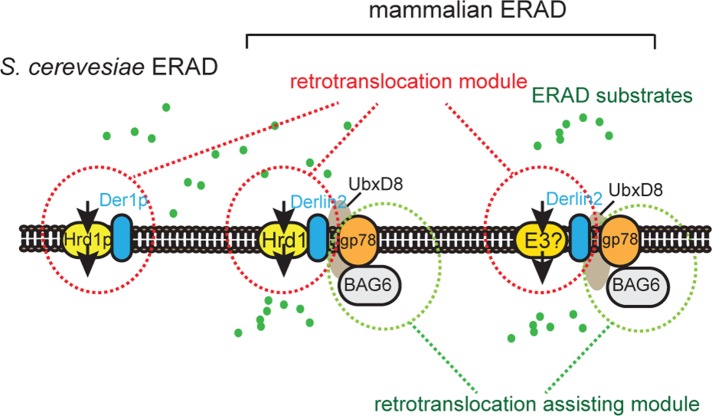
The functional relationship between mammalian ubiquitin ligase gp78 and Hrd1 was studied. Hrd1 is one of the essential retrotranslocation regulators conserved in yeast and mammalian cells, whereas gp78 serves an assisting role downstream of Hrd1 and possibly other ubiquitin ligases in mammalian cells.
Abstract
Eukaryotic cells eliminate misfolded proteins from the endoplasmic reticulum (ER) via a conserved process termed ER-associated degradation (ERAD). Central regulators of the ERAD system are membrane-bound ubiquitin ligases, which are thought to channel misfolded proteins through the ER membrane during retrotranslocation. Hrd1 and gp78 are mammalian ubiquitin ligases homologous to Hrd1p, an ubiquitin ligase essential for ERAD in Saccharomyces cerevisiae. However, the functional relevance of these proteins to Hrd1p is unclear. In this paper, we characterize the gp78-containing ubiquitin ligase complex and define its functional interplay with Hrd1 using biochemical and recently developed CRISPR-based genetic tools. Our data show that transient inactivation of the gp78 complex by short hairpin RNA–mediated gene silencing causes significant stabilization of both luminal and membrane ERAD substrates, but unlike Hrd1, which plays an essential role in retrotranslocation and ubiquitination of these ERAD substrates, knockdown of gp78 does not affect either of these processes. Instead, gp78 appears to act downstream of Hrd1 to promote ERAD via cooperation with the BAG6 chaperone complex. We conclude that the Hrd1 complex forms an essential retrotranslocation module that is evolutionarily conserved, but the mammalian ERAD system uses additional ubiquitin ligases to assist Hrd1 during retrotranslocation.
INTRODUCTION
In the secretory pathway, nascent polypeptides entering the endoplasmic reticulum (ER) frequently fold improperly due to either macromolecular crowding in the ER lumen, insufficient levels of chaperoning, or genetic lesions in the polypeptides. Accumulation of misfolded proteins can cause ER stress, which will induce cell death if not rectified. ER stress–induced cell death has been linked to the pathogenesis of many human diseases (Marciniak and Ron, 2006; Ron and Walter, 2007; Liu and Ye, 2011).
ER-associated degradation (ERAD) is an evolutionarily conserved protein quality-control mechanism that eliminates misfolded proteins from the ER in eukaryotic cells (Vembar and Brodsky, 2008; Christianson and Ye, 2014; Ruggiano et al., 2014). Aberrant polypeptides in the ER lumen or on the ER membrane are recognized by ERAD machinery proteins and subsequently exported into the cytosol via a process termed retrotranslocation (Tsai et al., 2002). During retrotranslocation, misfolded polypeptides are conjugated with ubiquitin chains by ER-associated ubiquitin ligases on the cytosolic side of the ER membrane (Hirsch et al., 2009). Polyubiquitinated substrates are then extracted from the membrane by the conserved AAA (ATPase-associated with diverse cellular activities) ATPase complex p97-Ufd1-Npl4 and delivered to the 26S proteasome for degradation (Ye et al., 2001; Bays and Hampton, 2002; Jarosch et al., 2002; Rabinovich et al., 2002).
The retrotranslocation system in the ER membrane consists of several multiprotein complexes. Some of these complexes appear to operate in parallel to eliminate misfolded proteins of different classes. Each retrotranslocation complex is assembled around a ubiquitin ligase in the membrane (Christianson and Ye, 2014). Given that many ERAD ubiquitin ligases contain multiple transmembrane domains (Hirsch et al., 2009), and also because a retrotranslocation substrate was detected in proximity to the transmembrane domains of the ubiquitin ligase Hrd1p, it was proposed that ubiquitin ligases may form several protein-conducting channels in the membrane to export diverse misfolded polypeptides (Carvalho et al., 2010).
In Saccharomyces cerevisiae, there are two well-defined ubiquitin ligases in ERAD: Hrd1p and Doa10p (Bays et al., 2001; Swanson et al., 2001; Sato et al., 2009; Foresti et al., 2013). Biochemical and genetic studies support a simple “dichotomy of lesion” model, in which the location of the misfolded domains dictates the substrate specificity of these ligases (Vashist and Ng, 2004; Carvalho et al., 2006): In cooperation with cytosolic chaperones, the Doa10 complex ubiquitinates substrates with misfolded cytosolic domain (ERAD-C), whereas the Hrd1 complex primarily targets substrates carrying misfolded domains in the ER lumen (ERAD-L) or in membranes (ERAD-M). However, this conceptually straightforward model is based on studies using a limited number of model substrates. Thus exceptions to these rules may exist as more substrates are analyzed. Indeed, a recent report showed that Doa10p also recognizes a newly established ERAD substrate bearing a “degron” in the transmembrane region (Habeck et al., 2015).
The division of labor rule is also not applicable to the mammalian ERAD system, which has been studied with more model substrates. The mammalian ERAD system is also more complex, as it consists of far more regulators than the yeast system. In addition to homologues of the Hrd1 and Doa10 complexes, the mammalian ERAD system uses a large number of additional ubiquitin ligases. It seems unlikely that each of these ligases would form an exit tunnel for misfolded proteins, because many of them contain either no transmembrane domain or a limited number of transmembrane segments. Thus it is more likely that many of these ubiquitin ligases need to act in conjunction with either Hrd1 or other retrotranslocation channel candidates. Cooperation between ubiquitin ligases during ERAD has been reported (Younger et al., 2006; Morito et al., 2008; Jo et al., 2011a), but how ubiquitin ligase partners collaborate in ERAD has remained unclear.
Among the identified mammalian ubiquitin ligases, gp78 is unique, because it is homologous to Hrd1 at the amino acid–sequence level, yet it has an interaction network that is largely distinct from that of Hrd1 (Christianson et al., 2011): while the mammalian Hrd1 complex contains homologues of the yeast Hrd1p complex members such as SEL1L, OS-9, Derlin1/2/3, and HERP (Lilley and Ploegh, 2005; Ye et al., 2005; Mueller et al., 2008; Christianson et al., 2011; Huang et al., 2013b), gp78 interacts with UbxD8, UBAC2, Ube2g2, and Derlin1 (Christianson et al., 2011). This is probably caused by sequence variations in the membrane domains. Moreover, a few unique protein interaction motifs in gp78 may also contribute to its specific networking property. Functionally, gp78 appears to use a distinct mechanism to assemble ubiquitin chains, because it can stably interact with its cognate ubiquitin-conjugating enzyme Ube2g2 to assemble ubiquitin chains on its active site. This is owing to the presence of a specific high-affinity ubiquitin-conjugating enzyme-binding motif G2BR and a ubiquitin-binding CUE domain in gp78 (Das et al., 2009; Li et al., 2009). At least in vitro, preformed ubiquitin chains can be transferred en bloc to a substrate (Li et al., 2007), whereas such activity has not been reported for Hrd1. Despite these distinctions, the two enzymes share partially overlapping substrate specificity: certain ERAD substrates require both gp78 and Hrd1, whereas others depend on only one but not the other for degradation (Fang et al., 2001; Song et al., 2005; Ishikura et al., 2010; Bernasconi et al., 2010; Christianson et al., 2011). For those substrates whose degradation requires both gp78 and Hrd1, it is unclear whether these enzymes act in a sequential manner or in parallel.
In this paper, we report that mammalian Hrd1 and gp78 are not functionally redundant. Instead, these enzymes act in sequence to promote misfolded protein turnover. Hrd1 is involved in retrotranslocation and ubiquitination of ERAD substrates, whereas gp78 provides an accessory function downstream of Hrd1, probably through interaction with the cytosolic chaperone BAG6.
