression patterns (17). The DEGs were classified into specific biological processes (GO terms) and KEGG pathways, which were represented in heat maps. To be specific, DEG expression values were used in the hierarchical clustering analysis using gplots software (18).
GO and KEGG pathway analysis
The R packages GO.db (19) and KEGGREST (20) were used to detect GO categories and KEGG pathways that were significantly over represented in the DEGs compared with the whole genome. P<0.01 indicated significantly enriched GO terms, whereas P<0.05 indicated significantly enriched KEGG pathways.
Construction of biological networks
Protein-protein interaction (PPI) databases were downloaded from the Human Protein Reference Database (HPRD; http://www.hprd.org/) (21), the Biological General Repository for Interaction Datasets (BioGRID; http://thebiogrid.org/) (22), and the Human Protein-Protein Interaction Prediction database (PIPs; http://www.compbio.dundee.ac.uk/www-pips/) (23). The pair interactions, which were included in all three databases, were selected to be included in the curated PPI database. A total of 56,1405 pair interactions were identified in the PPI database. Cytoscape version 3.1.1 (http://www.cytoscape.org/) was used to construct interaction networks. Interacting gene pairs from the curated PPI database were imported as a stored network. Following functional enrichment analysis, the DEGs specified in significantly enriched GO terms and KEGG pathways were mapped to corresponding networks in order to analyze the interaction relationships.
Results
Analysis of DEGs
A comparison of the gene expression levels between metastatic and non-metastatic OS tumor samples was conducted using microarray analysis. A log2 fold-change >1.5 and an adjusted P<0.01 indicated that a gene was significantly differentially expressed. A total of 282 DEGs were obtained, of which 212 were upregulated and 70 were downregulated (Table I). The top 50 upregulated and downregulated DEGs are presented in Table II. Of the DEGs, at least eight genes may have been associated with OS metastasis to the lungs, including the homeobox only protein (HOPX), lysosomal-associated membrane protein-3 (LAMP3), chemokine (C-C motif) ligand-18 (CCL18), carcinoembryonic antigen-related cell adhesion molecule-6 (CEACAM6), keratin-19 (KRT19), prostaglandin-endoperoxide synthase-2 (PTGS2), clusterin (CLU), and nucleoside diphosphate kinase-1 (NME1).
Table I.
Statistical distribution of differentially expressed genes.
| Probes and genes | Probes | Genes |
|---|---|---|
| Alla | 10,348 | 7,323 |
| Differentially expressedb | 347 | 282 |
| Upregulated | 265 | 212 |
| Downregulated | 82 | 70 |
Affymetrix Human Genome U133A Array;
[log(fold change)] >1.5 and adjusted P<0.01.
Table II.
The top 50 upregulated and downregulated differentially expressed genes (DEGs) in the metastatic osteosarcoma (OS) tissue samples, as compared with the non-metastatic OS tissue samples.
| Gene symbol | Log2 (fold-change) | P-value | Adjusted P-value |
|---|---|---|---|
| Upregulated | |||
| SFTPC | 10.13 | 2.93E-22 | 3.26E-18 |
| SFTPA2 | 9.46 | 1.58E-17 | 3.50E-14 |
| SFTPB | 8.35 | 2.01E-16 | 3.72E-13 |
| SCGB1A1 | 6.43 | 3.23E-07 | 6.93E-05 |
| SFTPD | 6.27 | 1.63E-09 | 1.21E-06 |
| CYP4B1 | 5.46 | 2.33E-08 | 8.91E-06 |
| TACSTD2 | 5.44 | 4.06E-09 | 2.60E-06 |
| HOPX | 5.22 | 4.46E-11 | 5.50E-08 |
| LAMP3 | 5.16 | 6.76E-09 | 3.58E-06 |
| SERPINA1 | 5.01 | 1.73E-10 | 1.60E-07 |
| CLDN18 | 5.00 | 1.70E-10 | 1.60E-07 |
| CCL18 | 4.83 | 2.48E-08 | 8.91E-06 |
