Abstract
Genes and microRNAs (miRNAs) are considered to be key biological factors in human carcinogenesis. To date, considerable data have been obtained regarding genes and miRNAs in cancer; however, the regulatory mechanisms associated with the genes and miRNAs in cancer have yet to be fully elucidated. The aim of the present study was to use the key genes and miRNAs associated with laryngeal cancer (LC) to construct three regulatory networks (differentially expressed, LC-related and global). A network topology of the development of LC, involving 10 differentially expressed miRNAs and 55 differentially expressed genes, was obtained. These genes exhibited multiple identities, including target genes of miRNA, transcription factors (TFs) and host genes. The key regulatory interactions were determined by comparing the similarities and differences among the three networks. The nodes and pathways in LC, as well as the association between each pair of factors within the networks, such as TFs and miRNA, miRNA and target genes and miRNA and its host gene, were discussed. The mechanisms of LC involved certain key pathways featuring self-adaptation regulation and nodes without direct predecessors or successors. The findings of the present study have further elucidated the pathogenesis of LC and are likely to be beneficial for future research into LC.
Keywords: transcription factors, microRNA, target genes, network, laryngeal cancer
Introduction
Laryngeal cancer (LC) is one of the most common types of head and neck cancer. A large number of protein-coding and non-coding genes are known to be differentially expressed in LC cells, indicating a functional switch (1). This gene expression is regulated at the post-transcriptional level, and is involved in the regulation of cellular differentiation and development and metabolic processes (2).
Transcription factors (TFs) and microRNAs (miRNAs) have crucial roles in the regulation of gene expression (3). TFs are proteins that bind to cis-regulatory elements located in the upstream regions of genes in order to activate or suppress transcription, thereby acting to regulate gene expression at the transcriptional level. TFs may function alone or in combination with other proteins (4). miRNAs are evolutionarily conserved, endogenous, non-coding RNAs that post-transcriptionally modulate gene expression and are associated with tumorigenesis (5). Differentially expressed genes and miRNAs are significant factors in the development, metastasis and therapy of LC. miR-21, for example, has been suggested to play an oncogenic role in the cellular processes of LC (6), and c-Myc is highly expressed at the protein level in LC (7).
miRNAs target thousands of human genes, known as target genes; knowledge of these target genes is important for the analysis of the biological functions of miRNAs (8). A number of databases, including those that are computationally (9) and experimentally (10,11) validated, provide an adequate resource to facilitate an understanding of the associations between miRNAs and their targets. Certain miRNAs are located within genes known as host genes, and these miRNAs are transcribed in parallel with their host transcripts. Two different transcription classes of miRNAs have been identified: Exonic and intronic (12). Baskerville and Bartel (13) indicated that a close association exists between intronic miRNA and its host gene. Intronic miRNA and its host gene are usually expressed in a coordinated manner in biological progression, and typically work in combination to effect a biological function and the alteration of signaling pathways (14). The host gene is considered in the network of differentially expressed factors when its corresponding miRNA is differentially expressed. The findings of the above studies suggest that miRNAs can either work together with or separately from their host genes in the progression of cancer.
The aim of the present study was to focus on the associations among the elements in LC by collecting information on the differentially expressed and cancer-related genes and miRNAs involved in LC from databases and literature searches. Three types of association were considered: miRNAs targeting their targets, TFs regulating their miRNAs and miRNAs locating within their host genes. Three networks (differentially expressed, LC-related and global) were built with the obtained data. Following network construction, comparisons were made to find the similarities and differences among the three networks, and the regulatory pathways involving the differentially expressed elements and common TFs were separately extracted. We hypothesized that the pathways involving the differentially expressed elements would have the most significant influence on the progression of LC, with the abnormal modulation of these pathways promoting LC development.
Materials and methods
Dataset of experimentally validated target associations between miRNAs and target genes
The experimentally validated dataset of the gene targets of each specific miRNA was extracted based on the data provided by Tarbase 5.0 (10) and miRTarBase (11). The official symbols used in this study to unify all miRNAs and genes were obtained from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/gene/). The inclusion of these experimental data provides strong evidence to support the study. The collected results are referred to as dataset U1.
Dataset of experimentally validated regulatory associations between TFs and corresponding miRNAs
The dataset of the interaction between the TFs and miRNAs was collected from TransmiR (15), which is a manually extracted database. The results from this process are referred to as dataset U2.
Dataset of miRNAs and host genes
The host genes of respective miRNAs were manually established based on the data obtained from miRBase (16) and the NCBI. Official symbols and IDs were used to represent each host gene and miRNA. The collected results are referred to as dataset U3.
