Abstract
Telomere DNA-binding proteins protect the ends of chromosomes in eukaryotes. A subset of these proteins are constructed with one or more OB folds and bind with G+T-rich single-stranded DNA found at the extreme termini. The resulting DNA-OB protein complex interacts with other telomere components to coordinate critical telomere functions of DNA protection and DNA synthesis. While the first crystal and NMR structures readily explained protection of telomere ends, the picture of how single-stranded DNA becomes available to serve as primer and template for synthesis of new telomere DNA is only recently coming into focus. New structures of telomere OB fold proteins alongside insights from genetic and biochemical experiments have made significant contributions towards understanding how protein-binding OB proteins collaborate with DNA-binding OB proteins to recruit telomerase and DNA polymerase for telomere homeostasis. This review surveys telomere OB protein structures alongside highly comparable structures derived from replication protein A (RPA) components, with the goal of providing a molecular context for understanding telomere OB protein evolution and mechanism of action in protection and synthesis of telomere DNA.
Keywords: Telomere structural biology, single-stranded DNA-binding proteins, OB fold, x-ray crystallography, NMR, intrinsically disordered protein regions, molecular recognition, allostery
Introduction
Organization of the genome as linear DNA chromosomes capped by telomeres is a defining feature of Eukarya. Linear chromosomes present challenges with respect to stability and replication, and special handling procedures are required to prevent degradation, end-to-end fusion, and attrition with each cell division. Telomere specific proteins stabilize linear genomes through two principal functions: DNA protection and DNA synthesis (Verdun and Karlseder, 2007; de Lange, 2009). For DNA protection, telomere proteins bind with telomere DNA and ensure that natural chromosome ends are not recognized as DNA breaks. For DNA synthesis, telomere proteins recruit two polymerase enzymes: telomerase and DNA polymerase α/primase. Telomerase adds short tandem repeats of a G+T-rich sequence to the 3′-terminus using instructions encoded in a constitutive RNA template. DNA polymerase α/primase initiates and continues synthesis of the C+A-rich strand using the G+T-rich strand as a template. In all organisms investigated, the 3′-terminal G+T-rich strand ends with a portion that is single stranded, a structural feature that is consistent with biochemical requirements of telomerase, but which imparts risks and dangers since this form of DNA is more reactive chemically, is easily hydrolyzed by nucleases, looks like damaged DNA, and promotes recombination. OB fold proteins bind with the single-stranded G+T-rich strand and thus protect this highly sensitive DNA. The DNA-protective role that OB fold proteins play has been well appreciated. Recent results point also to a role for OB fold proteins in DNA synthesis by coordinating interactions among telomere components, including telomerase and DNA polymerase. The structures of these telomere OB proteins are the subject of this review.
OB fold proteins are widely distributed among all domains of life and participate in several aspects of single-stranded nucleic acid metabolism including DNA replication, DNA repair, protein translation, as well as telomere protection. Several excellent reviews examine the evolution and function of OB fold proteins (Theobald et al., 2003b; Agrawal and Kishan, 2003; Arcus, 2002; Flynn and Zou, 2010). The aim here is to survey DNA-binding and protein-binding OB proteins found at telomeres with a particular emphasis on three-dimensional molecular structure. This work is motivated by newly reported structures of telomere associated OB proteins and the realization that telomere OB protein complexes are more widely conserved than previously thought. The hope is that patterns (and exceptions) identified through structural comparison will generate expectations and testable ideas for newly discovered telomere OB proteins.
Overview of telomere and RPA OB proteins for which structures are available
The first structures of OB folds derived from telomere proteins and replication protein A (RPA) subunits focused on those proteins and domains with high affinity DNA-binding activity. RPA was isolated as an essential multi-subunit complex required for cell-free SV40 DNA replication reactions (Fairman and Stillman, 1988; Wold and Kelly, 1988; Brill and Stillman, 1991). Replication protein A is conserved across Eukarya, and is considered the central single-stranded DNA-binding component of DNA replication forks and DNA repair centers (Wold, 1997; Fanning et al., 2006). Constructed with large, medium, and small subunits, replication protein A closely resembles subcomplexes found at telomeres. Telomere DNA synthesis shares biochemical requirements with DNA replication, and the telomere OB proteins and replication protein A were likely preceded by a common ancestor that could protect single-stranded DNA and coordinate DNA exchange with polymerases and DNA repair enzymes. The telomere OB protein family includes protein-binding members, and structures of protein-interacting telomere OB proteins demonstrate structural homology with protein-interacting RPA subunits. The idea emerging is that replication protein A-like complexes were duplicated and rapidly acquired several specialized functions that all fall in the general category of genome guardians (Flynn and Zou, 2010).
Figure 1 provides an overview of telomere and RPA OB-containing proteins for which three-dimensional structures are available. Stn1 and Ten1 were first characterized as proteins which interact with Cdc13 to regulate telomere length in budding yeast (Grandin et al., 1997; Grandin et al., 2001). The Cdc13–Stn1–Ten1 (CST) complex appears to be conserved across eukaryotes with homologs of Stn1 and Ten1 identified in fission yeast (Martin et al., 2007), plants (Song et al., 2008), and vertebrates (Miyake et al., 2009; Wan et al., 2009), and an analog of Cdc13 (called Ctc1) identified in plants and mammals (Surovtseva et al., 2009; Miyake et al., 2009) [reviewed in (Wellinger, 2009; Price et al., 2010)]. Mammalian Stn1 and Ctc1 had previously been characterized as accessory factors that complex with and stimulate polymerase α/primase (Goulian et al., 1990; Goulian and Heard, 1990; Casteel et al., 2009). The analogous Stn1 and Cdc13 proteins in yeast, similarly interact with polymerase α/primase (Qi and Zakian, 2000; Hsu et al., 2004), and also with telomerase (Qi and Zakian, 2000; Pennock et al., 2001).
Figure 1.
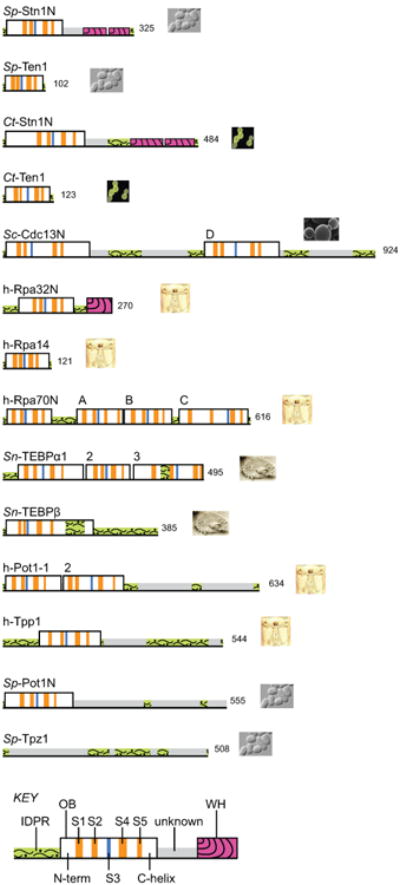
Overview of OB folds in telomere OB proteins and RPA. See on-line version for color; print version lacks color. Protein regions for which three-dimensional structures have been determined by x-ray crystallography or NMR are outlined as tall boxes; regions not yet structurally characterized are low profile. Numbers at the C-terminus indicate the number of residues in that protein. OB units are labeled to indicate the source organism, the protein name, and a unique identifier if the protein contains more than one OB unit or folded domain (see also Table 1 for names of OB units). Icons provide visual cues for the protein source organisms, which are listed here in order of appearance: Schizosaccharomyces pombe, Candida tropicalis, Saccharomyces cerevisiae, Homo sapiens, and Sterkiella nova. Within each OB fold, the conserved structural elements are highlighted with color as follows: S3, blue; S1, S2, S4, and S5, orange (see Fig 2 for further explanation of these elements). Winged helix-turn-helix (WH) motifs found at the C-terminus of Rpa32 and Stn1 are colored magenta with curved stripes. Sp-Ten1 represents the smallest OB unit with very short loops connecting each of the conserved S1 – S5 elements. Segments with a high probability of adopting intrinsically disordered structure are colored green with a wavy pattern. Note that in some cases an intrinsically disordered protein region (IDPR) adopts a fixed structure upon association with a binding partner, as is the case for the IDPR contained within the Sn-TEBPβ OB unit (tall box). Structurally uncharacterized regions within Sc-Cdc13, Sp-Pot1, Sp-Tpz1 and h-Pot1 with low IDPR scores (grey and no pattern) likely contain additional OB units as previously predicted (Theobald and Wuttke, 2004; Miyoshi et al., 2008). Amino acid sequence profiling to score for intrinsically disordered regions was accomplished with the program DISPro (Hecker et al., 2008) as implemented with the SCRATCH protein structure prediction server (Cheng et al., 2005).
In this review, each OB protein unit will be labelled with a short name indicating source organism, name of the complete protein, and either a letter or a number differentiating one OB unit from others in those proteins that have multiple parts (see Table 1 for a complete list of OB unit names used in this review). Thus, Sp-Stn1N is the N-terminal OB unit derived from Schizosaccharomyces pombe Stn1, and Sc-Cdc13D is the DNA-binding OB unit derived from Saccharomyces cerevisiae Cdc13 protein. The N-terminal domain of Stn1 is an OB fold (box with vertical stripes in Fig 1) and the structures of Sp-Stn1N and Ct-Stn1N have been determined in complex with species-specific homologs of Ten1 from Schizosachharomyces pombe and Candida tropicalis (Sun et al., 2009). The C-terminal domain of Stn1 consists of two winged helix-turn-helix (WH) motifs as deduced from crystal structures of the corresponding domain from Saccharomyces cerevisiae (Sun et al., 2009; Gelinas et al., 2009).
Table 1. Structurally characterized telomere protein and RPA-derived OB units.
| name | description (residue range; chain id pdb id*) |
|---|---|
| Sp-Stn1N | N-terminal OB unit found in Stn1, a ubiquitous telomere protein that closely resembles the medium-sized subunit of replication protein A, Rpa32. Stn1N forms a stable heterodimer with Ten1. Source: Schizosaccharomyces pombe. (16-154; chain A 3kf6) |
| Ct-Stn1N | ibid. Source: Candida tropicalis. (19-214; chain A 3kf8) |
| h-Rpa32N | N-terminal OB unit found in Rpa32, the DNA-replication and repair factor most closely related with telomere protein Stn1. Rpa32N forms a stable heterodimer with Rpa14 and a stable heterotrimer with Rpa14 and Rpa70C. Source: human. (41-176; chain A 2pi2, 1quq, also chain B 1l1o) |
| Sp-Ten1 | The single OB unit comprising Ten1, a ubiquitous telomere protein closely resembling the small subunit of replication protein A, Rpa14. Ten1 forms a stable heterodimer with Stn1. Source: Schizosaccharomyces pombe. (1-102; chain B 3kf6, also chain A 3k0x) |
| Ct-Ten1 | ibid. Source: Candida tropicalis. (2-121; chain B; 3kf8) |
| h-Rpa14 | The single OB unit comprising Rpa14, the DNA-replication and repair factor most closely related with telomere protein Ten1. Rpa14 forms a stable heterodimer with Rpa32 and a heterotrimer with Rpa32 and Rpa70C. Source: human. (2-118; chain E 2pi2, chain A 1l1o, chain B 1quq) |
| Sc-Cdc13N | OB unit from the cell division control protein Cdc13, a telomere protein with multiple OB units. Cdc13 appears related, by analogy or paralogy, to the large subunit of replication protein A (Rpa70) and to Ctc1. Cdc13N is the N-terminal OB unit and forms a stable homodimer which binds the catalytic unit of the DNA polymerase α/primase complex. Dimerization is additionally linked to modest binding activity for long single-stranded DNA fragments. Source: Saccharomyces cerevisiae. (13-224; chain A 3oiq; 3oip; 3nws; 3nwt) |
| Sc-Cdc13D | ibid. Cdc13D binds single-stranded DNA, with ultra-high affinity to the sequence d(GTGTGGGTGTG). Source: Schizosaccharomyces pombe. (500-686; chain A 1s40; 1kxl) |
| h-Rpa70N | OB unit from Rpa70, the large subunit of replication protein A. Rpa70N is the N-terminal OB unit and binds with the tumor suppressor and DNA-damage checkpoint protein p53. Source: human. (1-128; chain A 2b3g) |
| h-Rpa70C | ibid. Rpa70C is the C-terminal OB unit which forms a heterotrimer with Rpa32 and Rpa14. Source: human. (439-616; chain C 1l1o) |
| h-Rpa70A | ibid. Rpa70A and Rpa70B bind single-stranded DNA. Source: human. (183-298; chain A 1jmc, 1fgu) |
| h-Rpa70B | ibid. (299-420; chain A 1jmc, 1fgu) |
| h-Pot1-1 | OB unit from protection of telomere protein 1, a ubiquitous telomere protein with dual roles in protection and synthesis of telomere DNA. Pot1-1 and Pot1-2 form a stable single-stranded DNA-binding domain that is highly comparable in structural form with the DNA-binding domain of Sn-TEBPα and that exhibits high affinity and specificity for the sequence d(TTAGGGTTAG). Source: human. (6-145; chain A 1xjv, 3kjo, 3kjp) |
| h-Pot1-2 | ibid (149-299; chain A 1xjv, 3kjo, 3kjp) |
| Sp-Pot1N | N-terminal DNA-binding OB unit in Pot1 from fission yeast. The complete DNA-binding domain probably includes a second OB unit, the structure of which is unavailable. The structure of Sp-Pot1N complexed with d (GGTTAC) is highly comparable with structures of h-Pot1-1 and Sn-TEBPα1 seen as portions of DNA-complexed proteins. Source: Schizosaccharomyces pombe. (5-174; chain A 1qzh, 1qzg) |
| h-Tpp1 | A Pot1-interacting protein containing at least one OB unit that is highly comparable to Sn-TEBPβ in terms of structure and function. Tpp1 enhances DNA-binding affinity, DNA versus RNA discrimination, and telomerase processivity activity measured for Pot1. Source: human. (96-241; chain A 2i46) |
| Sn-TEBPα1 | OB unit from the telomere end-binding protein alpha subunit. TEBPα1 and α2 form the N-terminal DNA-binding domain which binds d(TTTTGGGG) with modest affinity and sequence specificity. The complete TEBPα protein forms a complex with TEBPβ that is stable only in the presence of DNA. Source: Sterkiella nova. (36-204; chain A 2i0q, 1otc, 1jb7, 1kix, 1k8g, 1ph1, 1ph2, and several others) |
| Sn-TEBPα2 | ibid. (205-313; chain A 2i0q, 1otc, 1jb7, 1kix, 1k8g, 1ph1, 1ph2, and several others) |
| Sn-αN | The N-terminal DNA-binding domain of TEBPα, synonymous with TEBPα1 + α2. Source: Sterkiella nova. (36-313; chain A 2i0q, 1otc, 1jb7, 2kix, 1k8g, 1ph1, 1ph2, and several others) |
| Sn-TEBPα3 | The C-terminal OB unit from the telomere end-binding protein alpha subunit which interacts with TEBPβ, the second telomere end-binding subunit. TEBPα–TEBPβ association is DNA-dependent. Source: Sterkiella nova. (325-495; chain A 2i0q, 1otc, 1jb7, 1ph1, etc.) |
| Sn-TEBPβ | The second telomere end-binding protein subunit from Sterkiella nova, which appears related by paralogy with human Tpp1 and with the fission yeast protein Tpz1. TEBPβ contains one OB unit, an internal IDPR insertion (residues 165-200), and a C-terminal IDPR tail (residues 234-385). TEBPβ associates with the TEBPα-DNA complex, extending the DNA-binding interface to include d(TTTTGGGGTTTTGGGG), and enhancing DNA affinity. Source: Sterkiella nova. (9-223; chain B 2i0q, 1otc, 1jb7, 1ph1, etc.) |
| Sn-αNβΔ | Chimera protein constructed with two DNA-binding OB units of TEBPα fused to the OB unit of TEBPβ. αNβΔ binds d(TTTTGGGGTTTTGGGG), and a crystal structure shows DNA interactions not seen in previous structures. Source: Sterkiella nova. (35-316 of α, 1-155+201-231 of β, [156-200, including IDPR of β deleted]) |
Certain crystal space groups contain multiple copies of the same protein. In these cases only the first instance of a particular chain is identified. For OB units that have been structurally determined multiple times multiple pdb ids are listed, the first of which corresponds with structures used in analyses presented in this review.