RESULTS
Characterization of the gp78-UBAC2-UbxD8 complex
The interaction networks of gp78 and Hrd1 have recently been revealed by biochemical pull-down and mass spectrometry analyses (Christianson et al., 2011; Jo et al., 2011b; Wang et al., 2011). Among gp78 interactors, UbxD8 and UBAC2 were known to form a stable subcomplex (Figure 1A; Olzmann et al., 2013), whereas Derlin2 stably interacts with Hrd1, Sel1L, and HERP (Huang et al., 2013a, b). However, it is not entirely clear whether gp78 forms stable interactions with both the Hrd1 complex and the UbxD8-UBAC2 complex in a stoichiometric manner. To address this question, we first performed coimmunoprecipitation experiments using cells expressing FLAG-tagged gp78 or FLAG-tagged Hrd1. We compared endogenous interactors that were coprecipitated with these ligases by immunoblotting. As expected, all the previously described interactors such as p97, BAG6, Sel1L, HERP, OS9, Derlin2, UBAC2, and UbxD8 could be coprecipitated by each of these ligases, but the relative amounts were different: gp78 preferentially bound to UBAC2, BAG6, and p97, whereas Sel1L, HERP, and OS9 were enriched in Hrd1 pull-down (Figure 1B). Although Derlin2 was similarly present in gp78 and Hrd1 pull-down samples, the interaction of Derlin2 with gp78 is sensitive to detergent (Christianson et al., 2011). Immunoprecipitation of FLAG-gp78 also pulled down a small amount of endogenous Hrd1 and vice versa (unpublished data). Importantly, immunoprecipitation of endogenous Hrd1 pulled down a small amount of gp78 and vice versa (Figure 1, C and D). These results support the notion that gp78 and Hrd1 form two distinct complexes: gp78-UBAC2-UbxD8 and Hrd1-Sel1L-HERP-Derlin2-OS9, which communicate with each other, probably via a transient link established by UbxD8 and Derlin2. It is noteworthy that all the components of the Hrd1 complex are conserved from budding yeast to humans, but the gp78 complex is only present in higher eukaryotes.
FIGURE 1:
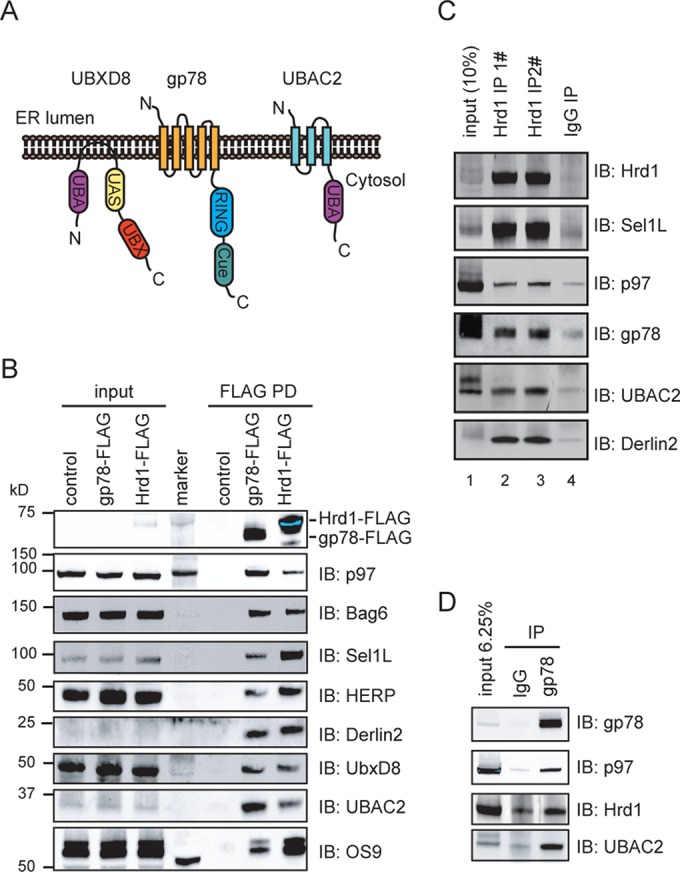
gp78 forms a transient interaction with Hrd1. (A) Schematic diagram of the domain structure of gp78, UbxD8, and UBAC2. (B) Hrd1 and gp78 bind to different interactors with different affinities. Control transfected HEK293T cells or cells overexpressing FLAG-Hrd1 or FLAG-gp78 were lysed in the CHAPS lysis buffer. Proteins immunoprecipitated with FLAG beads were analyzed by immunoblotting. (C and D) Endogenous interaction of Hrd1 with gp78. (C) 293T cells were lysed in a CHAPS-containing lysis buffer. The cell extracts were subject to immunoprecipitation by either control antibody or two affinity-purified Hrd1 antibodies. Where indicated, a fraction of the cell lysate was analyzed directly by immunoblotting. (D) 293T-derived cell extracts were subject to immunoprecipitation by the indicated antibodies.
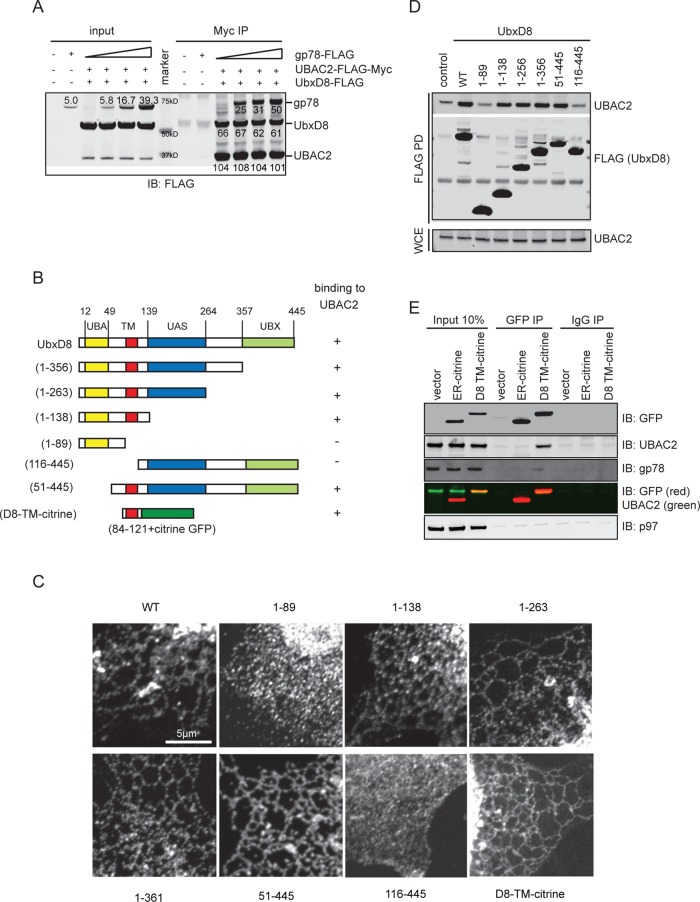
Because the molecular composition and the function of the Hrd1 complex had been extensively characterized in S. cerevisiae and in human cell lines (Lilley and Ploegh, 2005; Ye et al., 2005; Carvalho et al., 2006; Mueller et al., 2008; Christianson et al., 2011; Huang et al., 2013b), we decided to focus our study on the gp78-UbxD8-UBAC2 complex. We first wished to determine the protein stoichiometry within this complex. Because the level of endogenous proteins is too low to reliably measure these interactions, and also because of the difference in antibody sensitivity, we chose to reconstitute this complex in cells by coexpressing recombinant proteins that all carry the same FLAG tag. One of the proteins (the one expressed at a lower level) also contained a unique Myc epitope, which was used to pull down the complex. Immunoblotting analyses of purified complex showed that UbxD8 interacted stably with UBAC2, consistent with previous results (Olzmann et al., 2013). When gp78 was coexpressed, the interaction between UbxD8 and UBAC2 was not enhanced, but a significant amount of gp78 was coprecipitated with UbxD8 and UBAC2. On the basis of quantitative immunoblotting, we estimated that gp78, UbxD8, and UBAC2 were present in the precipitated complex in an approximate ratio of 1:1:2 (Figure 2A).
FIGURE 2:
Characterization of the gp78-UbxD8-UBAC2 complex. (A) Stoichiometry of the gp78-UbxD8-UBAC2 complex. The indicated proteins were overexpressed in HEK293T cells. Cells were lysed in NP40 lysis buffer, and proteins immunoprecipitated with anti-Myc antibodies were analyzed by quantitative immunoblotting. (B) Schematic diagram of the UBXD8 variants used in the pull-down study. All constructs contain a FLAG tag at the amino terminus. (C) Subcellular localization of the UbxD8 variants. COS7 cells expressing the indicated UbxD8 variants were stained by FLAG antibodies. Scale bar: 5 μm. (D) Cells overexpressing FLAG-tagged UBXD8 constructs were lysed in a NP40-containing lysis buffer. Proteins coprecipitated by FLAG beads were analyzed by immunoblotting. (E) The transmembrane domain of UbxD8 is sufficient to bind UBAC2.