Differentially expressed miRNAs in LC and LC-related miRNAs
The differentially expressed miRNAs (overexpressed, downregulated, and upregulated) in LC were mainly extracted from mir2Disease (17), which is a manually created database of differentially expressed miRNAs in various human diseases. In addition, relevant literature was mined using PubMed (http://www.ncbi.nlm.nih.gov/pubmed) to identify differentially expressed miRNAs associated with LC, the miRNAs involved in LC progression, and differentially expressed and non-differentially expressed miRNAs. The collection of LC-related miRNAs was completed in the same manner. The collected results are referred to as dataset U4.
Differentially expressed genes in LC and LC-related genes
The differentially expressed genes in LC were gathered from several sources, including Cancer Genetics Web (http://www.cancerindex.org/geneweb/), literature searches and the single nucleotide polymorphism (SNP) database of the NCBI (http://www.ncbi.nlm.nih.gov/snp/). Following the collection of the differentially expressed gene data, the LC-related genes were collected from the relevant literature and the GeneCards database (18). The P-Match algorithm (19), which operates on the concepts of pattern matching and weight matrix approaches to identify TF binding sites (TFBSs) in DNA sequences, was additionally employed. These TFs were considered to be LC-related genes and only the TFs that appeared in TransmiR (15) were focused on. Promoter region sequences [1,000 nucleotide (nt)] of the targets of differentially expressed genes were downloaded from the UCSC genome browser database (20). The P-Match algorithm was used to identify the TFBSs in the 1,000-nt promoter region sequences, and these TFBSs were mapped onto the promoter region of the targets. The matrix library utilized by P-Match contains sets of known TFBSs collected from the TRANSFAC database, and thus enables a wide range of TFBSs to be searched for. The vertebrate matrix and restricted high-quality criterion were used. The differentially expressed genes that were obtained were included in the LC-related gene set. The collected results are referred to as dataset U5.
Network construction at three levels
Three regulatory networks were constructed to discuss the regulatory interactions between the genes and miRNAs in LC. The regulatory associations for the TFs, miRNAs, targets and host genes from datasets U1, U2 and U3 were extracted. Following the combination of the associations in these datasets, the global network was derived. Differentially expressed and LC-related elements were separately extracted from U4 and U5, and then separately mapped onto the global network. The differentially expressed and LC-related networks were separately derived following the combination of the associations.
Results
Differentially expressed network in LC
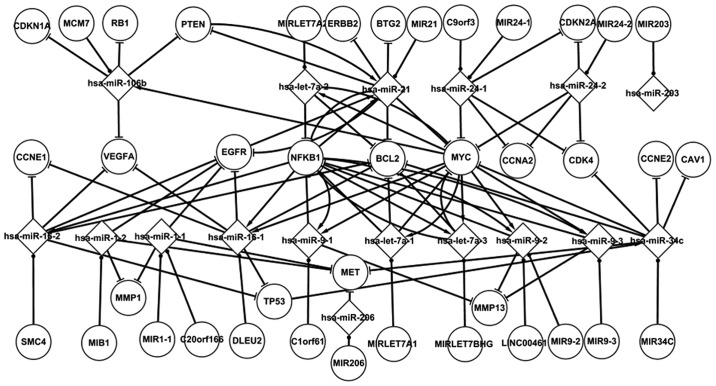
Fig. 1 shows numerous important regulatory interactions among the differentially expressed elements in LC. Each node displayed in the figure represents a differentially expressed gene or miRNA. This network is composed of five TFs (TP53, PTEN, MYC, EGFR and NFKB1), the targets of miRNAs, miRNAs and their host genes. With the exception of the host genes, all the other elements are differentially expressed in LC. There are three types of associations between the elements in LC: miRNAs targeting genes, host genes containing miRNAs and genes regulating miRNAs. Several types of the regulatory interactions between miRNAs and genes are shown in Fig. 1. Most notable are the five TF-related pathways. hsa-miR-106b and hsa-miR-21 target PTEN, which regulates hsa-miR-21. hsa-miR-21 targets EGFR in LC. It is suggested that PTEN could indirectly influence EGFR through hsa-miR-21. An miRNA may target one or several genes. hsa-miR-1–2, for example, targets EGFR, MMP1 and MET. Certain regulatory circuits can also be found in this network. For example, hsa-let-7a-3 regulates and targets NFKB1 at the same time. A number of special characteristics of the host genes and their miRNAs are highlighted in Fig. 1. A host gene may contain one or several miRNAs, and these miRNAs may target other genes. MIR21, for example, contains hsa-miR-21, which targets PTEN, MYC, BCL2, EGBB2, NFKB1, BTG2 and EGFR. The nodes in the differentially expressed network have already been confirmed by tests. The nodes were confirmed using numerous sources, including Cancer Genetics Web, the SNP database of NCBI, and mir2Disease (17). This differentially expressed network partly reveals the regulatory mechanism of LC.
Figure 1.
Regulatory interactions in the differentially expressed network in laryngeal cancer.