The structural domains of Stn1 are highly comparable with those found in the medium-sized RPA subunit, Rpa32 (Sun et al., 2009). Stn1 and Rpa32 each contain an N-terminal OB unit and a C-terminal WH motif. Tandem duplication of the WH unit appears to be a telomere-specific innovation since Rpa32 has one WH and Stn1 has two (Gelinas et al., 2009). The structure of the OB fold from human Rpa32, h-Rpa32N, has been determined as part of a heterodimer with the RPA small subunit, Rpa14 (Bochkarev et al., 1999), and as part of a heterotrimer with Rpa14 and the C-terminal OB unit derived from the RPA large subunit, Rpa70 (Bochkareva et al., 2002). The structure of a winged helix-turn-helix domain of human Rpa32 has been determined by NMR, in an unliganded form and as complexes with peptides derived from DNA-repair enzymes (Mer et al., 2000). These and other structures of RPA-derived subcomplexes illustrate the modular nature of OB units in forming OB-DNA and OB-protein complexes, a theme that also applies to telomere OB proteins.
In budding yeast, the Stn1–Ten1 subcomplex recruits Cdc13, a larger telomere protein with multiple OB units. Two OB units contained within Cdc13 have been structurally characterized (Mitton-Fry et al., 2002; Mitchell et al., 2010; Sun et al., 2011), and two others were predicted by sequence profile analysis (Theobald and Wuttke, 2004). A centrally located OB unit in Cdc13, Sc-Cdc13D, binds telomere single-stranded DNA with high affinity, and the solution structure of a Sc-Cdc13D-DNA complex has been determined by NMR (Mitton-Fry et al., 2004). Structures of the N-terminal OB unit of Cdc13, Sc-Cdc13N, are now available in an unliganded state (Mitchell et al., 2010; Sun et al., 2011) and as a complex with a peptide derived from Pol1 (Sun et al., 2011), the catalytic component of the DNA polymerase α/primase complex essential for initiation of semi-conservative DNA synthesis.
The large subunit of replication protein A, Rpa70, contains four OB units, each of which has been structurally characterized. A structure of the N-terminal OB unit, Rpa70N, was determined in complex with a peptide derived from the tumor suppressor and DNA-damage checkpoint protein p53 (Bochkareva et al., 2005). Structures of the tandem Rpa70A+Rpa70B DNA-binding OB units have been determined by x-ray crystallography, as an unliganded protein (Bochkareva et al., 2001) and in complex with single-stranded DNA (Bochkarev et al., 1997). A structure of the C-terminal OB unit, Rpa70C, was obtained as part of a higher-order assembly involving Rpa32N and Rpa14 (Bochkareva et al., 2002).
The Cdc13(or Ctc1)–Stn1–Ten1 complex works alongside a second telomere specific complex centered on Pot1 and its interacting partner Tpp1. OB units within the Pot1–Tpp1 subcomplex are structurally similar to OB components within the Sn-TEBPα–TEBPβ heterodimer from Sterkiella nova (formerly Oxytricha nova), consistent with the idea that this subcomplex has been conserved across evolution (Baumann and Cech, 2001; Xin et al., 2007; Wang et al., 2007). The Pot1–Tpp1 complex has a ying/yang relationship with telomerase, acting to sequester and protect single-stranded DNA so that access to telomerase is limited (Liu et al., 2004; Ye et al., 2004; Lei et al., 2005), and also serving to recruit telomerase and enhance telomerase processivity (Xin et al., 2007; Wang et al., 2007; Zaug et al., 2010; Latrick and Cech, 2010). Presumably these two diametrically opposed activities are regulated to meet requirements at different stages of the cell cycle.
Principle DNA-binding activity for Pot1–Tpp1 and TEBPα–TEBPβ resides with two N-terminal OB units (Pot1-1 and Pot1-2 in Pot1; TEBPα1 and TEBPα2 in Sn-TEBPα), and structures for these domains in complex with species-specific single-stranded telomere DNA are available (Lei et al., 2004; Classen et al., 2001; Peersen et al., 2002). Additional OB units contained within Pot1 and Sn-TEBPα mediate interaction with Tpp1 and Sn-TEBPβ, respectively. The structure of a Sn-TEBPα–TEBPβ heterodimer complexed with single-stranded DNA showed details for DNA interaction and protein-protein association (Horvath et al., 1998).
A crystal structure of the OB unit derived from Tpp1 showed structural features shared with Sn-TEBPβ but not seen in other OB proteins (Wang et al., 2007). Multiple regions in both Tpp1 and Sn-TEBPβ score high probability for intrinsically disordered protein regions (IDPR)s (see Fig 1), providing additional support for the idea that Tpp1 and Sn-TEBPβ are descendants of a common ancestral protein. Outside of the OB unit, structural information for Tpp1 is sparse and a detailed view of the Pot1–Tpp1 interface is unavailable. The C-terminal IDPR of Sn-TEBPβ comprises residues 231-385 which are similarly recalcitrant to structure determination (Buczek and Horvath, 2006a). As observed in crystal structures of a Sn-TEBPαβ–DNA complex (Horvath et al., 1998; Buczek and Horvath, 2006a) the TEBPα–TEBPβ interface is constructed by OB-OB contacts between the OB unit of Sn-TEBPβ and the middle (Sn-TEBPα2) and C-terminal (Sn-TEBPα3) OB unit of Sn-TEBPα. The OB-OB portion of the TEBPα–TEBPβ interface is augmented by 46 residues in Sn-TEBPβ (residues 165β-200β) which adopt relatively extended peptide structure that partially wraps around Sn-TEBPα3. 2D-NMR spectra recorded for Sn-TEBPβ and deletion variants of Sn-TEBPβ indicated residues 165β-200β are highly mobile prior to association with Sn-TEBPα and single-stranded DNA, meaning these residues are an IDPR insertion contained within the Sn-TEBPβ OB unit that co-fold upon assembly of the ternary complex, as inferred from inspection of the Sn-TEBPαβ–DNA crystal structure (Horvath et al., 1998).
The Pot1–Tpp1 and Sn-TEBPα–TEBPβ complexes are recapitulated in fission yeast as a Sp-Pot1–Tpz1 complex. The structure of the N-terminal OB unit resident in Sp-Pot1 has been determined in complex with single-stranded DNA (Lei et al., 2003). The DNA-binding Sp-Pot1N OB unit is thought to collaborate with a second, immediately adjacent OB unit (Trujillo et al., 2005; Croy et al., 2006); however, the two-OB Sp-Pot1 DNA-binding domain was difficult to express and a structure for the putative second OB fold is currently unavailable. At the time this review was prepared, a structure for the S. pombe Tpp1 homolog, Tpz1 was also unavailable. Sequence profiling shows C-terminal regions of Sp-Tpz1 score a high probability of adopting intrinsically disordered structure (Fig 1), meaning IDPRs are likely a conserved feature important for some aspect of h-Tpp1/Sn-TEBPβ/Sp-Tpz1 function.
Structural architecture of the OB fold
Structural similarity among OB proteins was first described by Murzin (Murzin, 1993). Elements defining the OB fold comprise five beta strands arranged as two anti-parallel beta sheets that form a partially closed structure reminiscent of a barrel (Fig 2). Previously, OB folds were reliably identified only by structure determination. With advances in sequence profile searches and a now robust collection of OB structures, it is currently possible to pull out OB proteins from genome databases with a fair degree of confidence (Baumann and Cech, 2001; Theobald and Wuttke, 2004; Martin et al., 2007; Gao et al., 2007; Miyake et al., 2009). Sequence matching is challenging because the core beta strand elements support diverse insertions and embellishments at connecting segments as well as N-terminal and C-terminal extensions. Plasticity in structure likely makes the OB protein especially well suited for divergent evolution and establishment of new functions. For this review we are most interested in three principle OB functions: binding single-stranded DNA, binding extended peptide or helical segments, and binding other OB fold units. Figure 2 illustrates the conserved core OB structure engaged in each of these three activities.
Figure 2.
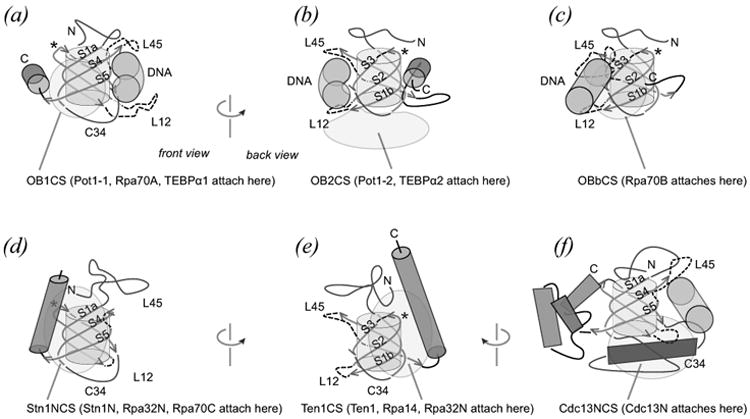
Architecture of RPA and telomere protein OB folds. See on-line version for color; print version lacks color. Conserved beta strand elements (orange arrows, labeled) wrap around the OB fold core (grey cylinder). Loops (dashed) connect the beta strands in a characteristic topology so that beta strands S1a-S4-S5 form one anti-parallel beta sheet and strands S1b-S2-S3 form a second sheet. From the vantage point chosen here, the N-terminal region (blue, labeled N) and S3-S4 connecting portion (blue, labeled C34) cap the OB beta barrel from “above” and “below”. The view shown in panels (a), (d) and (f) correspond with the “front” of the OB fold, and the view shown in panels (b), (c) and (e) correspond with the “back”. Contact surfaces typically seen in telomere OB protein structures are indicated with transparent pink shapes. (a-c) Examples of single-stranded DNA-binding OB folds show DNA (green cylinder) at the classical oligo-binding surface defined by L45 and L12 from above and below. For DNA-binding OB units, the C-terminal helix (blue, labeled C) lies along the equator of the OB fold (a, b) or is absent (c) as observed for Rpa70A. Each of these DNA-binding OB folds collaborates with a second OB unit that makes intramolecular contact at one of three contact surfaces, labeled OB1CS (a) OB2CS (b) and OBbCS (c). (d-f) Examples of protein-binding OB folds. For these protein-binding OB units, the C-helix aligns in an axial manner and participates in defining two contact surfaces used in OB fold inter-molecular association, labeled Stn1NCS (d) and Ten1CS (e) on the front and back of the structure. (f) A third OB protein association interface is defined by the dimer interface of Cdc13N, which is constructed with symmetry equivalent C34-helices (dark blue). In this (Sc-Cdc13N)2 structure, the classical oligo-binding site is altered so that C34-helices assume the role normally played by L12 in contacting an oligopeptide ligand (green cylinder).
The naming of structural elements and contact surfaces will simplify further analysis and discussion. As labeled in Figure 2, the five beta strands are called S1a, S1b, S2, S3, S4, and S5. Strand S1 forms the outer edge of both beta sheets, with topology S1a-S4-S5 in one beta sheet, and S1b-S2-S3 in the second beta sheet. Connecting loops are L12, L23, and L45, with the name indicating the two connected strands. S3 connects to S4 with either extended peptide or helical structure or a mixture of both, and this structural element is labeled C34 to indicate a different, more elaborate connection compared with the Lij loops. C34 seals the “bottom” of the OB barrel along the otherwise open S1b/S5 seam. In some cases L45 serves a similar role and folds so as to connect the two beta sheets at the “top” of the OB barrel along the S1a/S3 seam. The N-terminal region consistently forms some kind of elaboration that caps the OB barrel. Most OB folds end with a C-terminal helix (C-helix). The C-helix is characteristically embedded between or alongside L23 and the C-terminal portion of C34. In most telomere OB proteins, the C-helix is oriented along the equator of the OB structure or parallel to the barrel's axis of radial symmetry. For some telomere protein and RPA-derived OB units (e.g. Sn-TEBPα3) the C-helix is missing, and in two OB proteins (Sn-TEBPβ and h-Tpp1) the C-helix is removed from its normally observed location and is found instead towards the “back” and “top” of the OB structure interacting with residues of the N-terminal element.
OB fold units generally work in pairs, either covalently tethered in one protein or as individual subunits of a heterodimer. For example, DNA-binding domains of the S. nova telomere protein Sn-TEBPα and its human homolog h-Pot1 each comprise two OB folds. In both of these DNA-binding domains, OB1 fits onto the “front” of OB2 as defined by the S1aS4S5 sheet (Fig 2(a)). For this review, contact surfaces will be named according to the prototypical OB unit that fits into each surface. Accordingly, the OB1-associated contact surface is designated OB1CS. OB2 fits onto the “bottom” of OB1 as defined by the C34 connector and L12, and this OB2-associated contact surface is called OB2CS (Fig 2(b)).
Contact surfaces connecting two OB units may associate in response to binding of DNA, or these may stably associate in both DNA-bound and ligand-free states. The N-terminal DNA-binding domain of Sn-TEBPα, Sn-TEBPαN, serves as an example of the latter category. The crystal structure of Sn-TEBPαN has been determined in unbound and DNA-bound states, and the two OB units (Sn-TEBPα1 and Sn-TEBPα2) make the same interactions with each other in both states, meaning intramolecular OB-OB contacts do not change in response to DNA-binding for this protein (Classen et al., 2001; Peersen et al., 2002).
Different from the situation described for Sn-TEBPα, the principal DNA-binding OB units of Rpa70 (Rpa70A and Rpa70B) respond dramatically to binding of single-stranded DNA (Bochkarev et al., 1997; Bochkareva et al., 2001). In the unliganded state, Rpa70A and Rpa70B are loosely tethered and make very modest contact with each other. In the DNA-bound state, Rpa70 OB units are rotated by 45 ° and make more significant interaction. Rpa70A contacts Rpa70B at OB1CS very similarly to the arrangement seen for DNA-binding OB units in Sn-TEBPα and Pot1, but Rpa70B contacts Rpa70A not at OB2CS but at a different surface, OBbCS, defined by a highly extended L45 and the outer edge of the S1aS2S3 sheet on the “back” of the OB structure (Fig 2 (c)).
The path taken by single-stranded DNA is either straight or bent depending on the particular contact surface connecting two DNA-binding OB units. Each of the structurally characterized single-stranded DNA-binding OB units cradles ssDNA at the classical oligo-binding site defined by L45 and L12 (Fig 2(a-c)). In Rpa70, with front-to-back arrangement of OB1CS and OBbCS, single-stranded DNA continues from the oligo-binding site on Rpa70A to that of Rpa70B in a straight path without changing direction. In the DNA complexes observed for h-Pot1 and Sn-TEBPα, the telomere single-stranded DNA must bend sharply to maintain contact with the oligo-binding sites at two OB units because these are arranged front-to-bottom with use of OB1CS and OB2CS. Bending of single-stranded DNA may be a telomere-specific innovation, as previously noted (Croy and Wuttke, 2006).
These examples show that structural organization of OB units within an OB protein has consequences for DNA structure. Further levels of organization and new contact surfaces are apparent from examination of how telomere proteins associate with each other. Telomere OB proteins often assume asymmetrical quarternary structure through association of unequal protein subunits. For example, the telomere OB proteins from S. nova were isolated from ciliates as a DNA-bound TEBPα–β heterodimer (Gottschling and Zakian, 1986; Price and Cech, 1989). In this case, subunit association is DNA-dependent (Fang and Cech, 1993; Fang et al., 1993; Buczek et al., 2005) and involves OB-OB contacts, OB-oligopeptide contacts, as well as OB-DNA contacts.
The telomere proteins Stn1 and Ten1 form a stable heterodimer, which binds single-stranded DNA and other telomere components, such as Sc-Cdc13 in budding yeast and Ctc1 in plants and vertebrates, with functional consequences for telomere homeostasis (Pennock et al., 2001; Gao et al., 2007; Martin et al., 2007; Song et al., 2008; Wan et al., 2009; Surovtseva et al., 2009; Miyake et al., 2009) [also reviewed in (Linger and Price, 2009)]. Recently determined structures of the Stn1 N-terminal OB fold (Stn1N) complexed with Ten1 from two yeast species show that the Stn1N–Ten1 interface relies heavily on C-helices contributed from both OB proteins (Sun et al., 2009). The C-helix of Stn1 lies along a surface on Ten1 defined by the S1aS4S4 beta sheet and C-helix of Ten1, (Stn1CS in Fig 2(d)). The C-helix of Ten1 lies along a different surface on Stn1 defined by the opposite S1bS2S3 beta sheet and C-helix, (Ten1CS in Fig 2 (e)). Each of these contact surfaces is highly analogous with those found at the Rpa32–Rpa14 heterodimer interface, with Rpa14 fitting into the Ten1CS on Rpa32N and Rpa32N fitting into the Stn1CS on Rpa14. At the interface of Rpa70C and the Rpa32–Rpa14 subcomplex, Rpa32N provides the Stn1CS which interfaces with Ten1CS provided by Rpa70C.