We next performed mapping experiments to define the UbxD8 domain that is involved in UBAC2 binding. To this end, we created a series of UbxD8 truncated mutants (Figure 2B). Immunofluorescence imaging experiments confirmed that UbxD8 mutants bearing the transmembrane hairpin were all correctly localized to the ER membrane, whereas those lacking the transmembrane domain (1–89 and 116–445) were localized largely in the cytosol (Figure 2C). Coimmunoprecipitation experiments showed that deletion of the carboxyl-terminal cytosolic domain did not affect its interaction with UBAC2. However, further deletion of a segment between residue 89 and 116, which comprised the hairpin-shaped transmembrane domain, disrupted the interaction with UBAC2 (Figure 2, B and D). Likewise, removal of the amino-terminal UBA domain of UbxD8 did not affect binding to UBAC2, but further deletion of the transmembrane segment diminished this interaction (Figure 2, B and D). Together these results suggested that UbxD8 might use its transmembrane domain to bind UBAC2.
To further confirm the interaction of the transmembrane domain of UbxD8 with UBAC2, we generated a construct that expressed the UbxD8 transmembrane domain in fusion with a monomeric yellow fluorescence protein (UbxD8 TM-citrine). When expressed in cells, this protein was localized to the ER membrane (Figure 2C), suggesting that the transmembrane domain of UbxD8 is sufficient for ER targeting. We then performed a coimmunoprecipitation experiment. We used ER-citrine that was localized to the ER by the transmembrane domain of Sec61β as a negative control. Indeed, UbxD8 TM-citrine could be coprecipitated with endogenous UBAC2, whereas ER-citrine did not bind. UbxD8 TM-citrine also bound weakly to gp78 but not to p97. Thus we concluded that the transmembrane domain of UbxD8 is involved in forming the stable UbxD8-UBAC2 complex, which interacts with gp78. Because gp78 overexpression does not enhance the interaction of UbxD8 with UBAC2 (Figure 2A), the interaction between UbxD8 and UBAC2 does not seem to be mediated by gp78.
gp78 and Hrd1 are required for ERAD of both luminal and membrane substrates
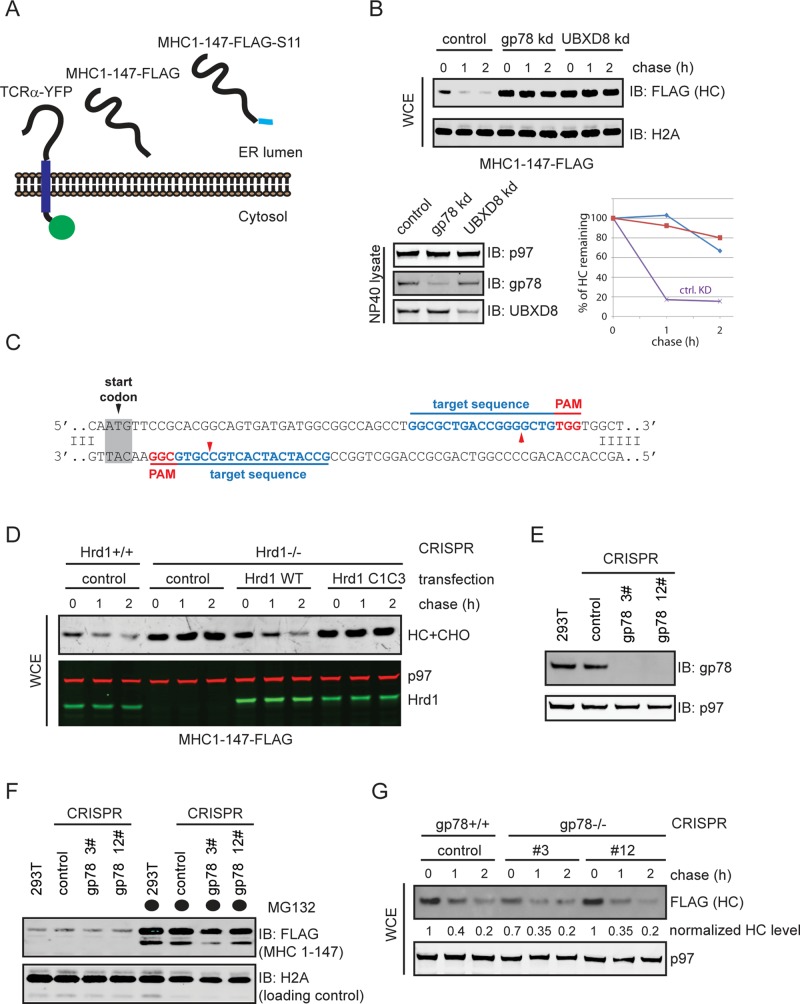
The physical interaction between the gp78 and the Hrd1 complex (Figure 1) suggested that these enzymes might cooperate with each other in ERAD. Conceptually, the two ligases might act in a linear pathway, or they might function in parallel. To study the functional interplay between gp78 and Hrd1, we sought model ERAD substrates that require both Hrd1 and gp78 for degradation. As a representative of membrane substrates, we chose TCRα (Figure 3A), a type I membrane protein that is mostly unassembled when overexpressed in tissue culture cells. As a result, overexpressed TCRα is rapidly degraded by a mechanism dependent on both gp78 and Hrd1 (Ishikura et al., 2010). Because previously reported luminal ERAD substrates do not require gp78 for efficient degradation, we engineered a truncated major histocompatibility complex (MHC) class I heavy-chain molecule (hereinafter referred to as “MHC 1-147”) that contains only a small luminal segment of the human leukocyte antigen (HLA) A2 allele (Figure 3A). A similar MHC class I mutant was previously established as a luminal ERAD substrate whose degradation requires the Hrd1 complex (Burr et al., 2013). We used short hairpin RNAs (shRNAs) to knock down either gp78 or UbxD8 and tested whether the half-life of MHC 1-147 was affected using a translation shutoff assay. The result suggested that gp78 and UbxD8 are both required for efficient degradation of MHC 1-147 (Figure 3B).
FIGURE 3:
Both gp78 and Hrd1 are required for ERAD of luminal and membrane substrates. (A) Diagram illustrating the model ERAD substrates used in this study. (B) Both gp78 and UbxD8 are required for degradation of MHC 1-147. Cells cotransfected with indicated shRNA constructs and a plasmid expressing MHC 1-147 were treated with cycloheximide for the indicated time points. Cells were directly lysed in the Laemmli buffer, and the whole-cell extracts were analyzed by immunoblotting. Graph on the right represents the quantification of the experiment. (C) Guiding sequence used to create hrd1 knockout CRISPR cell. The PAM sequence and the target sequence are colored in red and blue, respectively. Red arrow indicates the predicted Cas9 D10A cutting site. (D) Hrd1 is required for the degradation of MHC 1-147. Cycloheximide chase was performed in control CRISPR and hrd1 knockout CRISPR cells. Where indicated, plasmids expressing WT Hrd1 or a catalytically inactive Hrd1 (C1C3) mutant were cotransfected with MHC 1-147. Whole-cell extracts were analyzed by immunoblotting. (E) Verification of the gp78 CRISPR cells by immunoblotting. (F and G) gp78-deficient CRISPR cells do not have ERAD defects. The steady-state level of MHC 1-147 in either the parental HEK293T cells or the indicated CRISPR clones was analyzed by immunoblotting. Where indicated, cells were treated with the proteasome inhibitor MG132 (10 μm, 15 h). (G) The indicated CRISPR cells transfected with a plasmid expressing MHC 1-147 were treated with cycloheximide for the indicated time points. Cells were directly lysed in the Laemmli buffer, and the whole-cell extracts were analyzed by immunoblotting.
We used the recently developed CRISPR technology to generate HEK293T cells deficient for key components of these enzyme complexes (Ran et al., 2013). Hrd1 was first disrupted by expression of the CAS9 D10A mutant together with a pair of guide RNAs (Figure 3C). Because DNA double strain break is generated only when both guide RNAs bind to the same region within the target gene, this strategy was reported to greatly reduce the off-target effect (Ran et al., 2013). When we examined the degradation kinetics of MHC 1-147 in either control or Hrd1 CRISPR cells, MHC 1-147 was rapidly degraded in control cells that still had a normal level of Hrd1 expression, but the degradation was significantly inhibited in hrd1 CRISPR cells. The MHC 1-147 degradation could be largely restored by overexpression of wild-type (WT) but not by a catalytically inactive Hrd1 mutant (Figure 3D). When a similar strategy was used to knock out the Hrd1 partner Sel1L, it also resulted in significant stabilization of MHC 1-147 (unpublished data). Together these results suggested MHC 1-147 as a substrate of Hrd1. Moreover, the results also indicated that whether or not a substrate contains a transmembrane domain cannot predict its reliance on either gp78 or Hrd1 for degradation in mammalian cells.
We next created gp78 null CRISPR cells (Figure 3E) and examined the expression of both MHC 1-147 and TCRα in control and gp78 CRISPR knockout cells. We found that the steady-state levels of MHC 1-147 and TCRα were similar between control and gp78 null cells, suggesting that the degradation of these substrates was not affected by gp78 deletion (Figure 3F; unpublished data). This was indeed confirmed for MHC 1-147 using the translation shutdown assay (Figure 3G). These results were surprising, because transient knockdown of gp78 dramatically affected the degradation of the same substrates (Figure 3B; Ishikura et al., 2010; Liu et al., 2014). We concluded from these data that, for substrates whose degradation requires both Hrd1 and gp78, these ligases play distinct roles; the Hrd1 complex is essential, whereas the gp78 complex serves a function that could be compensated for when the gp78 complex is permanently inactivated. CRISPR cells might adapt to gp78 deficiency during the clonal expansion process (see Discussion).
