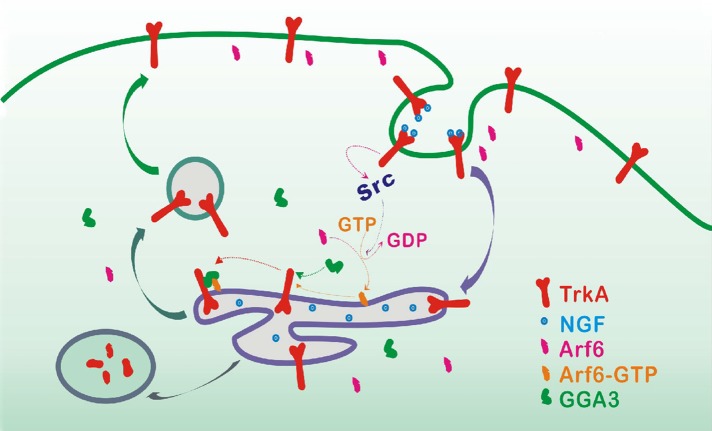
GGA3 binds directly to the TrkA internal DXXLL motif and mediates TrkA endocytic recycling. This effect is dependent on the activation of Arf6. GGA3 is a key player in a novel DXXLL-mediated recycling machinery for TrkA, where it prolongs the activation of Akt signaling and survival responses.
Abstract
Although TrkA postendocytic sorting significantly influences neuronal cell survival and differentiation, the molecular mechanism underlying TrkA receptor sorting in the recycling or degradation pathways remains poorly understood. Here we demonstrate that Golgi-localized, γ adaptin-ear–containing ADP ribosylation factor-binding protein 3 (GGA3) interacts directly with the TrkA cytoplasmic tail through an internal DXXLL motif and mediates the functional recycling of TrkA to the plasma membrane. We find that GGA3 depletion by siRNA delays TrkA recycling, accelerates TrkA degradation, attenuates sustained NGF-induced Akt activation, and reduces cell survival. We also show that GGA3’s effect on TrkA recycling is dependent on the activation of Arf6. This work identifies GGA3 as a key player in a novel DXXLL-mediated endosomal sorting machinery that targets TrkA to the plasma membrane, where it prolongs the activation of Akt signaling and survival responses.
INTRODUCTION
Nerve growth factor (NGF) plays critical roles in the development and maintenance of the vertebrate nervous system. NGF promotes neuronal survival and differentiation via binding to TrkA, which initiates receptor phosphorylation and activates downstream signal transduction cascades, including the Ras/mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K)/Akt, and phospholipase Cγ/protein kinase C (PKC) signaling pathways (Klesse et al., 1999; Pierchala et al., 2004; Zweifel et al., 2005). NGF also induces the ubiquitination and subsequent internalization of TrkA into endocytic vesicles (Geetha and Wooten, 2008). Once internalized into early endosomes, TrkA may either be recycled back to the plasma membrane (PM), leading to functional resensitization and prolongation of cell surface–specific signaling events, or sorted to the lysosomes for degradation, producing a prolonged attenuation of cellular signaling (Chen et al., 2005; Yu et al., 2011). Different biological responses to NGF are controlled by signaling from different intracellular locations. Neuronal differentiation is promoted by active TrkA localized to endosomes, whereas survival is accomplished by TrkA signaling at the cell surface (Zhang et al., 2000). Thus the process that directs the sorting of TrkA to diverse pathways has a significant effect on TrkA-mediated biological functions, and its alteration may lead to human diseases.
Endocytic trafficking of TrkA to degradative or recycling pathways is a complex and highly regulated process. A role for ubiquitin chains, proteasomes, and small GTPase Rab7 activity is critical in regulating TrkA delivery to lysosomes (Saxena et al., 2005a; Geetha and Wooten, 2008). In contrast, TrkA recycling to the cell surface requires a specific sequence located in the juxtamembrane region and the activity of the small GTPase Rab11 (Chen et al., 2005; Huang et al., 2013). However, in contrast to other receptor tyrosine kinases (RTKs), such as epidermal growth factor receptor (EGFR), the molecular mechanisms of TrkA endosomal sorting remain largely uncharacterized.
The Golgi-localized, γ adaptin-ear–containing ADP ribosylation factor–binding proteins (GGA1, GGA2, and GGA3) are a family of monomeric adaptors associated with the trans-Golgi network (TGN) and endosomes that function in the intracellular trafficking of receptors (Bonifacino, 2004). GGA proteins consist of four segments: a VHS domain that binds the acidic dileucine sorting signals (DXXLL, X = any amino acid) present in the cytoplasmic tail of various transmembrane proteins, such as mannose-6-phosphate receptor (MPR); a GAT domain, which interacts with the GTP-bound form of Arfs and ubiquitin; an unstructured hinge region, which binds clathrin; and a GAE domain, which interacts with a number of accessory factors (Bonifacino, 2004). The GGAs are recruited from the cytosol to membranes by Arf GTPase to promote the polymerization of clathrin for the sorting of cargo proteins (Bonifacino, 2004). GGAs are mainly localized at the TGN and are involved in the sorting and transport of receptors from the TGN to the endosome (Puertollano et al., 2001a). However, an additional pool of GGAs has also been localized on endosomes and is involved in endocytic sorting to the lysosome or PM. Indeed, GGA3 has been shown to be crucial for the sorting of EGFR and β-site amyloid precursor protein cleaving enzyme 1 (BACE1) to lysosomes through interactions with ubiquitin (Puertollano and Bonifacino, 2004; Kang et al., 2010) and for the recycling of the Met receptor to the PM through an interaction with the adaptor protein Crk (Parachoniak et al., 2011).
In this study, we investigated the role of GGAs in TrkA trafficking and signaling. We determined that GGA3 interacts directly with the TrkA cytoplasmic tail through an internal DXXLL motif and mediates TrkA functional recycling to the PM, sustained NGF-induced Akt activation, and cell survival. NGF/TrkA stimulation upregulates Arf6 activity via Src activation, which recruits GGA3 on endosomes to mediate TrkA recycling. These results identify a novel key role for GGA3 in TrkA recycling and signaling.
RESULTS
GGA3 interacts and colocalizes with TrkA on endosomes
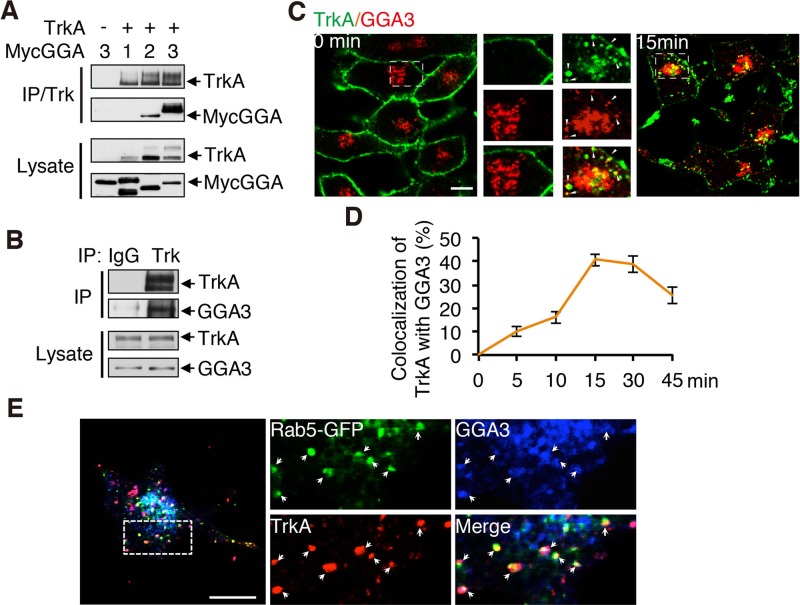
GGAs are found on endosomes and participate in the endocytic sorting of various tyrosine kinase receptors. To investigate the relevance of GGAs to TrkA trafficking, we first examined whether GGAs interact with TrkA using coimmunoprecipitation (coIP) assays of HEK293 cells expressing TrkA together with Myc-GGA1, Myc-GGA2, or Myc-GGA3. Among the three GGA proteins, GGA3 showed the strongest interaction with TrkA (Figure 1A). The binding capacity of TrkA with endogenous GGA3 was then confirmed by coIP experiments using the PC12 (615) cells stably expressing TrkA (Figure 1B; Hempstead et al., 1992). Using confocal microscopy, we next examined whether GGA3 and TrkA colocalized on endosomes. Serum-starved PC12 (615) cells were incubated at 4°C with anti-TrkA antibody (5C3) and then shifted to 37°C for different periods of time (0–45 min) to initiate endocytosis. 5C3 binds specifically to the ectodomain of TrkA and mimics NGF, inducing TrkA internalization and signaling (LeSauteur et al., 1996). Little colocalization was observed between GGA3 and TrkA in unstimulated cells (Figure 1C). However, in cells shifted to 37°C, colocalization in punctate patterns typical of early endosomes was observed between internalized TrkA and GGA3 (Figure 1C). Maximal colocalization was induced by 15 min of stimulation (Figure 1D), further suggesting that the codistribution of internalized TrkA with GGA3 occurs in early endosomes. Indeed, TrkA is present in EEA1- and transferrin-positive endosomes after 15 min of internalization (Supplemental Figure S1). Furthermore, a 1.9-fold increase in localization of GGA3 to early endosome was observed after 15 min of NGF stimulation (Supplemental Figure S2). To identify more precisely the sorting compartment in which TrkA and GGA3 colocalize, we compared their distribution with that of early endosome marker Rab5. After 15 min of internalization, cell surface–labeled TrkA (5C3) partially localized with GGA3 to rab5-labeled endosomes (Figure 1E), consistent with the recruitment of GGA3 to TrkA internalized in sorting endosomes. Taken together, these results support a possible role for GGA3 as an endocytic adaptor for TrkA.
FIGURE 1:
GGA3 interacts and colocalizes with TrkA on endosomes. (A) IP of lysates from HEK293T cells transfected with TrkA together with Myc-tagged GGA1, GGA2, or GGA3 using Trk antibody and immunoblotted as shown. (B) Confirmation of the interaction between TrkA and endogenous GGA3 by IP of lysates from PC12 (615) cells using TrkA antibody. (C) Confocal microscopy images comparing the distribution of endogenous GGA3 and cell surface–labeled TrkA receptors (5C3 antibodies) internalized for 0 or 15 min in PC12 (615) cells. Insets, regions of higher magnification. Arrowheads indicate colocalization between internalized TrkA and GGA3. Scale bar, 10 μm. (D) Quantification of the degree of overlap between GGA3 and TrkA internalized for different periods of time (as described in C). Manders coefficients were obtained using Olympus FluoView, version 1.6b, colocalization software after background subtraction (n = 30). (E) Confocal microscopy images comparing the distribution of endogenous GGA3, the early endosome marker Rab5, and cell surface–labeled TrkA receptors (5C3 antibodies) internalized for 15 min in PC12 (615) cells expressing GFP-tagged Rab5. Insets, regions of higher magnification; arrowheads indicate colocalization. Scale bar, 10 μm.
