Abstract
Heme oxygenase-1 (HO-1) is an inducible isoform of HO that is activated in response to oxidative stress and has anti-apoptotic and pro-proliferative effects on leukemia cells. RET, a tyrosine kinase receptor; its expression levels are associated with the differentiation degree of acute myelocytic leukemia (AML) cells. The promyelocytic leukemia (PML) gene inhibits cell proliferation and tumor growth, participates in the differentiation of hematopoietic progenitor cells and induces cell apoptosis. However, the association between the expression levels of HO-1, RET and PML genes and the risk stratification of AML and prognosis have not previously been reported. In the present study, HO-1 was expressed in the human AML Kasumi-1, HL-60 and THP-1 cell lines, and HO-1 expression was regulated by Hemin (20 µmol/l) and ZnPPIX (10 µmol/l). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis demonstrated that expression of RET and PML were positively and negatively correlated with HO-1 expression, respectively. Bone marrow samples (18 favorable, 55 intermediate, 15 adverse and 2 unknown karyotype AML cases and 20 healthy donors) were collected from 90 randomly selected AML patients upon their first visit. The mRNA and protein expression of HO-1, RET and PML in samples was detected by RT-qPCR and western blot analysis. At the mRNA level, the adverse group expressed significantly higher levels of HO-1 and RET compared with the levels in the favorable and normal groups. The PML mRNA expression levels in adverse patient samples was lower compared with those of the intermediate group and favorable group. Western blot analysis demonstrated that the expression levels of HO-1, RET and PML proteins in all risk groups exhibited the same pattern of expression as was observed for the mRNA levels. The overall survival and relapse-free survival rates were shortest in AML patients with high HO-1 expression (Kaplan-Meier; log-rank, P<0.01). The results of the present study therefore indicate that HO-1, RET and PML may be critical in the risk-stratification and prognosis of AML. However, additional samples and clinical data should be collected and analyzed in order to provide stronger evidence for this hypothesis.
Keywords: AML, HO-1, PML, RET, risk stratification, prognosis
Introduction
Acute myelocytic leukemia (AML) is a malignant, clonal disorder of the highly heterogeneous myeloid progenitor cells in the hematopoietic system (1). Treating patients with different prognoses upon first visit by various protocols may dramatically prolong survival and improve the quality of life. Risk stratification based on cytogenetic characteristics has been proposed to divide AML patients into 3 subgroups based on their cytogenetics: Favorable, intermediate and unfavorable risk, with the 5-year overall survival (OS) rates of 36, 22 and 5%, respectively (2). In the last few decades, markers for determining prognosis have been well studied, but performing accurate risk stratification upon diagnosis remains difficult (3). Meanwhile, molecular mechanisms for the etiology and progression of AML remain unclear, and AML genomes are expected to identify additional prognostic markers (4).
AML risk stratification and prognosis are affected by abnormal gene expression and gene mutation. Heme oxygenase-1 (HO-1) is an inducible isoform of HO-1 that is activated in response to oxidative stress, disease or other physiological stresses. The activation of HO-1 may result in a reduction of cell apoptosis and promotion of cell proliferation. In a previous study of 43 AML patients, 33 expressed high levels of HO-1 (77%) (5). In addition, higher levels of HO-1 are expressed in chronic lymphocytic leukemia (CLL) cells compared with normal lymphocytes (6). In a previous study, inducing high expression of HO-1 gene increased the expression of multidrug-resistance gene, accompanied by elevated IC50 values in a number of types of tumor cells (7). In another previous study, silencing the HO-1 gene facilitated the apoptosis of leukemia cells or tumor cells by reducing the antioxidant capacity (8). Moreover, inducing high HO-1 expression inhibited the expression of anti-oncogene P53 and increased the cell proliferation capability 1-fold compared with that of the control group (9). RET is a tyrosine kinase receptor that has been associated with papillary thyroid carcinoma (10), multiple endocrine neoplasia type 2 and gastric cancer (11); its expression level is associated with the degree of differentiation of AML cells (12). The expression of promyelocytic leukemia (PML) protein was first observed in patients with acute PML. The PML protein shares similar characteristics with numerous tumor inhibitors by suppressing cell proliferation and tumor growth, by participating in the differentiation of hematopoietic progenitor cells and by inducing cell apoptosis (13). Although PML is expressed in the majority of cell lines, it is only expressed in myeloid cells of human bone marrow (14). It has previously been reported that PML protein expression was weakened or even vanished when colon and breast cancer cells locally invaded and metastasized (15). Dynamic expression of PML gene is indicative of tumor progression, and has been used in the diagnosis and treatment of acute promyelocytic leukemia; therefore, it may be employed as a diagnostic marker in AML (16,17). To the best of our knowledge, the association between the expression levels of HO-1 and RET and AML and the risk stratification and prognosis of AML have not previously been reported. The present study aimed to endogenously regulate the expressions levels of HO-1 in Kasumi-1, HL-60 and THP-1 cells, and to investigate whether RET and PML genes were associated with HO-1 expression using RT-qPCR. In addition, bone marrow samples were collected from 90 randomly selected AML patients to analyze the association between abnormal expression levels of HO-1, RET and PML and the AML risk stratification and prognosis by using RT-qPCR and western blot analysis.