| C4BPA | 4.52 | 6.24E-07 | 1.05E-04 |
| GPRC5A | 4.43 | 9.93E-08 | 2.56E-05 |
| SLC34A2 | 4.32 | 1.43E-09 | 1.13E-06 |
| ADH1B | 4.21 | 5.01E-06 | 4.19E-04 |
| AGER | 4.19 | 6.63E-05 | 2.18E-03 |
| CEACAM6 | 4.13 | 2.23E-07 | 5.17E-05 |
| HPGD | 3.95 | 5.12E-07 | 9.01E-05 |
| MUC1 | 3.87 | 2.49E-08 | 8.91E-06 |
| CXCL2 | 3.74 | 1.76E-04 | 3.94E-03 |
| CDH1 | 3.71 | 4.49E-09 | 2.62E-06 |
| TMEM100 | 3.56 | 6.11E-05 | 2.09E-03 |
| TNS1 | 3.52 | 3.32E-07 | 6.96E-05 |
| SLPI | 3.43 | 4.91E-04 | 7.11E-03 |
| FOS | 3.33 | 3.80E-05 | 1.59E-03 |
| MFAP4 | 3.33 | 1.01E-08 | 4.47E-06 |
| MARCO | 3.27 | 1.69E-04 | 3.85E-03 |
| CAV1 | 3.18 | 3.83E-07 | 7.87E-05 |
| KRT19 | 3.17 | 3.03E-06 | 3.01E-04 |
| MEST | 3.15 | 1.95E-04 | 4.13E-03 |
| IGJ | 3.11 | 3.34E-04 | 5.65E-03 |
| AQP3 | 3.10 | 6.90E-07 | 1.09E-04 |
| KRT7 | 3.09 | 4.65E-07 | 8.85E-05 |
| EMP2 | 3.09 | 3.16E-06 | 3.06E-04 |
| ICAM1 | 3.08 | 6.29E-08 | 1.84E-05 |
| FCN3 | 3.05 | 3.34E-04 | 5.65E-03 |
| C1orf116 | 3.05 | 4.00E-05 | 1.61E-03 |
| PTGS2 | 3.02 | 2.48E-06 | 2.62E-04 |
| CD52 | 3.01 | 8.12E-06 | 5.94E-04 |
| CLIC5 | 3.00 | 1.37E-04 | 3.50E-03 |
| CLIC3 | 2.99 | 8.67E-05 | 2.67E-03 |
| VWF | 2.98 | 1.78E-04 | 3.95E-03 |
| CAV2 | 2.97 | 1.92E-06 | 2.21E-04 |
| CLU | 2.92 | 1.15E-04 | 3.13E-03 |
| APOC1 | 2.89 | 1.98E-04 | 4.18E-03 |
| CTSH | 2.88 | 3.47E-05 | 1.51E-03 |
| INHBA | 2.87 | 2.96E-05 | 1.37E-03 |
| TPSAB1 | 2.87 | 2.02E-05 | 1.09E-03 |
| PTGDS | 2.87 | 1.17E-05 | 7.63E-04 |
| Downregulated | |||
| MMP13 | -4.21 | 1.61E-06 | 2.00E-04 |
| PRAME | −3.72 | 5.97E-05 | 2.07E-03 |
| UCHL1 | −3.68 | 1.85E-04 | 4.01E-03 |
| RRM2 | −2.78 | 3.09E-06 | 3.01E-04 |
| KIAA0101 | −2.72 | 3.62E-06 | 3.34E-04 |
| ITGA4 | −2.71 | 3.65E-11 | 5.06E-08 |
| ADAM12 | −2.60 | 2.71E-04 | 4.96E-03 |
| CCNB1 | −2.57 | 4.13E-04 | 6.38E-03 |
| GOLM1 | −2.55 | 5.02E-08 | 1.51E-05 |
| ADAMTS5 | −2.44 | 2.66E-07 | 6.02E-05 |
| SLC6A15 | −2.43 | 1.78E-04 | 3.95E-03 |
| MAD2L1 | −2.39 | 2.95E-04 | 5.19E-03 |
| NEK2 | −2.35 | 6.61E-05 | 2.18E-03 |
| NCALD | −2.32 | 5.23E-04 | 7.38E-03 |
| CDK1 | −2.28 | 5.58E-04 | 7.68E-03 |
| BUB1 | −2.27 | 5.15E-07 | 9.01E-05 |
| ACP1 | −2.15 | 2.27E-05 | 1.16E-03 |
| TRIP13 | −2.15 | 3.25E-07 | 6.93E-05 |
| BCAT1 | −2.14 | 5.19E-05 | 1.92E-03 |
| RAD51AP1 | −2.09 | 2.95E-05 | 1.37E-03 |
| IGF2BP3 | −2.07 | 7.05E-04 | 8.83E-03 |
| CCNB2 | −2.05 | 6.36E-04 | 8.31E-03 |
| RFC5 | −2.04 | 2.75E-08 | 9.24E-06 |
| SRPK1 | −2.03 | 6.66E-04 | 8.53E-03 |
| AURKA | −2.03 | 3.16E-05 | 1.44E-03 |
| HIST3H2A | −2.00 | 1.02E-04 | 2.94E-03 |
| TYMS | −1.99 | 2.26E-04 | 4.48E-03 |
| PHB | −1.99 | 2.06E-07 | 4.97E-05 |
| KIF20A | −1.91 | 7.55E-04 | 9.22E-03 |
| RMDN1 | −1.88 | 2.41E-04 | 4.62E-03 |
| NME1 | −1.87 | 5.56E-06 | 4.50E-04 |
| MRPL42 | −1.86 | 7.13E-08 | 2.03E-05 |
| DNAJC12 | −1.86 | 6.06E-04 | 8.07E-03 |
| NPM3 | −1.86 | 1.08E-04 | 3.04E-03 |
| MRPS16 | −1.85 | 1.21E-06 | 1.65E-04 |
| PFKM | −1.85 | 2.62E-04 | 4.85E-03 |
| NDUFS1 | −1.85 | 7.34E-08 | 2.04E-05 |
| HSPE1 | −1.84 | 3.04E-05 | 1.39E-03 |
| NUSAP1 | −1.83 | 4.10E-04 | 6.38E-03 |
| RPP40 | −1.81 | 1.22E-04 | 3.21E-03 |
| ALG13 | −1.80 | 8.54E-05 | 2.64E-03 |
| EEF1E1 | −1.80 | 1.48E-05 | 8.69E-04 |
| MRPL35 | −1.79 | 3.93E-08 | 1.26E-05 |
| HMMR | −1.79 | 1.08E-06 | 1.51E-04 |
| UQCRFS1 | −1.77 | 2.74E-04 | 4.97E-03 |
| TWF1 | −1.77 | 2.42E-06 | 2.58E-04 |
| GGH | −1.71 | 5.79E-05 | 2.06E-03 |
| OIP5 | −1.70 | 4.83E-05 | 1.83E-03 |
| MCTS1 | −1.69 | 8.91E-08 | 2.36E-05 |
| CEP55 | −1.67 | 2.94E-06 | 2.96E-04 |
DEGs were selected if they had a log2(fold change) >1.5 and an adjusted P<0.01.