GGA3 is required for TrkA sorting to the recycling pathway
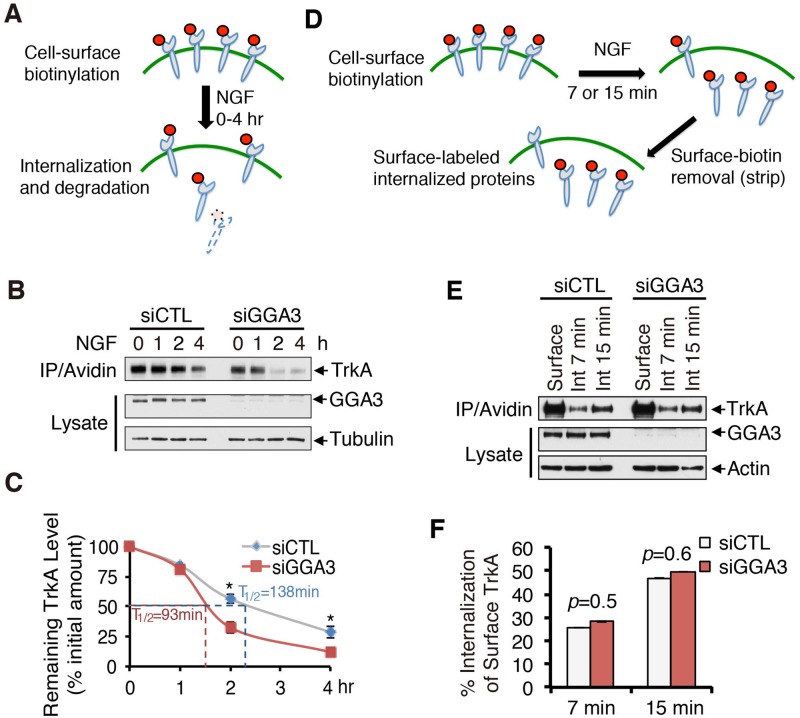
To investigate the functional role of GGA3 in TrkA trafficking, we examined the endocytic fate of TrkA in GGA3-depleted cells. We first compared the kinetics of TrkA degradation in PC12 (615) cells transfected with control or GGA3 small interfering RNA (siRNA) using the biotinylation assay schematized in Figure 2A. After cell-surface biotinylation, cells were treated with NGF for 0, 1, 2, or 4 h, lysed, pulled down with streptavidin beads, and immunoblotted for TrkA, allowing the assessment of proteolysis of endocytosed biotin-labeled TrkA receptors (Figure 2, A and B). A stronger reduction of the amount of biotin-labeled TrkA was detected in GGA3-depleted cells compared with control cells after 2 and 4 h of NGF stimulation (Figure 2B). Quantitative analysis indicated that the turnover of biotinylated TrkA increased by 33% in GGA3-depleted cells (T1/2 ≈ 93 min vs. T1/2 ≈ 138 min in control cells; Figure 2C). These results support the enhanced entry of TrkA into the degradative pathway in the absence of GGA3. This increase in TrkA degradation rate is not due to a lower initial amount of cell-surface TrkA, because quantitative analysis indicated that cell-surface TrkA levels under unstimulated conditions (NGF 0) were not significantly different in GGA3-depleted versus control cells (Supplemental Figure S3), suggesting that GGA3 depletion did not alter TrkA biosynthesis or constitutive trafficking of newly synthesized TrkA to the cell surface. In concordance, confocal microscopy analysis of cycloheximide chase experiments did not indicate any difference in TrkA intracellular distribution in control and GGA3-depleted cells (unpublished data), arguing against a role for GGA3 in TrkA transport from the TGN to the PM.
FIGURE 2:
GGA3 knockdown alters TrkA degradation but not its internalization. (A) Schematic of degradation assay. PC12 (615) cells were biotinylated at 4°C to label cell-surface proteins and stimulated with 10 ng/ml NGF for 0, 1, 2, or 4 h at 37°C to allow for internalization and degradation of biotinylated receptors. The remaining biotinylated proteins were collected with avidin and immunoblotted with TrkA antibodies. (B) Representative Western blots of the TrkA degradation assay performed in control and GGA3-depleted PC12 (615) cells. (C) Quantification of the degree of TrkA degradation from three independent experiments (as described in B). The amount of degraded TrkA is expressed as the percentage of the initial pool of cell surface–biotinylated TrkA (time 0). Student’s t test, *p < 0.05. (D) Schematic of internalization assay. PC12 (615) cells were biotinylated at 4°C to label cell-surface proteins and stimulated with NGF for 7 or 15 min at 37°C to allow for internalization. Any remaining biotin on cell-surface receptors was removed with glutathione treatment to assess only the internalized proteins and then collected with avidin and immunoblotted with TrkA antibodies. (E) Representative Western blots of the TrkA internalization assay performed in control and GGA3-depleted PC12 (615) cells. “Surface” refers to the total biotinylated cell-surface TrkA receptors in unstimulated cells not treated with glutathione; Int 7 min and Int 15 min refer to the internalized biotinylated receptors after stimulation with NGF for 7 and 15 min, respectively, and glutathione treatment. (F) Quantification of the degree of TrkA internalization from three independent experiments (as described in D and E). The amount of internalized TrkA is expressed as the percentage of the initial pool of cell-surface biotinylated TrkA (referred to as Surface in F). Student’s t test, *p < 0.05.
We hypothesized that the differences in degradation rates may be due to alterations in TrkA receptor trafficking at the initial internalization step and/or the endocytic sorting in the recycling pathway. Using a cleavable biotinylation assay, we first compared the internalization rate of TrkA in control and GGA3-depleted PC12 (615) cells. As outlined in Figure 2D, cells were surface labeled with sulfo-NHS-SS-biotin at 4°C, and internalization was initiated by incubating cells with NGF for 7 and 15 min at 37°C. The cells were next treated with glutathione, which cleaves biotin from proteins at the PM, allowing selective isolation of internalized biotinylated receptors that remained protected from cleavage. No obvious changes in the TrkA internalization ratio were observed in GGA3-knockdown cells (Figure 2, E and F), suggesting that GGA3 does not regulate the internalization rate of TrkA in response to NGF.
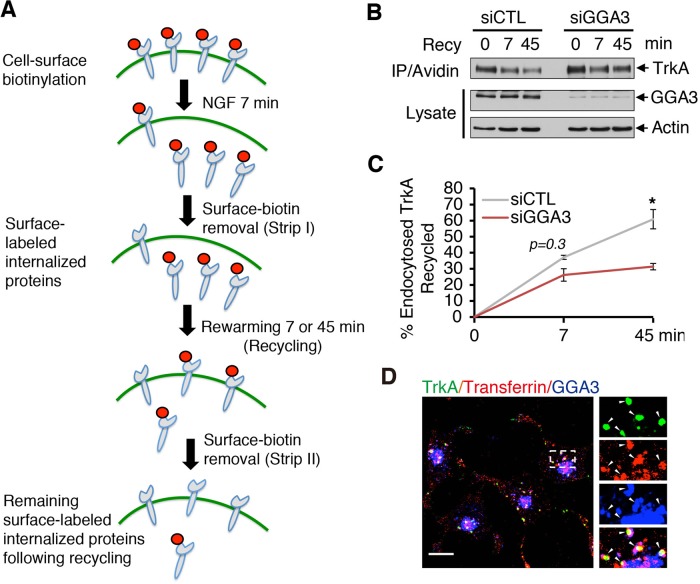
To assess whether GGA3 participated in the postendocytic recycling of TrkA, we performed a cleavable biotinylation assay (schematized in Figure 3A) in which the biotin-labeled cell surface receptors were internalized after 7 min of NGF treatment, followed by stripping with glutathione. The cells were then returned to 37°C for 7 or 45 min to allow for recycling, and any reappearing cell surface–biotinylated receptors were stripped again (Figure 3A). The TrkA signal lost in the second stripping procedure was considered the fraction of receptors that had been recycled. In GGA3-depleted cells, only 31% of TrkA had returned to the PM by 45 min, whereas in control cells, >60% had returned (Figure 3, B and C). However, GGA3 knockdown did not significantly alter the TrkA recycling ratio at 7 min (Figure 3C), a time point representing rapid recycling (Goh and Sorkin, 2013). Thus these biotinylation data indicate that GGA3 knockdown leads to slower recycling of TrkA, suggesting that in the absence of GGA3, TrkA traverses the endosomal sorting pathway to lysosomes.
FIGURE 3:
GGA3 is required for TrkA recycling. (A) Schematic of internalization assay. PC12 (615) cells were biotinylated at 4°C and stimulated with NGF for 7 min at 37°C to allow for internalization. After glutathione stripping of the remaining cell-surface biotin, cells were reincubated at 37°C for another 7 or 45 min to allow for the recycling of internalized receptors. Cells were then stripped again with glutathione to remove biotin from the surface-exposed biotinylated receptors recycled back to the cell surface. The remaining biotinylated proteins were then collected with avidin and immunoblotted with TrkA antibodies. The TrkA signal lost in the second stripping procedure was considered the fraction of recycled receptors. (B) Representative Western blots of the TrkA recycling assay performed in control and GGA3-depleted PC12 (615) cells. Recy 0 refers to the internalized biotinylated receptors before rewarming; Recy 7 and 45 refer to the remaining biotinylated receptors after rewarming. (C) Quantification of the degree of TrkA recycling from three independent experiments (as described in A and B). The amount of recycled TrkA is expressed as the percentage of the pool of biotinylated TrkA following the 7-min internalization period and before rewarming. Student’s t test, *p < 0.05. (D) Confocal microscopy images comparing the distribution of endogenous GGA3 with that of cell surface–labeled TrkA (5C3 antibodies) and transferrin receptor (Alexa Fluor 594–conjugated transferrin) internalized for 15 min in PC12 (615) cells. Insets, regions of higher magnification; arrowheads indicate colocalization. Scale bar, 10 μm.
Taken together, our results support a role for GGA3 as an adaptor involved in TrkA recycling. In concordance, GGA3 localized on vesicles labeled with both internalized TrkA and transferrin after 15 min of incubation at 37°C (Figure 3D), consistent with the recruitment of GGA3 to TrkA internalized in recycling endosomes.