Hrd1, but not gp78, is involved in substrate retrotranslocation and ubiquitination
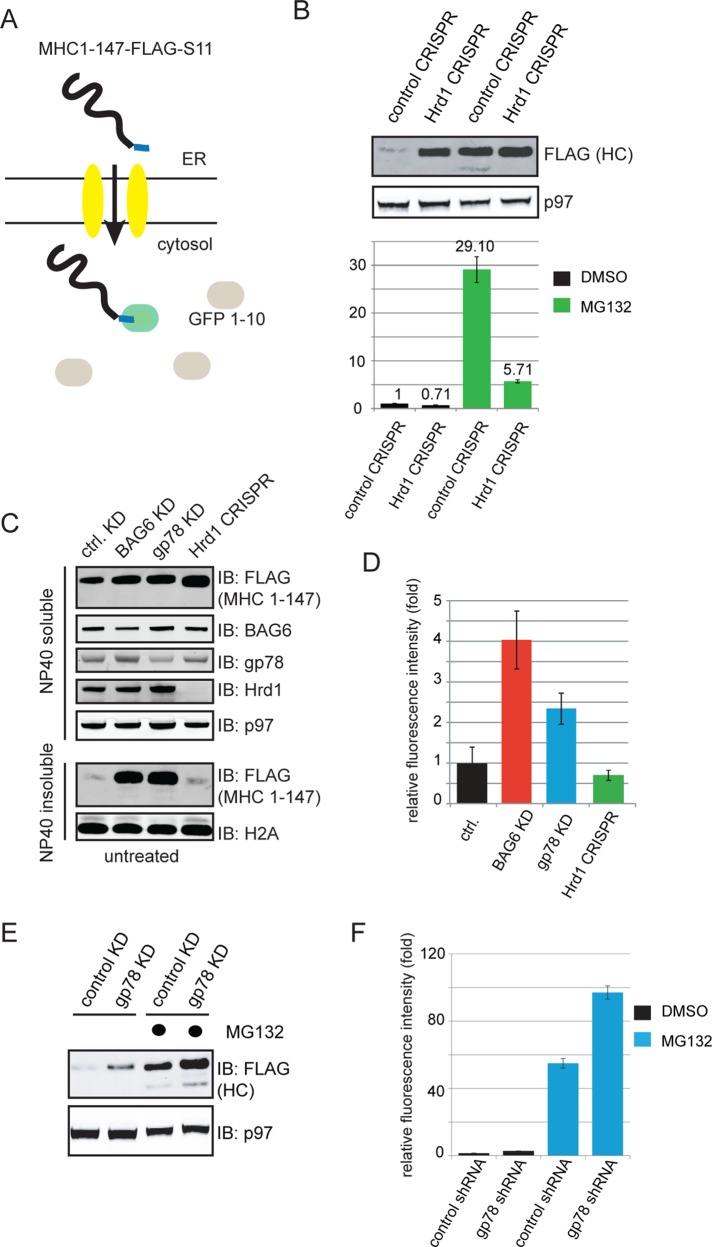
Next we tested whether both gp78 and Hrd1 were required for substrate polyubiquitination. We first expressed MHC 1-147 in control and Hrd1 CRISPR cells together with hemagglutinin (HA)-tagged ubiquitin. We then treated these cells with DMSO as a control or the proteasome inhibitor MG132 and immunoprecipitated MHC 1-147 from cell extracts under denaturing conditions. Immunoblotting with FLAG antibodies showed that inhibition of the proteasome caused accumulation of MHC 1-147 in both glycosylated and deglycosylated forms (Figure 4A, bottom panel, lane 3 vs. lane 1). The deglycosylated MHC 1-147 must have arrived in the cytosol, because the N-glycanase that processes the sugar chain is cytosolic (Blom et al., 2004). The glycosylated form accumulated mostly in the ER due to a retrotranslocation backup, although a fraction might have reached the cytosolic side of the ER membrane, but the glycan had not been removed. Knockout of hrd1 caused MHC 1-147 to accumulate only in the glycosylated form (Figure 4A, lane 2). When control and hrd1 CRISPR cells were treated with MG132, glycosylated MHC 1-147 was accumulated similarly, but the amount of deglycosylated MHC 1-147 was significantly less in hrd1 knockout cells than in control cells (Figure 4A, lane 4 vs. lane 3). These observations are consistent with the proposed function of Hrd1 in retrotranslocation. Immunoblotting with HA antibodies detected ubiquitinated MHC 1-147 in control cells, and as expected, ubiquitinated MHC 1-147 was increased when degradation was blocked by the proteasome inhibitor MG132 (Figure 4A, lane 3 vs. lane 1). However, the level of ubiquitinated MHC 1-147 was significantly reduced in Hrd1 CRISPR cells compared with control cells under both untreated and MG132-treated conditions (Figure 4A, lane 2 vs. lane 1 and lane 4 vs. lane 3). Similar results were obtained with the membrane ERAD substrate TCRα (Figure 4B). We next examined the role of gp78 in these ERAD processes by transiently knocking down the gp78 expression. Surprisingly, knockdown of gp78 neither reduced the level of deglycosylated MHC 1-147 nor inhibited MHC 1-147 ubiquitination (Figure 4, C, lane 4 vs. lane 3, and D). Likewise, ubiquitination of TCRα was also not inhibited by gp78 knockdown (Figure 4E). Thus, for substrates whose degradation requires both Hrd1 and gp78, only Hrd1 but not gp78 is essential for ubiquitination.
FIGURE 4:
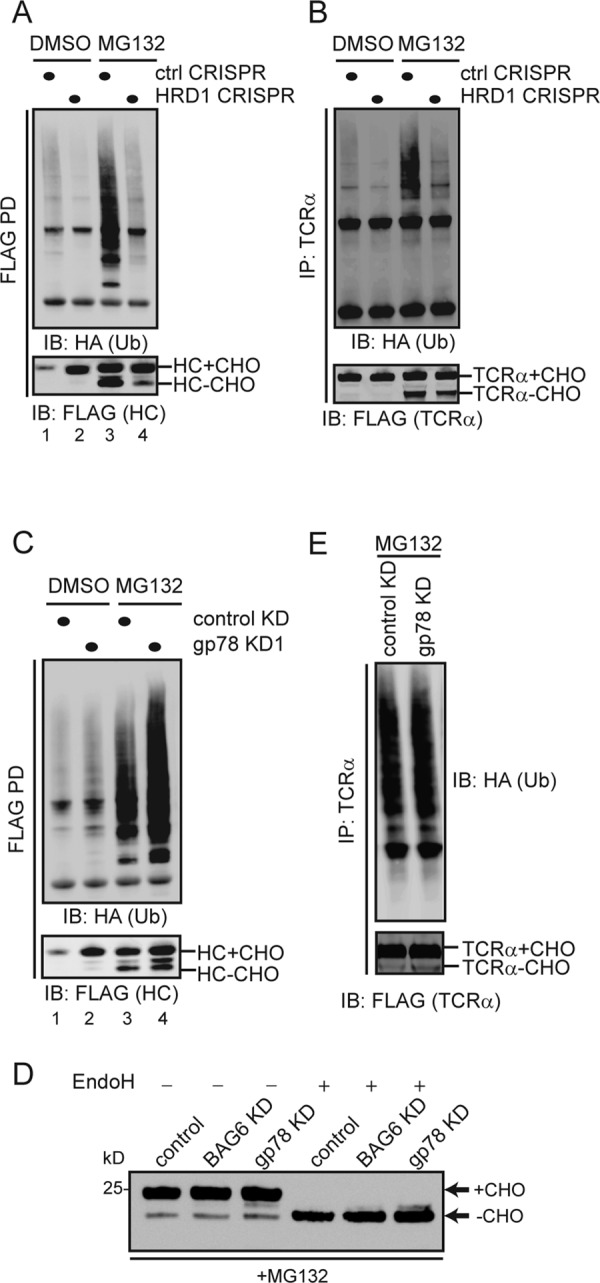
Hrd1 but not gp78 is required for ubiquitination of ERAD substrates. (A and B) The model ERAD substrates MHC 1-147-FLAG (A) and TCRα-YFP-FLAG (B) were cotransfected with a construct expressing HA-tagged ubiquitin in either control or hrd1 CRIPSR cell. The cells were treated with either DMSO as a control or with the proteasome inhibitor MG132 (10 μM, 15 h). Substrates immunoprecipitated from the cell extracts under denaturing conditions were analyzed by immunoblotting. The anti-HA blot reveals ubiquitinated substrate and the anti-FLAG blot shows the nonubiquitinated glycosylated and deglycosylated substrates. (C) As in A, except that cells treated with the indicated shRNA constructs were used. (D) As in B, except that cells treated with the indicated shRNA constructs were used. (E) Endoglycosidase H treatment of the indicated cell extract reveals the glycosylation pattern of MHC 1-147 in the indicated CRISPR cells that have been exposed to a proteasome inhibitor.