Construction of biological networks
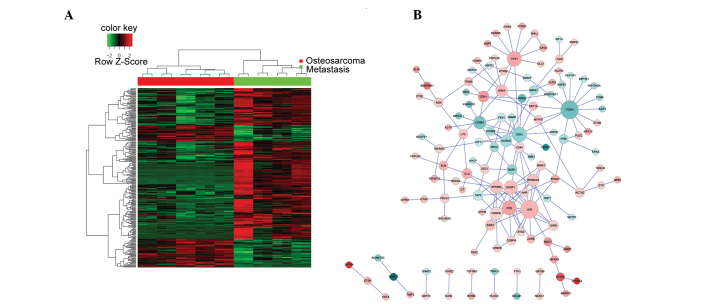
The non-metastatic and metastatic OS tumor samples had different gene expression profiles, as demonstrated by the heatmap representing the hierarchical clustering of all DEGs (Fig. 1A). Biological networks for upregulated and downregulated DEGs were constructed according to the protein-protein interactions identified in the HPRD, BIOGRID, and PIP databases (Fig. 1B). The majority of proteins were involved in >1 sub-networks. A total of four genes: JUN, CAV1, NFKB1A and ITGA4, were consistently over-represented in the networks; thus suggesting that they may contribute to the OS metastatic process. Of the four genes, JUN, CAV1 and NFKBIA were upregulated in the metastatic OS tumor samples, as compared with the non-metastatic OS tumor samples. Conversely, ITGA4 was downregulated in the metastatic OS tumor samples, as compared with the non-metastatic OS tumor samples. The network of all the DEGs was too complex to allow elucidation of the various functions of the sub-networks; thus requiring further analysis.
Figure 1.
A heat map of all the DEGs and their corresponding biological networks. (A) Heat map of hierarchical clustering of all DEGs (five conventional OS and four OS lung metastases tumor samples). Red and green in the heat map indicate high and low relative expression levels, respectively. (B) A biological network was constructed in order to visualize the interactions of all DEGs. The red/pink and blue coloring in the network indicate upregulated and downregulated expression levels, respectively. The darker the color, the greater the gene is differentially expressed between the metastatic and non-metastatic OS tissue samples. DEGs, differentially expressed genes; OS, osteosarcoma.
GO and KEGG pathway analysis
A GO and KEGG pathway analysis was performed for the DEGs. A total of 529 GO terms and 10 KEGG pathways were over-represented in the metastatic OS tumor samples, as compared with the non-metastatic OS tumor samples (Table III). P<0.01 indicated significantly enriched GO terms, whereas P<0.05 indicated significantly enriched KEGG pathways. The five most significantly enriched GO terms included: Cell proliferation; response to external stimulus; positive regulation of biological process; cell migration; and cellular component organization or biogenesis (Table IV). The most significantly enriched KEGG pathways included: Extracellular matrix (ECM)-receptor interaction; cell adhesion molecules; complement and coagulation cascades; and osteoclast differentiation (Table V). Heat maps and biological networks of the five GO terms and the top KEGG pathways are presented in Figs. 2 and 3, respectively. The network analysis identified four genes that may be involved in the molecular events associated with the metastasis of OS cells to the lungs: JUN, CAV1, NFKBIA and ITGA4; thus suggesting that these genes may be potential targets for the treatment of patients with metastatic OS.
Table III.
Over-represented GO biological processes and KEGG pathways.
| Analysis | P-value | Number |
|---|---|---|
| GO BP | <0.01 | 529 |
| KEGG pathways | <0.05 | 10 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological processes.
Table IV.
Significantly altered Gene Ontology (GO) terms.
| GO-ID | P-value | Count | Term |
|---|---|---|---|
| GO:0008283 | 2.74E-09 | 63 | Cell proliferation |
| GO:0009605 | 6.13E-09 | 66 | Response to external stimulus |
| GO:0048518 | 1.75E-08 | 110 | Positive regulation of biological process |
| GO:0016477 | 7.80E-08 | 41 | Cell migration |
| GO:0071840 | 8.03E-08 | 125 | Cellular component organization or biogenesis |
Table V.
Significantly altered Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
| KEGG-ID | P-value | Count | Term |
|---|---|---|---|
| 04512 | 3.30E-04 | 8 | Extracellular matrix-receptor interaction |
| 04514 | 1.59E-03 | 9 | Cell adhesion molecules |
| 04610 | 2.82E-03 | 6 | Complement and coagulation cascades |
| 04380 | 1.61E-02 | 7 | Osteoclast differentiation |
Figure 2.