Materials and methods
Cell lines and cell culture
Human AML cell lines Kasumi-1, HL-60 and THP-1 were purchased from the American Type Culture Collection (Manassas, VA, USA). HO-1 is highly expressed in U937 cells but expressed at lower levels in HL-60 and Kasumi-1 cells. Hemin (20 µmol/l; Sigma-Aldrich, St. Louis, MO, USA) and ZnPPIX (10 µmol/l; Sigma-Aldrich) were used to upregulate and downregulate HO-1 expression, respectively. The cell lines were cultured in RPMI 1640 media with 10% fetal bovine serum (Gibco Life Technologies, Carlsbad, CA, USA) with 100 U/ml penicillin and 100 mg/ml streptomycin (Solarbio Science & Technology, Beijing, China) in a 37°C humidified atmosphere containing 5% CO2/95% air.
Patients and samples
The 90 AML patients (mean age, 36 years; range, 13–75 years) comprised 55 males and 35 females. A total of 20 healthy volunteers were also selected (mean age, 25 years; range, 18–56 years), consisting of 12 males and 8 females: They were either hospitalized patients or bone marrow donors who visited Hospital Affiliated to Guiyang Medical College from February 2011 to August 2014. According to the Southwest Oncology Group classification system (18), the patients were divided into 1 case of M1, 45 cases of M2, 24 cases of M4 and 20 cases of M5. The patients were divided into a low-risk group (18 cases), an intermediate-risk group (55 cases) and a high-risk group (15 cases) based on the guidelines of National Comprehensive Cancer Network (19,20). In addition, chromosome karyotype was unknown in 2 cases. Bone marrow (BM) samples were collected from clinically examined patients and normal donors. All of the patients and volunteers signed written informed consent forms for their samples to be stored and used for research purposes in advance. All data were collected and analyzed anonymously. The present study was approved by the Ethics Committee of the Hospital Affiliated to Guiyang Medical College. The clinical features of AML patients are listed in Table I.
Table I.
Clinical features of 90 AML pretreatment patients.
| Characteristic | No. patients | |
|---|---|---|
| Gender | ||
| Male | 55 | |
| Female | 35 | |
| Age | ||
| <60 | 78 | |
| ≥60 | 12 | |
| FAB subtype | ||
| M1 | 1 | |
| M2 | 45 | |
| M4 | 24 | |
| M5 | 20 | |
| Chromosome karyotype | ||
| Favorable | 18 | |
| Intermediate | 55 | |
| Adverse | 15 | |
| Not obtained | 2 | |
| Type of AML | ||
| De novo | 90 | |
| Secondary | 0 | |
| PB blasts, %a | 46 (0–97) | |
| BM blasts, %a | 60 (22–95) | |
| WBC count, g/la | 43.4 (0.4–420) | |
| Hb concentration, g/la | 80 (56–156) | |
| PLT count, g/la | 65 (30–119) | |
| Induction | ||
| DA | 25 | |
| IA | 45 | |
| TA | 14 | |
| Mortality before induction | 6 | |
| CR following induction therapy, (%) | 60 (73.1) | |
| Consolidation | ||
| MDAC/HDAC | 80 | |
| Mortality before CR/lost before consolidation | 10 | |
| HSCT | ||
| Auto-HSCT | 6 | |
| Allo-HSCT | 28 |
Median (Range).PB, peripheral blood; BM, bone marrow; WBC, white blood cells; Hb, hemaglobin; PLT, platelet; DA, daunorubicin and cytarabine; IA, idarubicin and cytarabine; TA, pirarubicin and cytarabine; CR, complete remission; MDAC, medium dose cytarabine; HDAC, high dose cytarabine; HSCT, hematopoietic stem cell transplantation.
Total RNA extraction and cDNA synthesis
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) was applied to extract total RNA from Kasumi-1, HL-60, THP-1 cells and patient samples. Ultraviolet spectroscopy was performed using an Eppendorf BioPhotometer (Eppendorf, Hamburg, Germany) to determine the RNA concentration and purity. Total RNA (2 µg) was reverse-transcribed into first-strand cDNA using the first-strand PrimeScript™ RT Reagent kit with gDNA Eraser (Takara Bio, Inc., Tokyo, Japan) according to the manufacturer's instructions. The final reaction volume was 20 µl.
