Abstract
Emerging evidence indicates that microRNA control and modulate immunity. MicroRNA have not been investigated in acquired aplastic anemia, a T-cell-mediated immune disease. Analysis of 84 microRNA expression levels in CD4+ and CD8+ T cells of patients with aplastic anemia revealed concurrent down-regulation of miR-126-3p, miR-145-5p, miR-223-3p, and miR-199a-5p (>3-fold change, P<0.05) in both T-cell populations, which were unique in aplastic anemia compared to other hematologic disorders. MiR-126-3p and miR-223-3p were down-regulated in CD4+ T effector memory cells, and miR-126-3p, miR-145-5p, and miR-223-3p were down-regulated in CD8+ T effector memory and terminal effector cells. Successful immunosuppressive therapy was associated with restoration to normal expression levels of miR-126-3p, miR-145-5p, and miR-223-3p (>2-fold change, P<0.05). In CD4+ and CD8+ T cells in aplastic anemia patients, MYC and PIK3R2 were up-regulated and proved to be targets of miR-145-5p and miR-126-3p, respectively. MiR-126-3p and miR-145-5p knockdown promoted proliferation and increased interferon-γ and granzyme B production in both CD4+ and CD8+ T cells. Our work describes previously unknown regulatory roles of microRNA in T-cell activation in aplastic anemia, which may open a new perspective for development of effective therapy. Clinicaltrials.gov identifier: NCT 01623167
Introduction
Aplastic anemia (AA) is an acquired bone marrow disease characterized by trilineage marrow hypoplasia, a paucity of hematopoietic stem and progenitor cells, and pancytopenia of the peripheral blood due to immune cell attack on the bone marrow. The responsiveness of AA to immunosuppressive therapies remains the best evidence of an underlying immune pathophysiology: the majority of patients show hematologic improvement after only transient T-cell depletion by antithymocyte globulins.1 Although immunity to hematopoietic progenitors by activated T cells has been considered to be responsible for the pathogenesis of AA, little is known about the molecular basis of T-cell activation.
Effector cells in AA have been identified by immunophenotyping as activated cytotoxic T cells expressing Th1 cytokines, especially γ-interferon (IFN-γ);2 by oligoclonal expansion of CD8+ CD28− T cells, defined by flow cytometry for T-cell receptor (TCR) Vβ subfamilies; and using spectratyping to detect skewing of CDR3 length and sequencing of the CDR3 region to define a molecular clonotype.3 Polymorphisms in cytokine genes, associated with an increased immune response, are also more prevalent in AA.4 Constitutive expression of T-bet, a transcriptional regulator that is critical to Th1 polarization, is present in the majority of AA patients.5 Genome-wide transcriptional analysis of T cells from AA patients has implicated some components of innate immunity in AA, including Toll-like receptors and natural killer cells.6 However, the precise mechanisms underlying activation of T cells in AA are still unclear.
MicroRNA (miRNA, miR) are a group of small, conserved, non-coding RNA molecules that primarily modulate gene expression at the post-transcriptional level by hybridization to complementary sequences in the 3′ untranslated region (3′UTR) of their corresponding mRNA.7 miRNA bind to the ribonucleoprotein complex RNA-induced silencing complex, which in addition also binds to the 3′ UTR of complementary mRNA.8 The double-stranded complex between miRNA and mRNA is then degraded, which leads to decreased protein translation.9
Approximately 30% of the human genome is estimated to be regulated by miRNA, and a single miRNA can potentially regulate hundreds of proteins.10 More than 1,000 miRNA have been identified in mammals and implicated in a wide range of biological functions.11 MiRNA contribute to the pathophysiology of a number of important human diseases such as cancer,12 cardiovascular disease, and neurodegenerative disorders.13,14 There is emerging evidence that miRNA play crucial roles in controlling and modulating immunity.15 Dysregulation of miRNA can lead to autoimmune diseases, such as rheumatoid arthritis, multiple sclerosis, and inflammatory bowel disease.16–18 Normalization of dysregulated miRNA can be therapeutic in murine disease models. MiRNA thus represent novel molecular diagnostic markers and potential targets for therapeutics.19
We hypothesized that dysregulated miRNA expression might lead to aberrant T-cell activation in AA. In this work, we used quantitative reverse transcriptase polymerase chain reaction (RT-qPCR)–based approaches to assess miRNA expression in CD4+ and CD8+ T cells from AA patients. We demon strate that down-regulation of miR-126-3p and miR-145-5p promotes CD4+ and CD8+ T-cell activation by increasing MYC and PIK3R2 expression levels and T-cell proliferation, of potential importance in the pathogenesis of AA. Our results provide a pharmacological rationale for the potential use of synthetic miRNA mimics to limit disease.