The crystal structures of the N-terminal OB fold from Sc-Cdc13 revealed a dimeric quarternary structure with C2 rotational symmetry (Mitchell et al., 2010; Sun et al., 2011). Symmetrical dimers and tetramers are familiar OB arrangements for single-stranded DNA-binding proteins in Eubacteria and Archaea [reviewed in (Horvath, 2008b)], but unusual for eukaryote OB proteins with a role in DNA metabolism which are generally constructed as asymmetrical hetero-oligomers. The Sc-Cdc13N OB unit contains a highly developed C34 region. A large helix in the N-terminal portion of C34 fits into a groove formed between the C34 helix and S5 of a symmetry related Sc-Cdc13N subunit (Cdc13NCS in Fig 2 (f)). As will be discussed further below, the Sc-Cdc13N dimer interface is functionally important for telomere DNA synthesis (Mitchell et al., 2010; Sun et al., 2011), and Cdc13 dimerization is evolutionarily conserved (Sun et al., 2011).
The examples discussed so far have served to partially introduce structural elements that differentiate telomere OB protein from each other and from replication protein A subunits. The following sections further analyze structural similarities and differences with the aim of identifying innovations that were functionally important for telomeres and also to detangle evolutionary relationships. Structure is especially important for this quest because much (but not all) of the historical record for OB proteins has been scrambled at the amino acid sequence level.
Structural conservation and diversity
Structures of telomere OB proteins have been instrumental in the hunt for homologous telomere OB proteins. The first crystal structure of telomere OB fold proteins from Sterkiella nova provided molecular details explaining DNA binding and recognition and revealed structural homology with replication protein A (RPA), another OB fold protein involved in DNA metabolism (Horvath et al., 1998). The next telomere OB protein to be described structurally was the single-stranded DNA-binding domain derived from Cdc13, the principle single-stranded telomere DNA-binding agent in budding yeast. The NMR structure of Sc-Cdc13D showed that OB folds would be a conserved feature of telomere end protection (Mitton-Fry et al., 2002). The N-terminal OB fold unit in S. nova TEBPα facilitated identification of a homologous telomere DNA-binding OB protein, Pot1, (Baumann and Cech, 2001), and structural similarity for S. pombe Pot1, human Pot1 and Sn-TEBPα derived DNA-binding domains support an evolutionary relation (Lei et al., 2003; Lei et al., 2004).
Figure 3 shows three-dimensional models of OB units grouped according to function and structural criteria. Each unit is either derived from a telomere OB protein or from replication protein A. Each example is viewed in the same orientation, with the S1aS4S5 sheet toward the viewer, the C-helix on the left (if present), and loops L12 and L45 on the right. Root mean squared deviations calculated for representative pairwise comparisons ranges from 0.9 to 2.6 Å (mean rmsd = 2.0 Å, n = 36) for 44-49 overlapping Cα positions (Table 2). Aligned in this way, the five conserved beta strands are recognizable in each OB unit (contrasting ribbon color in Fig 3). While OB conserved elements have been preserved nearly intact over millions of years of evolution, slight truncations and insertions in the beta strands are nevertheless apparent. For example, several members, especially those belonging to the DNA-binding group (Fig 3(a)), have one extra residue in S4 that is not found in S4 of the Stn1 group (Fig 3(c)). Also, the N-terminal portion of S1 is shorter by two residues in the Sn-TEBPβ/Tpp1 group (Fig 3(b)). Conserved element S2 terminates with a tight turn configured with an Asp-Gly-Thr/Ser-Gly motif, and deviations from this pattern are evident in newly determined structures of a Candida tropicalis Ct-Stn1N–Ten1 heterodimer (Sun et al., 2009) and structures of the Saccharomyces cerevisiae (Sc-Cdc13N)2 homodimer (Mitchell et al., 2010; Sun et al., 2011).
Figure 3.
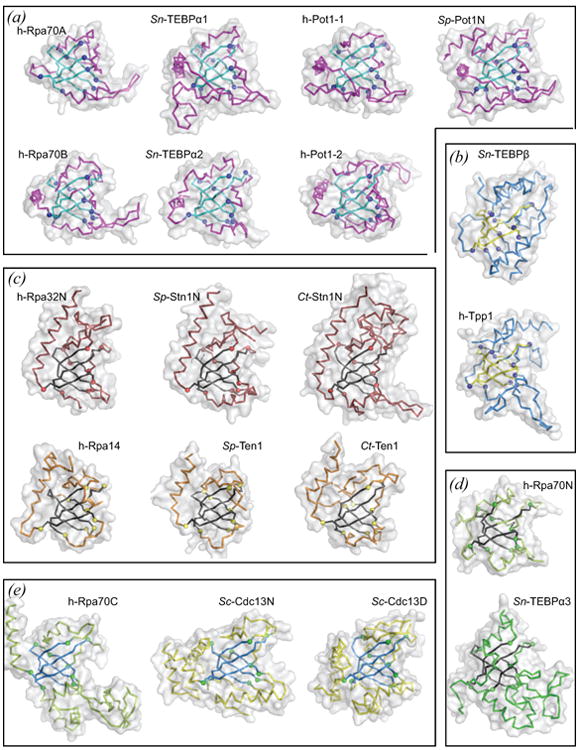
Structures of telomere protein and RPA-derived OB folds. See on-line version for color; print version lacks color. Each OB unit is depicted as an alpha-carbon trace inside a semi-transparent accessible surface (grey) oriented as in Fig 2(a) with the classical oligo-binding surface on the right, N-terminal extension on top and C34 connecting element on bottom. Each OB fold was aligned on the basis of 43-49 equivalent alpha carbon positions contained within the structurally conserved core comprising strands S1 – S5 (contrasting color). Representative pairwise root mean squared deviations are reported in Table 2. Each panel groups OB structures according to shared binding function and shared structural features. (a) The single-stranded DNA-binding group includes two OB units from each of h-Rpa70, Sn-TEBPα, and h-Pot1, and the N-terminal OB unit from Sp-Pot1. In addition to DNA-binding function, six of the seven OB units in this group position the C-helix along the equator of the OB barrel. (b) Structures are shown for Sn-TEBPβ and its human homolog Tpp1, each of which collaborates with its respective DNA-binding partner (Sn-TEBPβ with Sn-TEBPα, h-Tpp1 with h-Pot1) to enhance DNA-binding affinity. Sn-TEBPβ and Tpp1 additionally position the C-helix at the back of the OB unit, interacting with the N-terminal extension. (c) Members of the Rpa32N/Stn1N group each dimerize with a smaller subunit from the Rpa14/Ten1 group, and all members of these two groups feature a C-helix that is positioned axially along the OB barrel. (d) Rpa70N and Sn-TEBPα3 each bind polypeptide ligands and lack a C-helix. (e) The three OB units grouped here feature a highly elaborate C34 element connecting S3 and S4. Note that Sc-Cdc13D could be placed with the group containing Sn-TEBPα1 and h-Pot1-1 on the basis of DNA-binding function; however, recently discovered structural similarity with Sc-Cdc13N and the phylogeny presented in Figure 4 argue for placement together with h-Rpa70C and Sc-Cdc13N. Structures were rendered with PyMOL [PyMOL Molecular Graphics System, Version 0.99, Schrödinger, LLC] using atomic coordinates obtained from the protein data bank (PDB) listed here by PDB identifier: h-Rpa70A and h-Rpa70B, 1jmc (Bochkarev et al., 1997); Sn-TEBPα1, Sn-TEBPα2, Sn-TEBPα3, and Sn-TEBPβ, 2i0q (Buczek and Horvath, 2006a); h-Pot1-1 and h-Pot1-2, 1xjv (Lei et al., 2004); Sp-Pot1N, 1qzh (Lei et al., 2003); h-Tpp1, 2i46 (Wang et al., 2007); h-Rpa32N and h-Rpa14, 2pi2 (Deng et al., 2007); Sp-Stn1N and Sp-Ten1, 3kf6; Ct-Stn1N and Ct-Ten1, 3kf8 (Sun et al., 2009); h-Rpa70N, 2b3g (Bochkareva et al., 2005); h-Rpa70C, 1l1o (Bochkareva et al., 2002); Sc-Cdc13N, 3oiq (Sun et al., 2011); Sc-Cdc13D, 1s40 (Mitton-Fry et al., 2004).
Table 2. Root mean squared deviation for pairwise comparison of representative telomere OB units*.
| RMSD (Å) | Sp-Ten1 | Sn-α3 | Sc-Cdc13N | h-Tpp1 | h-Pot11 | h-Pot12 | Sn-α1 | Sn-α2 | min | max |
|---|---|---|---|---|---|---|---|---|---|---|
| Sp-Stn1N | 1.1 | 1.8 | 1.5 | 1.7 | 1.8 | 1.9 | 2.0 | 1.8 | 1.1 | 2.0 |
|
|
||||||||||
| Sp-Ten1 | 2.1 | 1.5 | 1.6 | 2.1 | 2.0 | 2.1 | 2.1 | 1.1 | 2.1 | |
|
|
||||||||||
| Sn-α3 | 1.7 | 2.3 | 2.4 | 2.6 | 2.5 | 2.5 | 1.7 | 2.6 | ||
|
|
||||||||||
| Sc-Cdc13N | 2.2 | 2.4 | 2.3 | 2.6 | 2.4 | 1.5 | 2.6 | |||
|
|
||||||||||
| h-Tpp1 | 2.3 | 2.3 | 2.2 | 2.2 | 1.6 | 2.3 | ||||
|
|
||||||||||
| h-Pot1-1 | 1.7 | 0.9 | 1.6 | 0.9 | 2.4 | |||||
|
|
||||||||||
| h-Pot1-2 | 1.6 | 1.5 | 1.5 | 2.6 | ||||||
|
|
||||||||||
| Sn-α1 | 1.7 | 0.9 | 2.6 | |||||||
|
|
||||||||||
| Sn-α2 | 1.5 | 2.5 | ||||||||
|
|
||||||||||
Only amino acid residues in the structurally conserved OB core (n = 42-49) were included in pairwise superpositions.
Structural diversity outside of the conserved beta core is readily apparent. For example, the N-terminal OB barrel “lid” comprises segments of extended structure plus a short helix in most OB units, but in Ct-Stn1N, this region constitutes a large subdomain sitting on top of the OB fold (Fig 3 (c)). Insertions are apparent in loops connecting beta strand core elements. The L12 loop connecting strands S1 and S2 normally comprises between 0-16 residues but contains 43 residues in the N-terminal OB fold of Rpa70 (Rpa70N in Fig 3(d)) and 33 residues in the C-terminal OB fold of Sn-TEBPα (Sn-TEBPα3 in Fig 3(d)). Insertions such as these in L12 of h-Rpa70N and Sn-TEBPα3 were likely fixed in the course of evolution because these provided some advantageous function. The structure of Rpa70N shows at least a portion of the L12 insertion making contact with a helical peptide element derived from the tumor suppressor protein p53, suggesting that a function of insertions may be to increase contact surface with other proteins active in DNA repair and cell cycle check points (Bochkareva et al., 2005). In the structure of Sn-TEBPα complexed with Sn-TEBPβ and single-stranded DNA, the L12 insertion in Sn-TEBPα3 contributes to interactions with an oligopeptide element derived from Sn-TEBPβ (Horvath et al., 1998).
Insertion in the region connecting L23 and S3 has occurred for at least three DNA-binding telomere OB proteins. In DNA-binding OB units derived from Sc-Cdc13, Sn-TEBPα and h-Pot1, the L23 insertion exceeds 10 residues (24 in Sc-Cdc13D, 13 in Sn-TEBPα1 and 28 residues in h-Pot1-2). As seen in the NMR structure of Sc-Cdc13D complexed with single-stranded DNA, the L23 insertion contributes a significant portion of the DNA-protein interface (Mitton-Fry et al., 2004). Analysis of the dynamic properties of Sc-Cdc13D by NMR showed that the L23 insertion is folded with similar topology in both unliganded and DNA-bound states (Eldridge and Wuttke, 2008). Motions in the slow (ms) and intermediate (μs) timescales respond to DNA-binding, however, indicating that L23 residues participate in an induced-fit type of mechanism for molecular recognition of telomere DNA (Eldridge and Wuttke, 2008).
Structural roles played by analogous L23 insertions found in Sn-TEBPα1 and h-Pot1-2 are less clear but apparently do not involve direct contact with DNA. The structure of h-Pot1 complexed with telomere DNA shows that the L23 insertion found in h-Pot1-2 fits over the S1bS2S3 beta sheet and thus occludes OBbCS (Lei et al., 2004), possibly so as to block interaction with other proteins at this contact surface. Comparison of several crystal structures containing Sn-TEBPα1 shows that the L12 insertion in that OB unit can adopt a range of positions (Horvath et al., 1998; Classen et al., 2001; Peersen et al., 2002; Theobald and Schultz, 2003; Buczek and Horvath, 2006a). In some of these, it appears as an appendage, and temperature factors in these cases are elevated, indicating L12 insertion residues are flexible and likely dynamic in solution.
Elaboration of the C34 element is apparent in several examples: in Sc-Cdc13N (described above), in the DNA-binding OB fold unit of Sc-Cdc13 (compare Sc-Cdc13N and Sc-Cdc13D, Fig 3 (e)), and also in the C-terminal OB folds of Rpa70C and Sn-TEBPα3 (Fig 3 (d, e)). In addition to constructing a dimer interface, C34 elaboration in Sc-Cdc13N contributes to binding of a helical peptide element derived from the catalytic subunit of the polymerase α/primase complex (Sun et al., 2011). The C34 elaboration in Rpa70C appears to provide structural reinforcement for an equally large L12 insertion. Concave surfaces and grooves apparent for this C34 + L12 elaboration in Rpa70C suggest a role in binding DNA or peptide elements in other proteins. In Sn-TEBPα3 at least a portion of the C34 + L12 elaboration serves to increase the Sn-TEBPα–β protein-protein interface (Horvath et al., 1998), which is critical for cooperative DNA binding (Fang et al., 1993; Buczek et al., 2005).
Evolutionary relationships
OB proteins likely evolved from a single common ancestor (Theobald and Wuttke, 2005); therefore, it is reasonable to consider evolutionary paths by which telomere OB units and RPA OB units were derived. Telomere and RPA OB units can be grouped into clades on the basis of homologous structural features (e.g. disposition of C-helix) and on the basis of analogous function (e.g. binding directly with single-stranded DNA). After completing the structural alignment of telomere and RPA OB folds (Fig 3), I wondered if evolutionary relations among the OB units could be inferred from amino acid character in the corresponding sequence alignment. Figure 4(a) shows the structure-based alignment of 21 OB fold amino acid sequences. This alignment is similar to a structure-based alignment of OB fold proteins involved in nucleic acid recognition (Theobald et al., 2003b) and a structure-based alignment of a more limited set of DNA-binding telomere OB fold proteins (Horvath, 2008a).
Figure 4.
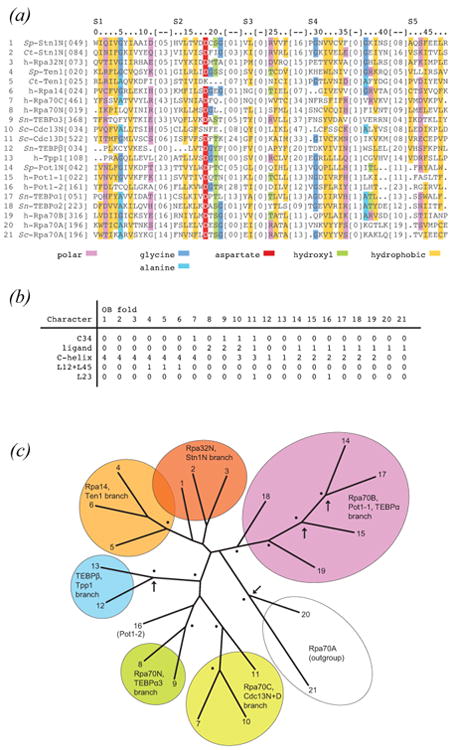
Evolutionary history of telomere OB proteins. See on-line version for color; print version lacks color. (a) Amino acid sequences corresponding with structurally conserved elements (S1 – S5, labeled) were aligned on the basis of alpha carbon structural superpositions. Indexes (top of alignment) reference positions within the sequence alignment beginning with zero and ending with 48. To calculate the position within any one OB protein simply add the alignment index and [inserted residues] to the [first position] provided. Sequence positions meeting chi-squared test criteria for significance are color coded as indicated. (b) Morphological and binding function characters of OB proteins are as follows: “C34” scored 1 if more than 30 residues connect strands S3 and S4, and scored 0 otherwise; “ligand” scored 0 if current structures do not contain a ligand at the classical oligo-binding site, scored 1 if this ligand is single-stranded DNA, and scored 2 if this ligand is polypeptide; “C-helix” scored 0 if the C-helix is missing (e.g. h-Rpa70A), scored 1 if the C-helix is displaced to the back of the OB fold (h-Tpp1 and Sn-TEBPβ), scored 2 for equatorially positioned C-helix (e.g. h-Pot1 and Sn-TEBPα1), scored 3 for a three-helix cluster (Sc-Cdc13N and Sc-Cdc13D), and scored 4 for axially positioned C-helix (e.g. Stn1N); “L12+L45” scored 1 if the combined number of residues in L12 and L45 is less than 10 residues (Ten1 and Rpa14), and scored 0 otherwise; L23 scored 1 if the number of residues in the L12 connection between S2 and S3 exceeded 20 residues (Sc-Cdc13D and h-Pot1-2), and scored 0 otherwise. (c) Tree obtained from analysis of combined sequence and morphological characters. The tree is deeply rooted with each clade containing OB members from unicellular and metazoan lineages, implying gene duplications probably occurred in a common ancestor prior to divergence of ciliates, yeast, and vertebrates. Analysis of mixed data types was accomplished with MrBayes [Bayesian Analysis of Phylogeny, version 3.1.2, (Altekar et al., 2004)]. A total of 10,000 trees were sampled during the course of parallel Metropolis coupled Markov chain Monte Carlo simulations which applied gamma-distributed LG rates of amino acid exchange (Le and Gascuel, 2008). Nodes which were observed in more than 50% of sampled trees are labeled with a dot; nodes with greater than 50% frequency for analysis of sequence characters alone (no morphological characters) are also marked with an arrow. h-Rpa70A was chosen as the outgroup in this analysis because of long branch lengths obtained in preliminary analyses with ProtTest [ProtTest, Selection of Models of Protein Evolution, version 2.4 (Abascal et al., 2005)].