Quantitative polymerase chain reaction (qPCR)
cDNA was subjected to qPCR using SYBR-Green PCR Master Mix (TianGen Biotech, Beijing, China) and the ABI Prism 7500 Sequence Detection System (Applied Biosystems Life Technologies, Foster City, CA, USA). Melting curve analysis was applied to guarantee the specificity of amplification. β-Actin was used as the internal control. The primer sequences were as follows: HO-1, sense, 5′-ACC CAT GAC ACC AAG GAC CAGA-3′ and antisense, 5′-GTG TAA GGA CCC ATC GGA GAA GC-3′, with a PCR product of 157 bp; RET, sense, 5′-TCG AGG GAT GCT TAC TGG GA-3′ and antisense, 5′-GTT GTT CTC ATG CAG CCG TG-3′, with a PCR product of 160 bp; PML, sense, 5′-CAG GAG CCC CGT CAT AGG AA-3′ and antisense, 5′-GAG GGC TGG AAG AGA CCGA-3′, with a PCR product of 143 bp; β-actin, sense, 5′-TCA CCC ACA CTG TGC CCA TCT ACGA-3′ and antisense, 5′-CAG CGG AAC CGC TCA TTG CCA ATGG-3′, with a PCR product of 195 bp. Briefly, qPCR (total volume, 20 µl) was performed with 0.5 µl cDNA, 0.75 µl forward primer, 0.75 µl reverse primer, 9 µl SYBR-Green PCR Master Mix and 9 µl dd H2O. The fluorescence was automatically measured using the Mastercycler® ep realplex (Eppendorf). The thermal cycling conditions were: 1 min at 94°C followed with 40 cycles at 94°C for 10 sec, and 60°C for 15 sec, then 72°C for 5 min and finally paused at 4°C. The relative gene expression was calculated relative to β-actin according to the following equations: i) ∆Ct = Ct (Target gene) - Ct (β-actin); ii) ∆∆Ct = ∆Ct (Treatment) - ∆Ct (Control); and iii) Gene expression = 2−ΔCt or Gene expression = 2−ΔΔCt.
Western blot analysis
BM cells were harvested from the clinically examined patients and normal donors. After washing twice in ice-cold PBS, the cells were lysed by sonication in RIPA buffer containing 1 mM Phenylmethanesulfonyl fluoride (Solarbio Science & Technology) for 20 min. The lysate was then centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was collected and mixed with loading buffer. The final solution was boiled for 10 min. An equal amount of protein (100–150 µg) was loaded and run on a 10% SDS-PAGE gel and transferred onto PVDF membranes (Millipore Corporation, Milford, MA, USA), which were then blocked in 5% non-fat milk in TBST buffer at 4°C overnight. The membranes were treated with corresponding primary antibodies at a dilution of 1:1,000 dilution (monoclonal mouse anti-human HO-1 32.8 kDa, cat no. ab12220; monoclonal rabbit anti-human Ret 124 kDa, cat no. ab134100; polyclonal rabbit anti-human PML 69 kDa, cat no. ab53773; monoclonal mouse anti-human β-actin 42 kDa, cat no. ab6276; Abcam, Cambridge, UK) for 2 h while shaking at room temperature. Subsequently, the membranes were washed in TBST 3 times (10 min each) and incubated with the corresponding conjugated secondary antibody at a dilution of 1:1,000 [horseradish peroxidase-labeled (HRP)goat anti-rabbit IgG (H+L), cat no. A0208; HRP-labeled goat anti-mouse IgG (H+L), cat no. A0216; Beyotime Institute of Biotechnology, Shanghai, China) for 2 h at room temperature. After washing in TBST three times (10 min each), protein bands were visualized on film by enhanced chemiluminescence (7sea Biotech, Shanghai, China) following the manufacturer's instructions. The expression levels of the target genes were normalized to that of β-actin.
Statistical analysis
Data were statistically analyzed using SPSS software, version 11.5 (SPSS Inc, Chicago, IL, USA). All data are presented as the mean ± standard deviation. Statistical significance among groups was determined by one-way analysis of variance and q-text. Optical density from western blotting was quantified with Quantity One software (version 4.5.2; Bio-Rad Laboratories, Inc., Hercules, CA, USA). P<0.05 was considered to indicate a statistically significant difference. Kaplan-Meier survival curves were plotted and analyzed with the log-rank test.
Results
Association between mRNA expression levels of RET and PML with HO-1
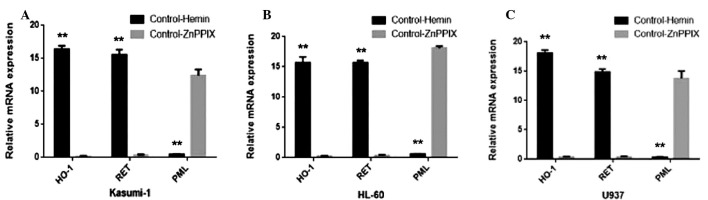
The association between the mRNA expression levels of RET and PML with HO-1 were quantitatively assessed using qPCR and the SYBR® Green I technique in human AML cell lines U937, HL60 and Kasumi-1. 2−ΔΔCt ≥1 was considered to indicate gene upregulation, and 2−ΔΔCt <1 was considered to indicate downregulation. Under the control of Hemin (20 µmol/l), RET and HO-1 was upregulated, and PML was downregulated. Under the control of ZnPPIX (10 µmol/l), PML was upregulated, and RET and HO-1 was downregulated in all 3 cell lines. Thus, regulating the expression of HO-1 was positively associated with RET and negatively associated with PML (Fig. 1).