LC-related network
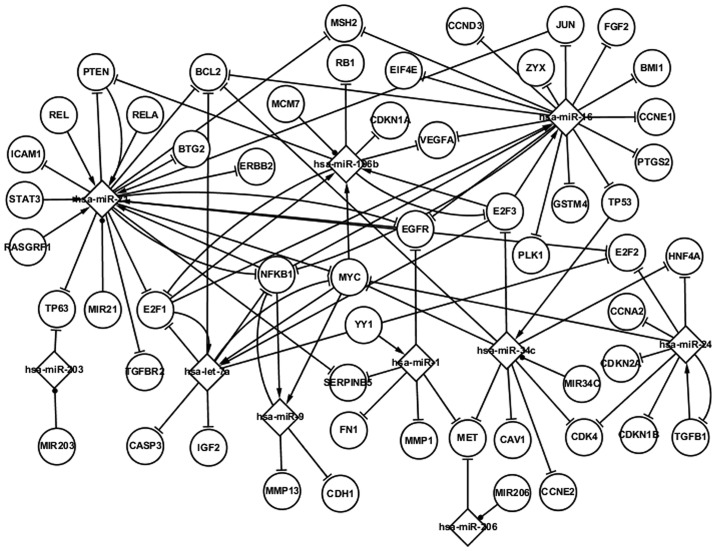
Fig. 2 shows numerous regulatory interactions among genes and miRNAs in LC. The network is composed of differentially expressed genes and miRNAs, LC-related genes and miRNAs, targets of miRNAs and host genes of miRNAs. The LC-related network can be regarded as an extension of the differentially expressed network, based on the fact that the differentially expressed genes and miRNAs are considered to be LC-related elements. Fig. 2 shows five differentially expressed TFs (TP53, PTEN, MYC, EGFR and NFKB1) and nine LC-related TFs (E2F1, E2F3, JUN, RASGRF1, REL, RELA, STAT3, TGFB1, YY1) in LC. A group of additional pathways can also be observed in Fig. 2; for example, STAT3 and RELA regulate hsa-miR-21, which targets ICAM1, and E2F3 and E2F1 regulate hsa-miR-106b, which in turn targets E2F3 and RB1. YY1 regulates hsa-miR-1, which targets FN1. The LC-related network shows more topological associations than the differentially expressed network and contributes to the understanding of the pathogenesis of LC.
Figure 2.
Regulatory interactions in the laryngeal cancer-related network.
Global network in LC
The global network is the aggregate of datasets U1, U2 and U3 and includes all the interactions. The global network accommodates the LC-related network, which additionally contains the differentially expressed network.
Analysis of host genes and their miRNAs in LC
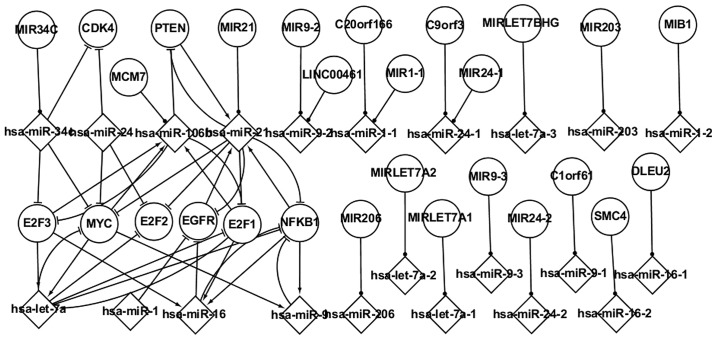
Host genes and their miRNAs exhibit certain important characteristics, and are key biological factors involved in the regulatory process. Fig. 3 shows a number of pathways involving host genes and their miRNAs. Although these host genes are not differentially expressed in LC, their miRNAs are subject to differential expression. Numerous important regulatory associations exist among the host genes, TFs, gene targets and the miRNAs contained within the host genes.
Figure 3.
Associations between host genes and their microRNAs in the differentially expressed network.
MIR34C is the host gene of hsa-miR-34c, which targets E2F3, MYC and CDK4. MCM7 is the host gene of hsa-miR-106b, which targets E2F3, E2F1 and PTEN. hsa-miR-106b is regulated by E2F3, E2F1 and MYC. C1orf61 contains hsa-miR-9-1, which does not target any gene. hsa-miR-21 is regulated by PTEN, NFKB1 and EGFR, and hsa-miR-21 and PTEN form a self-adaptation association. hsa-miR-16 is regulated by E2F3, E2F1 and NFKB1. It is suggested that an understanding of host genes and their miRNAs could further aid with the clarification of the pathogenesis underlying LC.