To further validate our conclusion, we adopted a recently established retrotranslocation assay that is based on the split–green fluorescent protein (GFP) technology (Zhong and Fang, 2012). GFP contains 11 β-strands that form a β-barrel with an embedded fluorophore. Deletion of the last β-strand (GFP1-10) abolishes the fluorescence of GFP, but when GFP1-10 is localized in the same subcellular compartment with another protein tagged with the 11th β-strand of GFP (s11), the GFP fluorophore can be reconstituted to regain fluorescence (Figure 5A). We focused our study on MHC 1-147, because retrotranslocation is a prerequisite for ubiquitination and degradation of luminal ERAD substrates. We appended the carboxyl-terminal 11 residues of GFP to the carboxyl-terminus of MHC 1-147 (MHC 1-147 S11). This construct was coexpressed with GFP1-10 in cells. Because GFP1-10 was cytosolically localized, fluorescence would be generated only when retrotranslocated MHC 1-147 S11 accumulated significantly in the cytosol. Because retrotranslocation and degradation are usually tightly coupled in WT cells, very little fluorescence was detected in control CRISPR cells. Knockout of hrd1 accumulated MHC 1-147 S11, but no green fluorescence was detected, suggesting that the substrate was mainly localized in the ER lumen. By contrast, treatment with MG132 uncoupled retrotranslocation from degradation, leading to accumulation of retrotranslocated MHC 1-147 S11 and enhancement of fluorescence intensity by ∼30-fold (Figure 5B), but under the same condition, the increase in fluorescence intensity was reduced to only approximately sixfold when the hrd1 gene was disrupted. These results are consistent with the notion that Hrd1 mediates retrotranslocation of MHC 1-147, but also indicated the presence of a Hrd1-independent route for export of MHC 1-147.
FIGURE 5:
Hrd1 but not gp78 is involved in substrate retrotranslocation. (A) Schematic illustration of the split-GFP–based retrotranslocation assay. (B) Hrd1 is required for the retrotranslocation of MHC 1-147. Plasmids expressing s11-tagged MHC 1-147 and GFP S1-10 were cotransfected into control or hrd1 CRISPR cells. Cells were treated with DMSO (control) or 10 μM proteasome inhibitor MG132 for 15 h. Top, NP40-soluble lysates were analyzed by immunoblotting. p97 was used as a loading control. Bottom, fluorescence intensity in the same cells was measured. The graph is from three independent experiments. (C and D) MHC 1-147 still undergoes retrotranslocation in BAG6 and gp78 knockdown cells. Similar to B, shRNA construct or CRISPR cell line was used to knockdown or knockout the indicated gene without MG132 treatment. Both NP40-soluble and NP40-insoluble fractions were analyzed. Bar graph represents the average of three independent experiments. (E and F) gp78 knockdown with MG132 treatment. Bar graph represents the average of three independent experiments.
We next determined the level of retrotranslocated MHC1-147 S11 in gp78 and BAG6 knockdown cells. We included BAG6 knockdown cells in the study because BAG6 is a recently identified chaperone holdase that maintains the solubility of retrotranslocated products to facilitate ERAD (Wang et al., 2011). Accordingly, its depletion should not block retrotranslocation. As expected, knockdown of either BAG6 or gp78 caused significant accumulation of MHC 1-147 S11 as detected by immunoblotting. Strikingly, a large fraction of MHC 1-147 S11 stabilized in either BAG6 or gp78 knockdown cells was resistant to extraction by the nonionic detergent Nonidet P-40 (NP40; Figure 5C). By contrast, MHC 1-147 S11 accumulated in Hrd1-deficient cells was largely soluble in NP40. When fluorescence intensity in these cells was measured, we noted that knockdown of BAG6 increased green fluorescence by approximately fourfold, whereas gp78 knockdown also consistently increased the green fluorescence intensity, albeit less dramatically than BAG6 depletion (Figure 5D). By contrast, Hrd1-deficient cells consistently showed a small reduction in basal fluorescence intensity. Another distinction between gp78 knockdown and Hrd1 CRISPR cells is that gp78 knockdown slightly increased rather than reduced the green fluorescence under MG132-treated conditions (Figure 5, E and F). Together these results indicate that, unlike Hrd1, gp78 is not required for retrotranslocation of MHC 1-147 S11. The results also suggest that gp78 may be functionally more relevant to the BAG6 complex, which assists retrotranslocation by maintaining the solubility of retrotranslocation products.
gp78 functions downstream of the Hrd1 complex
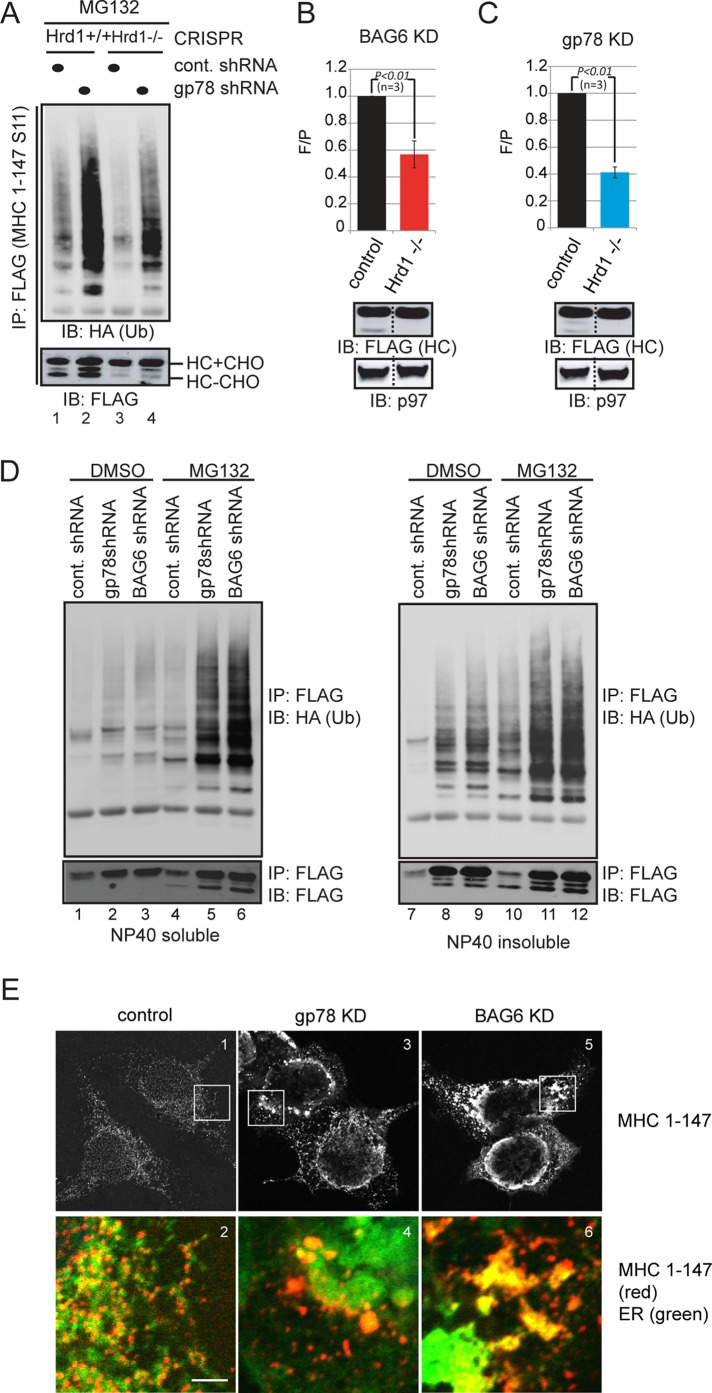
The observation that Hrd1 deficiency reduces retrotranslocation and ubiquitination of MHC 1-147 and that gp78 knockdown slightly increases MHC 1-147 ubiquitination suggests that these enzymes act at two distinct steps. To precisely determine the functional relationship between these ligases, we knocked down gp78 in Hrd1 CRISPR cells. Indeed, under the gp78 knockdown condition, Hrd1 depletion greatly reduced the level of ubiquitinated MHC 1-147 and retrotranslocated deglycosylated heavy chain molecules (Figure 6A). Moreover, the split-GFP assay showed that the gain in green fluorescence observed under BAG6 and gp78 knockdown condition was reversed upon depletion of Hrd1 (Figure 6, B and C). From these data, we concluded that gp78 assists Hrd1-mediated retrotranslocation of MHC 1-147 at a step downstream of retrotranslocation and ubiquitination.
