Abstract
MicroRNAs (miRNAs) are important gene regulators that participate in tumorigenesis. Previous studies have implicated that miR-214 is a tumor suppressor that is capable of inhibiting human hepatocellular carcinoma (HCC) cell growth. However, the mechanism by which miR-214 suppresses tumor development remains unknown. In the present study, miR-214 was observed to suppress tumor proliferation by directly targeting E2F transcription factor 3 (E2F3) in HCC cells. Colony formation, cell cycle and proliferation assays were employed to study the tumor suppressor role of miR-214 in cell proliferation. In addition, western blotting and dual-luciferase reporter assays were used to evaluate whether E2F3 was a target of miR-214. The results of these analyses revealed that E2F3 was a novel target of miR-214. Furthermore, enhanced expression of miR-214 or silencing of E2F3 inhibited the proliferation of HCC SMMC-7721 cells. These findings suggest that miR-214 suppresses HCC growth by targeting E2F3, and may provide a novel approach for the treatment of human HCC.
Keywords: miR-214, hepatocellular carcinoma, proliferation, E2F3
Introduction
Hepatocellular carcinoma (HCC) is one of the most common types of malignancy and the third leading cause of cancer-associated mortalities worldwide, due to its poor prognosis and frequent relapse and metastasis (1,2). Furthermore, HCC is difficult to treat, since it is usually diagnosed at advanced stages (3). Thus, studying the molecular mechanism of HCC in order to identify novel prognostic markers or therapeutic targets for the treatment of HCC is of value.
MicroRNAs (miRNAs) are small noncoding RNAs of ~22 nucleotides (nt) in length that regulate gene expression at the post-transcriptional level (4). miRNAs are initially transcribed as long primary precursor molecules named pri-miRNAs, which are processed in a number of endonuclease reactions to produce the mature effector 22-nt miRNA molecule (5). Through the specific targeting of the 3-untranslated regions (3′-UTRs) in the target genes of the miRNA, the translational repression and degradation of their target mRNA occurs (6). By targeting various transcripts, miRNAs are involved in a number of fundamental biological processes, including cell proliferation and differentiation, apoptosis, development and metabolism (7–9). Previous studies suggest that the expression of miRNAs is remarkably dysregulated in cancer, due to numerous epigenetic and genomic alterations (10,11). Several miRNAs have been demonstrated to act as tumor suppressor genes or oncogenes in tumors (12). As one miRNA may potentially regulate numerous mRNAs, it is clear that they are important regulatory molecules. Consistently, previous studies have considered miRNAs as important biomarkers and promising therapeutic targets for the treatment of various diseases (13). Recently, miR-214 was reported to be involved in certain types of cancer, including breast (14), adrenocortical (15), ovarian (16) and gastric cancer (17). Thus, the roles of miR-214 may be critical in the development of cancer.
In the present study, bioinformatics analysis was performed to assess E2F transcription factor 3 (E2F3) as a potential target of miR-214, and in vitro experiments were conducted to investigate the effect of overexpression of miR-214 or silencing of E2F3 in HCC proliferation. Furthermore, in HCC cells overexpressing miR-214, the cell cycle distribution was analyzed. The protein levels of cyclin D1 (CCND1) and cyclin-dependent kinase 6 (CDK6) were measured and dual-luciferase reporter assays were conducted in these cells.
Materials and methods
Cell lines
The human HCC cancer cell line SMMC-7721 and HEK293 cells were obtained from the Key Laboratory of Environmentally and Genetically Associated Diseases (Xi'an Jiaotong University College of Medicine, Xi'an, China), and cultured in Dulbeccos modified Eagles medium supplemented with 10% fetal bovine serum (PAA Laboratories GmbH), at 37°C in a humidified chamber with 5% CO2.