Transcriptional network of TFs and differentially expressed miRNAs
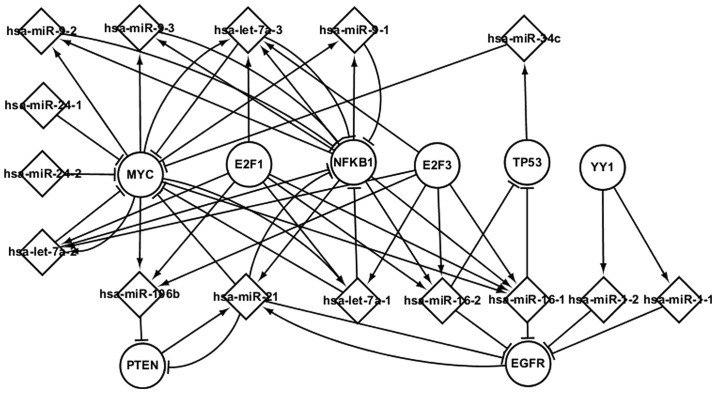
A further analysis of the differentially expressed miRNAs exhibiting regulatory interactions with common TFs was performed. These miRNAs and TFs influence their successors by targeting and regulating them, respectively. The elements involved in LC progression are shown in the predicted transcriptional network in Fig. 4.
Figure 4.
Regulatory interactions in the predicted transcriptional network in laryngeal cancer.
Fig. 4 shows the regulatory interactions among common TFs and miRNAs, as well as their targets, in LC. The most important TFs are MYC, PTEN, NFKB1 and EGFR. These four TFs and their differentially expressed miRNAs interact to affect the progression of LC. MYC and NFKB1 coregulate hsa-let-7a-3 and hsa-9-1, which target NFKB1. As shown in Fig. 4, a differentially expressed miRNA may target one or several TFs, and a TF may regulate one or several differentially expressed miRNAs. hsa-miR-16-1, for example, is regulated by four TFs (E2F3, NFKB1, E2F1 and MYC); five miRNAs (hsa-miR-21, hsa-miR-16-1, hsa-miR-16-2, hsa-let-7a-1 and hsa-let-7a-2) target EGFR; and YY1 regulates two miRNAs (hsa-miR-1-1 and hsa-miR-1–2), which target EGFR. The predicted transcriptional network showing the TFs and miRNAs is likely to further enhance the understanding of the pathogenesis of LC.
Comparison and analysis of the characteristics of the differentially expressed genes in the networks
In the dataset of the differentially expressed genes, nodes were classed according to the regulatory association of adjacent nodes in the three networks in order to compare and analyze the interaction characteristics of each differentially expressed gene.
With regard to the TFs, the first class of TF has six adjacent nodes (three successors and three predecessors). Three of these TFs, PTEN, TP53 and MYC, exhibit special characteristics, with both direct predecessors and successors. MYC, for example, is represented in the three networks, as shown in Table I. MYC is a notable tumor suppressor with distinct characteristics in the three networks. In the differentially expressed network, 11 miRNAs target MYC, which regulates 22 miRNAs; in the LC-related network, 12 miRNAs target MYC, which regulates 24 miRNAs; and in the global network, 18 miRNAs target MYC, which regulates 32 miRNAs. In the differentially expressed network, the predecessors indirectly influence the successors through MYC. As shown in Table I, hsa-miR-17 targets MYC, which in turn regulates hsa-miR-17, thus generating a self-adaptation association. This self-adaptation association may be crucial in the progression of LC. MYC indirectly influences the expression of other genes via certain miRNAs; for example, MYC regulates hsa-let-7a-1, which targets BCL2. A number of TFs also indirectly influence MYC via certain miRNAs; for example TP53 regulates hsa-miR-34c, which targets MYC. These interactions show that there are numerous complex associations among MYC, miRNAs and other genes. In the global network, the involvement of the six previously mentioned miRNAs (predecessors) and seven additional miRNAs (successors) in LC has not been validated. Despite this, the collected information is likely to be of use in the elucidation of the pathogenesis of LC.
Table I.
Regulatory interactions between microRNAs (miRNAs) and MYC in the three networks.