Pairwise comparison of representative sequences within the group considered here gave a range of percent identity from 4.7% (random) to 33.3% (homologous), with an average percent identity of 15% (Table 3). Universally conserved sequence motifs are absent. Consequently, more sensitive methods were applied to identify positions experiencing constraints during evolution (color coded in Fig 4(a)). Chi-squared statistical tests (with Yates correction for continuity) gave especially strong signals (p < 0.001) for positions 5 and 18, where the occurrence of Gly and Asp were significantly more frequent than expected by chance. Glycine at position 5 is thought to facilitate wrapping of S1 around the core, allowing this beta strand to form the outer edge of both SaS4S5 and SbS2S3 beta sheets. Ten of the 21 members use Gly at this position. Seven of the 21 OB proteins satisfy the same structural constraint with Ala, the second smallest amino acid residue, at position 5, but there are four examples that violate the Gly5/Ala5 guideline for constructing an OB fold.
Table 3. Percent identity for pairwise comparison of representative telomere OB sequences*.
| Identity (%) | Sp-Ten1 | Sn-α3 | Sc-Cdcl3N | h-Tppl | h-Pot1-1 | h-Pot1-2 | Sn-α1 | Sn-α2 | min | max |
|---|---|---|---|---|---|---|---|---|---|---|
| Sp-Stn1N | 10.2 | 16.3 | 12.8 | 17.0 | 20.0 | 11.1 | 13.3 | 21.3 | 10.2 | 21.3 |
| Sp-Ten1 | 12.2 | 8.5 | 21.3 | 13.3 | 17.8 | 11.1 | 14.9 | 8.5 | 21.3 | |
| Sn-α3 | 12.8 | 12.8 | 11.1 | 13.3 | 8.9 | 14.9 | 8.9 | 16.3 | ||
| Sc-Cdcl3N | 8.9 | 4.7 | 16.3 | 16.3 | 15.6 | 4.7 | 16.3 | |||
| h-Tppl | 14.0 | 30.2 | 11.6 | 13.3 | 8.9 | 30.2 | ||||
| h-Potl-1 | 13.3 | 33.3 | 17.8 | 4.7 | 33.3 | |||||
| h-Potl-2 | 13.3 | 24.4 | 11.1 | 30.2 | ||||||
| Sn-α1 | 22.2 | 8.9 | 33.3 | |||||||
| Sn-α2 | 13.3 | 24.4 |
Only amino acid residues included in the structurally conserved OB core (n = 42-49) were compared.
Aspartate at position 18 of the alignment followed by glycine and threonine comes closest to defining a conserved sequence motif. Although each of these positions was associated with high chi-squared signals (Asp18 has the highest score encountered) exactly zero of the 21 telomere OB folds has an exact match to the putative Asp18Gly19[Thr/Ser]20Gly21 motif, highlighting the difficulty associated with defining diagnostic sequence criteria for this protein family. Asp18 and residues 19-21 define a tight turn at the end of S2 which is followed either immediately or closely by S3. The most likely explanation for conservation at these positions is that the combination of Asp18, Ser20/Thr20, and either Gly19 or Gly21 stabilizes the OB structure or an otherwise rarely encountered folding intermediate. Constraints dictating relatively high conservation for these residues appear to have vanished for two newly reported structures (Ct-Ten1 and Sc-Cdc13N), which retain no vestige of the motif.
Sequence positions define alternating hydrophobic/polar positions (gold/magenta in Fig 4(a)), a pattern which has been noted previously for nucleic acid-binding OB proteins (Theobald et al., 2003b). Inspection of the telomere and RPA OB structures shows this pattern corresponds with the inside/outside disposition expected for beta sheet structure surrounding a hydrophobic core. Of the two types of positions, hydrophobic character has been more strongly preserved as judged by chi-squared scores (gold, p < 0.001). Positions marked as polar are apparently more tolerant to accepting a hydrophobic residue (magenta, p < 0.01). This situation likely reflects high thermodynamic penalties for burying a polar moiety and relatively low penalties for leaving a hydrophobic group on the outside of the OB fold. Another explanation is that hydrophobic patches close to the surface of the protein may be necessary to form functional interaction surfaces.
Attempts to define evolutionary relationships among OB proteins on the basis of the structure-aligned amino acid sequences consistently yielded trees (not shown) with comb-like structure and only two clearly supported clades: one defined by h-Tpp1 and Sn-TEBPβ, and the second defined by h-Pot1-1, Sp-Pot1N, and Sn-TEBPα1 (nodes marked with arrows in Fig 4(c)). The close evolutionary relationship for members in these two clades has been previously supported by functional and structural similarity (Baumann and Cech, 2001; Lei et al., 2003; Lei et al., 2004; Xin et al., 2007; Wang et al., 2007). Encouraged by this result, I attempted to further detangle the other branches by including morphological characters and binding function (Fig 4(b)) in addition to sequence information. The resulting unrooted tree (Fig 4(c)) exhibits topology congruent with previously published phylogenies (Theobald and Wuttke, 2005; Horvath, 2008a) and includes additional clades and inferences.
One expected branch includes Stn1N, Rpa32N, Ten1 and Rpa14. Structure determinations of the Rpa32N–Rpa14 heterodimer (Bochkarev et al., 1999) and two examples of a Stn1N–Ten1 complex (Sun et al., 2009) show clear similarity at multiple levels of organization. Each member possesses an axially oriented C-helix (Fig 3(b)) that is a central structural element of the heterodimer interface (Bochkarev et al., 1999; Sun et al., 2009). The Ten1 and Rpa14 members have ablated L12 and L45 loops, a trait that appears to be derived since most OB units have substantially longer L12 and L45 loops. Winged helix-turn-helix motifs are found in C-terminal domains particular to members of the Stn1 and Rpa32 clade (Mer et al., 2000; Sun et al., 2009; Gelinas et al., 2009), further supporting the close relationship of Stn1 with Rpa32 deduced from functional homology and sequence similarity (Gao et al., 2007).
Conservation of structure for Stn1N–Rpa32N and Ten1–Rpa14 subcomplexes suggests a conserved function that is similar for both telomere maintenance and for DNA replication and repair. Indeed, the Stn1N-derived OB fold can substitute for Rpa32N to restore function in rpa32−/− deletion mutants of S. cerevisiae (Gao et al., 2007). The principle role for the Rpa32–Rpa14 heterodimer is to contribute to single-stranded DNA binding and simultaneously coordinate protein-protein association and exchange events. The Rpa32–Rpa14 heterodimer recruits Rpa70 via OB-OB interactions with Rpa70C, as viewed in a crystal structure of the RPA trimerization core (Bochkareva et al., 2002), and the C-terminal winged helix-turn-helix (WH) domain of Rpa32 recruits an assortment of DNA repair proteins as observed by NMR (Mer et al., 2000). Genetic and biochemical tests suggest that the Stn1–Ten1 heterodimer functions at telomeres similarly with the protein recruitment functions provided by Rpa32–Rpa14 at DNA replication forks and repair centers. Stn1 and Ten1 each bind with single-stranded DNA (Gao et al., 2007), and S. cerevisiae Stn1–Ten1 delivers telomerase and the DNA polymerase α/primase complex to telomeres in a regulated manner (Pennock et al., 2001; Puglisi et al., 2008).
In S. cerevisiae, telomerase and polymerase α/primase recruitment by Sc-Stn1–Ten1 works through interactions with Cdc13 (Qi and Zakian, 2000; Chandra et al., 2001; Sun et al., 2011). Telomerase recruitment may work differently in other organisms since in S. pombe this function is executed by a Sp-Pot1-associated subcomplex that includes Tpz1 and Ccq1 (Miyoshi et al., 2008). Nevertheless, DNA polymerase α/primase recruitment by a Stn1-centered complex appears conserved. In plants and humans, Stn1 interacts with the conserved telomere component protein Ctc1 (Miyake et al., 2009; Surovtseva et al., 2009) with analogous functional consequence to Stn1 and Cdc13 in budding yeast; both Cdc13 and Ctc1 appear to deliver polymerase α/primase to telomere ends for synthesis of the C+A-rich strand.
OB units associated with high-affinity single-stranded DNA-binding protein domains were grouped in a single clade, but two expected members were placed in orphan branches: h-Pot1-2 and Sp-Cdc13D. The second DNA-binding OB unit in the human Pot1 protein, h-Pot1-2 is surprisingly located at the base of a branch defined by h-Rpa70N and Sn-TEBPα3, OB units which are known to function in protein-protein interactions. The relatively large L23 insert found in h-Pot1-2 may lead to long-branch attraction with Sc-Cdc13D which is located on a neighboring branch and also has a large L23 insertion. And it is certainly possible that divergent evolution has scrambled the protein pedigree for h-Pot1-2.
The telomere protein Sc-Cdc13 presents a challenge for the molecular evolutionist because character traits give conflicting conclusions regarding homology. On the basis of DNA-binding function, Sc-CDC13D would be placed together with the h-Pot1-1/Sp-Pot1N/Sn-TEBPα1 clade. Apparent structural similarity and inferred homology noted for Sc-Cdc13D and Sn-TEBPα (Mitton-Fry et al., 2002; Theobald et al., 2003a) may need reevaluation in light of a newly determined structure for the N-terminal OB unit of Cdc13, Sc-Cdc13N (Sun et al., 2011). Sc-Cdc13N possesses two structural characters that are shared with Sc-Cdc13D, but not commonly found among other telomere OB folds: (i) a highly elaborated C34 element connecting strands S3 and S4, and (ii) replacement of the prototypical single C-helix with a cluster of three or more helices. Dali-derived structure matching corroborate structural similarity between Sc-Cdc13N and Sc-Cdc13D (Mitchell et al., 2010; Sun et al., 2011). In addition to these structural similarities with Sc-Cdc13N, differences in DNA recognition that distinguish Sc-Cdc13D from other telomere single-stranded DNA-binding OB folds (to be discussed further below) support the idea that Sc-Cdc13D was derived by gene duplication within an ancestral Sc-Cdc13 protein and not through divergence from an ancestral DNA-binding OB fold (Sun et al., 2011).
Similar to the puzzle of placing Sc-Cdc13D within the telomere OB family, conflicting evidence exists for the relation between Cdc13 and Rpa70. The Rpa70–Rpa32–Rpa14 complex functions at DNA replication forks and during the course of DNA repair and recombination reactions [reviewed in (Wold, 1997; Fanning et al., 2006)]. The Cdc13–Stn1–Ten1 complex is similarly organized with large, medium and small subunits and functions to regulate DNA synthesis at telomeres. Argument by analogy leads to a conclusion that Rpa70 and Cdc13 are homologs; however, structural differences cast this Cdc13–Rpa70 relation in doubt. One difference involves the number of DNA-binding OB units. The DNA-binding domain of Cdc13 comprises a single OB unit; the high-affinity DNA-binding domain of Rpa70 comprises two OB units. Tandem DNA-binding OB units appears to be the ancestral state, since other telomere OB proteins from Sterkiella nova, fission yeast, and humans also use two OB units for binding single-stranded telomere DNA. Evolution may have replaced a primordial two-OB DNA-binding domain in Cdc13 so as to cope with a telomerase template “crisis” in the hemiascomycetes lineage –– a crisis that also lead to heterogeneous telomere DNA sequences and loss of double-stranded DNA binding factors such as TRF1 (Li et al., 2000; Teixeira and Gilson, 2005).
Although the DNA-binding domains of Sc-Cdc13 and Rpa70 are structurally different, the N-terminal and DNA-binding domains of Sc-Cdc13 share structural features also found in the C-terminal domain of Rpa70. The phylogram derived from Baysian analysis of telomere protein and RPA-derived OB units placed these three OB units (Sc-Cdc13N, Sc-Cdc13D, h-Rpa70C) in one clade (Fig 4). Structure determination of the remaining two predicted OB units within Sc-Cdc13 could help resolve the question of when and how Sc-Cdc13 and Rpa70 diverged during evolution. These questions also call attention to the possibility that certain OB units within otherwise homologous telomere and RPA proteins may have been lost or reinvented.
The human Sn-TEBPβ ortholog is h-Tpp1 (formerly PIP1, also PTOP) initially characterized as a protein that interacts with Pot1 and other telomere components (Ye et al., 2004; Liu et al., 2004; O'Connor et al., 2006). Homology with Sn-TEBPβ is based on distinguishing structural features shared by h-Tpp1 and Sn-TEBPβ that are rarely encountered or not apparent for other telomere OB proteins (Wang et al., 2007). For instance, the C-helix in both Sn-TEBPβ and h-Tpp1 is displaced from its prototypical location alongside the OB core and instead coalesces with L45 and elements of the N-terminal extension. The h-Tpp1 + Sn-TEBPβ branch stands out because of high probability confidence values (this node was observed in ∼90% of sampled trees) and also because it is the only branch in the tree that lacks an RPA-derived member. This last observation suggests the ancestral protein from which Sn-TEBPβ and h-Tpp1 descended may have been fixed in the eukaryotic lineage because it acquired a highly advantageous function at telomeres that was dispensable at DNA replication forks and DNA repair centers.
Given the critical importance of telomeres for genome stability one would expect h-Tpp1/Sn-TEBPβ orthologs in all extant taxa that maintain chromosome ends with telomeres and telomerase. A search for Sn-TEBPα and Sn-TEBPβ orthologs in Euplotes crassus (a distantly related marine cousin to Sterkiella nova) revealed two proteins with telomere single-stranded DNA-binding function and high sequence similarity to OB units derived from the N-terminal domain of Sn-TEBPα, but no match for Sn-TEBPβ (Price, 1990) [also reviewed in (Linger and Price, 2009)]. A similar situation appeared to hold for humans and fission yeast when Pot1 was first reported on basis of sequence similarity with Sn-TEBPα1 (Baumann and Cech, 2001). More recently, Sn-TEBPβ orthologs have been identified in humans (Xin et al., 2007; Wang et al., 2007), Schizosaccharomyces pombe (Miyoshi et al., 2008), Candida albicans (Yu et al., 2008), Saccharomyces cerevisiae (Lee et al., 2008; Lee et al., 2010), Candida parapsilosis and Lodderomyces elongisporus (Yen et al., 2011). Genetic tests for function in yeast clearly indicate that Sn-TEBPβ homologs function to coordinate telomere homeostasis (Miyoshi et al., 2008; Lee et al., 2010; Yen et al., 2011).
Structural character of OB interfaces
OB protein modules provide interaction surfaces for the recognition of diverse ligands including oligosaccharides, single-stranded nucleic acid, oligopeptide, and proteins. Understanding telomere OB protein function requires an appreciation for interactions leading to molecular recognition. With this goal in mind, this section examines interfaces observed in crystal and NMR structures of telomere OB proteins. Relevant RPA-derived structures provide perspective and hint at analogous assemblies likely at play for telomere protection and maintenance. The structures can be roughly divided into three categories according to ligand type and complexity: OB-oligomer, OB-OB, and composite OB. In the OB-oligomer category, telomere OB proteins bind with polypeptide or single-stranded DNA ligands (Fig 5). The OB-DNA interfaces have been expertly reviewed previously (Theobald et al., 2003b; Croy and Wuttke, 2006). The current analysis revisits some of this previous work so as to compare these OB-DNA interfaces with new OB-DNA structures and the second category of OB-OB interfaces (Fig 6). Composite OB structures constructed with both OB-oligomer and OB-OB interaction types feature more elaborate and inter-woven interfaces (Fig 7).