GGA3 mediates TrkA recycling through direct binding to TrkA internal DXXLL motifs
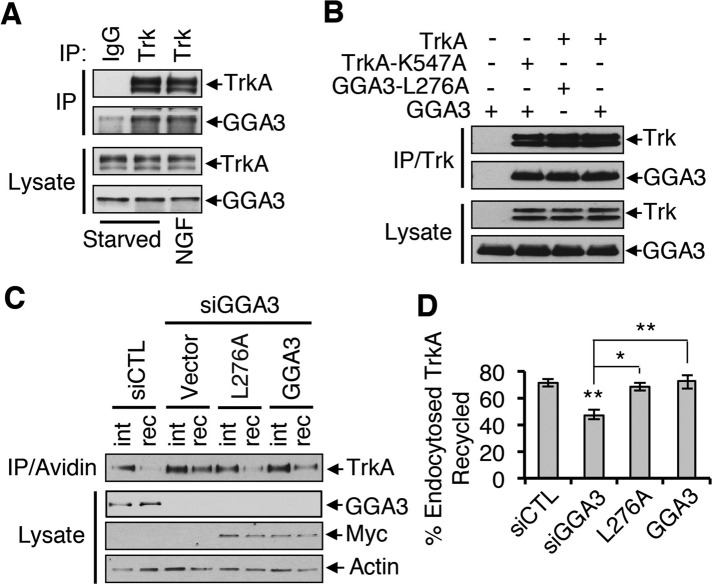
To establish the mechanism by which GGA3 regulates TrkA recycling, we characterized the interaction between TrkA and GGA3. Given that TrkA is ubiquitinated after NGF stimulation (Arevalo et al., 2006; Geetha and Wooten, 2008) and that GGA3 binds to ubiquitin and is involved in endosomal sorting of ubiquitinated cargo (Puertollano and Bonifacino, 2004; Kang et al., 2010), we first examined whether NGF treatment (10 min) increases the amount of endogenous GGA3 that associates with TrkA in PC12 (615) cells. The amount of TrkA that coimmunoprecipitated with GGA3 from NGF-treated cells was comparable to that from untreated cells (Figure 4A), suggesting that the interaction between TrkA and GGA3 is independent of TrkA activation and ubiquitination. This result was validated in two different manners. First, we observed interactions between GGA3 and a kinase-dead TrkA mutant (TrkA-K547A; Meakin et al., 1999) and between TrkA and the GGA3-L276A mutant, which cannot bind ubiquitin (Puertollano and Bonifacino, 2004; Figure 4B). Second, after reintroduction of GGA3-L276A into GGA3-depleted cells with an siRNA-resistant form of GGA3-L276A, the level of TrkA recycling was similar to that in cells rescued with siRNA-resistant wild-type GGA3 or in control cells (Figure 4, C and D), confirming that the ability of GGA3 to bind ubiquitin is not required for endosomal sorting of TrkA.
FIGURE 4:
The interaction between GGA3 and TrkA is independent of TrkA activation and ubiquitination. (A) Interaction of TrkA and GGA3 in PC12 (615) cells incubated with or without 10 ng/ml NGF for 10 min. Lysates were immunoprecipitated with GGA3 antibody and immunoblotted as shown. (B) Immunoprecipitation of TrkA from HEK293T cells transfected with a TrkA wild type, TrkA kinase-dead mutant (TrkA-K547A), GGA3 wild type, or GGA3 mutant unable to bind ubiquitin (GGA3-L276A) and immunoblotted as shown. (C) Analysis of TrkA recycling in GGA3-depleted PC12 (615) cells rescued with control cDNA (vector) or siRNA-resistant GGA3 wild type or mutant GGA3-L276A. Recycling assays were performed as described in Figure 3A. int, internalized biotinylated TrkA before rewarming; rec, remaining biotinylated receptors after 45 min of rewarming (recycling). (D) Quantification of the degree of TrkA recycling from three independent experiments (as described in C). The amount of recycled TrkA is expressed as the percentage of the pool of biotinylated TrkA after the 7-min internalization period and before rewarming. One-way ANOVA, *p < 0.05, **p < 0.01.
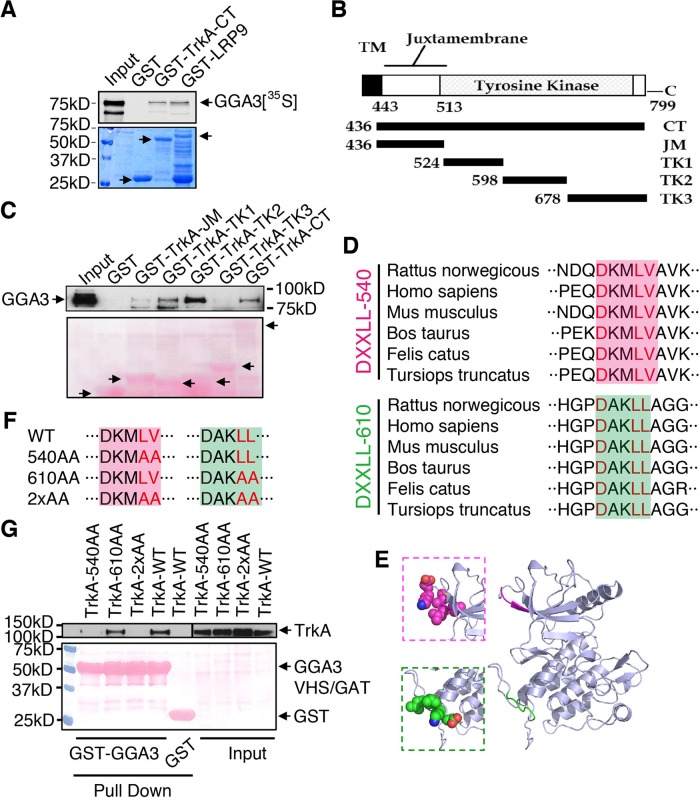
Previously identified GGA cargoes, such as MPR or LRP9, contain a consensus acidic cluster dileucine motif (DXXLL) at the extreme C-terminus of the cytoplasmic tail that directly interacts with the VHS domain of GGAs (Bonifacino, 2004; Boucher et al., 2008). Although TrkA lacks a putative DXXLL motif at the extreme C-terminus of its cytoplasmic tail, we next used an in vitro pull-down assay to investigate whether GGA3 binds directly to the C-terminus of TrkA. The 35S-labeled, in vitro–translated GGA3 bound to the glutathione S-transferase (GST)–TrkA-cytoplasmic tail in a similar way as to the GST-LRP9-cytoplasmic tail, which was used as a positive control (Figure 5A). In parallel, we confirmed the specificity of the GGA3 interaction, because neither GGA1 nor GGA2 interacted directly with TrkA (Supplemental Figure S4). To identify the specific region within TrkA that is required for the interaction with GGA3, we generated several TrkA C-terminal tail fragments (Figure 5B), and assessed their ability to interact with GGA3 using pull-down binding assays. GST-TrkA-TK1 and TK2 pulled down endogenous GGA3 from PC12 (615) cell lysate (Figure 5C). In contrast, no interaction with GGA3 was detected for GST-TrkA-JM, TK3, or GST alone (Figure 5C). Unexpectedly, we found that the two TrkA fragments that bound to GGA3, TK1 and TK2, contained putative DXXLL motifs (DKMLV in TK1, named DXXLL-540; DAKLL in TK2, named DXXLL-610; Figure 5D). Alignment of the amino acid sequences of TrkA C-terminal tails from several different mammals indicated that these two DXXLL motifs were conserved in several species (Figure 5D). To investigate the accessibility of both motifs for interaction with the GGA-VHS domain, we generated a homology model of the three-dimensional (3D) structure of the TrkA cytoplasmic tail (Figure 5E). According to the predicted 3D structure, the DXXLL-540 and the DXXLL-610 motifs are both exposed on the protein surface, but the aspartate of DXXLL-610 might be less accessible for interaction (Figure 5E).
FIGURE 5:
An internal DXXLL motif in the TrkA cytoplasmic domain is responsible for GGA3 binding. (A) GST pull-down experiment demonstrating that GGA3 interacts directly with TrkA. In vitro–translated, 35S-labeled GGA3 bound to GST-TrkA or LRP9 cytoplasmic tails. Bound proteins were resolved by SDS–PAGE and detected by autoradiography. The 5% input is shown. Bottom, Coomassie staining showing GST-tagged proteins. (B) Schematic diagram of TrkA fragments used for the pull-down assays. CT, C-terminus; JM, juxtamembrane region, TK, tyrosine kinase domain. (C) The deletion mutants shown in B were used to pull down endogenous GGA3 from PC12 (615) cell lysates. Bound proteins were detected with anti-GGA3 antibodies. Bottom, Ponceau staining showing GST-tagged proteins. (D) Alignment of two putative DXXLL motifs (DXXLL-540 and DXXLL-610) that are conserved in various mammalian TrkA cytoplasmic tails. (E) Computational modeling of the 3D structure of the TrkA cytoplasmic tail, showing the predicted locations of the DXXLL-540 (pink) and DXXLL-610 (green) motifs. Insets, atomic representations of the motifs. (F) Point mutations inserted into the DXXLL-540 and DXXLL-610 motifs. (G) The TrkA mutants shown in F were expressed in HEK293 cells, which were submitted to pull-down assays using GST or GST-GGA3 (VHS/GAT). Bound proteins were detected with anti-TrkA antibodies. Bottom, Ponceau staining showing GST-tagged proteins.
Because an inactivating mutation of the dileucine residues of the DXXLL sequence was previously reported to abrogate its interaction with GGA proteins (Puertollano et al., 2001a; Boucher et al., 2008), alanines were substituted for Leu-543 and Val-544 in the DXXLL-540 motif and for Leu-613 and Leu-614 in the DXXLL-610 motif or in both motifs (Figure 5F). Pull-down assays indicated that the interaction between TrkA and GGA3 was not affected by mutation of the DXXLL-610 motif (TrkA-610AA); however, the interaction was completely abolished by the DXXLL-540 (TrkA-540AA) or double DXXLL (TrkA-2xAA) mutations (Figure 5G). These results suggest that the DXXLL-540 motif is the key element contributing to the interaction between TrkA and GGA3 proteins.