Figure 1.
Correlation between mRNA expression of RET and PML with HO-1 in (A) Kasumi-1; (B) HL-60; and (C) U937 cells. HO-1 and RET exhibited higher expression following treatment with control-Hemin compared with control-ZnppIX, however, PML expression exhibited the opposite expression pattern in all 3 cell lines. Thus, RET was positively associated with HO-1 and PML was negatively associated with HO-1. The data is represented as the mean ± standard deviation from 3 independent experiments. **P<0.01 vs. control-ZnPPIX. HO-1, heme oxygenase-1; Hemin, the specific inductor of HO-1; ZnPPIX, the specific inhibitors of HO-1; Control, without any intervention.
mRNA expression levels of HO-1, RET and PML in favorable, intermediate and adverse AML patient groups
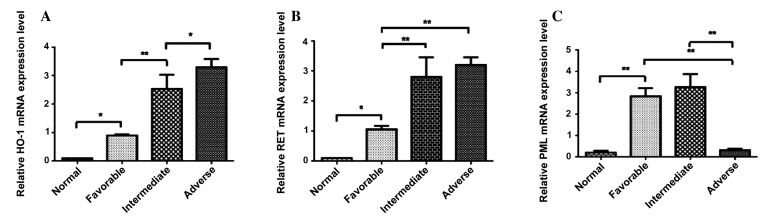
The adverse group (3.2241±0.3262) expressed significantly higher levels of HO-1 compared with the intermediate group (2.4356±0.6064)), intermediate group expressed significantly higher expression levels of HO-1 compared with the favorable group (0.8954±0.0200), and the favorable group expressed significantly higher levels of HO-1 compared with the normal group (0.7812) (Fig. 2A). RET mRNA was expressed at significantly higher levels in intermediate patients compared with those in the favorable group (1.1025±0.011, P<0.0001), RET mRNA was expressed at significantly higher levels in favorable patients compared with the normal group (0.0825, P<0.0001; Fig. 2B). No significant difference in the mRNA expression levels of RET were observed between the adverse and intermediate groups (Fig. 2B). By contrast, the PML mRNA expression level in the adverse patient group (0.2056±0.0122) was lower compared with those in the intermediate group (3.2017±0.9004, P<0.0001) and favorable group (2.8002±0.5201, P<0.0001). The PML mRNA expression level of the favorable group was higher compared with the normal group (0.7807±0.004, P<0.0001). No significant differences in the mRNA expression of PML were observed between the favorable and intermediate groups (Fig. 2C).
Figure 2.
(A) The expression of HO-1 mRNA in normal, favorable, intermediate and adverse risk patient groups. (B) The expression of RET mRNA in normal, favorable, intermediate and adverse risk patient groups. (C) The expression of PML mRNA in normal, favorable, intermediate and adverse risk patient groups. Statistical significance was determined by the one-way ANOVA analysis and q-text. The data is represented as the mean ± standard deviation from 3 independent experiments. HO-1, Heme oxygenase-1. RET, a tyrosine kinase receptor. PML, Promyelocytic leukemia gene. *P<0.05; **P<0.01.