Heat maps (left) and corresponding biological networks (right) for the five most significantly altered GO biological processes. (A) Cell proliferation (GO:0008283), (B) response to external stimulus (GO:0009605), and (C) positive regulation of biological process (GO:0048518). Red and green in the heat maps indicate high and low relative expression levels, respectively. The red/pink and blue coloring in the networks indicate upregulated and downregulated expression levels, respectively. The darker the color, the greater the gene expression levels were altered between the metastatic and non-metastatic osteosarcoma tissue samples. GO, gene ontology. (D) Cell migration (GO:0016477) and (E) cellular component organization or biogenesis (GO:0071840). Red and green in the heat maps indicate high and low relative expression levels, respectively. The red/pink and blue coloring in the networks indicate upregulated and downregulated expression levels, respectively. The darker the color, the greater the gene expression levels were altered between the metastatic and non-metastatic osteosarcoma tissue samples. GO, gene ontology.
Figure 3.
Heat map (left) and corresponding biological network (right) of significantly altered Kyoto Encyclopedia of Genes and Genomes pathways. (A) Extracellular matrix-receptor interaction, (B) cell adhesion molecules, and (C) osteoclast differentiation. Red, high relative levels of expression; green, low relative levels of expression in the heat map. Red/pink in the network, upregulated expression; blue in the network, downregulated expression. The darker the color, the greater the gene expression is altered.
Discussion
OS is the most common malignant bone tumor in children and adolescents. The prognosis for patients with OS is affected by whether or not metastasis to the lungs has occurred; thus highlighting the importance of developing novel therapeutic strategies for the treatment of patients with metastatic OS. Numerous genes associated with metastasis have previously been described for other types of cancer (24); however, the molecular mechanisms underlying the metastasis of OS are currently not well understood. The present study aimed to investigate the underlying metastatic processes of OS in order to identify potential biomarkers for the treatment of patients with metastatic OS.
The present study compared the gene expression profiles of non-metastatic and metastatic OS tissue samples, using microarray analysis. A total of 282 DEGs, including 212 upregulated and 70 downregulated DEGs, were identified, all of which had >1.5-fold change in gene expression levels. In addition, significantly enriched GO terms and KEGG pathways were analyzed, in order to identify significantly altered molecular events that were associated with metastasis, and biological networks were constructed to screen for candidate metastasis-associated genes, which may provide potential therapeutic targets for the treatment of patients with metastatic OS.
According to previous studies, a number of the DEGs identified in the present study may be associated with OS metastatic processes, including HOPX, LAMP3, CCL18, CEACAM6, KRT19, PTGS2, CLU and NME1. The majority of these genes, with the exception of NME1, were upregulated in the metastatic OS tissue samples, as compared with the non-metastatic OS tissue samples.
HOPX contains a homeobox-like domain lacking DNA binding properties due to the loss of required conserved residues. HOPX is a core regulator of epigenetics, and its aberrant expression has previously been associated with the progression of cancer (25). In addition, HOPX has been demonstrated to be involved in the metastatic process of sarcoma cells, and its knockdown decreased cell motility and metastasis formation in vitro and in vivo (26). The present study detected upregulation of HOPX in metastatic OS samples, as compared with in non-metastatic OS tissue samples; thus suggesting that upregulation of HOPX may accelerate metastasis in patients with OS.
LAMP3 is a member of the LAMP family of proteins and is recurrently upregulated in cancer cells (27). Previous studies have demonstrated an association between cell migration and LAMP3 expression in some solid tumors, including breast cancer (28) and cervical cancer (29). Furthermore, overexpression of LAMP3 has been associated with an enhanced metastatic potential in a cervical xenograft model (29). Alongside the findings of the present study, these results indicated that LAMP3 may promote the mobility of OS tumor cells.
It has previously been suggested that chemokines in the tumor microenvironment have an important role in tumor progression and metastasis (30). CCL18 is a small cytokine predominantly produced by the innate immune system. Li et al (31) suggested that CCL18 was able to induce breast cancer metastasis via phosphorylation of protein tyrosine kinase-2 and proto-oncogene tyrosine-protein kinase, and concomitant downstream signaling (31). In the present study, CCL18 was demonstrated to be overexpressed in the metastatic OS tumor samples, as compared with the non-metastatic OS tumor samples; thus suggesting that CCL18 may have a potential role in OS metastasis.
CEACAM6 belongs to a family of carcinoembryonic antigen cell adhesion molecules, and has functions in various biological processes, including cancer progression, inflammation, angiogenesis and metastasis (32). Previous studies have detected an association between CEACAM6 expression and poor prognosis of patients with primary cancers. As a marker of cancer progression and metastasis, the CEACAM protein family is considered to have high therapeutic value. Furthermore, specific monoclonal antibodies against CEACAM1 and CEACAM6 have been developed and have demonstrated their potential in the treatment of numerous types of cancer (33). The results of the present study demonstrated that CEACAM6 was upregulated in the metastatic OS samples, as compared with the non-metastatic OS samples; thus suggesting that CEACAM6 may be considered a potential therapeutic target for the treatment of patients with metastatic OS.