Figure 5.
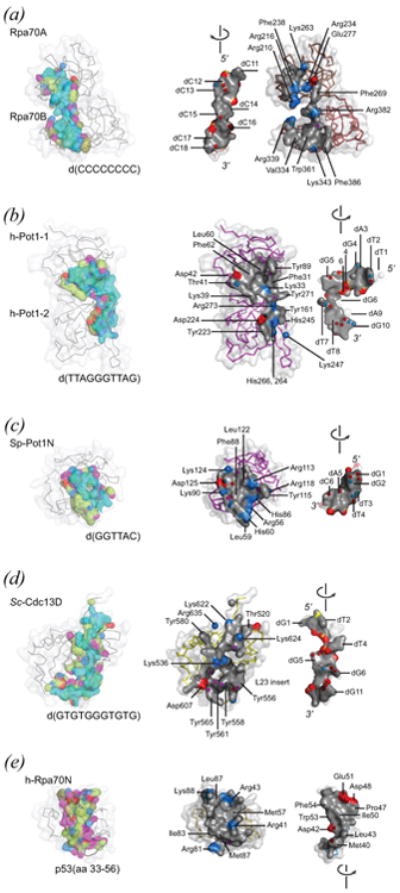
Simple OB-oligomer interfaces. See on-line version for color; print version lacks color. Each interface is depicted in two ways: as found in the complex (left-hand view) and in an exposed form generated by separating the DNA or peptide ligand from the OB component (right-hand view). In the complexed model, protein and DNA are both wrapped in a semi-transparent accessible surface. Protein is additionally rendered as an alpha carbon trace. DNA is colored cyan. Protein residues making contact with ligand (4 Å cutoff) are colored according to chemical type with aliphatic carbon atoms colored olive or lime, hydrogen bond acceptor groups colored red, hydrogen bond donor groups colored blue, groups ambivalent with respect to hydrogen bond donor/acceptor quality colored magenta. In the dissociated model, contact residues of the OB-oligomer interface are rendered as solid accessible surfaces (grey, with positively charged surface blue, and negatively charged surface red), and electronegative atoms making hydrogen bonds across the interface shown as spheres. Landmark residues are labeled. PDB identifiers are as follows: (a) 1jmc, major DNA-binding domain derived from Rpa70 (residues 183-420) complexed with d(C8); (b) 1xjv DNA-binding domain derived from h-Pot1 (residues 6-299) complexed with d(TTAGGGTTAG); (c) 1qzh, N-terminal OB unit derived from Sp-Pot1 (residues 5-174) complexed with d(GGTTAC); (d) 1s40, minimal DNA-binding domain derived from Sc-Cdc13 (residues 500-686) complexed with d(GTGTGGGTGTG); (e) 2b3g, N-terminal OB of h-Rpa70, h-Rpa70N (residues 1-128) complexed with p53-derived peptide (residues 33-56).
Figure 6.
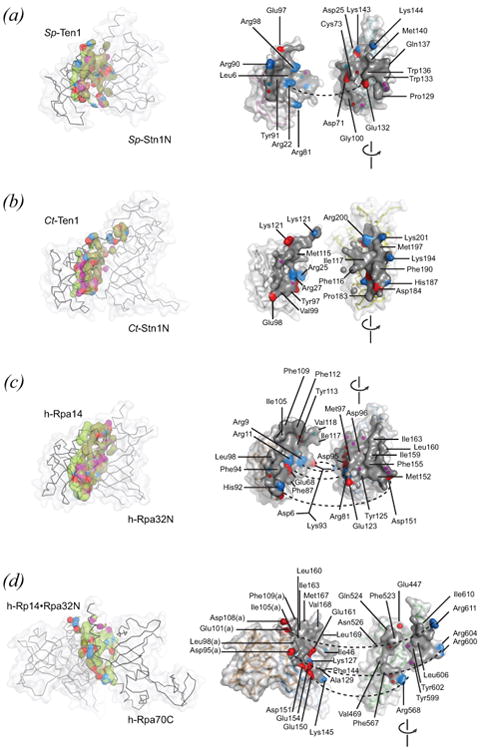
OB-OB interfaces as found in simple heteromeric telomere and RPA OB protein complexes. See on-line version for color; print version lacks color. The protein-protein interface is depicted in two ways: as found in the complex (left-hand view) and as exposed by separating protein components (right-hand view). In the model of the complex, aliphatic carbon atoms at the interface (4 Å cutoff) are colored lime for one component and olive for the second component. Groups with hydrogen-bonding potential are colored as in Figure 5 with hydrogen bond donor groups red, acceptor groups blue, and ambivalent (donor or acceptor) groups magenta. In the exposed view, interfaces are rendered as solid accessible surfaces (grey), with charged surfaces highlighted (red, negative; blue, positive), and electronegative atoms making hydrogen bonds across the interface depicted as spheres. Certain landmark residues are labeled. PDB identifiers are as follows: (a) 3kf6, Sp-Ten1 (full-length protein) associated with the N-terminal domain of Sp-Stn1 (residues 2-159); (b) 3kf8, Ct-Ten1 (full-length protein) associated with the N-terminal domain of Ct-Stn1 (residues 2-217); (c), 2pi2, h-Rpa14 (full-length protein, residues 2-118) associated with the N-terminal domain of h-Rpa32 (residues 41-176); (d) 1l1o, h-Rpa14 (full-length, residues 3-117) and h-Rpa32N (residues 44-171) associated with the C-terminal domain of h-Rpa70 (residues 439-616).
Figure 7.
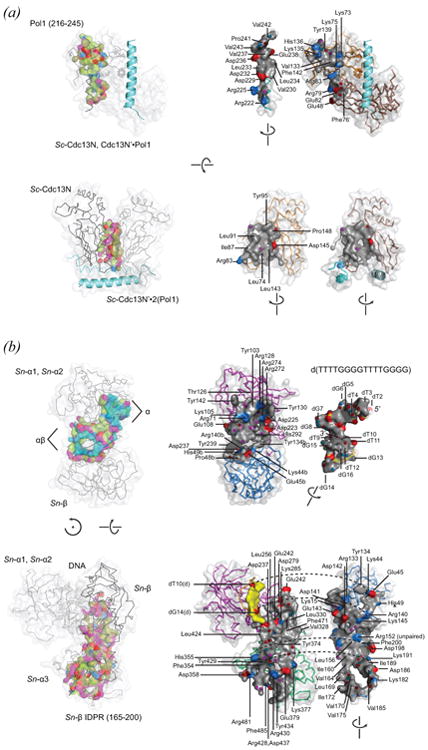
Composite OB interfaces as found in intricate multi-subunit OB protein complexes. See on-line version for color; print version lacks color. Interface surfaces are rendered and color-coded as described in Figures 5 and 6. (a) Multiple views are shown for homodimeric Sc-Cdc13N (residues 13-224) complexed with two copies of a Pol1-derived peptide (residues 216-245). One Pol1-derived peptide is shown as an accessible surface and a second is shown as a ribbon cartoon (blue). The (Sc-Cdc13N)2 OB-Pol1 interface involves residues contributed by each of the C2 symmetry-related Sc-Cdc13N subunits. The OB-peptide interface is highlighted in the top view, as found in the complex (left-hand view) and as separated components (right-hand view). The homodimer interface is highlighted in the bottom view of panel (a). Note that the two surfaces are nearly equivalent, and for this reason residue labels are only provided for one subunit. (b) Multiple views are shown for Sn-TEBPα and Sn-TEBPβ complexed with single-stranded telomere DNA. The DNA-protein interface formed between Sn-TEBPαβ and d(TTTTGGGGTTTTGGGG) is highlighted in the top view of panel (b). This structure contains a more complete view of the DNA-interface compared with previously published structures. It was determined to the 1.88 Å resolution limit (Rfree = 0.195, Rwork = 0.165) with use of an engineered protein constructed by fusing DNA-binding OB units of Sn-TEBPα and Sn-TEBPβ. The DNA occupies two connected binding surfaces – one comprising OB units Sn-TEBPα1 and Sn-TEBPα2 (labeled α), and a second involving Sn-TEBPβ and surfaces on Sn-TEBPα1 and Sn-TEBPα2 (labeled αβ). The protein-protein interface of Sn-TEBPα–β is highlighted in the bottom view. This structure was determined to the 1.91 Å resolution limit with use of full-length Sn-TEBPα and Sn-TEBPβ bound to d(GGGTTTTGGGG), which is a 5′-truncated form of the telomere single-stranded DNA. The protein-protein interface (grey with charged surfaces and hydrogen-bonded atoms highlighted) dwarfs contacts made by DNA with Sn-TEBPβ (yellow). PDB identifiers are as follows: (a) 3oiq, (Sun et al., 2011); (b) 2i0q, (Buczek and Horvath, 2006a).
The DNA-binding domain of Rpa70 bound with d(C8) is shown in Fig 5(a). DNA binds at the classical oligo-binding surface found at each of two OB units, and follows a straight path with 5′-proximal nucleotides contacting Rpa70A and 3′-proximal nucleotides contacting Rpa70B. The DNA strand orientation is the same for both Rpa70A and Rpa70B, with 5′→3′ pointed to the “back” when the OB unit is oriented as in Fig 2(a) with L45 on top, L12 below, and the oligomer binding surface on the right-hand side. The same DNA and OB unit polarity is observed in OB-DNA complexes of telomere proteins h-Pot1 (Fig 5(b)) and Sn-TEBPα (not shown), which also employ two OB units to contact a similar number of nucleotides. Different from the situation seen for Rpa70, DNA in these complexes bends sharply so as to follow a path defined by oligo-binding surfaces on the two OB units. OB-DNA interfaces seen for single-OB units derived from Sp-Pot1 (Fig 5(c)) and Sc-Cdc13 (Fig 5(d)) conserve the 5′→3′ DNA strand polarity. As pointed out previously, conservation in DNA strand polarity argues for descent by divergent evolution from a common OB ancestor (Theobald et al., 2003b).
Table 4 compares features of six OB-DNA interfaces for RPA and telomere proteins. The S. pombe Pot1 protein likely uses two OB units for binding DNA (Trujillo et al., 2005; Croy et al., 2006), meaning values such as surface area reported for the Sp-Pot1N–DNA complex reflect a portion of the total interface since the crystal structure was obtained for one OB unit. Also, the very large interface observed in a crystal structure of S. nova telomere-derived DNA complexed with DNA-binding portions of Sn-TEBPα and Sn-TEBPβ was divided into two parts so as to facilitate comparisons (the full DNA-TEBPαβ interface is summarized in Table 6). These parts correspond with 5′ and 3′-terminal halves of the 16-nucleotide S. nova telomere single-stranded DNA. Nucleotides of the 5′-half bind with a surface on Sn-TEBPα called the α-site; nucleotides of the 3′-half bind with a more intricate surface called the αβ-site that involves three OB units: the two DNA-binding OB units of Sn-TEBPα and portions of the OB unit of Sn-TEBPβ.
Table 4. Character of OB-DNA interfaces**.
| Character | Rpa70A+B | h-Pot1-1+2 | Sp-Pot1N | Sc-Cdc13D | Sn-TEBP α-site | Sn-TEBP αβ-site |
|---|---|---|---|---|---|---|
| OB units | 2 | 2 | 1* | 1 | 2 | 3 |
| PDB id | 1jmc | 1xjv | 1qzh | 1s40 | 1k8g | 2i0q |
| DNA sequence | CCCCCCCC | TTAGGGTTAG | GGTTAC | GTGTGGGTGTG | T2TTGGGG8 | T9TTTGGGG16 |
| SA% (Å2) | 1,387 | 1,610 | 875 | 1,666 | 1,303 | 1,678 |
| SA / OB (Å2) | 694 | 805 | 875 | 1,666 | 652 | 559 |
| DNA atoms (apolar)£ | 93 (13) | 124 (29) | 65 (11) | 124 (28) | 95 (12) | 98 (18) |
| Protein atoms (apolar)& | 96 (56) | 132 (72) | 90 (37) | 130 (77) | 98 (48) | 123 (59), α 86 (40), β 37 (19) |
| Salt bridge (bidentate)∞ | 3 | 2 | 1 | 0 | 3 | 4 (2) |
| Phosphate H-bond, not salt$ | 1 | 1 | 1 | 4 | 4 | 1 |
| H-bonds (bidentate)¢ | 7 | 15 (2) | 10 (2) | 8 | 14 (6) | 9 (2) |
| Base atoms | 50 | 65 | 44 | 57 | 59 | 54 |
| Aromatic (base)§ | 49 (34) | 60 (54) | 20 (14) | 38 (12) | 54 (45) | 53 (41) |
| C2′ atoms | 3 | 6 | 3 | 7 | 2 | 3 |
| C2′ contact (apolar) | 7 (7) | 10 (2) | 4 (1) | 19 (16) | 4 (3) | 7 (7) |
| Thymine C7 atoms | 0 | 3 | 0 | 1 | 2 | 4 |
| C7 contact (apolar) | 0 | 6 (5) | 0 | 1 (0) | 5 (0) | 7 (0) |
| Tyrosine / total residues (%) | 0 / 28 (0) | 5 / 31 (16.1) | 1 / 20 (5.0) | 7 / 28 (25.0) | 5 / 23 (21.7) | 9 / 47 (19.2) |
Values differ modestly from previously published values because these were computed here for protein and DNA components with first shell solvent molecules included. Also, the DNA-protein complex analyzed for Sn-TEBPα has clear electron density for nucleotides G2, G3 and G4, positions which were missing in previously published structures.
Sp-Pot1 DNA-binding domain is thought to comprise two OB units. Only the N-terminal OB unit has been structurally characterized, meaning these quantitative values likely reflect a portion of the full DNA-OB complex.
Surface area calculated by the program volume within the 3V package (Voss and Gerstein, 2010), using a 1.4 Å probe radius and 0.25 Å grid spacing. Note that results reported here include contributions from the first hydration shell and differ from literature reported values in some cases.
A DNA atom was counted if it was located within 4 Å of the DNA-protein interface. Apolar atoms of DNA (counted given in parentheses) were defined as aliphatic carbons of the deoxyribose moiety and the C5-methyl of the thymine base group.
Apolar atoms defined as any aliphatic carbon lacking aromatic character.
Salt bridge defined by phosphate oxygen less than 3.2 Å from positively charged hydrogen bond donor. Bidentate salt bridges involve two hydrogen bonds contributed by different N atoms of one arginine residue.
Phosphate H-bond (not salt) defined by non-bridging phosphate oxygens within 3.2 Å of a neutral (uncharged) hydrogen bond donor with appropriate geometry.
Hydrogen bond defined by complementary donor/acceptor pair within 3.2 Å with appropriate geometry that is not already included in the salt bridge count. Bidentate hydrogen bonds are constructed by two different atoms of one amino acid residue H-bonding with two different atoms of one DNA base moiety.
Aromatic contacts counted for atoms within 4.0 Å of DNA contributed by protein aromatic moieties, which includes the sidechains of Tyr, Phe, Trp, and His residues, and the guanidinium group of Arg residues. Numbers in parentheses count the number of aromatic protein atoms in close proximity with base moieties of the DNA.
Table 6. Character of composite OB interfaces.
| Character | Cdc13N2•Pol1 | Cdc13N(Pol1)2• Cdc13N′ | αNβΔ–DNA^ | (DNA)α–β‡ | α3–β162-195 | (DNA)α–βΔ |
|---|---|---|---|---|---|---|
| OB units | 2 | 2 | 3 | 4 | 1 | 3 |
| PDB id | 3oiq | 3oiq | n.a. | 2i0q | 2i0q | 2i0q |
| Surface area% (Å2) | 1,280 | 1,514 | 2,996 | 4,176 | 1,874 | 2,232 |
| Protein 1 (apolar)& | Cdc13N, 50 (24) | Cdc13N, 75 (36)# | α, 178 (86) | β, 261 (116) | β162-195, 116 (56) | βΔ, 145 (60) |
| Protein 2 (apolar) | Cdc13N′, 27 (5) | Cdc13N′, 62 (26) | β, 37 (19) | α, 237 (88) | α3, 102 (36) | α, 138 (53) |
| Component 3 (apolar) | Pol1, 84 (43) | n.a. | DNA, 193 (30) | n.a. | n.a. | n.a. |
| Salt bridges (bidentate)∞ | 7 (2) | 0 | 7 (2) | 3 | 0 | 3 |
| Ion pairs@ | 3 | 2 | 0 | 3 | 2 | 1 |
| H-bonds¢ | 7 | 3 | 21 | 28 | 14 | 13 |
| Tyrosine / total residues (%) | 2 / 34 (5.9) | 2 / 31 (6.5) | 9 / 47 (19.1) | 7 / 96 (7.3) | 4 / 48 (8.3) | 3 / 54 (5.6) |
Although the two Sc-Cdc13N subunits are symmetrical, the number of contact atoms differ in number because first-shell waters were assigned to the first subunit.