Protein expression levels of HO-1, RET and PML in favorable, intermediate and adverse AML patient groups
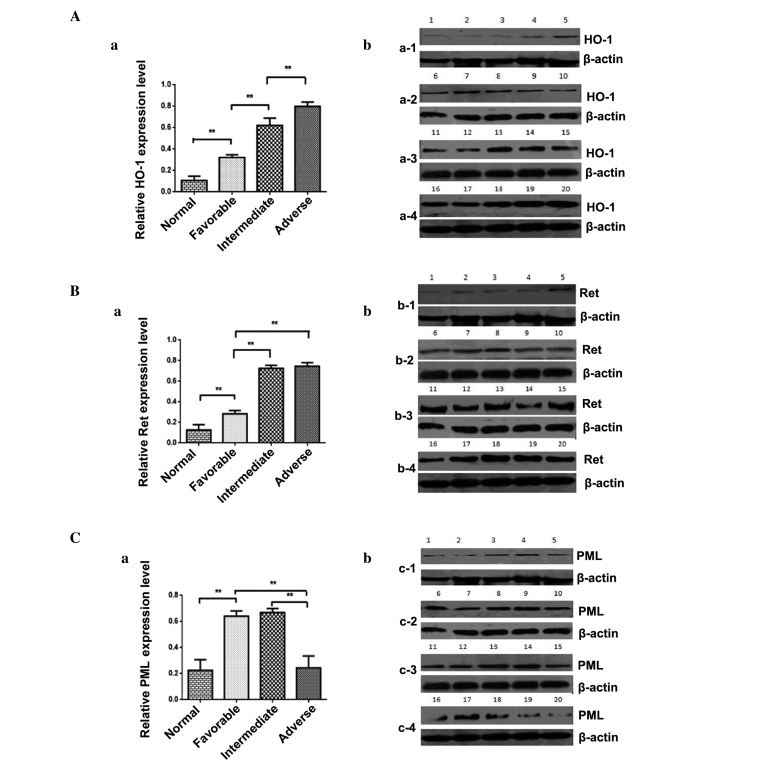
The protein level of HO-1 in the adverse group was notably higher compared with the intermediate group (P<0.01); the protein level of HO-1 in the intermediate group was notably higher compared with the favorable group (P<0.01); the protein level of HO-1 in the favorable group was notably higher compared with the normal group (P<0.01). No significant difference in the protein expression levels of PML were observed between the adverse group and intermediate group (P>0.05; Fig. 3A). Significantly higher Ret protein levels were expressed in intermediate patients compared with those in the favorable group (P<0.01) and significantly increased levels of Ret protein were expressed in favorable patients compared with the normal group (P<0.01), No significant differences in RET protein expression levels were observed between the intermediate and adverse groups (P>0.05) (Fig. 3B). The PML protein expression level of adverse patients was reduced compared with the intermediate group (P<0.01). The PML protein expression level was increased in the favorable group compared with the normal group (P<0.01). No significant differences in the mRNA expression of PML were observed between the favorable and intermediate groups (Fig. 3C).
Figure 3.
(a) Quantification of western blot analysis and (b) original blots of (A) HO-1; (B) Ret; (C) PML protein expression in normal, favorable, intermediate and adverse risk patients. Data is represented as the mean ± SD from 3 independent experiments. **P<0.01. HO-1, Heme oxygenase-1. RET, a tyrosine kinase receptor. PML, Promyelocytic leukemia gene.
Survival analysis
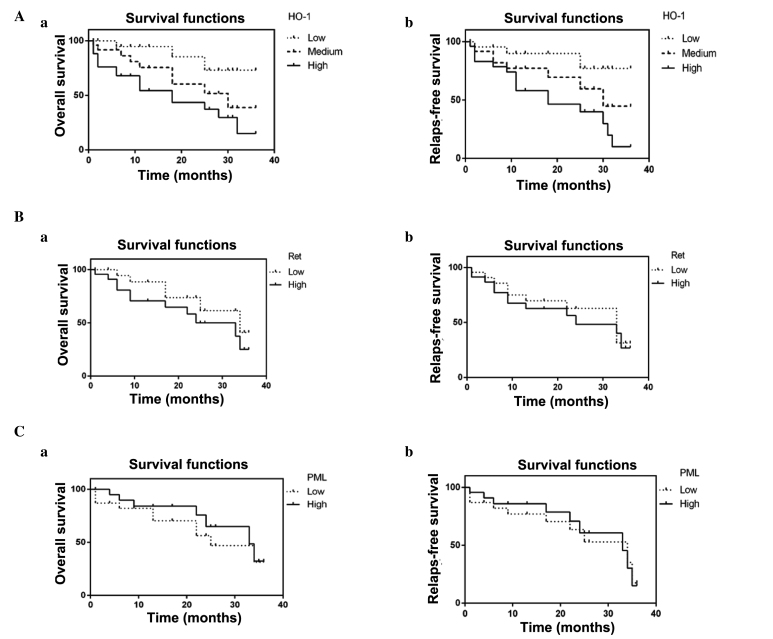
Survival curves were obtained using Kaplan-Meier analysis and the log-rank test was used to compare differences in survival among the 3 groups. According to the survival analysis, it was determined that the 3-year overall survival (OS) rate of the group with low levels of HO-1 protein expression was increased in comparison with the group with high levels of HO-1 protein expression (log-rank=9.517; P=0.0086; Fig. 4Aa), and the difference between each group was significant (P<0.05 for medium level vs. high level; P<0.05 for medium levels vs. low level). The 3-year relapse-free survival (RFS) rate of the group with low levels of HO-1 protein expression was higher compared with the group with low levels of HO-1 protein expression (log-rank=8.861; P=0.0119; Fig. 4Ab); the RFS rate of the group with medium levels of HO-1 protein expression was higher compared with the group with high levels of HO-1 protein expression (P<0.05; Fig. 4Ab); and the RFS rate of the group with low levels of HO-1 protein expression was higher compared with the group with medium levels of HO-1 protein expression (P<0.05; Fig. 4Ab). However, OS and RFS were not influenced by high vs. low Ret and PML expression levels in AML [Ret, OS: hazard ratio (HR) 1.833; 95% confidence interval (CI), 0.7227–4.839; P=0.2109 (Fig. 4Ba); RFS: HR, 1.200; 95% CI, 0.5296–2.818; P=0.6546 (Fig. 4Bb). PML, OS: HR, 1.714; 95% CI, 0.7112–4.297; P=0.2374 (Fig. 4Ca); RFS: HR, 1.111; 95% CI, 0.4564–2.757;P=0.8106) (Fig. 4Cb)].