| A, miRNAs targeting MYC | ||
|---|---|---|
| Differentially expressed network | LC-related network | Global network |
| hsa-let-7a-1 | hsa-let-7a-1 | hsa-let-7a-1 |
| hsa-let-7a-2 | hsa-let-7a-2 | hsa-let-7a-2 |
| hsa-let-7a-3 | hsa-let-7a-3 | hsa-let-7a-3 |
| hsa-let-7g | hsa-let-7g | hsa-let-7g |
| hsa-miR-145 | hsa-miR-145 | hsa-miR-145 |
| hsa-miR-17 | hsa-miR-17 | hsa-miR-17 |
| hsa-miR-26a-1 | hsa-miR-26a-1 | hsa-miR-26a-1 |
| hsa-miR-34a | hsa-miR-34a | hsa-miR-34a |
| hsa-miR-34b | hsa-miR-34b | hsa-miR-34b |
| hsa-miR-34c | hsa-miR-34c | hsa-miR-34c |
| hsa-miR-24-1 | hsa-miR-21 | hsa-miR-21 |
| hsa-miR-24-1 | hsa-miR-24-1 | |
| hsa-miR-20a | ||
| hsa-miR-24-2 | ||
| hsa-miR-378a | ||
| hsa-miR-98 | ||
| hsa-miR-26a-2 | ||
| hsa-miR-24-2 | ||
| B, miRNAs regulated by MYC | ||
| Differentially expressed network | LC-related network | Global network |
| hsa-let-7a-1 | hsa-let-7a-1 | hsa-let-7a-1 |
| hsa-let-7a-2 | hsa-let-7a-2 | hsa-let-7a-2 |
| hsa-let-7a-3 | hsa-let-7a-3 | hsa-let-7a-3 |
| hsa-let-7b | hsa-let-7b | hsa-let-7b |
| hsa-let-7d | hsa-let-7d | hsa-let-7d |
| hsa-let-7e | hsa-let-7e | hsa-let-7e |
| hsa-let-7g | hsa-let-7g | hsa-let-7g |
| hsa-miR-106 | hsa-miR-106 | hsa-miR-106 |
| hsa-miR-141 | hsa-miR-141 | hsa-miR-141 |
| hsa-miR-15a | hsa-miR-15a | hsa-miR-15a |
| hsa-miR-17 | hsa-miR-17 | hsa-miR-17 |
| hsa-miR-19a | hsa-miR-19a | hsa-miR-19a |
| hsa-miR-221 | hsa-miR-221 | hsa-miR-221 |
| hsa-miR-23b | hsa-miR-23b | hsa-miR-23b |
| hsa-miR-26a-1 | hsa-miR-26a-1 | hsa-miR-26a-1 |
| hsa-miR-29b-1 | hsa-miR-29b-1 | hsa-miR-29b-1 |
| hsa-miR-29b-2 | hsa-miR-29b-2 | hsa-miR-29b-2 |
| hsa-miR-29c | hsa-miR-29c | hsa-miR-29c |
| hsa-miR-34a | hsa-miR-34a | hsa-miR-34a |
| hsa-miR-9-1 | hsa-miR-9-1 | hsa-miR-9-1 |
| hsa-miR-9-2 | hsa-miR-9-2 | hsa-miR-9-2 |
| hsa-miR-9-3 | hsa-miR-9-3 | hsa-miR-9-3 |
| hsa-let-7c | hsa-let-7c | |
| hsa-miR-195 | hsa-miR-195 | |
| hsa-let-7f-1 | ||
| hsa-let-7f-2 | ||
| hsa-miR-106b | ||
| hsa-miR-16-1 | ||
| hsa-miR-18a | ||
| hsa-miR-19b-1 | ||
| hsa-miR-19b-2 | ||
The second class of TF, which includes EGFR, has five adjacent nodes (two successors and three predecessors). Six miRNAs target EGFR, and EGFR regulates hsa-miR-21 in three networks.
The remaining genes, i.e. those that do not regulate any miRNAs, can be grouped into one of three classes. In the first class of gene, each gene has three adjacent nodes (three predecessors). These genes include CDKN1A, ATM, MMP2 and MMP1, which are targeted by certain miRNAs but do not regulate any miRNAs. In the second class of gene, each gene has four types of adjacent nodes in the three networks (one successor and three predecessors). These genes include CDKN2A, RB1 and CDK4. In the third class of gene, each gene again has three adjacent nodes, but in this case they are three successors. An example of a class-three gene is FOXM1. It is suggested that the FOXM1 has less of an impact, as compared with other differentially expressed genes.
Comparison and analysis of the characteristics of the differentially expressed miRNAs
The first class of miRNA, which includes hsa-miR-34c, hsa-let-7a-1, hsa-let-7a-2 and hsa-let-7a-3, has six adjacent nodes (three predecessors and three successors). These four differentially expressed miRNAs (hsa-miR-34c, hsa-let-7a-1, hsa-let-7a-2 and hsa-let-7a-3) and their corresponding genes form four self-adaptation associations. Using hsa-let-7a-1 as an example, Table II lists the precursors and successors of the miRNA in the differentially expressed, LC-related and global networks. In the differentially expressed network, MYC regulates hsa-let-7a-1, which targets four differentially expressed genes; in the LC-related network, four genes regulate hsa-let-7a-1, which targets 13 genes; in the global network, 10 genes regulate hsa-let-7a-1, which targets 36 genes. Table II shows that MYC indirectly influences MYC, CASP8, NRAS and KRAS expression by regulating hsa-let-7a-1 in the differentially expressed network. MYC and hsa-let-7a-1 and E2F1 and hsa-let-7a-1 separately form self-adaptation associations. hsa-let-7a-1 indirectly influences other miRNAs via certain TFs; for example, hsa-let-7a-1 targets NFKB1, which regulates hsa-miR-16-1, hsa-miR-16-2, hsa-miR-21 and hsa-let-7a-3. A number of miRNAs also indirectly influence hsa-let-7a-1 via certain TFs; for example, hsa-miR-24-1 targets MYC, which regulates hsa-let-7a-1. These interactions show that hsa-let-7a-1 has numerous regulatory associations with genes and other miRNAs.