The DNA-protein interface contained in this complex is also analyzed in Table 4 as TEBPα–DNA and TEBPαβ–DNA subcomplexes
The TEBPα–β interface is analyzed as a complete protein-protein interface and also as OB-peptide [α3–β162-195] and OB-OB [(DNA)α–βΔ] subcomplexes (see columns to the right).
Surface area buried as a consequence of DNA binding with protein ranges from 875 (Sp-Pot1N) to 1678 (αβ-site) Å2, and the average value is 1,420 Å2 (with Sp-Pot1N excluded it is 1,530 Å2). These surface area calculations were obtained by assuming the solvent accessible surface of individual dissociated components can be estimated by structures seen in the associated complexes. This assumption introduces some systematic error since single-stranded DNA is likely to become more or less extended when free in solution. Even so, the values have some meaning and are useful for making comparisons within the OB-DNA category and with OB-protein interfaces described below.
When normalized by the number of OB units, the Sc-Cdc13 interface with DNA, measuring 1,666 Å2 OB−1, is the largest of this group. As explained previously (Mitton-Fry et al., 2004), the large contact surface of Sc-Cdc13D results from a large L23 insertion. Normalized in this way, the Sp-Pot1N interface with DNA, measuring 875 Å2 OB−1, is also somewhat larger than the median value of 750 Å2 OB−1. Consistent larger than average surface area per OB unit suggests that additional surfaces are accessible to DNA when single OB units are not involved in contacts with neighboring OB units. This OB versus DNA competition for limited surfaces on any one OB unit explains why the normalized DNA-buried surface area is so small (560 Å2 OB−1) for the three DNA-binding OB units comprising the Sn-TEBP αβ-site.
On average, a DNA-OB interface is constructed with 100 DNA atoms and 110 protein atoms. Generally, more than half of the atoms contributed by DNA belong to base groups (average, 55 atoms). Significantly fewer apolar atoms are contributed by DNA (average, 19 atoms). The number of apolar DNA atoms is also low by comparison with apolar protein atoms (average, 58 atoms). In this atom counting analysis, an atom was considered part of the interface if it came within 4 Å of an atom contained within the partner molecule, and apolar atoms were defined as aliphatic carbons with sp3 molecular orbital centers. Few apolar atoms contributed by DNA is likely a consequence of the fact that most atoms in DNA are either polar or heteroaromatic and only ∼25% are aliphatic.
The low proportion of hydrophobic contact surface characteristic of single-stranded DNA has consequences for the thermodynamic balance of enthalpy and entropy driving DNA-protein association. Large favorable entropy terms characterize processes that bury hydrophobic surfaces such as folding of globular, water soluble proteins. Studies of the thermodynamic character of Sn-TEBPαN binding with single-stranded d(T4G4) DNA showed that this process was entropy neutral (ΔS = 0) and entirely enthalpy driven (ΔH < 0) at 20 °C (Buczek and Horvath, 2006b). At higher temperatures entropy opposed DNA binding and enthalpy compensated by becoming larger in magnitude. This same pattern of favorable enthalpy and unfavorable entropy terms was also measured for Sp-Pot1N binding with d(GGTTAC) and single-nucleotide substitution variants (Croy et al., 2008). These trends indicate that single-stranded telomere DNA binding is not strongly driven by the hydrophobic effect, consistent with few apolar atoms contributed by single-stranded DNA at the OB-DNA interface.
DNA fragments longer than eight nucleotides appear to behave differently in OB-binding reactions. Reactions of d(G3T4G4) DNA binding with Sn-TEBPαN and Sn-TEBPα were favored by both enthalpy and entropy terms at 20 °C. As DNA length increased further, the enthalpy term diminished in magnitude and entropy became a more dominant favorable term. As is the case for many telomere sequences, these S. nova sequences contain G-rich portions which probably form Hoogsteen base pairs when not in contact with binding proteins. We interpreted the more favorable net entropy change and lower reliance on net enthalpy change to mean that these longer telomere DNAs must give up intra-molecular base pairing and base stacking interactions upon forming interactions with protein surfaces (Buczek and Horvath, 2006b; a).
The DNA-binding surfaces of telomere OB proteins have more positively charged residues compared with negatively charged residues (Fig 5, see also Fig 8). Nevertheless, the number of salt bridges is low, ranging from 0 to 3 salt-bridged phosphates per complex (average, 2), a circumstance which contributes to the salt-resistant nature of these DNA-protein complexes (Price and Cech, 1987). In cases where DNA-binding affinity has been measured at several salt concentrations, the contribution to DNA-binding affinity is approximately 85% non-electrostatic (Lei et al., 2002; Buczek and Horvath, 2006b).
Figure 8.
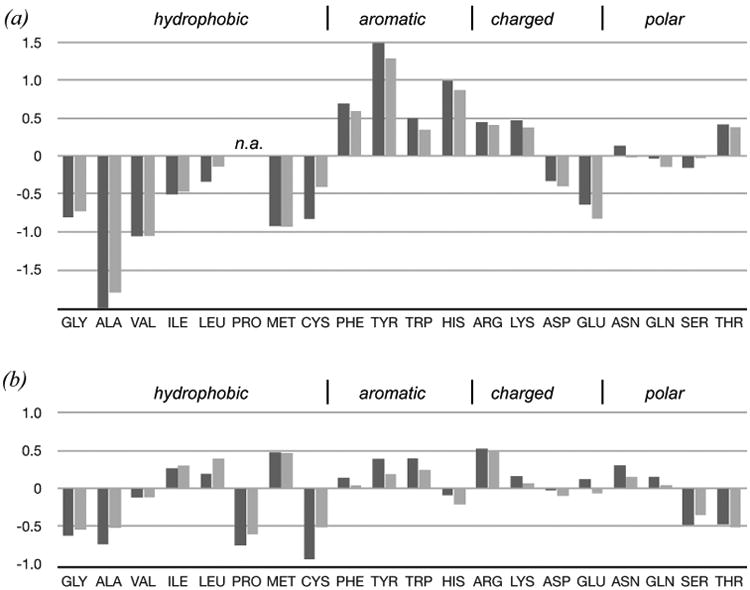
Propensities of amino acid residues at OB-DNA (a) and OB-protein interfaces (b). Propensity was quantified as ln(νi/νoi), where νi is the frequency of observing the ith amino acid residue type at an interface and νoi is the frequency of that amino acid type in a general population. The general population is either total amino acid composition of all OB proteins including regions of unknown structure (darkly shaded) or amino acid composition of surface exposed residues for OB units of known structure (lightly shaded). Residues enriched at an interface score above zero and residues depleted at an interface score below zero. The positive propensity observed for Tyr at OB-DNA interfaces is associated with strong statistical power especially by comparison with the total amino acid population (p < 0.001), but also with comparison with surface positions of OB proteins (p < 0.02). All other propensities failed to meet statistical tests for significance, including negative propensities observed for residues at OB-DNA interfaces. These negative propensities are subject to large uncertainties introduced by discontinuous sampling and few expected occurrences. Propensity for Pro at OB-DNA interfaces could not be computed because no Pro residues were observed (labeled n.a.).
What are these non-electrostatic contributions to DNA-binding affinity? Hydrogen bonds are relatively abundant as are contacts formed with aromatic protein residues. The number of hydrogen bonds involving DNA base moieties ranges from 7 to 15 (Table 4). Combining these numbers with those for hydrogen bonds with phosphate groups yields 10-21 hydrogen bonds per interface (average, 15), which are significantly larger numbers compared with the situation observed for protein-protein interfaces (see Table 5).
Table 5. Character of simple OB-protein interfaces.
| Character | Sp-Stn1N–Ten1 | Ct-Stn1N–Ten1 | Rpa32N–Rpa14 | Rpa70C–Rpa32/14 | Rpa70N–p53 |
|---|---|---|---|---|---|
| OB units | 2 | 2 | 2 | 3 | 1 |
| PDB id | 3kf6 | 3kf8 | 2pi2 | 1l1o | 2b3g |
| Surface area% (Å2) | 1,818 | 1,553 | 2,271 | 1,619 | 1,222 |
| Protein 1 (apolar)& | Sp-Stn1N, 102 (46) | Ct-Stn1N, 82 (43) | Rpa32N, 112 (45) | Rpa14/32, 97 (47) | Rpa70N, 75 (27) |
| Protein 2 (apolar)& | Sp-Ten1, 97 (37) | Ct-Ten1, 81 (39) | Rpa14, 184 (90) | Rpa70C, 79 (34) | p53 helix, 78 (38) |
| Salt bridges∞ | 2 (bidentate) | 2 | 2 (bidentate) | 2 | 2 (bidentate) |
| Ion pairs@ | 2 | 3 | 1 | 2 | 1 |
| Hydrogen bonds¢ | 8 | 2 | 10 | 5 | 2 |
| Tyrosine / total residues (%) | 1 / 34 (2.9) | 1 / 39 (2.6) | 3 / 57 (5.3) | 2 / 42 (4.8) | 0 / 29 (0) |
Ion pairs are oppositely charged groups within a distance range spanning 3.2 - 5 Å.
Aromatic contacts counted for aromatic atoms within 4.0 Å of protein partner. Atom count in parentheses includes that subset of aromatic contacts involving two aromatic moieties across the interface. Aromatic moieties are defined as the sidechains of Tyr, Phe, Trp, and His residues, and the guanidinium group of Arg residues.
Hydrogen bonds are augmented by van der Waals contacts, which often involve aromatic protein residues and base groups. In this analysis, aromatic protein moieties were defined as side chains of Tyr, Phe, Trp and His, and the guanidinium moiety of Arg residues. In many of the OB-DNA complexes, the number of aromatic atoms contributed by protein residues at the DNA-interface is approximately equal to the number of DNA base atoms, and a substantial fraction of aromatic, DNA-contacting protein residues make face-to-face stacking interactions with DNA bases. Sc-Cdc13D and Sn-TEBPαβ provide two notable exceptions to this trend. Aromatic protein residues lining the DNA-interface in these two cases interact principally with the deoxyribose backbone and make few (Sn-TEBPαβ) or no (Sc-Cdc13D) face-to-face stacking interactions with DNA bases.
OB-DNA interfaces are generally considered rich in aromatic residues (Theobald et al., 2003b; Croy and Wuttke, 2006). Are these types of residues over-represented at surfaces contacted by DNA? If the answer is yes, then surface aromatic residues could be a diagnostic test for identifying DNA-binding surfaces of OB proteins. To address this question I compared the frequency of encountering an aromatic residue at the DNA-contact surface (νaromatic = 0.37, nDNA-contact = 134) with aromatic residue frequencies computed for (i) surface exposed residues of all telomere OB proteins (νoaromatic = 0.17, nsurface = 2640), and (ii) all residues of all telomere OB proteins, including buried residues, surface exposed residues, and residues of undetermined structure (νoaromatic = 0.15, nall_residues = 7046). The sample rate for aromatic residues at DNA-contact surfaces is more than twice that observed for general surface exposed residues or all residues in this class of proteins. Although encouraging, this difference in sample rate does not pass chi-squared tests of significance (p = 0.2-0.5), meaning a diagnostic test for DNA-binding surfaces based on aromatic residues would give false-positives about 20-50% of the time.
I repeated the same sampling rate analysis for several categories of amino acid residues, including charged residues, positively charged residues, strongly hydrophobic residues, and each individual amino acid residue. Results for individual amino acid residues are summarized in Figure 8, which plots propensity of encountering each amino acid residue at an OB-DNA interface (and OB-protein interface), and is comparable to analyses of protein-protein interfaces presented by Janin (Janin et al., 2008). As with aromatic residues, these categories failed to differentiate DNA-contact surfaces from general surface exposed residues or the total amino acid population, with one exception: tyrosine. The frequency of encountering tyrosine at OB-DNA interfaces (νTyr = 0.14, nDNA-contact = 134) was 3.5-fold elevated above the average rate measured for surface exposed residues (νoTyr = 0.041, nsurface = 2640, p < 0.02), and 4.4-fold greater than the frequency of encountering tyrosine at any position in any telomere OB protein (νoTyr = 0.032, nall_residues = 7046, p < 0.001).
Tyrosine has special qualities that make it especially useful in molecular recognition (Koide and Sidhu, 2009; Kossiakoff and Koide, 2008). Tyrosine possesses a large surface area with few internal degrees of motion, meaning large favorable enthalpy and low entropy costs for making tyrosine contacts at macromolecular interfaces. Tyrosine can satisfy multiple interacting partners, including aromatic stacking partners, hydrophobic partners, and hydrogen bond donor and acceptor partners. The amphipathic nature of tyrosine residues allows for favorable interactions with solvent when the residue is not buried at a macromolecular interface. These physiochemical properties make tyrosine a logical choice for construction of OB-DNA interfaces, especially since the planar geometry of this residue fits easily into face-to-face stacking positions with heteroaromatic DNA bases.
Tyrosine frequencies observed for OB-DNA interfaces suggest that a diagnostic test constructed with tyrosine would have a much lower false positive rate compared with tests looking at all aromatic residues or any other category. Table 4 reports the number of tyrosine residues and total number of residues encountered at OB-DNA interfaces. Tyrosine is strongly over-represented in four cases (h-Pot1, Sc-Cdc13, Sn-TEBP α-site, and Sn-TEBP αβ-site, νTyr = 0.20, nDNA-contact = 129) and under-represented in two cases (Rpa70 and Sp-Pot1N, νTyr = 0.021, nDNA-contact = 48). The DNA-binding domain of Rpa70 appears to compensate for an absence of tyrosine by placing two phenylalanine and two tryptophan residues in contact with DNA. Sp-Pot1N has just one tyrosine (Tyr115) for 20 DNA-contacting residues; between 2-4 are expected on the basis of tyrosine frequencies for all DNA-binding OB surfaces. A second surface-exposed tyrosine residue (Tyr70) is located on the S1bS2S3 sheet of Sp-Pot1N and would probably make contact with a longer DNA ligand. Also, as noted above, the DNA-binding domain of Sp-Pot1 comprises two OB folds, and we have structural information for only one of these. Additional DNA-contacting tyrosine residues are probably contained within the second, structurally uncharacterized OB fold.
Summarizing these trends and patterns leads to an average character for DNA-contacting OB surfaces: 1500 Å2, constructed with 110 protein atoms contributed by 26 residues, 5 of which are tyrosine, 2 salt bridges, and 13 non-salt hydrogen bonds. How do these values compare with protein-protein interfaces observed for telomere protein and RPA-derived OB folds? Characteristics for simple OB-protein interfaces are provided in Table 5, and these interfaces are illustrated in Figure 5(e) and Figure 6. Surface areas calculated for simple OB-peptide and OB-OB interfaces range from 1,220 to 2,270 Å2, with an average value of 1,700 Å2. The average number of protein atoms is 200, about twice the count for DNA-protein interfaces, as would be expected for similarly sized interfaces since both surfaces are constructed with protein atoms in OB-protein complexes. A large fraction of these atoms are apolar (average, 0.44), and another significant portion are aromatic (average, 0.18). The simple OB-peptide and OB-OB examples have two or four oppositely charged residues engaged in 2 salt bridges across the interface. The number of non-salt hydrogen bonds ranges from 2 to 10, with an average of 5.4 such hydrogen bonds per interface.
These parameters compare well with surveys of protein-protein complexes (Xu et al., 1997; Janin et al., 2008), except that the number of hydrogen bonds found at OB-peptide and OB-OB protein interfaces is lower than expected. One trivial explanation, accounting for about 20% of the missing hydrogen bonds, is that my criteria for hydrogen bonds and salt bridges were more stringent. I counted hydrogen bonds and salt bridges if electronegative atoms were within 3.2 Å; surveys generally included examples with distances up to 3.4 Å for hydrogen bonds and 4.0 Å for salt bridges. The remaining hydrogen bond deficit (2-3 hydrogen bonds per interface) may reflect functional constraints working at OB-protein interfaces that have kept the number of hydrogen bonds as low as necessary for specificity, an idea that will be revisited in the section devoted to molecular recognition.
The frequency of encountering a tyrosine residue at a protein-protein interface ranges from 0 to 0.053, values which are not significantly different from the tyrosine frequency measured at surface exposed positions or for the total amino acid population. Aromatic residues as a category are likewise encountered at normal rates, and all of the tests described for DNA-contacting surfaces failed to detect a category or residue type that is significantly over-represented at OB-protein interfaces (see Fig 8).