Arf6 is required for GGA3-mediated TrkA recycling
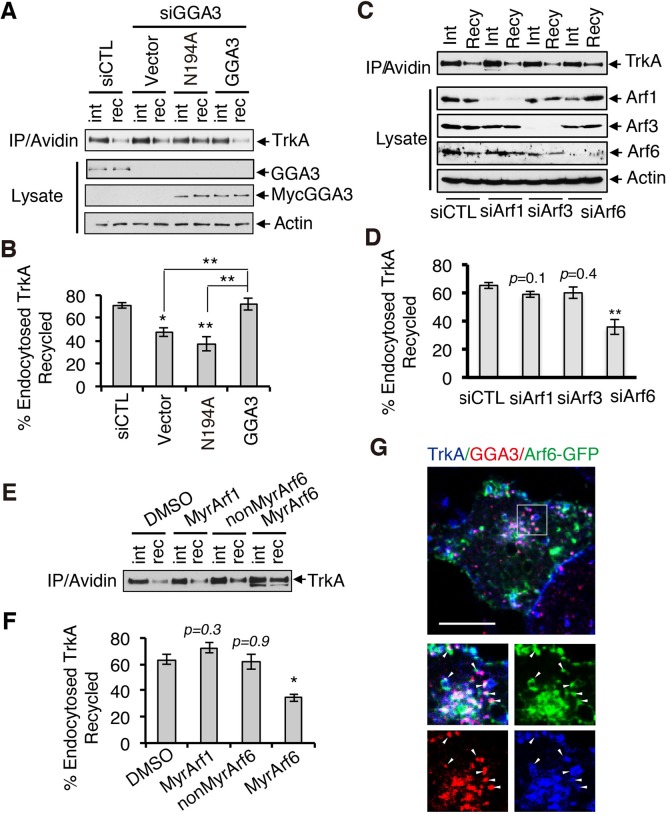
Having found that GGA3 binds directly to TrkA, we next investigated the mechanism by which GGA3 regulates the sorting of TrkA into the recycling pathway. Given that GGAs are recruited to the membrane by activated Arfs, we then examined the role of Arfs in GGA3-mediated TrkA recycling by rescue experiments using GGA3 N194A, a mutant that specifically uncouples GGA from interactions with Arf-GTP proteins (Puertollano et al., 2001b). Expression of siRNA-resistant GGA3, but not GGA3 N194A, restored TrkA recycling to the PM in GGA3- depleted PC12 (615) cells (Figure 6, A and B), suggesting that an interaction between GGA3 and Arf-GTP is required for the sorting of TrkA to the recycling pathway.
FIGURE 6:
GGA3-mediated TrkA recycling requires Arf6. (A) Analysis of TrkA recycling in GGA3-depleted PC12 (615) cells rescued with control cDNA (vector) or siRNA-resistant GGA3 wild type or GGA3 mutant unable to bind Arf-GTP (GGA3-N194A). Recycling assays were performed as described in Figure 3A. int, internalized biotinylated TrkA before rewarming; rec, remaining biotinylated receptors after 45 min of rewarming (recycling). (B) Quantification of the degree of TrkA recycling from three independent experiments (as described in A). The amount of recycled TrkA is expressed as the percentage of the pool of biotinylated TrkA after the 7-min internalization period and before rewarming. One-way ANOVA, *p < 0.05, **p < 0.01. (C) Analysis of TrkA recycling in PC12 (615) cells treated with control, Arf1, Arf3, and Arf6 siRNA. Recycling assays were performed as described in Figure 3A. int, internalized biotinylated TrkA before rewarming; rec, remaining biotinylated receptors after 45 min of rewarming (recycling). (D) Quantification of the degree of TrkA recycling from three independent experiments (as described in C). The amount of recycled TrkA is expressed as the percentage of the pool of biotinylated TrkA after the 7-min internalization period and before rewarming. One-way ANOVA, **p < 0.01. (E) Analysis of TrkA recycling in PC12 (615) cells treated with dimethyl sulfoxide (vehicle), MyrArf1, MyrArf6, and NonMyrArf6 (nonpermeant Arf6 peptide control). Recycling assays were performed as described in Figure 3A. int, internalized biotinylated TrkA before rewarming; rec, remaining biotinylated receptors after 45 min of rewarming (recycling). (F) Quantification of the effects of MyrArf1, MyrArf6, and NonMyrArf6 peptides on the degree of TrkA recycling from three independent experiments (as described in A). One-way ANOVA, *p < 0.05. (G) Confocal microscopy images comparing the distribution of Arf6-GFP, endogenous GGA3, and cell surface–labeled TrkA receptors (5C3 antibodies) internalized for 15 min in PC12 (615) cells. Insets, regions of higher magnification; arrowheads indicate TrkA-labeled vesicles. Scale bar, 10 μm.
Of the various Arf family members, Arf1, Arf3, and Arf6 are the best characterized, and all have been associated with the endocytic recycling pathway (Parachoniak et al., 2011; Kondo et al., 2012). We thus investigated whether these three Arf proteins participate in TrkA recycling, using specific siRNA depletion of Arf1, Arf3, and Arf6 (Figure 6C). Arf6 depletion decreased TrkA recycling by 29%. In contrast, Arf1 and Arf3 depletion did not significantly alter TrkA recycling (Figure 6D). The specific involvement of Arf6 activity in postendocytic TrkA recycling was confirmed using synthetic N-terminal Arf peptides that have been reported to block Arf activity (Figure 6E). The myristoylated peptides MyrArf1 and MyrArf6 are cell permeant and have been shown to inhibit the activation of Arf1 and Arf6, respectively (Kahn et al., 1992; Caumont et al., 1998; Choi et al., 2006). We found that MyrArf6 specifically inhibited the recycling of TrkA (Figure 6, E and F). In contrast, MyrArf1 and the nonmyristoylated Arf6 (NonMyrArf6) peptide, which served as a nonpermeant Arf6 peptide control, did not affect TrkA recycling (Figure 6, E and F). The concentration-dependent inhibitory effect of the MyrArf6 peptide on Arf6 activation (Arf6-GTP) was confirmed (Supplemental Figure S5). In summary, MyrArf6 not only confirmed the role of Arf6 in TrkA recycling, as shown using Arf6 siRNA, but also revealed the importance of Arf6 activation in TrkA recycling. Furthermore, supporting a role for Arf6 in TrkA recycling, cell surface–labeled TrkA did not colocalize with Arf1 or Arf3 (Supplemental Figure S6) but colocalized with both Arf6 and GGA3 after 15 min of internalization (Figure 6G).
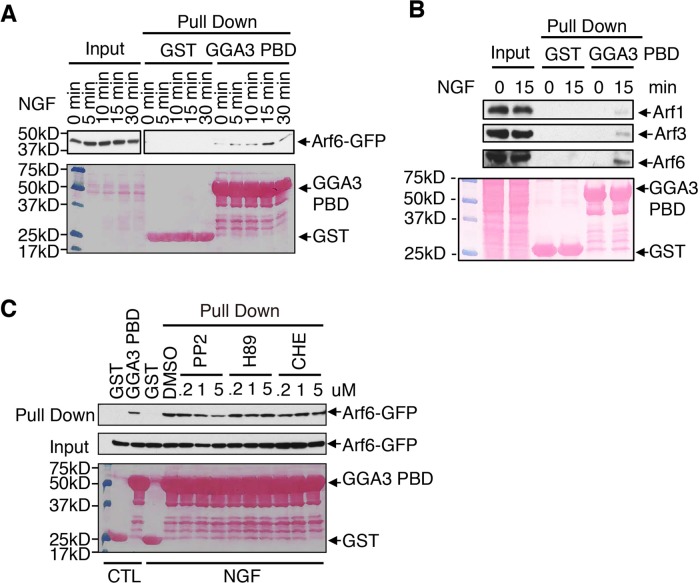
Arf6 activity can be regulated after the stimulation of cell surface receptors (Palacios and D’Souza-Schorey, 2003; Zhu et al., 2012). We therefore investigated whether NGF-induced TrkA could activate Arf6, using GST-GGA3 PBD pull-down assays that specifically bound the GTP-bound (active) form of Arf (Hafner et al., 2006). On stimulation with NGF, the levels of active GFP-tagged Arf6 (Arf6-GTP) were increased, with peak activity at 15 min (Figure 7A). Of interest, this time point coincided with TrkA localization at sorting endosomes (Figure 1C). To avoid the potential artifactual effects of the GFP tag on Arf6 activation, we analyzed the activation status of endogenous Arf6 in PC12 (615) cells after TrkA stimulation (Figure 7B). As observed for Arf6-GFP, the endogenous Arf6-GTP level increased upon stimulation with NGF for 15 min (Figure 7B). Endogenous Arf1 and Arf3 were also activated upon stimulation with NGF but at a much lower level than Arf6 (Figure 7B). These results suggest that Arf6 is activated in response to NGF stimulation in PC12 (615) cells and that Arf6 activity is required for TrkA sorting to the recycling pathway from early endosomes.
FIGURE 7:
Arf6 is activated in response to NGF stimulation. (A) Analysis of activation of Arf6-GFP after TrkA stimulation. Lysates from PC12 (615) cells expressing Arf6-GFP and stimulated with NGF for the indicated periods of time were incubated with GST or GST-GGA3 PBD, which specifically binds the GTP-bound form of Arf proteins. Bound proteins were analyzed by immunoblotting with anti-GFP antibodies. Bottom, Ponceau staining showing GST-tagged proteins. (B) Comparison of endogenous Arf1, Arf3, and Arf6 activation in response to NGF. Lysates from PC12 (615) cells stimulated with NGF for 15 min were incubated with GST or GST-GGA3 PBD. Bound proteins were analyzed by immunoblotting with anti-Arf1, Arf3, and Arf6 antibodies. The 5% input is shown. Bottom, Ponceau staining showing GST-tagged proteins. (C) Effect of Src, PKC, and PKA inhibitors on NGF-mediated Arf6 activation. The GST-GGA3 PBD pull-down assay were performed as described in A, except that the cells were preincubated for 1 h with specific inhibitors for Src (PP2), PKC (CHE), or PKA (H89) before NGF stimulation for 15 min. Bottom, Ponceau staining showing GST-tagged proteins.
To further understand the underlying mechanism by which TrkA activates Arf6, we examined the involvement of cytoplasmic kinases Src, PKC, and PKA, known downstream effectors of TrkA (Wooten et al., 2001; Pierchala et al., 2004; Herbst et al., 2011), on the activation of Arf6. PC12 (615) cells preincubated with 0.2, 1, or 2 μM of specific inhibitors for Src (PP2), PKC (CHE), or PKA (H89) were stimulated with NGF for 15 min, and the activity of Arf6 was assessed using GST-GGA3 PBD pull-down assays (Figure 7C). The inhibition of Src blocked the activation of Arf6 in a concentration-dependent manner, whereas the inhibitors of PKC and PKA had no effect (Figure 7C). Taken together, these data suggest that NGF stimulation up-regulates Arf6 activity via Src activation, which then recruits GGA3 from the cytosol to mediate TrkA recycling.
