Cancer has become a significant, predominate focus in world medicine over the past century. In 2008, a total of 7.6 million deaths internationally were attributed to the disease, giving cancer the dubious honor of being the leading cause of death worldwide.[1] The disease itself is actually a diverse subset of multiple genetic disorders, which leads to the generation of various rogue proteins and tumor suppressor knockouts that interrupt cellular homeostasis. Treatment success is highest when the specific genetic disorder causing a patient’s tumor is targeted through a personalized medicine approach, but this is often difficult because of issues with druggability of the desired target.
The most commonly mutated oncogene, at present deemed undruggable, is Ras (rat sarcoma), which has been identified in about 30 % of cancers.[2, 3] Ras comprises a family of guanosine-5′-triphosphate (GTP) binding proteins (K-Ras, N-Ras, and H-Ras) localized at the inner cell membrane, and these proteins serve as a molecular switch controlling cell survival, differentiation, and proliferation.[3] Many cancers utilize Ras proteins as survival pathways to drive and propagate the disease. Efforts thus far to directly target Ras at the GTP binding site have been met with much disappointment, as Ras proteins bind GTP with picomolar affinity.[4] Adding to this difficulty, GTP is present intracellularly at micromolar concentrations,[4] making the GTP pocket on Ras realistically unamenable to small-molecule inhibition. The druggability issue is made even more difficult because Ras does not contain any likely pockets where a small molecule can bind with high affinity.[4] Because of these problems, tumors bearing Ras mutations are difficult to treat.
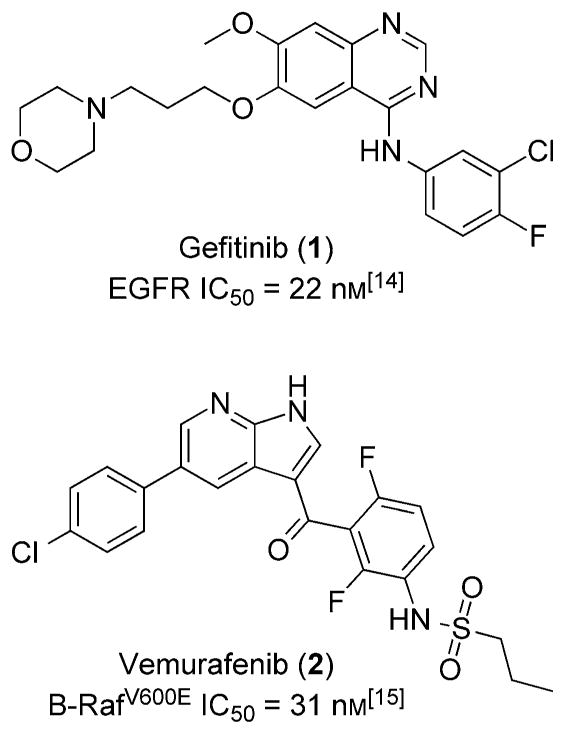
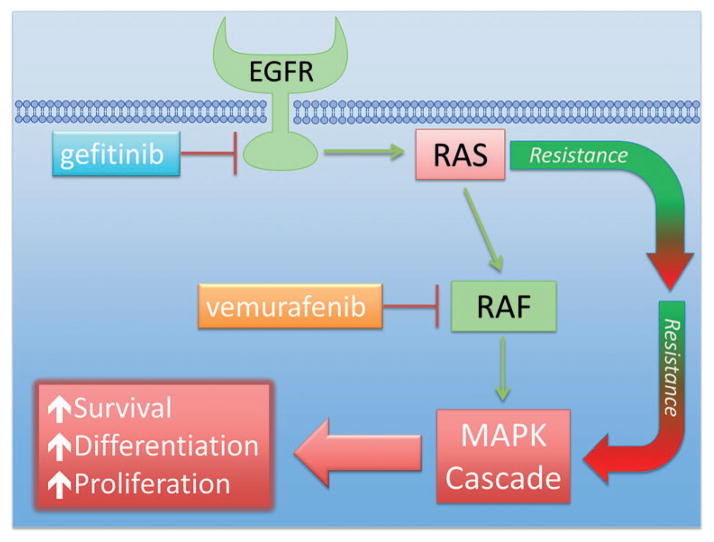
Current therapeutic avenues have been addressed and discussed in a comprehensive review by Wang et al.[5] Nevertheless, there have been a few transient success stories targeting the Ras protein, which primarily rely on inhibiting easily druggable kinases upstream and downstream in the Ras signal transduction pathway. This approach has been used to treat a certain subset of non-small-cell lung cancer (NSCLC). The NSCLC subset utilizes epidermal growth factor receptor (EGFR) as a survival pathway, which is upstream of Ras. Therapeutically, EGFR has been drugged with gefitinib (1), and significant tumor remission has been observed (Figure 1). However, efficacy is temporary, as NCSLC develops resistance to gefitinib over time. Interestingly, resistance has been shown to develop through activated Ras, where Ras is able to signal independent of EGFR stimulation.[6] In another example, vemurafenib (2) has been developed as a Raf (rapidly accelerated fibrosarcoma) inhibitor to treat late-stage melanoma, specifically inhibiting B-RafV600E. Raf is downstream of Ras, and treatment with vemurafenib initially blocks progression of the melanoma. However, treatment eventually becomes ineffective as a second Ras isoform (N-Ras) reactivates the mitogen-activated protein kinase (MAPK) signaling pathway and bypasses vemurafenib inhibition.[7] Therefore, all efforts to date to disrupt the Ras signaling pathway that show clinical promise have failed because of resistance conferred through the Ras protein (Scheme 1). Developing a strategy to target Ras directly has the potential to offer longer disease remission and overall better treatment options for Ras-driven tumors. However, the druggability of Ras at a clinically effective level still remains a major problem.