Comparison with OB-DNA interfaces shows that OB-protein interfaces are on average 13% larger as measured by buried surface area (but similarly sized as measured by number of atoms) and contain a similar number of salt bridges, about half the number of hydrogen bonds and one-third the number of tyrosine residues. Inspection of structural details shows that aromatic groups encountered at OB-protein interfaces make van der Waals contact, sometimes with other aromatic moieties, but aromatic face-to-face stacking interactions are absent. These comparisons suggest that switching from protein-binding to DNA-binding could be accomplished during evolution of OB folds with rather simple modifications such as the addition of three or more tyrosine residues and optimization of a few additional hydrogen bonding interactions. Such a switch likely occurred for the DNA-binding domain of Sc-Cdc13, which appears to be derived from a protein-binding lineage of OB folds (Sun et al., 2011).
Molecular recognition
The structures of OB-DNA, OB-peptide, and OB-protein interfaces suggest how these proteins distinguish between similar but different potential binding partners. Steric complementarity appears to play a major role. As illustrated in Figures 5-7, each binding surface provides a close, shape complementing fit to its binding partner. Contact surfaces at OB-OB interfaces have grooves, ridges, pits, and protuberances, and these steric details work to ensure that only the cognate partner matches and binds. The oligomer-binding surfaces of OB proteins are concave yet similarly detailed with unique features matching the surfaces presented by oligopeptide or DNA ligands. A search for defects (i.e. cavities) turned up few examples within the OB structures considered here, and these defects were not at binding interfaces with one exception: the OB-DNA interface of Sn-TEBPαβ. In the DNA–TEBPαβ complex, a large solvent-filled cavity (∼ 300 Å3) can be found close to the deoxyribose-phosphate moieties of nucleotides G13 and G14 and the bases of T10 and T11 (not shown). The role of this large cavity in the DNA-TEBPαβ structure is unknown, but speculative ideas are discussed in a later section on cooperativity and allostery.
In principle, oligomeric ligands should be more flexible and plastic, qualities that pose a challenge for establishing specificity by steric forces alone. Adaptability in a telomere DNA ligand has been captured as x-ray crystal structures of Sn-TEBPαβ in complex with non-cognate DNAs (Theobald and Schultz, 2003). DNA in these complexes extruded looped out nucleotides in order to avoid vacancies or clashes at the OB-DNA interface.
Hydrogen bonds and electrostatic terms likely resolve ambiguities for similarly shaped flexible ligands. Table 4 and Table 5 report the number of salt bridges and hydrogen bonds formed across simple OB-DNA and OB-protein interfaces. The number of these interactions seems small. Compared with the number of atoms at these interfaces an average of 14% are hydrogen-bonded across OB-DNA interfaces, and just 5% are hydrogen-bonded across OB-protein interfaces. The number of salt bridges ranges from 1-3 at both types of OB interface. These values are highly comparable with surveys of protein-protein interfaces (Xu et al., 1997; Janin et al., 2008), except that hydrogen-bond densities at OB-protein interfaces are lower.
The smaller than expected numbers may reflect a trade-off between specificity and affinity. Net contribution to stability for these hydrogen bonding and salt bridging interactions is determined as the balance of large desolvation costs and favorable enthalpic terms (Sheinerman and Honig, 2002). The distance and geometric requirements for the later means that only highly complementary surfaces can recoup the costs associated with forming a dehydrated interface, thus explaining how steric complementarity engenders molecular recognition and specificity. Each hydrogen bond and coulombic pair imposes a kinetic barrier to dissociation, which could be problematic for dynamic systems that must exchange binding partners. Evolution probably balances these constraints –– adding sufficient hydrogen-bonds and salt bridges to avoid confusion among binding partners and removing superfluous interactions so as to keep complexes dissociable and prevent kinetic traps that would otherwise interfere with telomere function.
The question of how telomere OB proteins distinguish among closely related DNA sequences has been considered carefully in a previous review (Croy and Wuttke, 2006). The general impression from binding studies is that most telomere DNA sequence positions are tolerant to substitution and only a few “hot spots” abrogate binding when replaced with a non-cognate nucleotide. We have examined nucleotide discrimination and selectivity for Sn-TEBPα and Sn-TEBPαβ with newly developed, more sensitive tools (Catherine Dy, 2010, PhD Dissertation Thesis, University of Utah, Biology). Previous work compared binding affinity for different DNA sequences in separate experiments (Classen et al., 2003; Lei et al., 2002; Anderson et al., 2003; Eldridge et al., 2006; Eldridge and Wuttke, 2008; Croy et al., 2008). The binding differences must necessarily be larger than experimental uncertainty to detect sequence specificity. In our new assay, different DNA sequences competed for the same binding sites in one experiment and the “winners” (and “losers”) were evaluated in a post-equilibrium analytic step. The competition assay thus directly measures sequence specificity, though apparent affinity for non-cognate sequences can be derived mathematically. Results showed the assay was very sensitive to even subtle nucleotide preferences, yet corroborated previously detected hot spots. The main findings of this work are summarized here.
The competition assay detected some level of nucleotide preference at every position along the S. nova telomere DNA, except at positions where the base makes no contact with protein in the crystal structure. Almost all sites tolerated substitutions with one or more of the three non-cognate nucleotides, consistent with true hot spots being rare. Mutational and structural analysis of residues contributing to nucleotide preference showed that it is possible to diminish and also switch nucleotide preference with single amino acid substitutions, but completely neutral sites were not attainable even if all potential hydrogen bond donors and acceptors had been removed.
Recent work in the Cech lab addressed a different recognition task: how telomere OB proteins distinguish DNA from structurally and chemically similar RNA molecules (Nandakumar et al., 2010). One important contact point identified in this work involved the h-Pot1 residue Phe62 and a C2′ atom of thymidylate, d(T), located at the fourth position of d(GGTTAGGGTTAG). The C5-methyl group of this nucleotide also makes close van der Waals contact with Phe62 in the crystal structure. Substitution of this position with ribothymidylate, a ribose analog which adds a 2′OH moiety at C2′, decreased binding affinity ∼2-fold when measured for h-Pot1 on its own and ∼14-fold when measured for h-Pot1 in the presence of h-Tpp1 (Nandakumar et al., 2010). Even stronger binding defects (∼4-fold with Pot1 and ∼119-fold with Pot1 and Tpp1) were detected when the position was replaced with uridylate, the ribose analog which adds a 2′OH at C2′ and additionally removes the C5-methyl moiety. The crystal structure of h-Pot1 with rU at this sensitive position shows that the 2′OH group (and absence of C5-methyl group) was accommodated through structural changes in DNA that perturb contacts with protein at adjacent nucleotides (Nandakumar et al., 2010).
The two chemical signatures distinguishing DNA from RNA are: (i) absence of a 2′OH group and (ii) presence of a C5-methyl group on thymine bases. Each of these makes DNA more hydrophobic compared with RNA, and the hydrophobic quality of Phe62 at the DNA-binding surface of h-Pot1 seems to explain dT versus rU preference (Nandakumar et al., 2010). To see if hydrophobic contact at C2′ and C5-methyl positions is general to DNA-binding OB proteins, I examined the chemical environment at C2′ and C5-methyl positions for telomere protein and RPA OB-DNA structures. Table 4 reports the number of C2′ and C5-methyl groups contacting each OB protein (defined by a 4.0 Å distance) and also the number and chemical type of protein atoms involved in making contacts with these moieties.
The number of C2′ atoms in contact with protein ranged from 2 – 7. On average, four C2′ atoms, corresponding with 47% of those available, came within range of making close van der Waals contact. At the DNA interfaces of h-Rpa70, Sc-Cdc13, and at the αβ-site of Sn-TEBP, each of the C2′ contact points is constructed with apolar aliphatic and aromatic carbon atoms and would, therefore, likely contribute to exclusion of RNA by a hydrophobic mechanism. With the exception of the Phe62–C2′ contact already described for h-Pot1, all C2′ contact points in h-Pot1 and its homolog Sp-Pot1N were constructed with hydrophilic polar groups. The α-site of Sn-TEBPα contacted two C2′ positions, the environments of which were polar (a tightly bound water molecule) in one case and hydrophobic (aromatic carbon atoms of Y293) in the other case.
A mixture of polar and apolar environments is similarly observed for C5-methyl groups. Compared with C2′ groups, there are fewer C5-methyl groups making contact with OB protein surfaces (average = 2), a situation that reflects the fact that thymine makes up only a portion of each telomere DNA sequence. In the DNA complex of Sp-Pot1N, the two C5-methyl groups participate in base-base stacking and do not closely contact protein. At the DNA interface with h-Pot1, three C5-methyl groups make close van der Waals contact with protein surfaces. For two of these the contact points are constructed with apolar carbon atoms contributed by aromatic residues (Phe62 and Tyr271). In all instances observed for Sc-Cdc13D, Sn-TEBPα and Sn-TEBPαβ, the C5-methyl contact points are constructed with hydrophilic polar moieties (e.g. hydroxyl, carboxamide and guanidinium groups). These observations indicate that over the course of evolution only modest pressure has been exerted to select for hydrophobic groups at C2′ and C5-methyl contact points. Steric complementarity may be sufficient to exclude RNA regardless of hydrophobic nature, or other cellular mechanisms may prevent telomere OB proteins from entering unproductive RNA complexes.
Composite OB interfaces
Composite OB interfaces can be thought of as combinations of simpler OB-oligomer and OB-OB interfaces described above. Figure 7 shows multiple views of the two structurally characterized telomere OB protein complexes with composite interfaces. These composite interfaces each have one or more OB-oligomer interface as a consequence of OB-OB association of two or more OB subunits. In the (Sc-Cdc13N)2 structure (Fig 7(a)), the two OB subunits are identical and each contributes to two OB-oligomer interfaces, which bind with peptide derived from the polymerase α/primase catalytic subunit, Pol1 (Sun et al., 2011). At the OB-oligomer interface, L45 of the classical oligomer-binding surface makes contact with the Pol1-derived helical peptide, and this portion of the composite interface resembles simple OB-oligomer complexes, especially h-Rpa70N–p53 and Sc-Cdc13D–DNA, as has been previously noted (Sun et al., 2011). The Pol1-binding interface is augmented through contacts with a smaller surface constructed from the N-terminal portion of C34 and L12 in the second Sc-Cdc13N subunit. Cdc13N dimerization and Pol1 binding are thus structurally linked, making recruitment of the polymerase α/primase complex sensitive to dimer stability (Sun et al., 2011).
The OB-OB homodimer interface is unique to Sc-Cdc13N. As illustrated in Fig 2(f), the dimer interface comprises helices of C34, the element that connects S3 and S4, and which is highly elaborated for this OB fold. Cdc13 dimerization is conserved; however, the protein region responsible for construction of the dimer interface is located in different domains in different yeast species, indicating that evolution may have solved this puzzle more than once with different outcomes (Sun et al., 2011). The selective pressure resulting in Cdc13 dimers may relate to interactions with the semi-conservative DNA synthesis apparatus, which also has C2-symmetric quarternary structure.
In the Sn-TEBPαβ–DNA structure, four OB units contributed by two unequal subunits collaborate to construct the OB-DNA interface (Fig 7(b)). Ironically, this highly complicated example was the first to be structurally characterized (Horvath et al., 1998). The composite interface can be divided into four parts: two OB-DNA interfaces, and two OB-protein interfaces. The two OB-DNA interfaces were analyzed above, and correspond with the α-site and αβ-site, each contacting one 8-nucleotide telomere DNA repeat (Table 4). The combined OB-DNA interface is summarized in Table 6 as one portion of the composite interface (Table 6). Protein-protein contacts in the Sn-TEBPαβ–DNA structure can be divided into an OB-OB interface and an OB-peptide interface. The two interfaces are contiguous. At the OB-OB interface, two of the three OB units in Sn-TEBPα make contact with the Sn-TEBPβ OB unit. At the OB-peptide interface, the single OB unit Sn-TEBPα3 binds with an oligopeptide element (residues 162β - 195β) that extends from the Sn-TEBPβ OB core.
The TEBPα3–peptide interface partly resembles other OB-oligomer complexes in that the classical oligomer-binding surface of TEBPα3 participates in contacting residues 162β - 195β. However, the oligomer-binding surface is much larger for TEBPα3 compared with other OB folds, a consequence of elaborations in L12 and contributions made by the N-terminal element of TEBPα3. The larger surface accommodates oligopeptide at two anti-parallel binding grooves with the result that residues of Sn-TEBPβ form an out-and-back loop. Strand orientation for the portion of the peptide binding with the L45 portion of the classical oligomer-binding site (residues 180β - 195β) is such that N→C points towards the “back” when the OB unit is oriented as in Fig 2(a). This peptide strand polarity matches that seen in other OB-peptide complexes (e.g. Sc-Cdc13N and h-Rpa70N). The other half of the TEBPβ-derived loop (residues 162β - 177β) interacts with the elaborated L12 of TEBPα3, and is oriented oppositely with N→C pointed towards the “front”, a situation which contrasts with other OB-peptide complexes.
The OB-OB interface observed for the Sn-TEBPαβ–DNA structure involves three OB units, two from Sn-TEBPα and one from Sn-TEBPβ. What are the contact surfaces involved and how do these compare with contact surfaces seen for other OB-OB complexes? The TEBPα-contacting surface of this OB-OB interface comprises the N-terminal element, L45 and C-helix of Sn-TEBPβ. The C-helix is positioned above the OB barrel towards the “back” when Sn-TEBPβ is oriented as in Fig 2(a), a structural architecture shared only with h-Tpp1. This TEBPα contact surface is unique to Sn-TEBPβ, as far as can be inferred from currently available structures, and hints at a potentially similar contact surface connecting Tpp1 and Pot1.
The TEBPβ-contacting surfaces of the OB-OB interface are found on OB units Sn-TEBPα2 and Sn-TEBPα3. The portion contributed by TEBPα2 corresponds with the classical oligo-binding surface and contacts the N-terminal half of the Sn-TEBPβ C-helix in a manner highly analogous with a simple OB-peptide complex seen for Rpa70N–p53. The portion contributed by TEBPα3 makes contact with the C-terminal half of the Sn-TEBPβ C-helix with use of a concave surface comprising S1, L23 and the C-terminal portion of C34. In most other OB units, this concave surface is unavailable for OB-OB interaction because it is occupied by the OB unit's C-helix (Fig 3). As noted in an earlier section, the C-helix is missing in Sn-TEBPα3, a relatively rare structural character trait shared with Rpa70A and Rpa70N (Fig 4(b)). Removal of the C-helix in Sn-TEBPα3 appears to be an innovation that allows for OB-OB interactions with the C-helix of Sn-TEBPβ, a type of partial domain swapping (Bennett et al., 1994; Schlunegger et al., 1997) between TEBPα3 and TEBPβ.
Additional structural characteristics for these composite interfaces are reported in Table 6. The Sc-Cdc13N–Pol1 interface is analyzed apart from the Cdc13N–Cdc13N' dimer interface. Separated in this way, each interface character is within normal ranges observed for OB-peptide or OB-OB complexes, except that the Sc-Cdc13N–Pol1 interface is unusually rich in salt bridges (7 versus 2 salt bridges observed at simple OB-protein interfaces). The combined surface area is atypically large. With two symmetrically equivalent Pol1-binding surfaces, the total surface area buried as a consequence of forming the dimer and associating with Pol1 is 4,070 Å2, more than twice the average value observed for a simple OB-protein interface.
Surfaces buried at the protein-protein interfaces of the Sn-TEBPαβ–DNA complex are similarly large by comparison with simple OB-protein interfaces. The total surface area buried at the α–β interface measures 4,170 Å2. When divided into two parts, the OB-OB and OB-peptide portions each buries larger-than-average surfaces but falls within range of surface areas measured for simple OB-protein interfaces. The number of hydrogen bonds detected at the α–β interface is more than seen for simple OB-protein interfaces (13-14 versus 2-10) and more comparable to values reported in general surveys of protein-protein interfaces (Janin et al., 2008).
Cooperativity and allostery
Composite interfaces described above provide a structural basis for understanding cooperativity and allostery. Cooperativity and allostery are emergent properties not evident for component parts. These emergent properties, especially allostery, likely play important roles in making telomere structure responsive to changing biochemical requirements at different times during the cell cycle [reviewed in (Horvath, 2008a; b)]. One way to demonstrate emergent properties is by comparison with the behavior of isolated parts. For example, cooperative oxygen binding and release by hemoglobin is easily recognized by comparison with simpler behavior of myoglobin.