GGA3 knockdown attenuates TrkA-dependent Akt signaling and cell survival
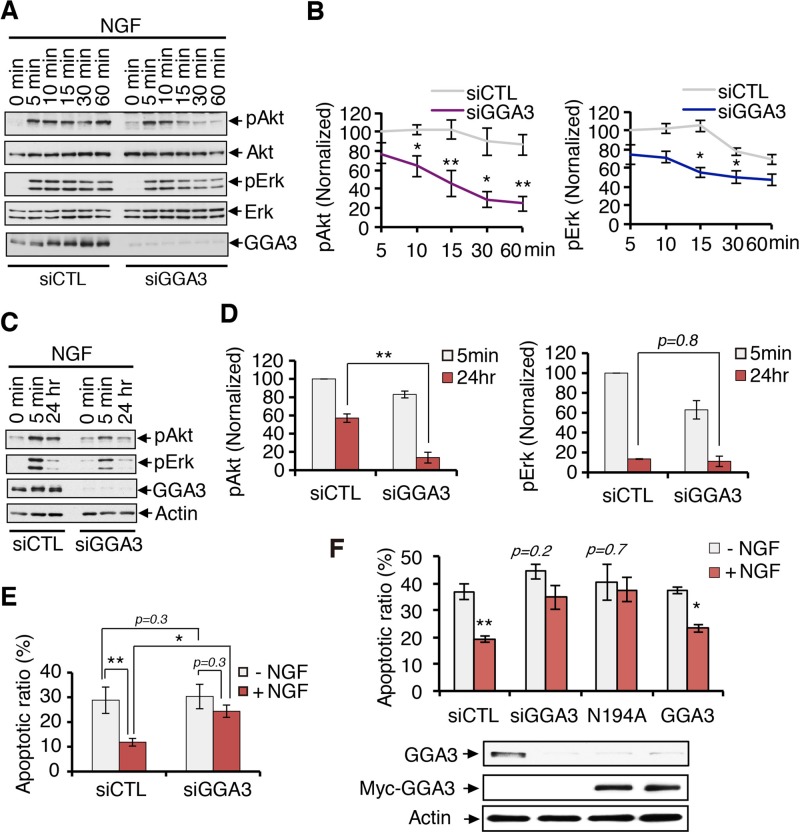
TrkA activates several signaling pathways required for the survival, differentiation, and maintenance of neurons. Therefore, the alteration of TrkA recycling by GGA3 knockdown may affect its downstream signaling. We thus examined its effect on the phosphorylation of Erk1/2 and Akt in response to NGF as readouts for MAPK and PI3K pathway activation, respectively (Figure 8A). GGA3 depletion attenuated the NGF signaling of both Akt and Erk1/2, with a stronger effect on Akt signaling (Figure 8A). Quantification indicated a 22% reduction of Erk1/2 activation and a 61% reduction of Akt activation in GGA3-knockdown cells compared with control cells after 60 min of stimulation with NGF (Figure 8B). These results suggest that GGA3 depletion may affect the sustained activation of TrkA-mediated signaling, which was confirmed by testing the levels of phosphorylated Akt and Erk1/2 using an established paradigm of prolonged 24-h stimulation with NGF (Figure 8C). In GGA3-knockdown cells, the level of phosphorylated Akt after 24 h of NGF stimulation was fourfold lower than that in control cells, whereas no significant difference in the level of phosphorylated Erk was observed (Figure 8D), indicating that GGA3 is involved in sustained Akt signaling downstream of TrkA. Because sustained Akt signaling is a prerequisite for NGF-induced cell survival, we next examined the effect of GGA3 depletion on the survival of PC12 (615) cells. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays were used to assess the proportions of apoptotic cells in nontransfected cells and cells treated with control siRNA or GGA3 siRNA after 24 h of incubation in the absence or presence of NGF (Figure 8E and Supplemental Figure S7). Although NGF treatment reduced the level of apoptosis in control cells by 60%, it did not prevent apoptosis of GGA3-depleted cells (Figure 8E). These data indicate that GGA3 activity is involved in NGF-dependent survival. Furthermore, the cell-surface TrkA levels were lower in GGA3-depleted cells then in control cells after 24 h of NGF stimulation (Supplemental Figure S8), which is consistent with previous findings showing that TrkA receptors that are localized at the cell surface after prolonged neurotrophin treatment elicit survival signaling (Zhang et al., 2000; Chen et al., 2005). Moreover, supporting a requirement for Arf in the recruitment of GGA3 for TrkA recycling and cell survival, expression of GGA3 N194A was unable to compensate for GGA3 depletion in preventing apoptosis in response to NGF (Figure 8F). These data thus demonstrate a requirement for Arf-GTP in GGA3-mediated TrkA recycling to the PM, sustained Akt activation, and cell survival.
FIGURE 8:
GGA3 knockdown alters NGF-induced, sustained Akt phosphorylation and cell survival. (A) Analysis of Akt and Erk phosphorylation in control and GGA3-depleted PC12 (615) cells stimulated with 10 ng/ml NGF for 0–60 min. Cells were collected with Laemmli buffer and immunoblotted as shown. (B) Quantification of the levels of phosphorylated Akt and Erk from three independent experiments (as described in A). The amounts of pAkt and pErk were normalized to total Akt and Erk levels, respectively. Student’s t test, *p < 0.05, **p < 0.01. (C) Analysis of Akt and Erk phosphorylation in control and GGA3-depleted PC12 (615) cells after 24 h stimulation with or without NGF. (D) Quantification of the levels of phosphorylated Akt and Erk from three independent experiments (as described in C). The amounts of pAkt and pErk were normalized to actin. One-way ANOVA, **p < 0.01. (E) Quantification of the level of apoptotic cells using TUNEL assays on control and GGA3-depleted PC12 (615) cells incubated with or without 10 ng/ml NGF for 24 h. The values are the means ± SEMs of three independent experiments. One-way ANOVA, *p < 0.05, **p < 0.01. (F) Quantification of the level of apoptotic cells in GGA3-depleted PC12 (615) cells rescued with control cDNA or siRNA-resistant GGA3 wild type or GGA3-N194A. The TUNEL assays are described in Figure 6E. Western blots confirmed the depletion of endogenous GGA3 and the expression of the siRNA-resistant form of GGA3 in the PC12 (615 cells). One-way ANOVA, *p < 0.05, **p < 0.01.
DISCUSSION
After NGF stimulation, internalized TrkA may either recycle to the PM or enter the degradative lysosomal pathway; however, the mechanisms that regulate entry of TrkA into these particular pathways are still poorly characterized. Our work demonstrates a key role for GGA3 in TrkA receptor recycling and signaling that promotes cell survival. We found that GGA3 depletion delays TrkA recycling and accelerates TrkA degradation. We also showed that GGA3 interacts directly with an internal DXXLL motif in the cytoplasmic tail of TrkA and colocalizes with TrkA and Arf6 in sorting endosomes. Furthermore, we observed that Arf6 is required for GGA3-mediated TrkA recycling and that Arf6 is activated by Src, a downstream effector of NGF-activated TrkA. On the basis of these findings, we propose the following model (Figure 9) for the function of GGA3 in TrkA endosomal recycling. After NGF stimulation, TrkA is internalized in early endosomes. In parallel, Arf6 is activated by TrkA through the Src kinase and recruits GGA3 to the endosomal membrane, where GGA3 directly binds to the DXXLL motif in TrkA. This interaction promotes the sorting of TrkA into the recycling pathway while decreasing its entry into the degradative pathway. The GGA3-dependent delivery of TrkA to the PM would then promote the cellular response to NGF, leading to sustained Akt activation and increased neuronal survival.
FIGURE 9:
Model for the GGA3-mediated recycling of TrkA. After NGF stimulation, TrkA is internalized in early endosomes. In parallel, Arf6 is activated by TrkA through Src and recruits GGA3 to the endosomal membrane, where GGA3 directly binds to a DXXLL motif in TrkA or aids in retaining a GGA3-TrkA complex in endosomal recycling membrane. This interaction promotes the sorting of TrkA to the recycling pathway, leading to sustained activation of Akt and cell survival.
GGA3 was first described as an adaptor protein that was localized at the TGN and involved in the sorting and transport of receptors, such as MPR, from the TGN to endosomes (Dell’Angelica et al., 2000). However, consistent with our results, accumulating evidence indicates an additional pool of GGAs that is localized to endosomes and involved in endocytic sorting of receptors either to the lysosome or the PM. GGA3 has been shown to be present on early endosomes and involved in EGFR degradation (Puertollano and Bonifacino, 2004). GGA3 has also been localized to Rab4-enriched endosomes, where it promotes the fast recycling of the Met receptor (Parachoniak et al., 2011). We observed a significant overlap of GGA3 with EEA1- or transferrin-labeled endosomes, consistent with its assignment as a recycling adaptor for TrkA. In contrast with Met GGA3–dependent recycling, our quantification data indicated that GGA3 is involved in the slow-recycling pathway of TrkA, which supports the fact that Trk receptors traffic through the slow Rab11-dependent endosomal recycling pathways in PC12 cells and neurons (Chen et al., 2005; Huang et al., 2013). The participation of GGA3 in both slow and rapid recycling of receptors may be due to the different methods by which GGA3 interacts with these receptors (direct/indirect interactions) or the involvement of other factors.
GGA3 controls postendocytic trafficking of transmembrane proteins through multiple mechanisms. GGA3 was found to interact with ubiquitin and be involved in the endosomal sorting of ubiquitinated receptors, such as EGFR and BACE1, to lysosomes (Puertollano and Bonifacino, 2004; Kang et al., 2010). It was also involved in the endosomal sorting of Met to the recycling pathway through an interaction with Crk (Parachoniak et al., 2011). Here we demonstrate that the direct interaction of GGA3 with a DXXLL motif in the cytoplasmic tail of TrkA controls the endosomal recycling of TrkA. Although the DXXLL GGA-binding motif is traditionally involved in anterograde transport from the TGN to the endosome, some studies reported its potential involvement in endosome-to-TGN retrograde transport (Wahle et al., 2005; Boucher et al., 2008). However, this work is the first evidence that DXXLL GGA-binding motifs may serve as recycling sorting signals. Of interest, the DXXLL motif found in TrkA is conserved among TrkA, TrkB, and TrkC, all of which can interact, with different affinity, with GGA3 (Supplemental Figure S9). Given that Trk receptors are known to recycle at different levels (Chen et al., 2005), the DXXLL GGA3-binding motif in their cytoplasmic domains may serve as a common recycling sorting signal, whereas other mechanisms, such as the previously identified TrkA juxtamembrane recycling motif (Chen et al., 2005), may regulate their recycling efficiency. In summary, even if the association of GGA3 with TrkA differs from its association with Met, this interaction supports a novel, GGA3-dependent step in receptor endosomal sorting to the recycling pathway.