Figure 4.
Correlation between low, medium and high (A) HO-1; (B) Ret; and (C) PML expression in AML patients and (a) 3-year overall survival and (b) relapse-free survival rates. HO-1, heme oxygenase-1; RET, a tyrosine kinase receptor. PML, Promyelocytic leukemia gene.
Discussion
Conducting accurate risk stratification for AML patients upon first visit may markedly prolong survival and improve the quality of life for the patient. Despite successful risk stratification and prognostic evaluation by examining gene mutation (21), cell genetic characteristics and age (22), AML remains difficult to treat successfully. In the present study, the association between expression levels of HO-1, RET and PML in AML cell lines and AML patients with different risks were studied. The HO-1 gene is located on human chromosome 22q12 and comprises 5 exons spanning 14 kb, and the molecular weight of HO-1 protein is 32.8 kDa. In general, HO-1 is expressed at higher levels in tumor cells, including cells from solid tumors, compared with normal tissue cells (23–25). When HO-1 is expressed at high levels, the expression of the tumor suppressor gene p53 is inhibited, and the proliferation capacity of tumor cells is notably enhanced; when HO-1 expression is inhibited, the expression of P53 is enhanced, and tumors cells proliferate at a reduced rate (9). In addition, HO-1 expression in CLL cells exceeds that in normal lymphocytes. In response to disease or stress, HO-1 expression results in resistance to cell apoptosis and promotes cell proliferation (26). Stuart et al (27) demonstrated that HO-1 was involved in the regulation of leukemia cell apoptosis, proliferation and differentiation through NF-κB, ROS and Nrf2. Mayerhofer et al (28) reported that inducing high HO-1 expression levels resulted in a reduced number of cells undergoing apoptosis and increased resistance to imatinib. However, there remains a lack of available studies regarding the expression status of HO-1 in the risk stratification and prognosis of AML. In the present study, RT-qPCR and western blot analysis were used to demonstrate that AML patients exhibited increased HO-1 expression levels compared with normal subjects. HO-1 expression levels increased with increasing AML risk. Moreover, high HO-1 expression levels were associated with a shorter OS and RFS rate in AML patients. This was consistent with the regulatory effects of HO-1 on the apoptosis, proliferation, differentiation and drug resistance of leukemia cells. The RET proto-oncogene is localized to chromosome 10 (10q11.2) and contains 21 exons spanning about 60 kb. The RET gene encodes Ret protein. Gattei et al (12) observed that RET expression in human AML was maturation-associated, which may indicate its involvement in developmental regulation during the differentiation of normal hematopoietic cells. In the present study, RT-qPCR demonstrated that RET expression may be correlated with HO-1, and the RT-qPCR and western blot analysis results indicated that the expression levels of RET gene increased with increasing risk amongst the AML patients, although no significant difference in the mRNA and protein expression levels of RET were observed between the adverse and intermediate groups. OS and RFS were not influenced by the level of Ret expression in AML patients. The PML gene is ~35 kB, is localized to chromosome 15 and contains 9 exons (29). The PML protein has been verified by in vivo and in vitro studies as a tumor growth inhibitory factor. Rego et al (30) demonstrated that deactivating the PML gene apparently triggered the onset of leukemia. Szendefi et al (31) were the first to report that PML expression was closely associated with increasing degrees of severity in cervical cancer. Subsequently, Gurrieri et al (15) reported that PML expression, which was not expressed in a number of types of tumor, including lung and breast cancer tumor tissues, was associated with tumor grade and stage. However, PML has not previously been associated with AML prognosis or risk stratification. In accordance with these previous findings, the present study demonstrated no association between PML expression and OS or RFS in AML patients. The PML protein expression level of adverse patients was reduced compared with those within the favorable group, whereas the PML expression levels of the intermediate-risk group were not lower than that of the low-risk group (32,33). The findings of the present study do not mimic the close association between PML expression and the degree of severity in cervical cancer (15,31). However, further studies based on larger sample sizes are required to investigate further.
In conclusion, the present study identified that there was a significant difference in the mRNA and protein expression levels of HO-1, RET and PML between low-risk and high-risk patients. In addition, high HO-1 expression was associated with shorter OS and RFS in AML patients. These results may be associated with AML risk stratification and prognosis, which may allow them to be used as suitable markers for evaluation in the future. However, in order to fully elucidate the exact association between HO-1, RET and PML with risk-stratification and prognosis, more samples need to be analyzed and further, larger studies are required.