Table II.
Regulatory interactions between hsa-let-7a-1 and genes in the three networks.
| A, Genes regulating hsa-let-7a-1 | ||
|---|---|---|
| Differentially expressed network | LC-related network | Global network |
| MYC | MYC | MYC |
| E2F1 | E2F1 | |
| E2F3 | E2F3 | |
| NFB1 | NFB1 | |
| EIF2C2 | ||
| FLI1 | ||
| FSH | ||
| LIN28 | ||
| LIN28B | ||
| TRIM32 | ||
| B, Genes targeted by hsa-let-7a-1 | ||
| Differentially expressed network | LC-related network | Global network |
| CASP8 | CASP8 | CASP8, KRAS |
| KRAS | KRAS | MYC, NRAS |
| MYC | MYC | APP, BCL2 |
| NRAS | NRAS | E2F1, E2F2 |
| APP | HRAS, IL6 | |
| BCL2 | ITGB3, NFKB1 | |
| E2F1 | THBS1, AMMECR1 | |
| E2F2 | CASP3, CASP9 | |
| HRAS | CCND2, DICER1 | |
| IL6 | EGR3, EIF2C4 | |
| ITGB3 | HMGA1, HMGA2 | |
| NFKB1 | HNRPDL, IGF2 | |
| THBS1 | LIN28A, MEIS1 | |
| NEFM, NF2 | ||
| NKIRAS2, PRDM1 | ||
| RAVER2, SLC20A1 | ||
| TRIM71, TUSC2 | ||
| UHRF2, ZFP36L1 | ||
The second class of miRNA, which includes hsa-miR-9-1, hsa-miR-9-2 and hsa-miR-9-3, has five adjacent nodes (two successors and three predecessors). In the differentially expressed network, MYC and NFKB1 regulate hsa-miR-9, and hsa-miR-9 targets MMP13 and NFKB1; in the LC-related network, MYC and NFKB1 regulate hsa-miR-9, and hsa-miR-9 targets CDH1, MYC and NFKB1.
The third class of miRNA, which includes hsa-miR-24-1, has four adjacent nodes (three successors and one predecessor). In the differentially expressed network, hsa-miR-24-1 is not regulated by a TF but targets CDK4, CDKN2A, MYC and CCNA2; in the LC-related network, TGFB1 regulates hsa-miR-24 and hsa-miR-24 target seven genes.
The fourth class of miRNA, which includes hsa-miR-203, has three adjacent nodes (two successors and one predecessor). In the LC-related network, hsa-miR-203 is not regulated by a TF but targets TP63.
Comparison and analysis of the characteristics of common TFs
Following the analysis of the differentially expressed network, the common TFs in the LC-related network were compared and analyzed using the same method as that used to compare the differentially expressed miRNAs. The first class of TF has six adjacent nodes (three successors and three predecessors). Six TFs (RUNX1, E2F1, E2F3, NFKB1, YY1 and CREB1) and their corresponding miRNAs form self-adaptation associations. The following section focuses on E2F1. Table III shows the predecessors and successors of E2F1, as well as the associated regulatory interactions. In the differentially expressed network, eight miRNAs target E2F1, which in turn regulates eight miRNAs; in the related network, nine miRNAs target E2F1, which regulates nine miRNAs; in the global network, 16 miRNAs target E2F1, which regulates 28 miRNAs. It can be noted from Table III that five miRNAs (hsa-let-7a-1, hsa-let-7a-2, hsa-let-7a-3, hsa-miR-106a and hsa-miR-17) and E2F1 separately form self-adaptation associations in the three networks. E2F1 is not differentially expressed in LC, but hsa-let-7a-1, hsa-let-7a-2 and hsa-let-7a-3 are subject to differential expression; thus, it can be inferred that the three miRNAs indirectly lead to changes in the expression of other miRNAs via E2F1. The pathways involving E2F1 and the differentially expressed miRNAs indicate that six miRNAs (hsa-miR-149, hsa-miR-23b, hsa-miR-34a, hsa-miR-15a, hsa-miR-15b and hsa-miR-19a) exhibit important differential expression in LC. These miRNAs not only are differentially expressed but also represent adjacent nodes of E2F1 that are frequently involved in transcription in cancer.
Table III.