DXXLL-type GGA sorting motifs were originally identified as situated one or two residues from the carboxy termini of transmembrane cargo proteins (Bonifacino, 2004). However, subsequent studies revealed that internal DXXLL sorting motifs also occur within the N- or C-terminal cytoplasmic domains of cargo molecules. Indeed, functional DXXLL sorting motifs were found situated 24 and 87 residues from the C-termini of LRP9 and LRP12 transmembrane receptors, as well as in the hinge regions of GGA1 and GGA3, which serve to autoregulate GGA function (Boucher et al., 2008; Doray et al., 2012). In this study, we identified two potential internal DXXLL motifs in the TrkA cytoplasmic tail, one situated 260 residues upstream of the extreme end of the cytoplasmic tail (DXXLL-540), and the other situated 190 residues upstream of the tail (DXXLL-610). However, substitution of LL with AA in these two DXXLL motifs indicated that only the DXXLL-540 motif was a functional site for GGA3 binding to TrkA. On analysis of the predicted 3D structure of TrkA cytoplasmic tail, both DXXLL motifs are exposed at the surface of the cytoplasmic domain of TrkA and could theoretically engage interaction with GGA3, but the aspartate of the DXXLL-610 motif seems slightly less accessible for interaction. Furthermore, the nature of the amino acids surrounding the internal DXXLL motif might also influence whether TrkA interacts with GGAs. Indeed, the importance of acidic residues immediately upstream of the DXXLL motif has been documented (Puertollano et al., 2001a; Kato et al., 2002; Doray et al., 2012). Of interest, an acidic residue (Asp-538; conserved in several species; Figure 5D) is found two residues upstream of DXXLL-540, but no acidic residue is observed upstream of DXXLL-610. Hence, although DXXLL-610 is accessible for interaction in the TrkA-TK2 fragment, the absence of interaction of this motif when located in the complete TrkA cytoplasmic tail might be explained by its reduced accessibility or the lack of appropriate surrounding residues. Of interest, we also noted that the cytoplasmic tail of TrkA preferentially binds GGA3 rather than GGA1 or GGA2. Significant differences in the relative affinity of DXXLL motifs for different GGAs have also been previously reported (Puertollano et al., 2001a; Boucher et al., 2008). It has been suggested that residues immediately upstream and downstream from the DXXLL motif, as well as the XX residues of this motif, may significantly affect the specificity of the interaction of GGA proteins with these motifs (Puertollano et al., 2001a; Doray et al., 2002; He et al., 2003). The level of autoinhibition of GGA1 and GGA3 in rat PC12 cells could also impair their binding to TrkA. Thus our findings not only identify a novel example of an internal DXXLL motif that specifically binds GGA3, they also reveal for the first time the presence of a functional DXXLL sorting motif in a RTK. Investigation of the importance of the residues surrounding the DXXLL-540 motif should provide new insights into the selectivity of specific interactions with GGA proteins and trafficking mechanisms associated with DXXLL-mediated sorting signals.
GGA3 can bind both ubiquitin and DXXLL motifs and can sort endosomal proteins into both degradation and recycling pathways. Given that TrkA is ubiquitinated after ligand binding, one can wonder how GGA3 preferentially uses the DXXLL motif to target TrkA to the recycling pathway. Phosphorylation has been shown to be a key factor in the regulation of GGA function toward different sorting pathways. For instance, phosphorylation of serine residues adjacent to or within the DXXLL signals of the MPRs and BACE1 increases the affinity of these signals for the VHS domain of the GGAs (Kato et al., 2002; He et al., 2003). Furthermore, the phosphorylation of GGAs themselves can regulate their function. GGA3 has been reported to be phosphorylated at S368 of the hinge region after EGFR stimulation, resulting in impaired interaction with ubiquitin without changes in its capacity to bind the DXXLL motif (Kametaka et al., 2005). Because there is no serine near the DXXLL-540 motif, it would be appropriate to examine whether NGF stimulation leads to the phosphorylation of GGA3 and changes its binding capacity to ubiquitin or the DXXLL-540 motif. The latter is less plausible because NGF stimulation did not alter GGA3–TrkA interactions. At first glance, this observation seems inconsistent with our model (Figure 9). By using IP assays, we observed that TrkA signaling activity was dispensable for the interaction with GGA3. In fact, we showed that TrkA interacted directly with GGA3 via an internal DXXLL motif. This observation would explain why the activation and subsequent phosphorylation/ubiquitination of TrkA were not needed for GGA3 association. Indeed, the solubilization of the proteins for the IP assay would allow the interaction of GGA3 with the DXXLL motif present on unstimulated/stimulated TrkA. Nevertheless, in the IF assay, the colocalization between TrkA and GGA3 was mainly observed in the endosomal compartment, suggesting that the interaction between GGA3 and internalized TrkA mainly occurs in this compartment in intact cells. According to the current sorting model for GGA proteins (Puertollano and Bonifacino, 2004), membrane-tethered Arf-GTP binds to the GAT domain of GGAs, which results in the recruitment of GGA to the membrane. The VHS domain of GGA binds DXXLL-type signals in the tails of receptors and other transmembrane cargo. However, the order of these steps has not been established. Of interest, we determined that the effect of GGA3 on TrkA sorting in the recycling pathway was dependent on Arf6. Arf6 is localized on endosomes and was previously associated with GGA3-dependent recycling of Met (Parachoniak et al., 2011). We showed that NGF/TrkA stimulation activates Arf6 through Src activity. In addition, Arf6 (Kobayashi and Fukuda, 2013) and GGA3 are recruited to recycling endosomes in response to NGF stimulation in PC12 cells. Relatedly, Arf6 depletion (siRNA) and inactivation (Myr-Arf6), as well as the uncoupling of GGA3 from Arf (N194A), impaired TrkA recycling to the PM. On the basis of these data, we thus proposed a model (Figure 9) in which the activation of Arf6 by TrkA recruits GGA3 to the endosomal membrane. Once on endosomes, GGA3 would directly bind to the DXXLL motif in TrkA and promote sorting into the recycling pathway. Alternatively, we cannot exclude that Arf6 could also aid in recruiting and sorting GGA3-TrkA complex formed earlier during TrkA transport to endosomes. Therefore these data support a model by which activation of Arf6 by RTKs such as Met or TrkA may aid in retaining and sorting GGA3-RTK complexes in endosomal-recycled membranes or serve to recruit other factors required for mediating RTK recycling.
Our study revealed that NGF/TrkA stimulation actives Arf6 through Src activity. The activation of Arf6 by Src was reported in other studies (Palacios et al., 2001; Heckel et al., 2009). However, the mechanism by which Src leads to Arf6 activation is poorly understood. Previous studies showed that Src down-regulated Arf6 activity via the ARF GTPase-activating protein GIT2 in osteoclasts (Heckel et al., 2009). Various Arf-GAPs interact with and are phosphorylated by Src (Brown et al., 1998; Donaldson and Jackson, 2011). Given that we observed that Src kinase inhibitor inhibited Arf6 activity after NGF stimulation, we suggest that Src could regulate the activity or localization of Arf6-GAP or even Arf6-GEF in PC12 cells. This potential mechanism needs to be assessed in future studies.
Postendocytic sorting of TrkA has significant effects on TrkA downstream signaling and on the physiological responses to NGF. Neuronal survival is maximally regulated by TrkA receptors localized at the PM via prolonged activation of the Akt signaling pathway, whereas neuronal differentiation is stimulated by sustained Erk activation by catalytically active TrkA in endosomes (Zhang et al., 2000). Our results show that GGA3 knockdown strongly attenuates the level of cell-surface TrkA, Akt sustained activation, and cell survival but has no significant effect on sustained Erk signaling. This supports a role for GGA3 in the endosomal sorting of TrkA to the recycling pathway, allowing TrkA delivery to the PM and prolonged Akt signaling, thereby leading to increased neuronal survival. Of interest, NGF and TrkA signaling have been shown to be involved in the pathogenesis of Alzheimer’s disease (AD). A major neuroanatomical feature of AD is the selective degeneration of basal forebrain cholinergic neurons (BFCNs), which express TrkA and whose normal function and survival require NGF trophic support. TrkA is reduced in BFCNs in AD, and failure of NGF-TrkA signaling has been considered the major cause of BFCN degeneration (Williams et al., 2006; Mufson et al., 2008). Of interest, the level of expression of GGA3 has also been reported to be reduced in the brains of AD patients (Tesco et al., 2007). Based on our findings, a reduced level of GGA3 may impair TrkA recycling and signaling, leading to a deficient trophic response and contributing to the pathogenesis of AD.
In conclusion, the results presented here identify GGA3 as a key player in a novel DXXLL-mediated endosomal sorting machinery that targets TrkA back to the PM. Consequently an alteration in GGA3 expression or activity may affect the proper recycling and signaling of TrkA and thereby contribute to the impaired neuronal survival that leads to pathologies such as AD.
MATERIALS AND METHODS
Antibodies, reagents, and plasmid constructions
Anti-Trk (c-14) polyclonal antibodies (pAbs) and Arf6 monoclonal antibodies (mAbs) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); anti-actin mAbs, tubulin mAbs, and anti-Flag M2 mAbs from Sigma-Aldrich (Saint Louis, MO); anti-EEA1 mAbs, GGA3 mAbs, Arf3 mAbs, and anti-Akt mAbs from BD Transduction Laboratories (Franklin Lakes, NJ); anti–phospho-Akt (Ser-473) pAbs, anti–p44/42 MAPK (Erk1/2) pAbs, anti–phospho-p44/42 MAPK (pErk1/2; Thr-202/Tyr-204) mAbs, and GGA3pAbs from Cell Signaling Technology (Beverly, MA); anti-hemagglutinin (HA) mAbs from Covance (Emeryville, CA); anti-Myc pAbs from Upstate (Temecula, CA); and anti-Arf1 goat pAbs from Abcam (Toronto, Canada). Affinity-purified anti-TrkA mAb 5C3 immunoglobulin G was provided by H. Uri Saragovi (Lady Davis Institute-Jewish General Hospital, Montreal, Canada). Alexa Fluor 594–transferrin and LysoTracker Red DND-99 were purchased from Life Technologies (Carlsbad, CA). MyrArf6(2-13), and non-MyrArf6(2-13) peptides were purchased from ANASPEC (Fremont, CA). MyrArf1(2-15) peptide was purchased from LifeTein (Hillsborough, NJ). pCR3.1-MycGGA1, pCR3.1-MycGGA2, and pCR3.1-MycGGA3 were provided by Juan Bonifacino (National Institutes of Health, Bethesda, MD). pcDNA3-HA-Arf6 was purchased from Addgene (Cambridge, MA). All of the mutants were generated by PCR and verified by DNA sequencing.