Figure 1.
Upstream (gefitinib) and downstream (vemurafenib) effectors of Ras signaling.
Scheme 1.
A schematic representation of the Ras signaling pathway. Gefitinib (1) and vemurafenib (2) block upstream and downstream of Ras, respectively. Resistance to both treatments is commonly observed through modification in Ras signaling. Therefore, drugging Ras directly could lead to better treatment outcomes.
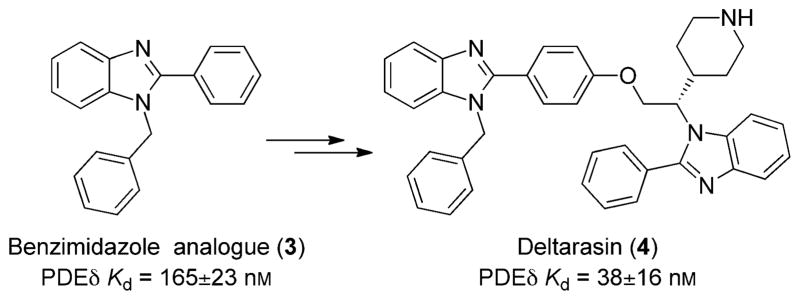
In an effort to reverse the uncertainty of drugging Ras, a recent breakthrough paper by Zimmerman and co-workers has revived the possibility of identifying a druglike inhibitor to block Ras activity.[8] Unlike other research efforts where Ras activity has been modified at high micromolar to millimolar drug concentrations by interrupting the Ras/SOS (son of sevenless) protein–protein interaction,[9,10] Zimmerman et al. managed to modify Ras activity at therapeutically relevant concentrations. The innovative technique involves interrupting a vital protein–protein interaction necessary for the localization of Ras to the cellular membrane by drugging the farnesyl binding pocket of photoreceptor cGMP phosphodiesterase δ subunit (PDEδ). Utilizing a high-throughput screening (HTS) assay, benzimidazole-based analogues 3 (Figure 2) were identified as hits that are capable of interrupting the binding of farnesylated K-Ras and prenyl binding protein PDEδ. This disruption obstructs membrane localization and therefore blocks K-Ras activity because the protein does not localize correctly. As with any cellular protein, correct location is essential for activity, and K-Ras is no exception. Therefore, Zimmerman and co-workers were able to circumvent Ras druggability issues by initiating a drug discovery campaign on a protein essential for Ras membrane localization.
Figure 2.
Novel compounds developed by Zimmerman and co-workers that disrupt K-Ras membrane localization.[8] The initial hit (3) was developed into deltarasin (4).
From the initial benzimidazole hit, crystal structure analysis of the analogue in PDEδ revealed that two benzimidazoles bind to a hydrophobic tunnel in the protein. Because of the proximity of the two benzimidazoles, a new compound was generated that linked the two ring systems, which displayed tight binding to PDEδ. The covalently linked molecule was chiral, and resolution of the two structures generated deltarasin (4). Deltarasin is biologically stable, penetrates cellular membranes, and binds to PDEδ with a Kd value of 38 ± 16 nM. Deltarasin is capable of interrupting the K-Ras/PDEδ protein–protein interaction at nanomolar concentrations and contains many druglike properties. The molecular weight of the compound is on the higher end (603 g mol−1), but medicinal chemistry efforts could easily optimize the structure to a more ideal druglike weight.
When dosed in cells, deltarasin can completely disrupt membrane localization of K-Ras. In K-Ras-sensitive cell lines and K-RasWT cell lines, deltarasin is selectively effective, only blocking proliferation of cell lines dependent on K-Ras. Ex vivo analysis revealed that complete inhibition of the K-Ras/PDEδ interaction occurs at a deltarasin concentration of 200 nM, while a measurable antiproliferative effect occurred at 3000 nM. The 15 times difference in activity was attributed to active efflux by ABC transporter proteins. An issue of concern is the possibility that deltarasin is targeted for active efflux by ABC transporters. Treatment with deltarasin will then lead to the selection of resistant cancer cells that express high levels of ABC genes, inevitably causing the development of a tumor mass resistant to treatment. Likely, the tumor mass will also be resistant to cytotoxic treatments like taxanes or vinca alkaloids, as resistance to natural-product-based chemotherapeutics is mediated through drug efflux proteins.[11] This problem should be at the forefront of medicinal chemistry efforts to develop future deltarasin analogues with clinical potential.