Acknowledgements
The authors are grateful for the help from all doctors in the Department of Hematology, Affiliated Hospital of Guiyang Medical College for the clinical sampling, and for the guidance from Dr. Ying Chen in Hematopoietic Stem Cell Transplantation Centre.
References
- 1.Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia (Review) Pediatr Clin North Am. 2008;55:21–51. doi: 10.1016/j.pcl.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 2.de Nully Brown P, Jurlander J, Pedersen-Bjergaard J, et al. The prognostic significance of chromosomal analysis and immunophenotyping in 117 patients with de novo acute myeloid leukemia. Leuk Res. 1997;21:985–995. doi: 10.1016/S0145-2126(97)00079-9. [DOI] [PubMed] [Google Scholar]
- 3.Tian Y, Huang Z, Wang Z, et al. Identification of novel molecular markers for prognosis estimation of acute myeloid leukemia: Over-expression of PDCD7, FIS1 and Ang2 may indicate poor prognosis in pretreatment patients with acute myeloid leukemia. PLoS One. 2014;9:e84150. doi: 10.1371/journal.pone.0084150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damm F, Heuser M, Morgan M, et al. Integrative prognostic risk score in acute myeloid leukemia with normal karyotype. Blood. 2011;117:4561–4568. doi: 10.1182/blood-2010-08-303479. [DOI] [PubMed] [Google Scholar]
- 5.Heasma SA, Zaiseva L, Bowles KM, et al. Protection of acute myeloid leukemia cells from apoptosis induced by front-line chemotherapeutics is mediated by haem oxyenase-1. Oncotarget. 2011;2:658–668. doi: 10.18632/oncotarget.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu RP, Hayashi T, Cottam HB, Jin G, Yao S, Wu CC, Rosenbach MD, Corr M, Schwab RB, Carson DA. Nrf2 responses and the theraputic selectivity of electrophilic compounds in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2010;107:7479–7484. doi: 10.1073/pnas.1002890107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishiya T, Kataoka H, Mori K, Goto M, Sugawara T, Furuhama K. Tienilic acid enhances hyperbilirubinemia in Eisai hyperbilirubinuria rats through hepatic multidrug resistance-associated protein 3 and heme oxygenase-1 induction. Toxicol Sci. 2006;91:651–659. doi: 10.1093/toxsci/kfj162. [DOI] [PubMed] [Google Scholar]
- 8.Hileman EO, Liu J, Albitar M, Keating MJ, Huang P. Intrinsic oxidative stress in cancer cells: A biochemical basis for therapeutic selectivity. Cancer Chemother Pharmacol. 2004;53:209–219. doi: 10.1007/s00280-003-0726-5. [DOI] [PubMed] [Google Scholar]
- 9.Fang J, Sawa T, Akaike T, Greish K, Maeda H. Enhancement of cheotherapeutic response of tumor cells by a heme oxygenase inhibitor, pegylated znic protoporphyrin. Int J Cancer. 2004;109:1–8. doi: 10.1002/ijc.11644. [DOI] [PubMed] [Google Scholar]
- 10.Zou M, Baitei EY, Alzahrani AS, BinHumaid FS, Alkhafaji D, Al-Rijjal RA, Meyer BF, Shi Y. Concomitant RAS, RET/PTC, or BRAF mutations in advanced stage of papillary thyroid carcinoma. Thyroid. 2014;24:1255–1266. doi: 10.1089/thy.2013.0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Tang JM, Wang L, Wu PP, Zhang M. Immunohistochemical detection of RET proto-oncogene product in tumoral and nontumoral mucosae of gastric cancer. Anal Quant Cytopathol Histpathol. 2014;36:128–136. [PubMed] [Google Scholar]
- 12.Gattei V, Degan M, Aldinucci D, De Iuliis A, Rossi FM, Mazzocco FT, Rupolo M, Zagonel V, Pinto A. Differential expression of the RET gene in human acute myeloid leukemia. Ann Hematol. 1998;77:207–210. doi: 10.1007/s002770050444. [DOI] [PubMed] [Google Scholar]
- 13.Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. Role of PML in cell grow and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 14.Gambaeorta M, Flenghi L, Fagioli M, Pileri S, Leoncini L, Bigerna B, Pacini R, Tanci LN, Pasqualucci L, Ascani S, et al. Heterogeneous nuclear expression of the promyelocytic leukemia (PML) protein in normal and neoplastic human tissues. Am Pathol. 1996;149:2023–2035. [PMC free article] [PubMed] [Google Scholar]
- 15.Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J NatL Cancer Inst. 2004;96:269–279. doi: 10.1093/jnci/djh043. [DOI] [PubMed] [Google Scholar]