Cell culture and transfections
HEK293T cells were obtained from Alexandra Newton (University of California, San Diego, La Jolla, CA) and grown in high-glucose DMEM (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS) supplemented with 100 U/ml penicillin–streptomycin. PC12 (615) cells were obtained from Moses V. Chao (New York University School of Medicine, New York, NY) and grown in DMEM containing 5% heat-inactivated horse serum and 10% FBS and supplemented with 100 U/ml penicillin–streptomycin and 200 μg/ml G418. HEK293T cells were transfected using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). PC12 (615) cells were electroporated using NucleofectorTM-II (Amaxa Biosystems, Cologne, Germany) according to the manufacturer’s instructions.
Coimmunoprecipitations, GST pull-down assays, and Western blot analysis
CoIP, pull-down, and immunoblotting assays were performed as described in Rosciglione et al. (2014). Briefly, GST fusion proteins were expressed in Escherichia coli BL21 cells and purified using glutathione–Sepharose 4B beads (Pharmacia, Piscataway, NJ) as previously described (Rosciglione et al., 2014). The 35S-labeled in vitro translation products were prepared using the TNT T7 rabbit reticulocyte Quick Coupled Transcription/Translation system (Promega, San Luis Obispo, CA) in the presence of [35S]EasyTag EXPRESS labeling mix (73% Met/22% Cys; 41,000 Ci/mmol; PerkinElmer, Waltham, MA). GST fusion protein (10 μg) was incubated with the in vitro–translated products, and all bound proteins were eluted with Laemmli buffer, resolved by SDS–PAGE, and visualized by autoradiography. For the GST pull-down assays using cell lysates, purified GST fusion proteins were incubated with 1 mg of lysate, and the bound proteins were separated by SDS–PAGE and detected by immunoblotting.
The Western blot quantification was performed using ImageJ software (National Institutes of Health, Bethesda, MD). Experiments were performed in triplicate, and the results are presented as the means ± SD. Statistical significance between two groups was assessed using the Student’s t test, and analysis of variance (ANOVA) was used for comparison with three groups or more. *p < 0.05 is considered significant.
Degradation assay
TrkA degradation assays were performed as described previously (Saxena et al., 2005b). Briefly, serum-starved cells were washed with cold phosphate-buffered saline and incubated for 30 min with 0.3 mg/ml sulfo-NHS-S-S-biotin at 4°C to biotinylate surface proteins. Unreacted biotin was quenched with ice-cold Tris-buffered saline (TBS), and then the cells were incubated at 37°C in DMEM containing 10 ng/ml NGF for 0–4 h to allow the biotinylated receptors to become internalized and degraded. Subsequently cells were washed and lysed with 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.1 mM EDTA (TNE) buffer. Biotinylated proteins were pulled down with streptavidin-conjugated Sepharose beads, washed, and subjected to SDS–PAGE and immunoblotted using TrkA antibodies. This assay detected surface-labeled TrkA receptors at different time points that had not been degraded, regardless of whether the proteins had been internalized or returned to the plasma membrane. These experiments were repeated at least three independent times.
Internalization assay
For TrkA internalization assays, serum-starved cells were incubated with the recycling inhibitor monensin (20 μM) and the protease inhibitors leupeptin (100 μg/ml), pepstatin A (100 μM), E64 (20 μg/ml), and MG132 (50 μM) for 30 min at 37°C. Cell-surface proteins were then subjected to biotinylation with sulfo-NHS-S-S-biotin (0.3 mg/ml), followed by free biotin quenching with TBS, as described for the degradation assay. Cells were then incubated at 37°C in DMEM with or without 10 ng/ml NGF for 7 or 15 min to induce internalization of biotinylated surface proteins. Afterward, the remaining cell-surface biotin was cleaved by glutathione treatment (50 mM glutathione, 75 mM NaCl, 75 mM NaOH, 0.01 g/ml bovine serum albumin, 10 mM EDTA) for 15 min at 4°C to assess only the internalized proteins. One sample of cell incubated without NGF was not treated with stripping solution in order to assess the initial pool of surface- biotinylated TrkA. Cells were washed and lysed with TNE buffer. Biotinylated proteins were pulled down with streptavidin-conjugated Sepharose beads, washed, and subjected to SDS–PAGE and immunoblotted using TrkA antibodies. These experiments were repeated at least three independent times.
Recycling assay
The recycling assay was performed as previously described (Huang et al., 2009). After serum starvation, cells were incubated with the protease inhibitors leupeptin (100 μg/ml), pepstatin A (100 μM), E64 (20 μg/ml), and MG132 (50 μM) for 30 min at 37°C. These inhibitors were included in all subsequent steps to inhibit proteolysis. Cell-surface proteins were biotinylated with sulfo-NHS-S-S-biotin and quenched with TBS, as described for the degradation assay, followed by incubation with 10 ng/ml NGF for 7 min at 37°C to induce internalization of biotinylated cell-surface TrkA. Afterward, cells were cooled on ice, and the remaining cell-surface biotin was cleaved by glutathione treatment, as described for the internalization assay. Internalized biotinylated receptors were thus protected from this biotin stripping. Subsequently cells were reincubated at 37°C in DMEM for 7 or 45 min to allow recycling of internalized receptors, followed by a second round of stripping with glutathione to remove biotin from the surface-exposed biotinylated receptors that had recycled back to the cell surface during the previous rewarming period. The TrkA signal lost in the second stripping procedure was considered the fraction of recycled receptors. The biotinylated proteins were then collected with avidin and immunoblotted with TrkA antibodies. These experiments were repeated at least three independent times.
Immunofluorescence
Cells were grown on coverslips were fixed with 3% paraformaldehyde in 100 mM phosphate buffer, pH 7.4, for 30 min, permeabilized with 0.1% Triton X-100 for 10 min or 0.1% saponin plus 5% FBS when cells were fed with Alexa Fluor 594–transferrin and LysoTracker Red DND-99, blocked with 10% FBS for 30 min at room temperature, and then incubated with primary antibodies for 1 h after Alexa Fluor–conjugated secondary antibodies (Molecular Probes, Eugene, OR) at room temperature. The cells were visualized using an inverted confocal laser scanning microscope (FV1000; Olympus, Tokyo, Japan) equipped with a PlanApo 60×/1.42 oil immersion objective. Images were acquired using Olympus FluoView, version 1.6a. The degree of colocalization between GGA3 and TrkA or EEA1 was quantified by calculating the Manders coefficient using Olympus FluoView v1.6b colocalization software, as described in Rosciglione et al., 2014. The quantification analysis was performed on 30 size-matched cells for each experimental condition, and the experiments were performed twice.
MyrArf peptides
Myristoylated (MyrARF6-(2–13)) and nonmyristoylated (ARF6-(2–13)) N-terminal ARF6 peptides (GKVLSKIFGNKE) were purchased from ANASPEC. Myristoylated (MyrARF1-(2–17)) N-terminal ARF1 peptide (GNIFANLFKGLFGKKE) was purchased from LifeTein. These synthetic peptides were added to PC12 (615) cells simultaneously with NGF to block the increase in Arf activity that occurs quickly after NGF stimulation and to avoid the effects of a longer incubation with these peptides on endocytosis or other cellular functions mediated by the basal Arf activity.
RNA interference and rescue
Nonspecific (CTL) siRNA scrambled II duplexes and siRNA against rat GGA3 (360658), Arf1 (64310), Arf3 (140940), and Arf6 (79121) were purchased from Dharmacon (Lafayette, CO). For RNA interference, 5 nM siRNA was electroporated using NucleofectorTM-II (Amaxa Biosystems), and the cells were analyzed 72 h after the transfection. Reversal of phenotype (rescue) was performed by transfecting PC12 (615) cells with cDNA encoding siRNA-resistant forms of wild-type or mutant (L276A, N194A) GGA3 24 h after the initial siRNA transfection using Lipofectamine 2000 and then analyzing the cells after 48 h.
Survival assays
PC12 (615) cells were plated at a density of 4.5 × 105 cells/ml on six-well plates coated with poly-l-lysine. Culture medium was replaced with DMEM with or without 10 ng/ml NGF, incubated for 24 h, and collected following the manufacturer’s instructions. The apoptotic cell ratio was assessed using the Guava TUNEL Kit assay and EasyCyte Mini System (Millipore, Billerica, MA).
Homology modeling
The I-TASSER (Zhang, 2008) server was used to generate a homology model of the cytoplasmic domain of TrkA. The model selected (model 1) had the highest confidence (C) and template modeling (TM) scores and a very low root-mean-square deviation. Such excellent statistics were expected because the crystal structure of the C-terminal domain of human TrkA was previously solved (Bertrand et al., 2012) and the percentage of identity between the human and rat primary structure is very high (78%).
Supplementary Material
Acknowledgments
We are grateful to Horacio Uri Saragovi, Juan Bonifacino, and Moses V. Chao for generous gifts of antibodies, cDNA, and cell lines. We thank Véronique Blais for technical help with the GUAVA EasyCyte flow cytometer. This work was supported by grants from the Canadian Institutes for Health Research and a Canada Research Chair to C.L.L.
Abbreviations used:
- Arf
ADP ribosylation factor
- BACE
β-site amyloid precursor protein cleaving enzyme
- coIP
coimmunoprecipitation
- CT
C-terminus
- EGFR
epidermal growth factor receptor
- GFP
green fluorescent protein
- GGA3
Golgi-localized, γ adaptin-ear–containing ADP ribosylation factor–binding protein 3
- GST
glutathione S-transferase
- GTP
guanosine triphosphate
- JM
juxtamembrane region
- LRP9
LDL-related protein 9
- MAPK
mitogen-activated protein kinase
- MPR
mannose-6-phosphate receptor
- NGF
nerve growth factor
- PBD
protein-binding domain
- PI3K
phosphatidylinositol 3-kinase
- PKA
protein kinase A
- PKC
protein kinase C
- PM
plasma membrane
- RTK
tyrosine kinases
- siRNA
small interfering RNA
- TGN
trans-Golgi network
- TK
tyrosine kinase domain
- TUNEL
terminal deoxynucleotidyl transferase nick end labeling.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-02-0087) on October 7, 2015.