FIGURE 6:
gp78 functions downstream of Hrd1 to promote substrate solubility in ERAD. (A) Depletion of Hrd1 under gp78 knockdown condition reduces substrate ubiquitination. The indicated CRISPR cells were cotransfected with MHC 1-147-FLAG-S11–expressing plasmid together with the indicated shRNA constructs and then treated with MG132 (10 μM, 15 h). MHC 1-147-FLAG-S11 was immunoprecipitated under denaturing conditions and analyzed by immunoblotting. Both ubiquitinated (HA blot) and deglycosylated forms of MHC class I heavy-chain 1-147 (HC) were reduced upon further knockout of Hrd1 in gp78 knockdown cells (lanes 2 and 4). (B and C) Fluorescence intensity in BAG6 or gp78 knockdown cells was reduced upon further depletion of Hrd1 (p < 0.01, n = 3). F/P, fluorescence intensity normalized against protein levels. Cell extracts prepared from a fraction of the cells were analyzed by immunoblotting to verify the protein level (bottom panels). (D) gp78 acts at a postubiquitination step in degradation of MHC 1-147. Cells expressing MHC 1-147 together with HA-tagged ubiquitin and the indicated shRNA constructs were lysed in a NP40-containing lysis buffer. After both the NP40-soluble and NP40-insoluble fractions were obtained, MHC 1-147 was immunoprecipitated from these fractions under denaturing conditions and analyzed by immunoblotting. (E) Both gp78 and BAG6 knockdown causes MHC 1-147 to aggregate in cells. All images were acquired with the same laser setting using a Zeiss LSM 780 confocal microscope. Scale bar: 5 μm.
Because BAG6 is a gp78-interacting chaperone that facilitates the transfer of retrotranslocation substrates from the ER membrane to the proteasome for degradation, and also because we observed accumulation of MHC 1-147 in an NP40-resistant form in both gp78 and BAG6 knockdown cells, we reasoned that gp78 may function together with the BAG6 complex to assist Hrd1 in retrotranslocation. To test this possibility, we further compared the ERAD phenotypes that are caused by knockdown of these factors. Consistent with our hypothesis, knockdown of gp78 and BAG6 gave rise to similar ERAD phenotypes with regard to retrotranslocation and degradation of MHC 1-147: under these knockdown conditions, both ubiquitinated and deglycosylated MHC 1-147 were detected in the NP40-insoluble fraction from gp78 and BAG6 knockdown cells, even without proteasome inhibition, suggesting that retrotranslocation still takes place (Figure 6D, lanes 7–9). MHC 1-147 also accumulated in the glycosylated form in NP40-insoluble fraction, which might be due to retrotranslocation backup or inefficient deglycosylation of retrotranslocated MHC 1-147 that has formed aggregates. Importantly, in MG132-treated cells, depletion of gp78 or BAG6 did not abolish accumulation of either ubiquitinated or deglycosylated MHC 1-147 in the NP40-insoluble fraction. To the contrary, a small increase was observed (Figure 6D, lanes 2 and 3 vs. lane 1, and lanes 5 and 6 vs. lane 4). These results are consistent with the notion that both gp78 and BAG6 function downstream of ubiquitination and retrotranslocation. Immunofluorescence experiments showed that MHC 1-147 was expressed at a low level in control COS7 cells (Figure 6E, panel 1), and it was localized to fine punctate structures that overlapped with an ER marker (Figure 6E, panel 2). When gp78 was knocked down, the level of MHC 1-147 was significantly increased, and the substrate accumulated in large puncta that appeared to be protein aggregates (Figure 6E, panels 3 and 4). Many of these MHC 1-147–containing protein aggregates were not colocalized with the ER marker (Figure 6E, panel 4), suggesting that it was probably formed after retrotranslocation. It is noteworthy that the ER network was significantly disrupted in gp78 knockdown cells as ER appeared to form aggregates. This was probably due to the accumulation of the aggregation-prone retrotranslocated proteins on the cytosolic surface of the ER membrane, which when aggregated, are expected to cause deformation of the ER. Similar phenotypes were observed in BAG6 knockdown cells, except that both the substrate-aggregation and ER-deformation phenotypes were more severe compared with gp78 knockdown cells (Figure 6E, panels 5 and 6). Together these results strongly suggest the possibility that gp78 may be required to maintain the functionality of the BAG6 chaperone complex to facilitate ERAD.
DISCUSSION
In this study, we characterized the gp78 complex with emphasis on its functional relationship with the Hrd1 complex in mammalian ERAD. Previous studies have shown that Hrd1 forms a stable complex with Derlin1, Derlin2, Sel1L, and HERP. We showed that gp78 forms a complex with UbxD8 and UBAC2 in a stoichiometric manner. This finding is consistent with the reported stable interaction between UbxD8 and UBAC2 (Olzmann et al., 2013). On the other hand, under similar conditions, immunoprecipitation of endogenous gp78 only coprecipitated a small fraction of Hrd1, and likewise, immunoprecipitation of endogenous Hrd1 pulled down a small amount of gp78. These results suggest that the two ligase complexes form only transient interactions. We also showed that gp78 interacts with the Hrd1 complex component Derlin2, consistent with a previous report (Christianson et al., 2011). However, unlike other subunits of the Hrd1 complex such as Sel1L, a large fraction of endogenous Derlin2 was coprecipitated with gp78 although the interaction was unstable in the presence of the nonionic detergent NP40 (Christianson et al., 2011). These observations suggest that the Derlin2 molecules bound to gp78 and Hrd1 may not originate from the same cellular pool. Given the reported oligomerization property of Derlins (Ye et al., 2005), Derlin2 might provide a link between Hrd1 and gp78 when the ligase-associated Derlin2 forms a homo-oligomer.
We previously showed that gp78 binds UbxD8 through its transmembrane domains (Xu et al., 2013). We now demonstrate that the UbxD8 transmembrane domain is also crucial for interaction with UBAC2. UbxD8 also interacts with the p97 ATPase by its UBX domain (Lee et al., 2008) and with the chaperone holdase BAG6 via a UBA domain (Xu et al., 2013). Through these interactions, UbxD8 helps recruit these chaperones to gp78 to promote ERAD of both membrane and soluble substrates. UbxD8 is also involved in lipid droplet (LD) metabolism, as its trafficking to LD stabilizes adipose triglyceride lipase, an enzyme that catalyzes the rate-limiting hydrolysis step of triglycerides (Olzmann et al., 2013). The function of UbxD8 in LD formation also requires its interaction with p97, but little is known about the role of UBAC2 in either ERAD or LD metabolism except that it is a rhomboid pseudoprotease that regulates the distribution of UbxD8 between ER and LD (Olzmann et al., 2013). The tight association of gp78 with UbxD8 and UBAC2 suggests a potential role for gp78 and UBAC2 in LD metabolism.
Although it is well established that ubiquitin ligases play important roles in ERAD, the precise function of these enzymes in this process is not completely defined. It is assumed that these enzymes are involved in ubiquitination of misfolded ERAD substrates, given the obvious requirement of ubiquitination in substrate delivery to the proteasome. Our study confirms that mammalian Hrd1 is essential for ubiquitination of both luminal and membrane ERAD substrates. However, our results do not support a similar role for gp78 in retrotranslocation. This is in line with a previous report showing that the transmembrane domains of gp78 were largely dispensable for ERAD (Tsai et al., 2007).
If gp78 can promote ERAD independent of substrate ubiquitination, what is its function in ERAD? Our recent work demonstrated that gp78 interacts with the cytosolic chaperone holdase complex BAG6-Ubl4A-Trc35. The latter promotes ERAD by maintaining the solubility of retrotranslocated polypeptides (Wang et al., 2011). We also showed that gp78 regulates ubiquitination of Ubl4A. Although hyperubiquitination of Ubl4A does not change its stability, it can negatively regulate BAG6 activity by controlling its integrity and its interaction with the cochaperone SGTA (Liu et al., 2014). The proper function of BAG6 seems to require the activity of the deubiquitinating enzyme USP13, which antagonizes gp78-mediated ubiquitination to ensure ERAD efficiency (Liu et al., 2014). The fact that gp78, although dispensable for retrotranslocation and ubiquitination, is still required for ERAD suggests that gp78-dependent ubiquitination of Ubl4A may also positively regulate BAG6 function. One possible explanation for these observations is that gp78-dependent ubiquitination and USP13-mediated deubiquitination form a cycle that regulates ordered interactions of BAG6 with distinct effectors during each round of retrotranslocation. The proposed regulation of BAG6 by gp78 is consistent with the observation that MHC 1-147 is accumulated in an NP40-insoluble form in both gp78 and BAG6 knockdown cells, and with the report that gp78 can also modulate the solubility of the ERAD substrate α1-antitrypsin to facilitate its degradation (Shen et al., 2006).
Recent studies suggest that ubiquitin-dependent regulation of ERAD machinery may be a common theme. Hrd1p also undergoes auto-ubiquitination, which may regulate either the oligomerization state or the activity of Hrd1p during retrotranslocation (Stein et al., 2014). Another component of the Hrd1 complex, HERP, could also be ubiquitinated by gp78, but in this case, the modification appears to only regulate the HERP stability (Yan et al., 2014). Future studies may elucidate more regulations of similar kinds, but it is noteworthy that identification of ERAD machinery factors as gp78 substrates does not necessarily rule out the possibility that gp78 may also be responsible for ubiquitination of certain ERAD substrates, as suggested previously (Lee et al., 2006; Chen et al., 2012).