Plasmid construction
In order to construct a vector for expression of miR-214, EcoRI and HindIII sites were inserted into the multiple cloning site of the pcDNA 6.2-GW/EmGFP-miR vector (Invitrogen Life Technologies, Carlsbad, CA, USA). The precursor of miR-214 was obtained by polymerase chain reaction using the following primers: Forward, 5′-CCG GAA TTC AAG GAA CCG CGA AGG AG-3′ and reverse, 5′-CCC AAG CTT TCA TT CAG GCT GGG TTG −3′. Next, miR-214 was inserted into the pcDNA 6.2-GW/EmGFP-miR vector, between the EcoRI and HindIII sites. RegRNA 2.0, a regulatory RNA motifs and elements finder software (regrna2.mbc.nctu.edu.tw), was used to predict genes associated with specific miRNAs. Bioinformatic analysis predicted E2F3 to be the target gene of miR-214. Thus, the specific fragments of E2F3 were synthesized. The luciferase-UTR reporter constructions were generated by inserting the wild-type (WT) or mutant (Mut)-E2F3 3′-UTR, carrying the predicted binding site for miR-214, into the pmirGLO Dual-Luciferase miRNA Target Expression vector (Promega Corporation, Madison, WI, USA), between the XhoI and SacI sites. The sequence of the miR-214 inhibitor was as follows: 5′-ACT GCC TGT CTG TGC CTG CTGT-3. The small interfering (si)RNA targeting E2F3 and the corresponding control (ctrl) siRNA were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China), and displayed the following sequences: si-E2F3 sense, 5′-GAC UUC AUG UGU AGU UGA UUU-3, and antisense, 5′-AUC AAC UAC ACA UGA AGU CUU-3; si-ctrl sense, 5′-UUC UCC GAA CGU GUC ACG UUU-3, and antisense, 5′-ACG UGA CAC GUU CGG AGA AUU-3.
Luciferase reporter assay
PmirGLO-E2F3-3′-UTR-WT or pmirGLO-E2F3-3′-UTR-Mut vectors were cotransfected with miR-214 into HEK293 cells, using Lipofectamine 2000 (Invitrogen Life Technologies). Firefly luciferase activity was measured at 48 h post-transfection, using a Dual-Luciferase Reporter Assay System (Promega Corporation), according to the manufacturers instructions. The results were normalized with Renilla luciferase. Each reporter plasmid was transfected ≥3 times, and each sample was assayed in triplicate.
Cell proliferation (MTT) and colony forming assays
For the MTT assay, SMMC-7721 cells were seeded at a density of 5,000 cells/well into 96-well cell culture plates, and transfected with miR-214 inhibitor, si-E2F3 or the corresponding control, using Lipofectamine 2000. Cells were incubated for 24, 48 or 72 h at 37°C in a humidified incubator with 5% CO2, followed by 4-h incubation in the presence of 20 µl MTT (Sigma-Aldrich, St. Louis, MO, USA). Next, the supernatant was removed, and 150 µl dimethyl sulfoxide (Guangdong Guanghua Sci-Tech Co., Ltd., Shenzhen, China) was added to the cells, prior to measurement of their absorbance at 490 nm in a FLUOstar OPTIMA microplate reader (BMG Labtech GmbH, Ortenberg, Germany). For the colony forming assay, SMMC-7721 cells were seeded at a density of 500 cells/well into 6-well plates, and transfected with miR-214, miR-ctrl, miR-214 inhibitor, miR-214 inhibitor-ctrl, si-E2F3 or si-ctrl. The cells were incubated for 14 days at 37°C in a humidified incubator with 5% CO2, and subsequently stained with a crystal violet solution (Sigma-Aldrich), and imaged with a GelDoc 2000 system and Quantity One software (version 4.3.1; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Western blot