Regulatory interactions between microRNAs (miRNAs) and E2F1 in the three networks.
| A, miRNAs targeting E2F1 | ||
|---|---|---|
| Differentially expressed network | LC-related network | Global network |
| hsa-let-7a-1 | hsa-let-7a-1 | hsa-let-7a-1 |
| hsa-let-7a-2 | hsa-let-7a-2 | hsa-let-7a-2 |
| hsa-let-7a-3 | hsa-let-7a-3 | hsa-let-7a-3 |
| hsa-miR-106a | hsa-miR-106a | hsa-miR-106a |
| hsa-miR-149 | hsa-miR-149 | hsa-miR-149 |
| hsa-miR-17 | hsa-miR-17 | hsa-miR-17 |
| hsa-miR-23b | hsa-miR-23b | hsa-miR-21 |
| hsa-miR-34a | hsa-miR-34a | hsa-miR-23b |
| hsa-miR-21 | hsa-miR-34a | |
| hsa-miR-106b | ||
| hsa-miR-126 | ||
| hsa-miR-20a | ||
| hsa-miR-223 | ||
| hsa-miR-330 | ||
| hsa-miR-93 | ||
| hsa-miR-98 | ||
| B, miRNAs regulated by E2F1 | ||
| Differentially expressed network | LC-related network | Global network |
| hsa-let-7a-1 | hsa-let-7a-1 | hsa-let-7a-1 |
| hsa-let-7a-2 | hsa-let-7a-2 | hsa-let-7a-2 |
| hsa-let-7a-3 | hsa-let-7a-3 | hsa-let-7a-3 |
| hsa-miR-106a | hsa-miR-106a | hsa-miR-106a |
| hsa-miR-15a | hsa-miR-15a | hsa-miR-15a |
| hsa-miR-15b | hsa-miR-15b | hsa-miR-15b |
| hsa-miR-17 | hsa-miR-17 | hsa-miR-17 |
| hsa-miR-19a | hsa-miR-19a | hsa-miR-195 |
| hsa-miR-195 | hsa-miR-19a | |
| hsa-miR-106b | ||
| hsa-let-7i | ||
| hsa-miR-16-1 | ||
| hsa-miR-16-2 | ||
| hsa-miR-18a | ||
| hsa-miR-18b | ||
| hsa-miR-19b-1 | ||
| hsa-miR-19b-2 | ||
| hsa-miR-20a | ||
| hsa-miR-20b | ||
| hsa-miR-223 | ||
| hsa-miR-25 | ||
| hsa-miR-363 | ||
| hsa-miR-449a | ||
| hsa-miR-449b | ||
| hsa-miR-449c | ||
| hsa-miR-92a-1 | ||
| hsa-miR-92a-2 | ||
| hsa-miR-93 | ||
The second class of TF, which includes RELA, has five adjacent nodes (two successors and three predecessors). In the differentially expressed network, RELA is not targeted by any differentially expressed miRNAs and it does not regulate any miRNAs; however, RELA regulates hsa-miR-21 in the LC-related network.
The third class of TF, which includes STAT1, NKX2–5 and E2F2, has three adjacent nodes (three successors or three predecessors). In the LC-related network, three differentially expressed miRNAs target E2F2, which does not, in turn, regulate any miRNAs.
The fourth class of TF, which includes REL and IRF1, has two adjacent nodes (two successors and no predecessors). In the LC-related network, no differentially expressed miRNAs target REL, but REL regulates hsa-miR-21.
In the final class of TF, which includes TFAP4 and NF1, each TF has one adjacent node (a predecessor).
Discussion
In the present study, three regulatory topological networks (differentially expressed, LC-related and global) were constructed in order to reveal the important pathways in LC and to establish a topological network regarding the development of LC. Following construction, the critical hubs and certain notable features within the networks, including self-adaptation regulation associations and nodes without direct predecessors or successors, were taken into consideration. In addition, certain significant regulatory associations involving differentially expressed genes, differentially expressed miRNAs and predicted TFs were identified.
The results of the study demonstrated that four differentially expressed miRNAs (hsa-let-7a-1, hsa-let-7a-2, hsa-let-7a-3 and hsa-miR-34c) and their corresponding genes form self-adaptation associations in LC, while six common TFs (E2F1, E2F3, NFKB1, RUNX1, YY1 and CRFB1) and their corresponding differentially expressed miRNAs form self-adaptation associations. There are numerous pathways involving interactions in which either one or multiple elements are differentially expressed. hsa-let-7a-1, for example, targets NFKB1, which in turn regulates hsa-miR-16-1, and hsa-miR-21 targets MYC, which regulates hsa-miR-16-1. These pathways perform a key function in LC.
Certain pathways have been found to affect the progression of LC. It has been suggested that hsa-mir-21 and PTEN play important roles in the progression of LC, with the two factors demonstrating a negative correlation (21). Some specific genes, miRNAs, and their interactions in LC have previously been identified. For example, the TP53Pro variant may contribute to the risk of LC development (22). Furthermore, Navarro et al (23) indicated that TP53 regulates hsa-miR-143 in resected non-small-cell lung cancer. The present study demonstrated that some pathways are not involved in LC; however, they influence the progression of numberous other carcinomas, for example hsa-miR-16 targets BCL2 in chronic lymphocytic leukemia (24). Although numerous functions of the identified pathways have not been elucidated in the present study, the biological functions of these pathways in other types of cancer may be transferable to LC. The remaining pathways that have not been detected in cancer may have some potential functions in LC, for example YY1 regulates hsa-miR-1-1. For those pathways of predicted TFs, some pathways have been detected in other carcinomas, for example miR-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis (25). In this way, information on the interactions among genes can be extrapolated from one type of carcinoma to another, and the blind spots of research on the pathology of LC can be highlighted.