KRT-19 is a member of the keratin family, which is responsible for maintaining the structural integrity of epithelial cells. The expression of KRT-19 has previously been associated with chemoresistance, and was demonstrated to confer an invasive potential on human hepatocellular carcinoma cells (HCC) (34). In the present study, KRT-19 was upregulated in the metastatic OS tissue samples, as compared with the non-metastatic OS tissue samples; thus suggesting that KRT-19 may have conferred an invasive ability on OS tumor cells.
PTGS2, which is also known as cyclooxygenase 2, catalyzes the conversion of arachidonic acid and O2 to prostaglandin H2, which is an important precursor in prostanoid biosynthesis. The overexpression of PTGS2 has been associated with the pathogenesis and cell mobility of various tumors, including Ewing sarcoma (35) and osteosarcoma (36,37). The selective PTGS2 inhibitors, celecoxib and meloxicam, have previously been demonstrated to inhibit cell proliferation and invasion in vitro and in vivo (35,38). In line with previous studies, upregulation of PTGS2 in the present study may have been associated with initiation of the OS metastatic process.
CLU is a 75–80 kDa heterodimeric protein, which is involved in the clearance of cellular debris and apoptosis (39). Furthermore, a role for CLU in tumor invasion has previously been reported for various cancer types; its downregulation via short hairpin RNA reduced migratory ability, whereas upregulation of CLU was associated with increased cell invasion in HCC (40). In the present study, CLU expression levels were upregulated in the metastatic OS tissue samples, as compared with the non-metastatic OS tissues samples; thus suggesting that CLU may increase the metastatic potential of OS.
NME1 exerts nucleoside diphosphate kinase and histidine protein kinase activities. It was initially identified as a suppressor of metastasis, and has been shown to be associated with numerous biological processes, including cell migration, proliferation and differentiation (41). Previous studies have suggested that the anti-metastatic effects of NME1 occur due to inhibition of the Ras/extracellular signal-regulated kinase signaling pathway in numerous types of human cancer, including melanoma, breast and stomach carcinomas (42–44). In addition, alterations in the expression profiles of genes regulated by NME1 have been demonstrated in melanoma and thyroid carcinomas (45). In the present study, NME1 expression levels were decreased in the metastatic OS tissue samples, as compared with the non-metastatic OS tissue samples; thus suggesting that cell migration and invasion may result from the downregulation of NME1 expression levels in the OS cells.
In the present study, the identified DEGs were clustered according to their functions by performing GO term and KEGG enrichment pathway analyses. GO term analysis was used to identify metastasis-associated biological processes that were over-represented in the OS metastatic tumor samples, as compared with the non-metastatic samples. The most significant biological processes included cell proliferation, response to external stimulus, positive regulation of biological process, cell migration, and cellular component organization or biogenesis. Notably, the majority of the DEGs were associated with cell proliferation and cell migration, which are key processes in metastasis. In addition, cellular responses to the tumor microenvironment were important upon the migration of the OS cells to the lungs. Metastasis is a complex cascade of events, which requires a precise gene regulatory network in order to overcome barriers that exist within the tumor microenvironment. The present GO analysis suggested that aberrant regulation of molecular events may contribute to metastasis of OS.
KEGG enrichment analysis demonstrated that the DEGs were involved in numerous pathways that have previously been associated with metastatic processes. Furthermore, the identification of pathways involving ECM-receptor interactions and cell adhesion molecules corroborated the results obtained from the GO analysis. It has previously been reported that cancer cell invasion and migration involves the degradation of ECM proteins, including matrix metalloproteinases and integrins (46). Based on the GO term and KEGG pathway analyses, the present study obtained a comprehensive understanding of the DEGs identified in the metastatic samples, including their functions, and upstream and downstream relationships.
In the present study, a signal network was constructed in order to identify the number of genes involved in significant biological processes and pathways. A total of four genes: JUN, CAV1, NFKBIA and ITGA4, were associated with numerous enriched biological pathways and formed the center of the network. All of the genes, with the exception of ITGA4, were upregulated in the metastatic OS samples, as compared with the non-metastatic OS samples. The JUN gene encodes the c-Jun protein, which may be activated via double phosphorylation in the c-Jun N-terminal kinase signaling pathway. It has previously been suggested that c-Jun may have roles in cell proliferation, the cell cycle, apoptosis prevention, and cancer progression (47); however, there is currently no direct evidence that associates c-Jun with metastasis. Sze et al (48) demonstrated that the C-terminal truncation of the hepatitis B virus X protein increased HCC cell migration via activation of c-Jun (48). Furthermore, activated c-Jun was predominantly expressed in invasive breast cancer cells and associated with proliferation and angiogenesis (49). CAV1 is a multi-functional scaffold protein associated with cell surface caveolae, which has previously been demonstrated to regulate numerous cancer-associated processes, including tumor growth, cell death and survival, and cellular transformation (50). In addition, numerous studies have associated CAV1 with metastasis. For example, upregulation of CAV1 has been associated with enhanced metastatic potential and exacerbated prognosis in HCC cells (51) and Ewing's sarcoma (52). NFKBIA is a member of the inhibitor of κB (IkB) proteins, which are able to inhibit the nuclear localization of nuclear factor-κB (53). There is currently no report supporting a role for NFKBIA in tumor metastasis; however, IkBg, another member of the IkB proteins, was previously shown to promote the metastatic progression of melanoma (54). The ITGA4 gene encodes the integrin alpha4 protein, which is involved in cell-cell and cell-ECM interactions (55). Previous studies have detected that over-expression of ITGA4 is associated with a reduction in the cell invasion of numerous types of cancer (56–58). In the present study, a network analysis enabled the identification of the most significantly enriched genes with the highest repetition frequency, which may be associated with OS metastasis.