Total protein content was isolated from SMMC-7721 cells transfected with miR-214, miR-ctrl, miR-214 inhibitor, miR-214 inhibitor-ctrl, si-E2F3 or siRNA-ctrl with RIPA buffer (Life Technologies, Grand Island, NY, USA) at 48 h post-transfection. Protein concentration was measured with a GeneQuant pro RNA/DNA calculator (GE Healthcare Life Sciences, Shanghai, China). Proteins were resolved on a 10% SDS-PAGE gel (Invitrogen Life Technologies) using SDS buffer (Sigma-Aldrich) for 2 h at 100 V, and transferred onto a polyvinylidene fluoride membrane (Merck Millipore, Beijing, China). Next, the membrane was blocked in Tris-buffered saline (pH 7.4; Sigma-Aldrich) with 0.05% Tween-20 (TBST; Sigma-Aldrich) and 5% non-fat dry milk (Yili Group, Hohhot, China) for 1 h, and incubated overnight at 4°C with rabbit anti-human E2F3 (cat. no. ab50917; dilution, 1:500; Abcam, Cambridge, UK), rabbit anti-human CDK6 (cat. no. 14052-1-AP; dilution, 1:1,000; ProteinTech Group, Inc., Chicago, IL, USA), rabbit anti-human CCND1 (cat. no. BS6532; dilution, 1:500; Bioworld Technology, Inc., St. Louis Park, MN, USA) and rabbit anti-human GAPDH (cat. no. 10494-1-AP; dilution, 1:2,000; ProteinTech Group, Inc.) polyclonal antibodies. Following 3 washes with TBST, the membrane was incubated with a goat anti-rabbit IgG secondary antibody (cat. no. 107616; dilution, 1:3,000; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 2 h. Following three washes with TBST, the proteins were scanned and the band density was measured using the ChemiDoc-It®415 Imager (Ultra-Violet Products Ltd., Cambridge, UK) and Immobilon Western Chemiluminescent HRP substrate (EMD Millipore, Billerica, MA, USA). Relative protein expression levels were normalized to the levels of GAPDH and Quantity One imaging software (Bio-Rad Laboratories, Inc.) was used to measure the relative protein expression.
Cell cycle analysis
SMMC-7721 cells were transfected with miR-214, miR-ctrl, miR-214 inhibitor, miR-214 inhibitor-ctrl, si-E2F3 or siRNA-ctrl. At 48 h post-transfection, the cells were harvested and fixed overnight at 4°C in ice-cold 70% ethanol. The cells were then washed in phosphate-buffered saline, and incubated at 4°C for 30 min with 1 ml staining solution, containing 20 µg/ml propidium iodide and 10 U/ml RNaseA (Sigma-Aldrich). Cell-cycle distributions were analyzed by fluorescence-activated cell sorting with a FACSort flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Statistical analysis
The statistical analyses were performed using SPSS software, version 13.0 (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± standard deviation of ≥3 samples, or as indicated. The Students t-test was used to analyze the differences between 2 groups. P<0.05 was considered to indicate a statistically significant difference.
Ethical approval
The present study was approved and supervised by the Medical Ethical Committee of the College of Medicine, Xi'an Jiaotong University (Xi'an, China).
Results
E2F3 is a direct target of miR-214
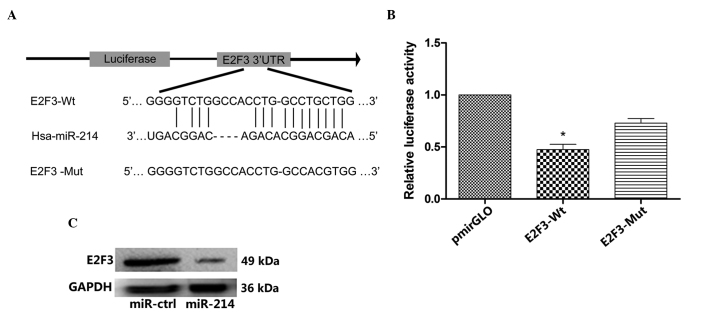
Among the large number of potential target genes of miR-214, E2F3 was selected for further analysis. Subsequently, a binding site for miR-214 was identified in the 3′-UTR of the E2F3 mRNA (Fig. 1A). In order to confirm whether miR-214 directly targets E2F3, 3′-UTR fragments of E2F3 were constructed, which contained a wild-type (WT) or mutant (Mut) binding site for miR-214. These binding sites were cloned into the region downstream of the pmiRGLO dual-luciferase reporter vector, and HEK293 cells were cotransfected with miR-214 and pmirGLO-E2F3-3′-UTR-WT or pmirGLO-E2F3-3′-UTR-Mut or pmirGLO. In the pmirGLO-E2F3-3′-UTR-WT group at 48 h, miR-214 significantly reduced the firefly luciferase activity, compared with pmirGLO (P<0.05; Fig. 1B), indicating that miR-214 may target directly the binding sequences at the 3′-UTR of E2F3. No significant difference was observed between the miR-214 and pmirGLO in the pmirGLO-E2F3-3′-UTR-Mut group. Furthermore, the transfection of miR-214 in the SMMC-7721 cells led to reduced expression of E2F3 at the protein level (Fig. 1C), demonstrating that miR-214 directly targeted E2F3.