- 16.Koken M, Linare-Cruz G, Quignon F, Viron A, Chelbi-Alix MK, Sobczak-Thépot J, Juhlin L, Degos L, Calvo F, de Thé H. The PML growth-suppressior has altered expression in human oncogenesis. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- 17.Cao TY, Duprez E, Borden KL, Freemont PS, Etkin LD. Ret finger protein is a normal component of PML nuclear bodies and interacts directly wity PML. J Cell Sci. 1998;111:1319–1329. doi: 10.1242/jcs.111.10.1319. [DOI] [PubMed] [Google Scholar]
- 18.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, Paietta E, Willman CL, Head DR, Rowe JM, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group study. Blood. 2000;96:4075–4083. [PubMed] [Google Scholar]
- 19.O'Donnell MR, Abboud CN, Altman J, et al. Acute myeloid leukemia. J Natl Compr Cancer Netw. 2012;10:984–1021. doi: 10.6004/jnccn.2012.0103. [DOI] [PubMed] [Google Scholar]
- 20.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European Leukemia Net. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez M, Levine RL, Rampal R. Integrating genomics into prognostic models for AML. Semin Hematol. 2014;51:298–305. doi: 10.1053/j.seminhematol.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juliusson G, Antunovic P, Derolf A, et al. Age and acute myeloid leukemia: Real world data on decision to treat and outcomes from the Swedish acute leukemia registry. Blood. 2009;113:4179–4187. doi: 10.1182/blood-2008-07-172007. [DOI] [PubMed] [Google Scholar]
- 23.Berberat PO, Dambrauskas Z, Gulbinas A, Giese T, Giese N, Künzli B, Autschbach F, Meuer S, Büchler MW, Friess H. Inhibition of heme oxygenase-1 increase responsiveness of pancreatic cancer cells to anticancer treatment. Clin Cancer Res. 2005;11:3790–3798. doi: 10.1158/1078-0432.CCR-04-2159. [DOI] [PubMed] [Google Scholar]
- 24.Mcalliister SC, Hansen SG, Ruhl RA, Raggo CM, DeFilippis VR, Greenspan D, Früh K, Moses AV. Kaposi sarcoma-associated herpesvirus (KSHV) induces heme oxygenase-1 expression and activity in KSHV-infected endothelial cells. Blood. 2004;103:3465–3473. doi: 10.1182/blood-2003-08-2781. [DOI] [PubMed] [Google Scholar]
- 25.Was H, Dulak J, Jozkowica A. Heme Oxygenase-1 in tumor biology and therapy. Curr Drug Targets. 2010;11:1551–1570. doi: 10.2174/1389450111009011551. [DOI] [PubMed] [Google Scholar]
- 26.Miyazaki T, Kirino Y, Takeno M, Samukawa S, Hama M, Tanaka M, Yamaji S, Ueda A, Tomita N, Fujita H, Ishigatsubo Y. Expression of heme oxygenase-1 in human leukemia cells and its regulation by transcriptional repressor Bach1. Cancer Sci. 2010;101:1409–1416. doi: 10.1111/j.1349-7006.2010.01550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rushworth SA, MacEwan DJ. HO-1 underlies resistance of AML cells to TNF-induced apoptosis. Blood. 2008;111:3793–3801. doi: 10.1182/blood-2007-07-104042. [DOI] [PubMed] [Google Scholar]
- 28.Mayerhofer M, Florian S, Krauth MT, Aichberger KJ, Bilban M, Marculescu R, Printz D, Fritsch G, Wagner O, Selzer E. Identification of heme oxygenase-1 as a novel BCR/ABL-dependent survival factor in chronic myeloid leukemia. Cancer Res. 2004;64:3148–3154. doi: 10.1158/0008-5472.CAN-03-1200. [DOI] [PubMed] [Google Scholar]
- 29.Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 30.Rego EM, Wang ZG, Peruzzi D, He LZ, Cordon-Cardo C, Pandolfi PP. Role of promyelocytic leukemia (PML) protein in tumor suppression. J Exp Med. 2001;193:521–529. doi: 10.1084/jem.193.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szendefi M, Walt H, Krasieva TB, Caduff R, Osann KE, LaMorte VJ. Association between promyelocyte protein and smallubiquitin-like modifier protein and the progression of cervical neoplasia. Obstet Gynecol. 2003;102:1269–1277. doi: 10.1016/S0029-7844(03)00845-7. [DOI] [PubMed] [Google Scholar]
- 32.Mu ZM, Chin KV, Liu JH, Lozano G, Chang KS. PML a growth suppressor disrupted in acute promyelocytic leukemia. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu JH, Mu ZM, Chang KS. PML suppresses oncogenic-transformation of NIH 3T3 cells by activated neu. J Exp Med. 1995;181:1965–1973. doi: 10.1084/jem.181.6.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]