Portions of each telomere OB protein composite interface can be isolated in the form of subcomplexes. For example, the (Sc-Cdc13N)2 dimer was stable even in the absence of Pol1-derived peptide, and the crystal structure of the ligand-free state was determined as part of the same work reporting the peptide-bound structure (Sun et al., 2011). Biochemical experiments and genetic tests demonstrate that designed alleles which influence Cdc13N dimer stability in vitro also change telomere homeostasis in vivo (Sun et al., 2011; Mitchell et al., 2010). Especially compelling is the observation that dimer instability correlates with severity of telomere length changes (Sun et al., 2011). The molecular mechanism responsible for linking Cdc13N dimerization with telomere homeostasis probably involves Pol 1 peptide binding and/or DNA-binding activity since dimerization defective alleles reduce or eliminate interaction with Pol1-derived peptide (Sun et al., 2011) and abrogate binding of long single-stranded DNA fragments (Mitchell et al., 2010). These results show that telomere length regulation can be optimized during evolution by increasing or decreasing Cdc13 dimer stability. On a shorter time scale, the DNA polymerase α/primase apparatus could be recruited and activated at telomeres by any mechanism that triggers Cdc13 dimerization during the cell cycle.
One of the two OB-DNA interfaces seen in the Sn-TEBPαβ–DNA structure can be prepared as a separate stable complex by combining Sn-TEBPα with single-stranded DNA without adding Sn-TEBPβ. Crystal structures containing the isolated Sn-TEBPα–DNA interface have been determined (Classen et al., 2001; Peersen et al., 2002). This DNA-TEBPα complex protects 8 nucleotides of DNA and likely corresponds with a stable intermediate encountered during the assembly of the DNA-TEBPαβ complex. The single-stranded portion of telomere DNA at the ends of macronuclear chromosomes of S. nova is exactly 16 nucleotides (Klobutcher et al., 1981), which contrasts with the situation in yeast, plants, and vertebrates, for which telomere single-stranded DNA is several hundred nucleotides and heterogeneous in length. All 16 nucleotides are protected by one Sn-TEBPαβ heterodimer. The OB-DNA interface of the Sn-TEBPαβ–DNA complex can be separated from much of the protein-protein interface by linking the DNA-binding portions of Sn-TEBPα and Sn-TEBPβ as a fusion protein (Buczek et al., 2005), designated as αNβΔ in Table 6. The crystal structure of αNβΔ complexed with 16-nucleotide single-stranded DNA provides a more complete view of the OB-DNA interface, but otherwise superimposes very closely with previous structures (Buczek et al., 2005; Buczek and Horvath, 2006a).
In contrast to the situation for OB-DNA subcomplexes, portions of the TEBPα–TEBPβ interface cannot be isolated as stable subcomplexes. Attempts to isolate the complex of Sn-TEBPα3 bound with a TEBPβ-derived peptide fragment failed (Pawel Buczek, personal communication), and crystals of TEBPα–TEBPβ did not grow except when DNA was added to reactions. Weak association of TEBPα and TEBPβ was detectable in the absence of DNA by ITC (Buczek and Horvath, 2006a). The DNA-dependence of TEBPα–TEBPβ association has been demonstrated in several contexts (Fang and Cech, 1993; Fang et al., 1993; Buczek et al., 2005), and binding linkage measured in this system amounts to ∼3 kcal of binding energy realized for the ternary complex that cannot be measured in combinations of simpler two-component binding reactions (Buczek and Horvath, 2006a). TEBPα–TEBPβ association is thus significantly more stable in the presence of DNA. Conversely, DNA-binding affinity increases 200-fold for the TEBPα–TEBPβ heterodimer compared with TEBPα alone (Buczek et al., 2005; Buczek and Horvath, 2006a). The excess binding energy derives partly from DNA-TEBPβ contacts (which are small at 400 Å2), and mostly from α–β contacts (which are substantial at 4,200 Å2) that are somehow not accessible except in the presence of DNA (Buczek and Horvath, 2006a).
A possible explanation for DNA-dependent α–β association relates to interface defects. As described earlier, such defects are rarely observed, yet the DNA–TEBPα–TEBPβ interface contains a large cavity that impinges on that point in the structure where all three components meet. In the absence of DNA, the size and extent of this defect would be expected to increase, possibly to the point of destabilizing the TEBPα–β heterodimer. The purpose of this cavity is unknown, and since comparable structures of DNA–Pot1–Tpp1 are unavailable, we also do not know if the cavity is conserved. One intriguing possibility is that this large unoccupied volume could bind with some other telomere factor, possibly a portion of telomerase or a telomerase-associated protein, and thus act as an effector site to trigger a DNA hand-off event through an allosteric mechanism.
Binding cooperativity in the Sn-TEBPαβ system is recapitulated in the human Pot1–Tpp1 complex since DNA-binding by Pot1 increases approximately 10-fold in the presence of Tpp1 (Xin et al., 2007; Wang et al., 2007). Cooperativity and binding linkage are biochemical behaviors associated with allosteric systems. Allostery could explain how telomere DNA-OB complexes switch from a DNA-protective state to a DNA-synthesis state (Buczek and Horvath, 2006a; Wang et al., 2007; Latrick and Cech, 2010). A regulated DNA protection ↔ DNA synthesis switch built around Sp-Pot1 and the Tpp1 homolog Sp-Tpz1 appears to be conserved in S. pombe, since the system shows rapid telomere defects if these proteins or interacting partners are removed (Miyoshi et al., 2008).
Conclusion and prospectus
Structures of telomere OB proteins have greatly expanded in number, making it possible to make meaningful structural comparisons. Structurally aligned amino acid sequences together with structural morphology characters provided a partially resolved view of deeply rooted evolutionary connections among telomere OB proteins and replication protein A. Comparison of OB-DNA and OB-OB interfaces led to general properties of each interface type and highlighted differences in the propensity of tyrosine residues, which is elevated for DNA-contacting surfaces. By comparison with these simple OB interfaces, architectures apparent for composite OB interfaces were significantly more complex. Structural complexity probably underlies emergent properties of specificity, cooperativity and allostery that are important for biological function of telomere OB proteins.
Structures of OB-DNA complexes characterized to date likely correspond with telomere DNA-protective states. Biochemical experiments and genetic analyses have demonstrated that telomere OB proteins can also promote telomere DNA synthesis, probably through allosteric mechanisms involving telomerase and the semi-conservative DNA synthesis apparatus. Evolutionary pressure selecting for allostery in these systems can be appreciated by considering alternative scenarios in which the system is unresponsive or stochastic. The DNA protective state requires high DNA affinity and slow DNA dissociation. The DNA synthesis apparatus has a limited time window in which to operate, and therefore cannot wait for such dissociation events to happen by chance. Additionally, an intermediate potentially encountered in a stochastic scenario is naked single-stranded DNA, which would be degraded, recombine with other parts of the genome, or elicit an inappropriate DNA damage repair response. Allosteric regulation avoids all these risky and dangerous possibilities. An important goal for future structural work will be to uncover the molecular form associated with DNA synthesis state(s) and to puzzle out exactly how putative allosteric mechanisms trigger switching between states. What do the moving parts look like? and how do they work? The answers will open new areas of knowledge with high dividends in terms of understanding telomere structure, evolution and biology.
Acknowledgments
I thank Isabel Gardett for constructive suggestions on organization and style, Kigen Curtice and Dr. Catherine Dy for reading of drafts, Dr. Douglas Theobald for explanations of Bayesian analysis, Dr. Gerda Saxer Quance for discussions about evolution, and Dr. Michael Horvath for suggestions regarding chi-squared significance tests.
Footnotes
Declaration of interest The NIH supported a portion of this work (GM067994).
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Agrawal V, Kishan KV. OB-fold: growing bigger with functional consistency. Curr Protein Pept Sci. 2003;4:195–206. doi: 10.2174/1389203033487207. [DOI] [PubMed] [Google Scholar]
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- Anderson EM, Halsey WA, Wuttke DS. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry. 2003;42:3751–3758. doi: 10.1021/bi027047c. [DOI] [PubMed] [Google Scholar]
- Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr Opin Struct Biol. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Choe S, Eisenberg D. Refined structure of dimeric diphtheria toxin at 2.0 A resolution. Protein Sci. 1994;3:1444–1463. doi: 10.1002/pro.5560030911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Belegu V, Korolev S, Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, Bochkarev A. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill SJ, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- Buczek P, Horvath MP. Structural reorganization and the cooperative binding of single-stranded telomere DNA in Sterkiella nova. J Biol Chem. 2006a;281:40124–40134. doi: 10.1074/jbc.M607749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczek P, Horvath MP. Thermodynamic characterization of binding Oxytricha nova single strand telomere DNA with the alpha protein N-terminal domain. J Mol Biol. 2006b;359:1217–1234. doi: 10.1016/j.jmb.2006.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczek P, Orr RS, Pyper SR, Shum M, Kimmel E, Ota I, Gerum SE, Horvath MP. Binding linkage in a telomere DNA-protein complex at the ends of Oxytricha nova chromosomes. J Mol Biol. 2005;350:938–952. doi: 10.1016/j.jmb.2005.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. A DNA polymerase-{alpha} {middle dot} primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284:5807–5818. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen S, Lyons D, Cech TR, Schultz SC. Sequence-specific and 3′-end selective single-strand DNA binding by the Oxytricha nova telomere end binding protein alpha subunit. Biochemistry. 2003;42:9269–9277. doi: 10.1021/bi0273718. [DOI] [PubMed] [Google Scholar]
- Classen S, Ruggles JA, Schultz SC. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein alpha subunit both uncomplexed and complexed with telomeric ssDNA. J Mol Biol. 2001;314:1113–1125. doi: 10.1006/jmbi.2000.5191. [DOI] [PubMed] [Google Scholar]
- Croy JE, Fast JL, Grimm NE, Wuttke DS. Deciphering the mechanism of thermodynamic accommodation of telomeric oligonucleotide sequences by the Schizosaccharomyces pombe protection of telomeres 1 (Pot1pN) protein. Biochemistry. 2008;47:4345–4358. doi: 10.1021/bi701778x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11-389. J Mol Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Habel JE, Kabaleeswaran V, Snell EH, Wold MS, Borgstahl GE. Structure of the full-length human RPA14/32 complex gives insights into the mechanism of DNA binding and complex formation. J Mol Biol. 2007;374:865–876. doi: 10.1016/j.jmb.2007.09.074. [DOI] [PubMed] [Google Scholar]
- Eldridge AM, Halsey WA, Wuttke DS. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry. 2006;45:871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge AM, Wuttke DS. Probing the mechanism of recognition of ssDNA by the Cdc13-DBD. Nucleic Acids Res. 2008;36:1624–1633. doi: 10.1093/nar/gkn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman MP, Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Cech TR. Oxytricha telomere-binding protein: DNA-dependent dimerization of the alpha and beta subunits. Proc Natl Acad Sci U S A. 1993;90:6056–6060. doi: 10.1073/pnas.90.13.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Gray JT, Cech TR. Oxytricha telomere-binding protein: separable DNA-binding and dimerization domains of the alpha-subunit. Genes Dev. 1993;7:870–882. doi: 10.1101/gad.7.5.870. [DOI] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit Rev Biochem Mol Biol. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- Gelinas AD, Paschini M, Reyes FE, Heroux A, Batey RT, Lundblad V, Wuttke DS. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc Natl Acad Sci U S A. 2009;106:19298–19303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Zakian VA. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- Goulian M, Heard CJ. The mechanism of action of an accessory protein for DNA polymerase alpha/primase. J Biol Chem. 1990;265:13231–13239. [PubMed] [Google Scholar]
- Goulian M, Richards SH, Heard CJ, Bigsby BM. Discontinuous DNA synthesis by purified mammalian proteins. J Biol Chem. 1990;265:18461–18471. [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- Hecker J, Yang JY, Cheng J. Protein disorder prediction at multiple levels of sensitivity and specificity. BMC Genomics. 2008;9(1):S9. doi: 10.1186/1471-2164-9-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath MP. Evolution of telomere binding proteins. In: Tomaska L, Nosek J, editors. Origin and Evolution of Telomeres. Austin, TX: Landes Bioscience; 2008a. pp. 83–99. [Google Scholar]
- Horvath MP. Single-stranded nucleic acid-binding proteins. In: Rice PA, Correll CC, editors. Protein-Nucleic Acid Interactions: Structural Biology. London: Royal Society of Chemistry; 2008b. pp. 91–128. [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- Hsu CL, Chen YS, Tsai SY, Tu PJ, Wang MJ, Lin JJ. Interaction of Saccharomyces Cdc13p with Pol1p, Imp4p, Sir4p and Zds2p is involved in telomere replication, telomere maintenance and cell growth control. Nucleic Acids Res. 2004;32:511–521. doi: 10.1093/nar/gkh203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q Rev Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Swanton MT, Donini P, Prescott DM. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci U S A. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide S, Sidhu SS. The importance of being tyrosine: lessons in molecular recognition from minimalist synthetic binding proteins. ACS Chem Biol. 2009;4:325–334. doi: 10.1021/cb800314v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossiakoff AA, Koide S. Understanding mechanisms governing protein-protein interactions from synthetic binding interfaces. Curr Opin Struct Biol. 2008;18:499–506. doi: 10.1016/j.sbi.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lee J, Mandell EK, Rao T, Wuttke DS, Lundblad V. Investigating the role of the Est3 protein in yeast telomere replication. Nucleic Acids Res. 2010;38:2279–2290. doi: 10.1093/nar/gkp1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol. 2008;15:990–997. doi: 10.1038/nsmb.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci U S A. 2007;104:14038–14043. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- Mitchell MT, Smith JS, Mason M, Harper S, Speicher DW, Johnson FB, Skordalakes E. Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation. Mol Cell Biol. 2010;30:5325–5334. doi: 10.1128/MCB.00515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J Mol Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc Natl Acad Sci U S A. 2010;107:651–656. doi: 10.1073/pnas.0911099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci U S A. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peersen OB, Ruggles JA, Schultz SC. Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA. Nat Struct Biol. 2002;9:182–187. doi: 10.1038/nsb761. [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Price CM. Telomere structure in Euplotes crassus: characterization of DNA-protein interactions and isolation of a telomere-binding protein. Mol Cell Biol. 1990;10:3421–3431. doi: 10.1128/mcb.10.7.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–3165. doi: 10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Cech TR. Telomeric DNA-protein interactions of Oxytricha macronuclear DNA. Genes Dev. 1987;1:783–793. doi: 10.1101/gad.1.8.783. [DOI] [PubMed] [Google Scholar]
- Price CM, Cech TR. Properties of the telomeric DNA-binding protein from Oxytricha nova. Biochemistry. 1989;28:769–774. doi: 10.1021/bi00428a053. [DOI] [PubMed] [Google Scholar]
- Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27:2328–2339. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Schlunegger MP, Bennett MJ, Eisenberg D. Oligomer formation by 3D domain swapping: a model for protein assembly and misassembly. Adv Protein Chem. 1997;50:61–122. doi: 10.1016/s0065-3233(08)60319-8. [DOI] [PubMed] [Google Scholar]
- Sheinerman FB, Honig B. On the role of electrostatic interactions in the design of protein-protein interfaces. J Mol Biol. 2002;318:161–177. doi: 10.1016/S0022-2836(02)00030-X. [DOI] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yang Y, Wan K, Mao N, Yu TY, Lin YC, Dezwaan DC, Freeman BC, Lin JJ, Lue NF, Lei M. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell Res. 2011;21:258–274. doi: 10.1038/cr.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Gilson E. Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res. 2005;13:535–548. doi: 10.1007/s10577-005-0999-0. [DOI] [PubMed] [Google Scholar]
- Theobald DL, Cervantes RB, Lundblad V, Wuttke DS. Homology among telomeric end-protection proteins. Structure. 2003a;11:1049–1050. doi: 10.1016/s0969-2126(03)00183-7. [DOI] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003b;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Schultz SC. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 2003;22:4314–4324. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS. Divergent evolution within protein superfolds inferred from profile-based phylogenetics. J Mol Biol. 2005;354:722–737. doi: 10.1016/j.jmb.2005.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Bunch JT, Baumann P. Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of heterogeneous fission yeast telomeres. J Biol Chem. 2005;280:9119–9128. doi: 10.1074/jbc.M414511200. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- Voss NR, Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38:W555–562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Qin J, Songyang Z, Liu D. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem. 2009;284:26725–26731. doi: 10.1074/jbc.M109.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ. The CST complex and telomere maintenance: the exception becomes the rule. Mol Cell. 2009;36:168–169. doi: 10.1016/j.molcel.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wold MS, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- Xu D, Tsai CJ, Nussinov R. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng. 1997;10:999–1012. doi: 10.1093/protein/10.9.999. [DOI] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WF, Chico L, Lei M, Lue NF. Telomerase and Retrotransposons: Reverse Transcriptases That Shaped Genomes Special Feature Sackler Colloquium: Telomerase regulatory subunit Est3 in two Candida species physically interacts with the TEN domain of TERT and telomeric DNA. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1017855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Wang F, Lei M, Lue NF. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol. 2008;15:985–989. doi: 10.1038/nsmb.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]