Figure 1.
miR-214 directly targets E2F3. (A) Binding sequence for miR-214 in the 3′-UTR of E2F3. A Mut was introduced in the 3′-UTR of WT E2F3, in the complementary site to the binding region of miR-214. (B) Luciferase assay in HEK293 cells: WT or Mut pmirGLO-E2F3-3′-UTR or pmirGLO was cotransfected with miR-214 in HEK293 cells. Following 48 h, relative luciferase values were measured and normalized by Renilla luciferase activity. The luciferase activity in pmirGLO-E2F3-3′-UTR-WT was observed to be significantly reduced by overexpression of miR-214. The data represent the mean ± standard deviation of 3 independent experiments (*P<0.05 vs. pmirGLO). (C) Protein expression levels of E2F3 were measured by western blot analysis. Hepatocellular carcinoma SMMC-7721 cells were transfected with miR-214 or miR-ctrl, and the expression levels of E2F3 were detected by western blotting at 48 h post-transfection. GAPDH was used as a reference gene. The results indicated that miR-214 represses the translation of the E2F3 protein in SMMC-7721 cells. E2F3, E2F transcription factor 3; UTR, untranslated region; Mut, mutation; WT, wild-type; ctrl, control; Hsa, Homo sapiens.
miR-214 suppresses the growth of SMMC-7721 cells through targeting E2F3. As indicated above, miR-214 was observed to directly target E2F3
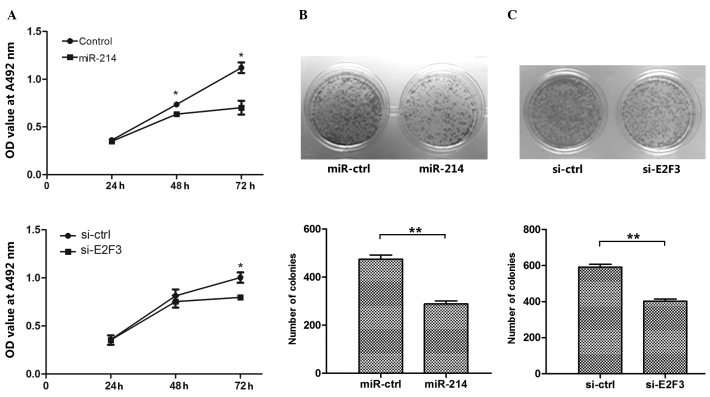
To further investigate the effects of E2F3-mediated miR-214 on the proliferation of SMMC-7721 cells, miR-214 or si-E2F3 were transfected into SMMC-7721 cells, with miR-ctrl or siRNA-ctrl as controls. The results of the MTT assay indicated that overexpression of miR-214 markedly inhibited the growth of SMMC-7721 cells 48 h and 72 h following transfection with miR-214 (P=0.044 and P=0.011, respectively; Fig. 2A). In addition, overexpression of miR-24 markedly inhibited the growth of SMMC-7721 cells 72 h after transfection with si-E2F3 (P=0.021; Fig. 2A). Similarly, a reduced number of colonies were observed in SMMC-7721 cells transfected with miR-214 or si-E2F3, compared with the controls (P<0.01; Fig. 2B and C). These findings suggest that miR-214 may reduce the growth of SMMC-7721 cells through targeting E2F3.