In conclusion, the present study identified a total of 282 DEGs in the metastatic OS tissue samples, as compared with the non-metastatic OS issue samples, of which 212 were upregulated and 70 were downregulated. GO term, KEGG pathway and network analyses identified numerous genes that may have a role in the metastasis of OS cells, and these may be considered as potential therapeutic targets in the treatment of patients with OS.
References
- 1.Whelan J, McTiernan A, Cooper N, Wong YK, Francis M, Vernon S, Strauss SJ. Incidence and survival of malignant bone sarcomas in England 1979-2007. Int J Cancer. 2012;131:E508–E517. doi: 10.1002/ijc.26426. [DOI] [PubMed] [Google Scholar]
- 2.Bielack SS, Carrle D, Hardes J, Schuck A, Paulussen M. Bone tumors in adolescents and young adults. Curr Treat Options Oncol. 2008;9:67–80. doi: 10.1007/s11864-008-0057-1. [DOI] [PubMed] [Google Scholar]
- 3.Hegyi M, Semsei AF, Jakab Z, Antal I, Kiss J, Szendroi M, Csoka M, Kovacs G. Good prognosis of localized osteosarcoma in young patients treated with limb-salvage surgery and chemotherapy. Pediatr Blood Cancer. 2011;57:415–422. doi: 10.1002/pbc.23172. [DOI] [PubMed] [Google Scholar]
- 4.Botter SM, Neri D, Fuchs B. Recent advances in osteosarcoma. Curr Opin Pharmacol. 2014;16:15–23. doi: 10.1016/j.coph.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Mialou V, Philip T, Kalifa C, Perol D, Gentet JC, Marec-Berard P, Pacquement H, Chastagner P, Defaschelles AS, Hartmann O. Metastatic osteosarcoma at diagnosis: Prognostic factors and long-term outcome-the French pediatric experience. Cancer. 2005;104:1100–1109. doi: 10.1002/cncr.21263. [DOI] [PubMed] [Google Scholar]
- 6.Flores RJ, Li Y, Yu A, Shen J, Rao PH, Lau SS, Vannucci M, Lau CC, Man TK. A systems biology approach reveals common metastatic pathways in osteosarcoma. BMC Syst Biol. 2012;28:50–67. doi: 10.1186/1752-0509-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Zhang L, Zhang G, Li S, Duan J, Cheng J, Ding G, Zhou C, Zhang J, Luo P, et al. Osteosarcoma metastasis: Prospective role of ezrin. Tumour Biol. 2014;35:5055–5059. doi: 10.1007/s13277-014-1799-y. [DOI] [PubMed] [Google Scholar]
- 8.Rao-Bindal K, Rao CK, Yu L, Kleinerman ES. Expression of c-FLIP in pulmonary metastases in osteosarcoma patients and human xenografts. Pediatr Blood Cancer. 2013;60:575–579. doi: 10.1002/pbc.24412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagao-Kitamoto H, Nagata M, Nagano S, Kitamoto S, Ishidou Y, Yamamoto T, Nakamura S, Tsuru A, Abematsu M, Fujimoto Y, et al. GLI2 is a novel therapeutic target for metastasis of osteosarcoma. Int J Cancer. 2015;136:1276–1284. doi: 10.1002/ijc.29107. [DOI] [PubMed] [Google Scholar]
- 10.Huang G, Nishimoto K, Zhou Z, Hughes D, Kleinerman ES. miR-20a encoded by the miR-17-92 cluster increases the metastatic potential of osteosarcoma cells by regulating Fas expression. Cancer Res. 2012;72:908–916. doi: 10.1158/0008-5472.CAN-11-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Cai X, Wang Y, Tang H, Tong D, Ji F. MicroRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol Rep. 2010;24:1363–1369. doi: 10.3892/or_00000994. [DOI] [PubMed] [Google Scholar]
- 12.Diao CY, Guo HB, Ouyang YR, Zhang HC, Liu LH, Bu J, Wang ZH, Xiao T. Screening for metastatic osteosarcoma biomarkers with a DNA microarray. Asian Pac J Cancer Prev. 2014;15:1817–1822. doi: 10.7314/APJCP.2014.15.4.1817. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Irizarry R, MacDonald J, Gentry J. Gcrma: Background adjustment using sequence information. R package. version 2.36.0. [Google Scholar]
- 14.Gentleman R, Carey V, Huber W, Hahne F. Genefilter: Methods for filtering genes from microarray experiments. R package. version 1.46.1. [Google Scholar]
- 15.Smyth GK. Limma: Linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 2005:397–420. doi: 10.1007/0-387-29362-0_23. [DOI] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 17.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 18.Warnes GR, Bolker B, Bonebakker L, et al. gplots: Various R programming tools for plotting data. R package version 2. 2009 [Google Scholar]
- 19.Carlson M. GOdb: A set of annotation maps describing the entire Gene Ontology. R package. version 2.14.0. [Google Scholar]
- 20.Tenenbaum D. KEGGREST: Client-side REST access to KEGG. R package. version 1.4.0. [Google Scholar]