Notably, previous studies and our current work have shown that small interfering RNA (siRNA)- or shRNA-mediated knockdown of gp78 inhibits the degradation of many ERAD substrates (Chen et al., 2012). However, our studies using the gp78 CRISPR cells have not revealed any significant ERAD defects in cells lacking gp78. We presume that the ERAD phenotypes observed in gp78 knockdown cells are not caused by an unknown off-target effect, because they were observed using siRNA/shRNAs with different targeting sequences. In addition, siRNA-mediated knockdown of each component of the gp78 complex (including gp78, UbxD8, and UBAC2) inhibited ERAD in all instances (Fang et al., 2001; Song et al., 2005; Shen et al., 2006; Tsai et al., 2007; Morito et al., 2008; Christianson et al., 2011; Jo et al., 2011a, b; Liu et al., 2014). By contrast, CRISPR cells lacking these proteins are functionally competent in ERAD, at least for the substrates tested here (Figure 3; unpublished data). Therefore, the inconsistency between the knockdown and CRISPR studies is probably due to an unknown mechanism that adapts the CRISPR cells to gene deficiency. In fact, as CRISPR knockout cell lines were each derived from a single cell, it took several weeks of clonal expansion to establish these lines. As a result, cells were given sufficient time to adapt to gene deficiency. If one function of gp78 is to regulate BAG6 via ubiquitination of Ubl4A, the proposed adaptation does not seem to involve a redundant chaperone, because shRNA-mediated silencing of BAG6 still caused significant stabilization of MHC 1-147 in gp78-deficient CRISPR cells (unpublished data). Thus the regulation of the BAG6 complex might be achieved by additional ubiquitin ligase(s) in the absence of gp78. Consistent with this view, we noticed previously that knockdown of gp78 only reduces rather than completely abolishes ubiquitinated Ubl4A, suggesting the existence of other ubiquitin ligases for Ubl4A (Liu et al., 2014). Regardless of the mechanism by which CRISPR cells adapt to gp78 deficiency in vitro, the observation that liver-specific ablation of gp78 in mice does inhibit lipid biosynthesis, in part due to stabilization of the gp78 substrate Insig-1 in vivo (Liu et al., 2012), indicates that adaptation to gene deletion observed in tissue culture cells may not occur at the organism level. Moreover, it is worth pointing out that although gp78 CRISPR cells did not have any apparent ERAD defects, the Hrd1 CRISPR cells generated under the same condition were impaired in ERAD of both luminal and membrane substrates. These observations further underscore the differential roles played by these ligases in ERAD.
In summary, our study demonstrates that gp78 has an accessory function both downstream and in parallel with Hrd1 in ERAD. A plausible explanation for this observation is that gp78 may cooperate with both Hrd1 and another ubiquitin ligase, each of which forms a route to export ERAD substrates (Figure 7). This model is supported by genetic evidence that knockout of the hrd1 gene does not completely abolish either retrotranslocation or ubiquitination of MHC 1-147, and that knockdown of gp78 in Hrd1 CRISPR cells can lead to further stabilization of this substrate. We propose that the gp78 complex acts as an assisting module downstream of these ubiquitin ligases. The requirement for gp78 may be largely confined to substrates that are prone to aggregation, which necessitates the involvement of a downstream chaperone holdase such as BAG6. The proposed modular organization of the mammalian ERAD system resolves the long-standing discrepancy between the budding yeast and the mammalian ERAD systems, and indicates that de novo origination of additional accessory modules accounts for the complexity of the mammalian ERAD system.
FIGURE 7:
The functional relationship between gp78 and Hrd1. Hrd1 is the essential retrotranslocation regulator conserved in yeast and mammalian cells, whereas gp78 serves an assisting role downstream of Hrd1 and possibly another ubiquitin ligase in mammalian cells. gp78 may promote ERAD by maintaining the functionality of the BAG6 complex.
MATERIALS AND METHODS
Cell lines, plasmids, and antibodies
The HEK293T cells were purchased from the American Type Culture Collection (Manassas, VA) and maintained in DMEM (Corning cellgro) containing 10% fetal bovine serum (FBS) and penicillin–streptomycin (10 U/ml). CRISPR cell lines were generated using the CAS9 D10A nickase following the published procedures (Ran et al., 2013). Specifically, for each gene to be targeted, we chose two guiding RNA sequences. For each targeting construct, a pair of oligos corresponding to the guiding RNA sequences were synthesized and annealed. The annealed oligos were ligated into pX330-U6-Chimeric_BB-CBh-hSpCas9n containing the D10A mutation using the BbsI ligation sites. The pX330-U6-Chimeric_BB-CBh-hSpCas9 was a gift from Feng Zhang (Addgene plasmid 42230). The D10A mutation was introduced by PCR-based site-directed mutagenesis. The two targeting plasmids were cotransfected into HEK293T cells. At 48 h posttransfection, genomic DNA was collected from a fraction of the cells to verify the efficiency of target gene interruption by Surveyor assay (Transgenomic, NE). The remaining cell pool was diluted and seeded at less than one cell in a single well in a 96-well plate. Single clones were collected and expanded. Positive clones were selected by immunoblot analyses of the target protein level.
Plasmids expressing UBXD8 constructs were described previously (Xu et al., 2013). The D8-TM-Citrine–expressing construct was made by fusing the transmembrane domain of UbxD8 (residues 84–121) and monomeric citrine YFP (mCitrine-C1 was a kind gift from Robert Campbell, Michael Davidson, Oliver Griesbeck, and Roger Tsien; Addgene plasmid 54587) using two rounds of PCR. The final product of the PCR was cloned to SalI and NotI sites of the pRK5 vector.
Plasmid expressing WT UBCA2 was purchased from Origene (Rockville, MD). Plasmids expressing gp78 shRNA and the corresponding control plasmid were a kind gift of Fang Shenyun (University of Maryland).
For construction of the pCMV-MHC1-147-FLAG-S11 plasmid, a DNA fragment encoding the first 147 amino acids of the HLA-A2 allele of the MHC class I heavy chain with a FLAG tag appended at the carboxy terminus was amplified by PCR using the following primers:
Forward: 5′-ACGCGGAAGATCTCACCATGGTACCGTGCACGC TGCTC-3′
Reverse: 5′-ACGCGTCGACCCTTTGTCATCATCGTCCTTGTAG TCCAGGGCGATGTAATCCTTGCC-3′
The DNA fragment was purified and digested by BglII and SalI and then ligated to the pCMV S11 GFP vector (Sandia BioTech). For creating pCMV-MHC1-147-FLAG, a stop codon was introduced right after the FLAG tag in the pCMV-MHC1-147-FLAG-s11 construct by site-directed mutagenesis. For generation of a pLNCX2-TCRα-YFP-FLAG construct, pLNCX2 TCRα-YFP plasmid was used as a template. A FLAG tag was inserted downstream of the YFP tag by PCR using the following primers:
Forward: 5′-ATGGACGAGCTGTACAAGGATTACAAGGATGAC GACGATAAGTGAGTCGACAGGCCTA-3′
Reverse: 5′-TAGGCCTGTCGACTCACTTATCGTCGTCATCCTTG TAATCCTTGTACAGCTCGTCCAT-3′
siRNA targeting gp78 was purchased from Dharmacon, GE Healthcare (Lafayette, CO): ON-TARGETplus SMARTpool siRNA targeting AMFR (J-006522-05, J-006522-06, J-006522-07, J-006522-08). siRNA targeting UBXD8 was purchased from Life Technologies (Grand Island, NY), siRNA ID s23260.
Antibodies for Hrd1 were generated using the following peptides as antigens (CDGEPDAAELRRRRLQKLE-amide and CHSVDLQSENPWDNKAVY-amide). The antibodies were affinity purified using resins immobilized with these peptides following a standard protocol from Thermo Scientific. gp78, GFP, BAG6, H2A, and p97 antibodies were previously described (Wang et al., 2011). Commercial antibodies used in the study include anti-FLAG M2 (Sigma-Aldrich F1804; 1:2000), anti-HA (Sigma-Aldrich H3663; 1:1000), anti-UBXD8 (Protein Tech 16251-1-AP; 1:1000), anti-ubiquitin P4D1 (Santa Cruz Biotechnology sc-8107; 1:1000), and anti-p97 (Fitzgerald 10R-2367; 1:1000). The anti-UBAC2 antibody is a kind gift of Peter Espenshade (Johns Hopkins University).
Transfection and gene silencing
On day 0, 0.5 × 106 control or Hrd1 CRISPR cells were seeded. shRNA constructs together with the plasmid expressing ERAD substrates were cotransfected using TransIT 293 on day 1. Cells were harvested 48 h posttransfection.