Figure 2.
Overexpression of miR-214 or silencing of E2F3 reduces hepatoma cell growth. (A) The proliferation capacity of hepatocellular carcinoma SMMC-7721 cells was measured by MTT assay at 24, 48 and 72 h post-transfection with miR-214, si-E2F3, miR-ctrl or siRNA-ctrl. The results indicated that overexpression of miR-214 or silencing of E2F3 reduced cell growth. (B and C) The ability to form colonies was used to measure cell proliferation in SMMC-7721 cells, transfected as indicated in panel A. The colony formation ability of these cells was observed to be significantly reduced following (B) overexpression of miR-214 or (C) silencing of E2F3. The data corresponds to the results of 3 independent experiments. *P<0.05, **P<0.01 vs. ctrl. E2F3, E2F transcription factor 3; si, silencing; ctrl, control; OD, optical density; A, absorbance.
miR-214 induces G1-S phase arrest and inhibits cell proliferation by suppressing E2F3-dependent cell cycle regulation
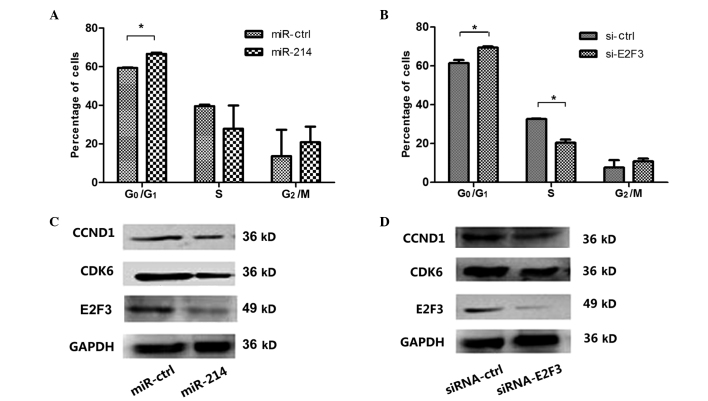
Overexpression of miR-214 led to a significant increase in the percentage of cells in G0/G1 phase, compared with the control (P<0.05; Fig. 3A). Similarly, si-E2F3 led to an increase in G0/G1 phase cells and a reduction in S phase cells, compared with the si-ctrl (P<0.05; Fig. 3B) at 48 h. Furthermore, overexpression of miR-214 reduced the expression levels of E2F3 in SMMC-7721 cells, compared with the control treatment (Fig. 3C), which was in agreement with the results of the treatment with si-E2F3 (Fig. 3D). These findings suggest that miR-214 may induce the G1-S phase arrest of SMMC-7721 cells and suppress cell growth by targeting E2F3.
Figure 3.
Overexpression of miR-214 or silencing of E2F3 induces G1-S arrest. (A and B) Hepatocellular carcinoma SMMC-7721 cells were transfected with (A) miR-214 or (B) si-E2F3 with corresponding controls, and the cell cycle distribution was assessed 48 h later. The histogram indicates the percentage of cells at the G0-G1, S and G2-M cell cycle phases (Students t-test, *P<0.05 vs. ctrl). The expression levels of E2F3, CDK6 and CCND1 in SMMC-7721 cells were measured by western blotting at 48 h post-transfection with (C) miR-214 or (D) si-E2F3. GAPDH was used as a reference control. The data corresponds to the results of 3 independent experiments. *P<0.05 vs. ctrl. E2F3, E2F transcription factor 3; si, silencing; ctrl, control; CDK6, cyclin-dependent kinase 6; CCND1, cyclin D1.
Inhibition of miR-214 contributes to proliferation of SMMC-7721 cells
To experimentally confirm the loss-of-function of miR-214, miR-214-inhibitor oligonucleotides were used to silence the expression of miR-214. The inhibition of miR-214 increased the proliferation of SMMC-7721 cells, as indicated by the MTT assay (Fig. 4A). The inhibitor of miR-214 promoted colony formation and induced G1-S phase transition in SMMC-7721 cells (P<0.05; Fig. 4B and C). Furthermore, cells transfected with miR-214-inhibitor oligonucleotides exhibited increased expression levels of E2F3, as determined by western blot analysis (Fig. 4D). Taken together, these results demonstrated the important contribution of endogenous miR-214 to the proliferation of SMMC-7721 cells in liver cancer progression.