In conclusion, the present study has partly uncovered the regulatory associations in the development of LC and supplied comprehensive data associated with LC. These data can be used to guide medical investigators and biologists undertaking further research in LC. Investigations into protein interactions and regulatory patterns (upregulation and downregulation) will enable the construction of a more comprehensive and extensive network for LC.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (no. 60973091) and the Science and Technology Development Plan of Jilin Province (no. 20130101166JC).
References
- 1.Zhang Y, Chen Y, Yu J, Liu G, Huang Z. Integrated transcriptome analysis reveals miRNA-mRNA crosstalk in laryngeal squamous cell carcinoma. Genomics. 2014;104:249–256. doi: 10.1016/j.ygeno.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Liu M, Wu H, Liu T, Li Y, Wang F, Wan H, Li X, Tang H. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009;19:828–837. doi: 10.1038/cr.2009.72. [DOI] [PubMed] [Google Scholar]
- 3.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 4.Zhu MH, Xu ZW, Wang KH, Wang N, Li Y. microRNA and gene networks in human pancreatic cancer. Oncol Lett. 2013;6:1133–1139. doi: 10.3892/ol.2013.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F, Song G, Liu M, Li X, Tang H. miRNA-1 targets fibronectin1 and suppresses the migration and invasion of the HEp2 laryngeal squamous carcinoma cell line. FEBS Lett. 2011;585:3263–3269. doi: 10.1016/j.febslet.2011.08.052. [DOI] [PubMed] [Google Scholar]
- 6.Ren J, Zhu D, Liu M, Sun Y, Tian L. Downregulation of miR-21 modulates Ras expression to promote apoptosis and suppress invasion of Laryngeal squamous cell carcinoma. Eur J Cancer. 2010;46:3409–3416. doi: 10.1016/j.ejca.2010.07.047. [DOI] [PubMed] [Google Scholar]
- 7.Sang J, Liu L, Tian F, Jin H, Yuan L, Lou W. The expression of c-myc in the tissues of human laryngeal squamous cell carcinoma and the effect of siRNA-mediated inhibition of c-myc on proliferation in laryngeal carcinoma Hep-2 cells. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2011;25:695–700. [PubMed] [Google Scholar]
- 8.Wang K, Xu Z, Wang N, Xu T, Zhu M. MicroRNA and gene networks in human diffuse large B-cell lymphoma. Oncol Lett. 2014;8:2225–2232. doi: 10.3892/ol.2014.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36(Database):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: A functional update of TarBase. Nucleic Acids Res. 2009;37(Database):D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu SD, Lin F-M, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39(Database):D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia W, Li J, Li S, Chen L, Ding H, et al. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun. 2010;396:978–982. doi: 10.1016/j.bbrc.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Lu M, Qiu C, Cui Q. TransmiR: A transcription factor-micro RNA regulation database. Nucleic Acids Res. 2010;38(Database):D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37(Database):D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al. GeneCards Version 3: The human gene integrator. Database (Oxford) 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chekmenev DS, Haid C, Kel AE. P-Match: Transcription factor binding site search by combining patterns and weight matrices. Nucleic Acids Res. 2005;33(Web Server):W432–W437. doi: 10.1093/nar/gki441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res. 2011;39(Database):D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Lei DP, Jin T, Zhao XN, Li G, Pan XL. Altered expression of miR-21 and PTEN in human laryngeal and hypopharyngeal squamous cell carcinomas. Asian Pac J Cancer Prev. 2011;12:2653–2657. [PubMed] [Google Scholar]
- 22.Zemleduch R, Lianeri M, Rydzanicz M, Gajecka M, Szyfter K, Jagodziński PP. Contribution of polymorphism in codon 72 of TP53 gene to laryngeal cancer in Polish patients. Oral Oncol. 2009;45:683–686. doi: 10.1016/j.oraloncology.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Navarro A, Diaz T, Gallardo E, Viñolas N, Marrades RM, Gel B, Campayo M, Quera A, Bandres E, Garcia-Foncillas J, et al. Prognostic implications of miR-16 expression levels in resected non-small-cell lung cancer. J Surg Oncol. 2011;103:411–415. doi: 10.1002/jso.21847. [DOI] [PubMed] [Google Scholar]
- 24.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shumizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok MT, Wang H, Chen J, Ng SS, Chen M, et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene. 2013;32:5078–5088. doi: 10.1038/onc.2012.526. [DOI] [PubMed] [Google Scholar]