REFERENCES
- Arevalo JC, Waite J, Rajagopal R, Beyna M, Chen ZY, Lee FS, Chao MV. Cell survival through Trk neurotrophin receptors is differentially regulated by ubiquitination. Neuron. 2006;50:549–559. doi: 10.1016/j.neuron.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Bertrand T, Kothe M, Liu J, Dupuy A, Rak A, Berne PF, Davis S, Gladysheva T, Valtre C, Crenne JY, Mathieu M. The crystal structures of TrkA and TrkB suggest key regions for achieving selective inhibition. J Mol Biol. 2012;423:439–453. doi: 10.1016/j.jmb.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS. The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Boucher R, Larkin H, Brodeur J, Gagnon H, Theriault C, Lavoie C. Intracellular trafficking of LRP9 is dependent on two acidic cluster/dileucine motifs. Histochem Cell Biol. 2008;130:315–327. doi: 10.1007/s00418-008-0436-5. [DOI] [PubMed] [Google Scholar]
- Brown MT, Andrade J, Radhakrishna H, Donaldson JG, Cooper JA, Randazzo PA. ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol Cell Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caumont AS, Galas MC, Vitale N, Aunis D, Bader MF. Regulated exocytosis in chromaffin cells. Translocation of ARF6 stimulates a plasma membrane-associated phospholipase D. J Biol Chem. 1998;273:1373–1379. doi: 10.1074/jbc.273.3.1373. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Ieraci A, Tanowitz M, Lee FS. A novel endocytic recycling signal distinguishes biological responses of Trk neurotrophin receptors. Mol Biol Cell. 2005;16:5761–5772. doi: 10.1091/mbc.E05-07-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Karim ZA, Whiteheart SW. Arf6 plays an early role in platelet activation by collagen and convulxin. Blood. 2006;107:3145–3152. doi: 10.1182/blood-2005-09-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B, Bruns K, Ghosh P, Kornfeld S. Interaction of the cation-dependent mannose 6-phosphate receptor with GGA proteins. J Biol Chem. 2002;277:18477–18482. doi: 10.1074/jbc.M201879200. [DOI] [PubMed] [Google Scholar]
- Doray B, Misra S, Qian Y, Brett TJ, Kornfeld S. Do GGA adaptors bind internal DXXLL motifs? Traffic. 2012;13:1315–1325. doi: 10.1111/j.1600-0854.2012.01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha T, Wooten MW. TrkA receptor endolysosomal degradation is both ubiquitin and proteasome dependent. Traffic. 2008;9:1146–1156. doi: 10.1111/j.1600-0854.2008.00751.x. [DOI] [PubMed] [Google Scholar]
- Goh LK, Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2013;5:a017459. doi: 10.1101/cshperspect.a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Schmitz A, Famulok M. Quantification of ARF-GTP in HepG2 by pulldown with GST-GGA3(1–316). Protoc Exchange. 2006 2006, doi:10.1038/nprot.2006.412. [Google Scholar]
- He X, Zhu G, Koelsch G, Rodgers KK, Zhang XC, Tang J. Biochemical and structural characterization of the interaction of memapsin 2 (beta-secretase) cytosolic domain with the VHS domain of GGA proteins. Biochemistry. 2003;42:12174–12180. doi: 10.1021/bi035199h. [DOI] [PubMed] [Google Scholar]
- Heckel T, Czupalla C, Expirto Santo AI, Anitei M, Arantzazu Sanchez-Fernandez M, Mosch K, Krause E, Hoflack B. Src-dependent repression of ARF6 is required to maintain podosome-rich sealing zones in bone-digesting osteoclasts. Proc Natl Acad Sci USA. 2009;106:1451–1456. doi: 10.1073/pnas.0804464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempstead BL, Rabin SJ, Kaplan L, Reid S, Parada LF, Kaplan DR. Overexpression of the trk tyrosine kinase rapidly accelerates nerve growth factor-induced differentiation. Neuron. 1992;9:883–896. doi: 10.1016/0896-6273(92)90241-5. [DOI] [PubMed] [Google Scholar]
- Herbst KJ, Allen MD, Zhang J. Spatiotemporally regulated protein kinase A activity is a critical regulator of growth factor-stimulated extracellular signal-regulated kinase signaling in PC12 cells. Mol Cell Biol. 2011;31:4063–4075. doi: 10.1128/MCB.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SH, Wang J, Sui WH, Chen B, Zhang XY, Yan J, Geng Z, Chen ZY. BDNF-dependent recycling facilitates TrkB translocation to postsynaptic density during LTP via a Rab11-dependent pathway. J Neurosci. 2013;33:9214–9230. doi: 10.1523/JNEUROSCI.3256-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SH, Zhao L, Sun ZP, Li XZ, Geng Z, Zhang KD, Chao MV, Chen ZY. Essential role of Hrs in endocytic recycling of full-length TrkB receptor but not its isoform TrkB.T1. J Biol Chem. 2009;284:15126–15136. doi: 10.1074/jbc.M809763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Randazzo P, Serafini T, Weiss O, Rulka C, Clark J, Amherdt M, Roller P, Orci L, Rothman JE. The amino terminus of ADP-ribosylation factor (ARF) is a critical determinant of ARF activities and is a potent and specific inhibitor of protein transport. J Biol Chem. 1992;267:13039–13046. [PubMed] [Google Scholar]
- Kametaka S, Mattera R, Bonifacino JS. Epidermal growth factor-dependent phosphorylation of the GGA3 adaptor protein regulates its recruitment to membranes. Mol Cell Biol. 2005;25:7988–8000. doi: 10.1128/MCB.25.18.7988-8000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang EL, Cameron AN, Piazza F, Walker KR, Tesco G. Ubiquitin regulates GGA3-mediated degradation of BACE1. J Biol Chem. 2010;285:24108–24119. doi: 10.1074/jbc.M109.092742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Misra S, Puertollano R, Hurley JH, Bonifacino JS. Phosphoregulation of sorting signal-VHS domain interactions by a direct electrostatic mechanism. Nat Struct Biol. 2002;9:532–536. doi: 10.1038/nsb807. [DOI] [PubMed] [Google Scholar]
- Klesse LJ, Meyers KA, Marshall CJ, Parada LF. Nerve growth factor induces survival and differentiation through two distinct signaling cascades in PC12 cells. Oncogene. 1999;18:2055–2068. doi: 10.1038/sj.onc.1202524. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Fukuda M. Arf6, Rab11 and transferrin receptor define distinct populations of recycling endosomes. Commun Integr Biol. 2013;6:e25036. doi: 10.4161/cib.25036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Hanai A, Nakai W, Katoh Y, Nakayama K, Shin HW. ARF1 and ARF3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct Funct. 2012;37:141–154. doi: 10.1247/csf.12015. [DOI] [PubMed] [Google Scholar]
- LeSauteur L, Maliartchouk S, Le Jeune H, Quirion R, Saragovi HU. Potent human p140-TrkA agonists derived from an anti-receptor monoclonal antibody. J Neurosci. 1996;16:1308–1316. doi: 10.1523/JNEUROSCI.16-04-01308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meakin SO, MacDonald JI, Gryz EA, Kubu CJ, Verdi JM. The signaling adapter FRS-2 competes with Shc for binding to the nerve growth factor receptor TrkA. A model for discriminating proliferation and differentiation. J Biol Chem. 1999;274:9861–9870. doi: 10.1074/jbc.274.14.9861. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, Counts SE, Perez SE, Ginsberg SD. Cholinergic system during the progression of Alzheimer’s disease: therapeutic implications. Expert Rev Neurother. 2008;8:1703–1718. doi: 10.1586/14737175.8.11.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios F, D’Souza-Schorey C. Modulation of Rac1 and ARF6 activation during epithelial cell scattering. J Biol Chem. 2003;278:17395–17400. doi: 10.1074/jbc.M300998200. [DOI] [PubMed] [Google Scholar]
- Palacios F, Price L, Schweitzer J, Collard JG, D’Souza-Schorey C. An essential role for ARF6-regulated membrane traffic in adherens junction turnover and epithelial cell migration. EMBO J. 2001;20:4973–4986. doi: 10.1093/emboj/20.17.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parachoniak CA, Luo Y, Abella JV, Keen JH, Park M. GGA3 functions as a switch to promote Met receptor recycling, essential for sustained ERK and cell migration. Dev Cell. 2011;20:751–763. doi: 10.1016/j.devcel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierchala BA, Ahrens RC, Paden AJ, Johnson EM., Jr Nerve growth factor promotes the survival of sympathetic neurons through the cooperative function of the protein kinase C and phosphatidylinositol 3-kinase pathways. J Biol Chem. 2004;279:27986–27993. doi: 10.1074/jbc.M312237200. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Aguilar RC, Gorshkova I, Crouch RJ, Bonifacino JS. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 2001a;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Randazzo PA, Presley JF, Hartnell LM, Bonifacino JS. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell. 2001b;105:93–102. doi: 10.1016/s0092-8674(01)00299-9. [DOI] [PubMed] [Google Scholar]
- Rosciglione S, Theriault C, Boily MO, Paquette M, Lavoie C. Galphas regulates the post-endocytic sorting of G protein-coupled receptors. Nat Commun. 2014;5:4556. doi: 10.1038/ncomms5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Bucci C, Weis J, Kruttgen A. The small GTPase Rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor TrkA. J Neurosci. 2005a;25:10930–10940. doi: 10.1523/JNEUROSCI.2029-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Howe CL, Cosgaya JM, Steiner P, Hirling H, Chan JR, Weis J, Kruttgen A. Differential endocytic sorting of p75NTR and TrkA in response to NGF: a role for late endosomes in TrkA trafficking. Mol Cell Neurosci. 2005b;28:571–587. doi: 10.1016/j.mcn.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Tesco G, Koh YH, Kang EL, Cameron AN, Das S, Sena-Esteves M, Hiltunen M, Yang SH, Zhong Z, Shen Y, et al. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle T, Prager K, Raffler N, Haass C, Famulok M, Walter J. GGA proteins regulate retrograde transport of BACE1 from endosomes to the trans-Golgi network. Mol Cell Neurosci. 2005;29:453–461. doi: 10.1016/j.mcn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Williams BJ, Eriksdotter-Jonhagen M, Granholm AC. Nerve growth factor in treatment and pathogenesis of Alzheimer’s disease. Prog Neurobiol. 2006;80:114–128. doi: 10.1016/j.pneurobio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Wooten MW, Vandenplas ML, Seibenhener ML, Geetha T, Diaz-Meco MT. Nerve growth factor stimulates multisite tyrosine phosphorylation and activation of the atypical protein kinase C’s via a src kinase pathway. Mol Cell Biol. 2001;21:8414–8427. doi: 10.1128/MCB.21.24.8414-8427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Calvo L, Anta B, Lopez-Benito S, Southon E, Chao MV, Tessarollo L, Arevalo JC. Regulation of trafficking of activated TrkA is critical for NGF-mediated functions. Traffic. 2011;12:521–534. doi: 10.1111/j.1600-0854.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moheban DB, Conway BR, Bhattacharyya A, Segal RA. Cell surface Trk receptors mediate NGF-induced survival while internalized receptors regulate NGF-induced differentiation. J Neurosci. 2000;20:5671–5678. doi: 10.1523/JNEUROSCI.20-15-05671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, London NR, Gibson CC, Davis CT, Tong Z, Sorensen LK, Shi DS, Guo J, Smith MC, Grossmann AH, et al. Interleukin receptor activates a MYD88-ARNO-ARF6 cascade to disrupt vascular stability. Nature. 2012;492:252–255. doi: 10.1038/nature11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.