Figure 4.
The inhibition of miR-214 contributes to the proliferation of hepatocellular carcinoma SMMC-7721 cells. (A) Cell proliferation ability was determined by MTT assay at 24, 48 and 72 h post-transfection with an inhibitor of miR-214 or control inhibitor. (B) The ability to form colonies was used to detect cell proliferation in SMMC-7721 cells, transfected as indicated in panel A. (C) The cell cycle was determined in SMMC-7721 cells at 48 h post-transfection with the inhibitor of miR-214 or inhibitor-ctrl. The histogram represents the percentage of cells in the G0-G1, S and G2-M cell cycle phases. (D) The expression levels of E2F3, CDK6 and CCND1 were measured by western blotting at 48 h post-transfection with the inhibitor of miR-214 or inhibitor-ctrl. GAPDH was used as a reference control. The data corresponds to the results of 3 independent experiments.*P<0.05 vs. inhibitor-ctrl. E2F3, E2F transcription factor 3; CDK6, cyclin-dependent kinase 6; CCND1, cyclin D1; ctrl, control; OD, optical density.
Discussion
HCC is the third leading cause of cancer-associated mortalities worldwide, resulting in >600,000 mortalities/year, and a 5-year survival rate of <5% when left untreated (18). However, the molecular mechanisms of HCC remain largely unknown. Recent evidence has suggested that miRNAs may be involved in the progression of human cancer, since numerous miRNAs have been observed to be deregulated during tumorigenesis in humans (19). Previous studies have demonstrated that miR-214 is downregulated in cervical cancer and upregulated in human pancreatic cancer (20,21). The present study has demonstrated that the expression of miR-214 was downregulated in HCC tissues and cell lines. Therefore, miR-214 may be a tumor suppressor miRNA.
E2F3 is a member of the E2F family of transcription factors, which participate in the regulation of diverse cellular processes, including cell differentiation and proliferation (22). E2Fs are involved in the regulation of the transcription activity required for the induction of the S phase of the cell cycle, during the transition from quiescence to proliferation (23). The proliferation of eukaryotic cells is divided into 4 phases: G1, S, G2 and M, as reviewed by Malumbres and Barbacid (24)and Yang et al (25). The activity of E2Fs is regulated by the retinoblastoma protein (pRB), a tumor suppressor that is functionally inactivated in the majority of human tumors. E2F transcription factors contain various heterodimers, each of which is composed of 1 E2F and 1 DP subunit (26). The family members E2F1, 2 and 3 predominantly associate with pRb, and each of these E2Fs are sufficient to induce quiescent cells to enter the S phase through a period of high transcriptional activity (27). Previous studies conducted in cells of higher eukaryotes have indicated that cyclin D-dependent kinases CDK4 and 6 possibly regulate the G1-S phase transition by phosphorylating pRb, and consequently releasing the E2F transcription factors. In the present study, miR-214 was able to suppress the growth of SMMC-7721 cells by targeting E2F3, and arrest the cell cycle progression at the G1 phase. In addition, the relative expression of the cell cycle regulatory protein E2F3 was reduced by miR-214. Furthermore, si-E2F3 and an inhibitor of miR-214 were used to evaluate the biological effects of miR-214 in HCC SMMC-7721 cells. The previously observed effects on cell proliferation due to the overexpression of miR-214 in these cells were consistent with the results obtained upon treatment of the cells with si-E2F3, and opposite to those observed when the cells were treated with the miR-214-inhibitor.