Immunoblotting and immunoprecipitation
Cells were lysed in NP40 lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM sodium chloride, 2 mM magnesium chloride, 0.5% NP40, 1 mM dithiothreitol [DTT], and a protease inhibitor mixture) unless indicated otherwise. Cell extracts were subject to centrifugation to separate soluble and insoluble fractions. For most experiments, the soluble fractions were analyzed. Where indicated in the figure legends, the NP40-insoluble pellet fractions were resolubilized by Laemmli buffer for immunoblotting. Immunoblotting was performed according to the standard protocol. Fluorescently labeled secondary antibodies (Rockland) were used for detection. The fluorescent bands were imaged and quantified on a Li-Cor Odyssey infrared imager using the software provided by the manufacturer. For immunoprecipitation, the soluble extract was incubated with FLAG-agarose beads (Sigma-Aldrich) or protein A–Sepharose CL-4B (GE Healthcare) bound with antibodies against specific proteins. After incubation, the beads were washed two times with NP40 wash buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM sodium chloride, 2 mM magnesium chloride, and 0.1% NP40. The proteins on beads were assayed by immunoblotting.
For detection of ubiquitin conjugates on ERAD substrates, HEK293T cells (∼2 × 106) were harvested 24 h posttransfection and lysed in 150 μl buffer D (1% SDS, 5 mM DTT). Cells were immediately heated at 95°C for 10 min to disrupt protein complexes. Cell extract was diluted 10-fold with the NP40 lysis buffer containing a protease inhibitor cocktail. After centrifugation at 16,100 × g for 10 min to remove insoluble materials, cleared cell lysates were subject to immunoprecipitation with FLAG beads.
All immunoblot images are representative of at least two independent experiments. Data shown in the bar figures are the average values from three independent experiments with the SDs. p Values were calculated using a two-tailed t test.
Immunofluorescence experiments
For detection of the subcellular localization of protein by fluorescence labeling, cells were seeded on fibronectin-coated coverglass and transfected. Cells were then fixed with phosphate buffer saline containing 2% paraformaldehyde for 20 min at room temperature, after which they were permeabilized with phosphate buffer saline containing 10% FBS and 0.2% saponin and stained with antibodies in the same buffer according to a standard protocol. Images were acquired with a Zeiss LSM780 confocal microscope.
Cycloheximide chase
Gene knockdown and plasmid transfection were performed as described above. Cells were harvested 48 h after transfection and were resuspended in 1.8 ml fresh DMEM containing 50 mM HEPES buffer (pH 7.5) and 50 μg/ml cycloheximide. Cells were then incubated at 37ºC. Equal numbers of cells were taken at 0, 1, and 2 h for immunoblot analysis.
The split-GFP assay
shRNA constructs or siRNA were cotransfected with pCMV-GFP 1-10 and MHC1-147 S11 plasmids into the same number of control or hrd1 CRISPR cells on day 1. At 48 h posttransfection, cells were harvested in 1 ml phosphate-buffered saline (PBS). Cells in PBS (100 μl) were added to a 96-well plate to measure the fluorescence intensity by the Victor 3 plate reader (Perkin Elmer-Cetus). PBS was used as a blank control. The remaining cells were lysed, and the extracts were analyzed by immunoblotting as described above.
Acknowledgments
We thank members of the Ye lab for discussions and suggestions and Nia Soetandyo for creating the pCMV-MHC1-147-FLAG-S11 construct. This research was supported by the Intramural Research Program of the National Institute of Diabetes, Digestive and Kidney Diseases.
Abbreviations used:
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HA
hemagglutinin
- HLA
human leukocyte antigen
- LD
lipid droplet
- MHC
major histocompatibility complex
- NP40
Nonidet P-40
- PBS
phosphate-buffered saline
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-06-0354) on September 30, 2015.
REFERENCES
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Bays NW, Hampton RY. Cdc48-Ufd1-Npl4: stuck in the middle with Ub. Curr Biol. 2002;12:R366–R371. doi: 10.1016/s0960-9822(02)00862-x. [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom D, Hirsch C, Stern P, Tortorella D, Ploegh HL. A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised. EMBO J. 2004;23:650–658. doi: 10.1038/sj.emboj.7600090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr ML, van den Boomen DJ, Bye H, Antrobus R, Wiertz EJ, Lehner PJ. MHC class I molecules are preferentially ubiquitinated on endoplasmic reticulum luminal residues during HRD1 ubiquitin E3 ligase-mediated dislocation. Proc Natl Acad Sci USA. 2013;110:14290–14295. doi: 10.1073/pnas.1303380110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Du S, Fang S. gp78: a multifaceted ubiquitin ligase that integrates a unique protein degradation pathway from the endoplasmic reticulum. Curr Protein Pept Sci. 2012;13:414–424. doi: 10.2174/138920312802430590. [DOI] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2011;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, Mc Feeters RL, Altieri AS, Ji X, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O, Ruggiano A, Hannibal-Bach HK, Ejsing CS, Carvalho P. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. eLife. 2013;2:e00953. doi: 10.7554/eLife.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeck G, Ebner FA, Shimada-Kreft H, Kreft SG. The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J Cell Biol. 2015;209:621. doi: 10.1083/jcb.20140808804292015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Huang CH, Chu YR, Ye Y, Chen X. Role of HERP and a HERP-related protein in HRD1-dependent protein degradation at the endoplasmic reticulum. J Biol Chem. 2013a;289:4444–4454. doi: 10.1074/jbc.M113.519561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Hsiao HT, Chu YR, Ye Y, Chen X. Derlin2 protein facilitates HRD1-mediated retro-translocation of sonic hedgehog at the endoplasmic reticulum. J Biol Chem. 2013b;288:25330–25339. doi: 10.1074/jbc.M113.455212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura S, Weissman AM, Bonifacino JS. Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem. 2010;285:23916–23924. doi: 10.1074/jbc.M110.127936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- Jo Y, Lee PC, Sguigna PV, DeBose-Boyd RA. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc Natl Acad Sci USA. 2011a;108:20503–20508. doi: 10.1073/pnas.1112831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y, Sguigna PV, DeBose-Boyd RA. Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 2011b;286:15022–15031. doi: 10.1074/jbc.M110.211326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JN, Song B, DeBose-Boyd RA, Ye J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J Biol Chem. 2006;281:39308–39315. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- Lee JN, Zhang X, Feramisco JD, Gong Y, Ye J. Unsaturated fatty acids inhibit proteasomal degradation of Insig-1 at a postubiquitination step. J Biol Chem. 2008;283:33772–33783. doi: 10.1074/jbc.M806108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TF, Tang JJ, Li PS, Shen Y, Li JG, Miao HH, Li BL, Song BL. Ablation of gp78 in liver improves hyperlipidemia and insulin resistance by inhibiting SREBP to decrease lipid biosynthesis. Cell Metab. 2012;16:213–225. doi: 10.1016/j.cmet.2012.06.014. [DOI] [PubMed] [Google Scholar]
- Liu Y, Soetandyo N, Lee JG, Liu L, Xu Y, Clemons WM, Jr, Ye Y. USP13 antagonizes gp78 to maintain functionality of a chaperone in ER-associated degradation. eLife. 2014;3:e01369. doi: 10.7554/eLife.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ye Y. Proteostasis regulation at the endoplasmic reticulum: a new perturbation site for targeted cancer therapy. Cell Res. 2011;21:867–883. doi: 10.1038/cr.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, Tanaka K, Iwai K, Nagata K. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRΔF508. Mol Biol Cell. 2008;19:1328–1336. doi: 10.1091/mbc.E07-06-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci USA. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc Natl Acad Sci USA. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich K, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014;204:869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Ballar P, Fang S. Ubiquitin ligase gp78 increases solubility and facilitates degradation of the Z variant of alpha-1-antitrypsin. Biochem Biophys Res Commun. 2006;349:1285–1293. doi: 10.1016/j.bbrc.2006.08.173. [DOI] [PubMed] [Google Scholar]
- Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell. 2014;158:1375–1388. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Mendoza A, Mariano JM, Zhou M, Kostova Z, Chen B, Veenstra T, Hewitt SM, Helman LJ, Khanna C, Weissman AM. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat Med. 2007;13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
- Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Liu Y, Soetandyo N, Baek K, Hegde R, Ye Y. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell. 2011;42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Liu Y, Lee JG, Ye Y. A ubiquitin-like domain recruits an oligomeric chaperone to a retrotranslocation complex in endoplasmic reticulum-associated degradation. J Biol Chem. 2013;288:18068–18076. doi: 10.1074/jbc.M112.449199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Liu W, Zhang H, Liu C, Shang Y, Ye Y, Zhang X, Li W. Ube2g2-gp78-mediated HERP polyubiquitylation is involved in ER stress recovery. J Cell Sci. 2014;127:1417–1427. doi: 10.1242/jcs.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. Inaugural article: recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Fang S. Live cell imaging of protein dislocation from the endoplasmic reticulum. J Biol Chem. 2012;287:28057–28066. doi: 10.1074/jbc.M112.381798. [DOI] [PMC free article] [PubMed] [Google Scholar]