Zimmerman and co-workers then went on to evaluate deltarasin in animal models using PancTu-I cells, a K-Ras-driven pancreatic cell line. The cells were injected subcutaneously (s.c.), and deltarasin was administered via intraperitoneal (i.p.) injection once (QD, 10 mg kg−1 and 15 mg kg−1) or twice (BID 10 mg kg−1) daily. A major side effect observed was a loss of approximately 15 % body weight in mice receiving deltarasin. The weight loss is an indication that deltarasin interferes with important metabolic pathways and is a forewarning for possible patient-related toxicities. Nevertheless, mouse weight did stabilize after two days and deltarasin did slow the growth of the tumors. Mice receiving the BID dose displayed statistically significant (P ≤ 0.01) tumor reduction, as well as the mice receiving the 15 mg kg−1 QD dose (P ≤ 0.05). Therefore, Zimmerman et al. were able to show for the first time the possibility of directly targeting oncogenic Ras in a mammalian system using a small molecule with dosing regimes that are therapeutically relevant.
A novel method for drugging Ras-driven tumors has been discovered and will likely lead to a new drug class. However, current research suggests that the utilized tumor model is not ideal for studying this type of pancreatic cancer.[12, 13] PancTu-I cells are derived from pancreatic ductal adenocarcinoma (PDA), and PDA xenografts respond to chemotherapy. In fact, gemcitabine is active in certain PDA xenograft models and pancreatic cell lines (not PancTu-I), yet minimally extends patient survival in the clinic.[12] New research shows that when PDA develops, the disease is fibrous with low vasculature. Therefore the disease is hard to drug, as blood vessels that facilitate the trafficking of chemotherapeutics into the tumor are practically nonexistent.[13] This is considerably different from the vascularized microenvironment of a s.c. tumor xenograft. The s.c. microenvironment contains blood vessels that deliver the drug into the tumor, which does not accurately model PDA and can display efficacy that does not translate into the clinic, as is the case with gemcitabine.[13] Model choice is essential to accurately predict clinical significance, and therefore deltarasin should be tested in PDA-engineered mouse models (GEMMs), which more closely resemble the PDA pathology observed in humans.[12]
Nevertheless, a strong proof of concept has been developed that indisputably rejuvenates the possibility for a successful drug discovery campaign addressing oncogenic Ras. At present, it is important to test deltarasin in cell lines with developed drug resistance initiated through Ras signaling. Cell lines refractory to gefitinib or vemurafenib could see a substantial rebound in treatment response when dosed with deltarasin, because deltarasin will block Ras-driven drug resistance. As the undruggable becomes druggable, new paradigms will surface yielding novel tools to advance the fight against cancers that at one point had minimally effective treatments. This should galvanize excitement and confidence that revolutionary cancer treatments can and will be uncovered, an idea usually met with hesitation and uncertainty. As we wield breakthrough tools to advance the war on cancer, our confidence in treating the disease must always be accompanied with circumspection.
References
- 1.The World Health Organization (WHO) Cancer: Fact sheet N°297. 2013. [Google Scholar]
- 2.Roberts PJ, Der CJ. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 3.Adjei AA. J Natl Cancer Inst. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 4.Gysin S, Salt M, Young A, McCormick F. Genes Cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Kaiser CE, Frett B, Li HY. J Med Chem. 2013;56:5219–5230. doi: 10.1021/jm3017706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin B, Ariyama H, Baba E, Tanaka R, Kusaba H, Harada M, Nakano S. Cancer Chemother Pharmacol. 2006;58:577–584. doi: 10.1007/s00280-006-0219-4. [DOI] [PubMed] [Google Scholar]
- 7.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PIH, Waldmann H. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 9.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJC, Bowman KK, Wu J, Giannetti AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang W, Fang G. Proc Natl Acad Sci USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. Angew Chem. 2012;124:6244–6247. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole SPC, Deeley RG. BioEssays. 1998;20:931–940. doi: 10.1002/(SICI)1521-1878(199811)20:11<931::AID-BIES8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 12.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, Frese KK, DeNicola G, Feig C, Combs C, Winter SP, Ireland-Zecchini H, Reichelt S, Howat WJ, Chang A, Dhara M, Wang L, Rückert F, Grützmann R, Pilarsky C, Izeradjene K, Hingorani SR, Huang P, Davies SE, Plunkett W, Egorin M, Hruban RH, Whitebread N, McGovern K, Adams J, Iacobuzio-Donahue C, Griffiths J, Tuveson DA. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pérez-Mancera PA, Guerra C, Barbacid M, Tuveson DA. Gastroenterology. 2012;142:1079–1092. doi: 10.1053/j.gastro.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen MW, Pedersen N, Ottesen LH, Poulsen HS. Br J Cancer. 2005;93:915–923. doi: 10.1038/sj.bjc.6602793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KYJ, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D’Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]