In conclusion, in the present study miR-214 was demonstrated to directly target E2F3 and induce cell cycle arrest at the G1 phase. Furthermore, miR-214 suppressed the growth of HCC SMMC-7721 cells through targeting E2F3. Therefore, these data suggest that miR-214 may be a tumor suppressor and potential therapy for the treatment of HCC in the future.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81171398) and The Key Science and Technology Major Program of Shaanxi Province, China (grant no. 2010ZDKG-50).
References
- 1.Avila MA, Berasain C, Sangro B, Prieto J. New therapies for hepatocellular carcinoma. Oncogene. 2006;25:3866–3884. doi: 10.1038/sj.onc.1209550. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Fuster J, Bruix J. Barcelona-Clínic Liver Cancer Group: The Barcelona approach: Diagnosis, staging and treatment of hepatocellular carcinoma. Liver transplantation. 2004;10(Suppl 1):S115–S120. doi: 10.1002/lt.20034. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan SH, Wu CW, LAF Y, Chi C-W, Lin WC. miR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 2008;28:907–911. [PubMed] [Google Scholar]
- 7.Wiemer EA. The role of microRNAs in cancer: No small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 9.Liu W, Mao SY, Zhu WY. Impact of tiny miRNAs on cancers. World J Gastroenterol. 2007;13:497–502. doi: 10.3748/wjg.v13.i4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Yao J, Shi X, Hu L, Li Z, Song T, Huang C. MicroRNA-302b suppresses cell proliferation by targeting EGFR in human hepatocellular carcinoma SMMC-7721 cells. BMC Cancer. 2013;13:448. doi: 10.1186/1471-2407-13-448. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Suzuki H, Maruyama R, Yamamoto E, Kai M. Epigenetic alteration and microRNA dysregulation in cancer. Front Genet. 2013;4:258. doi: 10.3389/fgene.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 13.Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol. 2010;10:543–550. doi: 10.1016/j.coph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Schwarzenbach H, Milde-Langosch K, Steinbach B, Müller V, Pantel K. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res Treat. 2012;134:933–941. doi: 10.1007/s10549-012-1988-6. [DOI] [PubMed] [Google Scholar]
- 15.Tömböl Z, Szabó PM, Molnár V, Wiener Z, Tölgyesi G, Horányi J, Riesz P, Reismann P, Patócs A, Likó I, et al. Integrative molecular bioinformatics study of human adrenocortical tumors: MicroRNA, tissue-specific target prediction and pathway analysis. Endocr Relat Cancer. 2009;16:895–906. doi: 10.1677/ERC-09-0096. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Kong W, He L, Zhao J-J, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al. MicroRNA expression profiling in human ovarian cancer: MiR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 17.Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013;13:68. doi: 10.1186/1475-2867-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirahata A, Fan W, Sakuraba K, Yokomizo K, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, Nemoto H, et al. MACC 1 as a marker for vascular invasive hepatocellular carcinoma. Anticancer Res. 2011;31:777–780. [PubMed] [Google Scholar]
- 19.Karp X, Ambros V. Developmental biology. Encountering microRNAs in cell fate signaling. Science. 2005;310:1288–1289. doi: 10.1126/science.1121566. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Chen S, Luan X, Li Y, Liu M, Li X, Liu T, Tang H. MicroRNA-214 is aberrantly expressed in cervical cancers and inhibits the growth of HeLa cells. IUBMB Life. 2009;61:1075–1082. doi: 10.1002/iub.252. [DOI] [PubMed] [Google Scholar]
- 22.Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- 23.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, Nevins JR. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 25.Yang L, Meng Y, Bao C, Liu W, Ma C, Li A, Xuan Z, Shan G, Jia Y. Robustness and backbone motif of a cancer network regulated by miR-17-92 cluster during the G1/S transition. PLoS One. 2013;8:e57009. doi: 10.1371/journal.pone.0057009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeGregori J. The Rb network. J Cell Sci. 2004;117:3411–3413. doi: 10.1242/jcs.01189. [DOI] [PubMed] [Google Scholar]
- 27.Lukas J, Herzinger T, Hansen K, Moroni MC, Resnitzky D, Helin K, Reed SI, Bartek J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]