- 21.Prasad Keshava TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database-2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O'Donnell L, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDowall MD, Scott MS, Barton GJ. PIPs: Human protein-protein interaction prediction database. Nucleic Acids Res. 2009;37:D651–D656. doi: 10.1093/nar/gkn870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Ma L. The emerging molecular machinery and therapeutic targets of metastasis. Trends Pharmacol Sci. 2015;36:349–359. doi: 10.1016/j.tips.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita K, Katoh H, Watanabe M. The homeobox only protein homeobox (HOPX) and colorectal cancer. Int J Mol Sci. 2013;14:23231–23243. doi: 10.3390/ijms141223231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovárová D, Plachy J, Kosla J, Trejbalová K, Čermák V, Hejnar J. Downregulation of HOPX controls metastatic behavior in sarcoma cells and identifies genes associated with metastasis. Mol Cancer Res. 2013;11:1235–1247. doi: 10.1158/1541-7786.MCR-12-0687. [DOI] [PubMed] [Google Scholar]
- 27.Ozaki K, Nagata M, Suzuki M, Fujiwara T, Ueda K, Miyoshi Y, Takahashi E, Nakamura Y. Isolation and characterization of a novel human lung-specific gene homologous to lysosomal membrane glycoproteins 1 and 2: Significantly increased expression in cancers of various tissues. Cancer Res. 1998;58:3499–3503. [PubMed] [Google Scholar]
- 28.Nagelkerke A, Bussink J, Mujcic H, Wouters BG, Lehmann S, Sweep FC, Span PN. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res. 2013;15:R2. doi: 10.1186/bcr3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanao H, Enomoto T, Kimura T, Fujita M, Nakashima R, Ueda Y, Ueno Y, Miyatake T, Yoshizaki T, Buzard GS, et al. Overexpression of LAMP3/TSC403/DC-LAMP promotes metastasis in uterine cervical cancer. Cancer Res. 2005;65:8640–8645. doi: 10.1158/0008-5472.CAN-04-4112. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Yao Y, Gong C, Yu F, Su S, Liu B, Deng H, Wang F, Lin L, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li HY, Cui XY, Wu W, Yu FY, Yao HR, Liu Q, Song EW, Chen JQ. Pyk2 and Src mediate signaling to CCL18-induced breast cancer metastasis. J Cell Biochem. 2014;115:596–603. doi: 10.1002/jcb.24697. [DOI] [PubMed] [Google Scholar]
- 32.Kuespert K, Pils S, Hauck CR. CEACAMs: Their role in physiology and pathophysiology. Curr Opin Cell Biol. 2006;18:565–571. doi: 10.1016/j.ceb.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32:643–671. doi: 10.1007/s10555-013-9444-6. [DOI] [PubMed] [Google Scholar]
- 34.Govaere O, Komuta M, Berkers J, Spee B, Janssen C, de Luca F, Katoonizadeh A, Wouters J, van Kempen LC, Durnez A, et al. Keratin 19: A key role player in the invasion of human hepatocellular carcinomas. Gut. 2014;63:674–685. doi: 10.1136/gutjnl-2012-304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barlow M, Edelman M, Glick RD, Steinberg BM, Soffer SZ. Celecoxib inhibits invasion and metastasis via a cyclooxygenase 2-independent mechanism in an in vitro model of Ewing sarcoma. J Pediatr Surg. 2012;47:1223–1227. doi: 10.1016/j.jpedsurg.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, Cai M, Ji F, Lou LM. The impact of COX-2 on invasion of osteosarcoma cell and its mechanism of regulation. Cancer Cell Int. 2014;14:27. doi: 10.1186/1475-2867-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee EJ, Choi EM, Kim SR, Park JH, Kim H, Ha KS, Kim YM, Kim SS, Choe M, Kim JI, Han JA. Cyclooxygenase-2 promotes cell proliferation, migration and invasion in U2OS human osteosarcoma cells. Exp Mol Med. 2007;39:469–476. doi: 10.1038/emm.2007.51. [DOI] [PubMed] [Google Scholar]
- 38.Naruse T, Nishida Y, Hosono K, Ishiguro N. Meloxicam inhibits osteosarcoma growth, invasiveness and metastasis by COX-2-dependent and independent routes. Carcinogenesis. 2006;27:584–592. doi: 10.1093/carcin/bgi240. [DOI] [PubMed] [Google Scholar]
- 39.Pucci S, Mazzarelli P, Nucci C, Ricci F, Spagnoli LG. CLU “in and out”: Looking for a link. Adv Cancer Res. 2009;105:93–113. doi: 10.1016/S0065-230X(09)05006-4. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Jiang K, Kang X, Gao D, Sun C, Li Y, Sun L, Zhang S, Liu X, Wu W, et al. Tumor-derived secretory clusterin induces epithelial-mesenchymal transition and facilitates hepatocellular carcinoma metastasis. Int J Biochem Cell Biol. 2012;44:2308–2320. doi: 10.1016/j.biocel.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 41.Marshall JC, Collins J, Marino N, Steeg P. The Nm23-H1 metastasis suppressor as a translational target. Eur J Cancer. 2010;46:1278–82. doi: 10.1016/j.ejca.2010.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takács-Vellai K. The metastasis suppressor Nm23 as a modulator of Ras/ERK signaling. J Mol Signal. 2014;9:4. doi: 10.1186/1750-2187-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarrett SG, Novak M, Dabernat S, Daniel JY, Mellon I, Zhang Q, Harris N, Ciesielski MJ, Fenstermaker RA, Kovacic D, et al. Metastasis suppressor NM23-H1 promotes repair of UV-induced DNA damage and suppresses UV-induced melanomagenesis. Cancer Res. 2012;72:133–143. doi: 10.1158/0008-5472.CAN-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouatas T, Salerno M, Palmieri D, Steeg PS. Basic and translational advances in cancer metastasis: Nm23. J Bioenerg Biomembr. 2003;35:73–79. doi: 10.1023/A:1023497924277. [DOI] [PubMed] [Google Scholar]
- 45.McCorkle JR, Leonard MK, Kraner SD, Blalock EM, Ma D, Zimmer SG, Kaetzel DM. The metastasis suppressor NME1 regulates expression of genes linked to metastasis and patient outcome in melanoma and breast carcinoma. Cancer Genomics Proteomics. 2014;11:175–194. [PMC free article] [PubMed] [Google Scholar]
- 46.Price JT, Thompson EW. Mechanisms of tumour invasion and metastasis: Emerging targets for therapy. Expert Opin Ther Targets. 2002;6:217–233. doi: 10.1517/14728222.6.2.217. [DOI] [PubMed] [Google Scholar]
- 47.Vogt PK. Fortuitous convergences: The beginnings of JUN. Nat Rev Cancer. 2002;2:465–469. doi: 10.1038/nrc818. [DOI] [PubMed] [Google Scholar]
- 48.Sze KM, Chu GK, Lee JM, Ng IO. C-terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology. 2013;57:131–139. doi: 10.1002/hep.25979. [DOI] [PubMed] [Google Scholar]
- 49.Vleugel MM, Greijer AE, Bos R, van der Wall E, van Diest PJ. c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum Pathol. 2006;37:668–674. doi: 10.1016/j.humpath.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 50.Burgermeister E, Liscovitch M, Röcken C, Schmid RM, Ebert MP. Caveats of caveolin-1 in cancer progression. Cancer Lett. 2008;268:187–201. doi: 10.1016/j.canlet.2008.03.055. [DOI] [PubMed] [Google Scholar]
- 51.Yu H, Shen H, Zhang Y, Zhong F, Liu Y, Qin L, Yang P. CAV1 promotes HCC cell progression and metastasis through Wnt/β-catenin pathway. PLoS One. 2014;9:e106451. doi: 10.1371/journal.pone.0106451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sáinz-Jaspeado M, Lagares-Tena L, Lasheras J, Navid F, Rodriguez-Galindo C, Mateo-Lozano S, Notario V, Sanjuan X, Del Garcia Muro X, Fabra A, Tirado OM. Caveolin-1 modulates the ability of Ewing's sarcoma to metastasize. Mol Cancer Res. 2010;8:1489–1500. doi: 10.1158/1541-7786.MCR-10-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crépieux P, Kwon H, Leclerc N, Spencer W, Richard S, Lin R, Hiscott J. I kappaB alpha physically interacts with a cytoskeleton-associated protein through its signal response domain. Mol Cell Biol. 1997;17:7375–7385. doi: 10.1128/mcb.17.12.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torabian SZ, de Semir D, Nosrati M, Bagheri S, Dar AA, Fong S, Liu Y, Federman S, Simko J, Haqq C, et al. Ribozyme-mediated targeting of IkappaBgamma inhibits melanoma invasion and metastasis. Am J Pathol. 2009;174:1009–1016. doi: 10.2353/ajpath.2009.080207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kummer C, Ginsberg MH. New approaches to blockade of alpha4-integrins, proven therapeutic targets in chronic inflammation. Biochem Pharmacol. 2006;72:1460–1468. doi: 10.1016/j.bcp.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 56.Park J, Song SH, Kim TY, Choi MC, Jong HS, Kim TY, Lee JW, Kim NK, Kim WH, Bang YJ. Aberrant methylation of integrin alpha4 gene in human gastric cancer cells. Oncogene. 2004;23:3474–3480. doi: 10.1038/sj.onc.1207470. [DOI] [PubMed] [Google Scholar]
- 57.Qian F, Vaux DL, Weissman IL. Expression of the integrin alpha 4 beta 1 on melanoma cells can inhibit the invasive stage of metastasis formation. Cell. 1994;77:335–347. doi: 10.1016/0092-8674(94)90149-X. [DOI] [PubMed] [Google Scholar]
- 58.Gosslar U, Jonas P, Luz A, Lifka A, Naor D, Hamann A, Holzmann B. Predominant role of alpha 4-integrins for distinct steps of lymphoma metastasis. Proc Natl Acad Sci USA. 1996;93:4821–4826. doi: 10.1073/pnas.93.10.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]