Methods
Patients and treatment
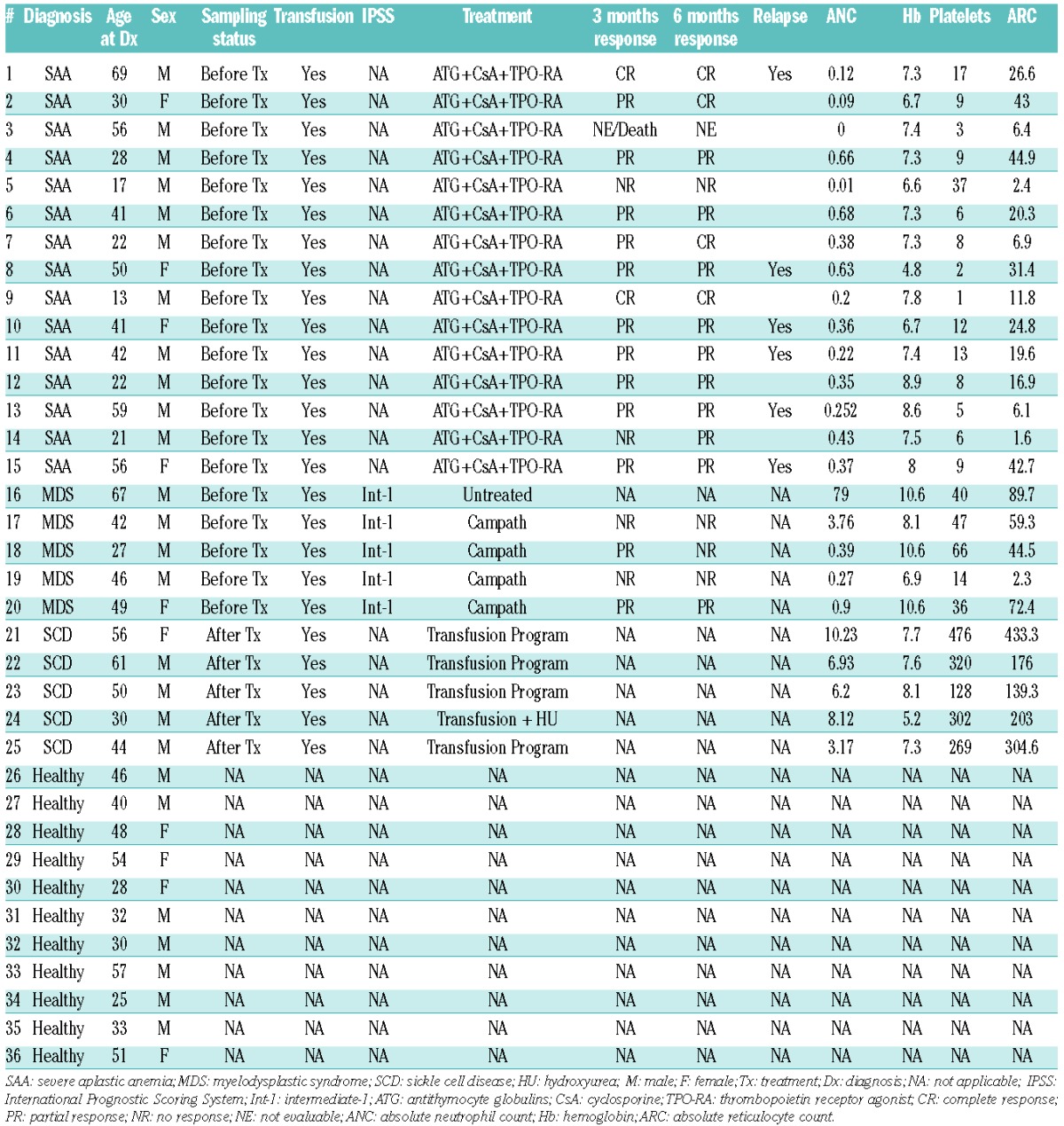
Blood samples were obtained after informed consent from 15 patients with severe AA and 11 age-matched healthy donors. The median age of AA patients was 41 years (range, 13–69 years). MiRNA expression levels of all 15 patients were analyzed at diagnosis. Standard criteria were used for the diagnosis of AA and the evaluation of disease severity.20 From 15 AA patients, 12 samples were used to determine miRNA expression in lymphocyte subsets and three samples were used to determine miRNA expression in T-cell subsets. From 11 healthy donors, eight samples were used to determine miRNA expression in lymphocyte subsets, and three samples were used to determine miRNA expression in T-cell subsets. Serial samples were collected before and after immunosuppressive therapy in six cases. Blood samples from five patients with low-risk myelodysplastic syndrome (MDS) at diagnosis and five patients with red blood cell transfusion-dependent sickle cell disease (SCD) were used for comparison. The demographic and clinical characteristics of the 25 patients are summarized in Table 1. All AA patients received horse anti-thymocyte globulin + cyclosporine + eltrombopag on a clinical research protocol (clinicaltrials.gov, #NCT01623167). All human subjects were enrolled on clinical protocols approved by the NHLBI Institutional Review Board.
Table 1.
Characteristics of the patients and healthy controls.
Flow cytometry and cell sorting
The gating strategies for sorting lymphocyte subsets in human or mouse samples, and T-cell subsets in human samples are summarized in Online Supplementary Figure S1.
RNA isolation
Total RNA, including small RNA, was isolated using the miRNeasy kit (QIAGEN, Valencia, CA, USA), according to the manufacturer’s instructions. RNA concentration was measured using a Nanodrop device (Peqlab, Erlangen, Germany).
T-cell and B-cell activation pathway-focused microRNA polymerase chain reaction-array analysis
Using miScript miRNA PCR Array Human or Mouse T-Cell and B-Cell Activation (QIAGEN, Sabiosciences) with 84 miRNA (Online Supplementary Table S1), miRNA PCR array focused on T-cell and B-cell activation pathways was performed, according to the manufacturer’s instructions. For each array, a minimum of 250 ng total RNA was retrotranscribed using the miScript II RT kit (QIAGEN) with HiSpec buffer, according to the manufacturer’s instruction. Data were median-centered and submitted to hierarchical clustering using the Cluster 3 program with Pearson correlation as similarity metrics and centroid linkage clustering. Results were displayed as heatmaps generated using the TreeView program.21
Custom microRNA targets polymerase chain reaction-array analysis
To examine target genes of four selected miRNA, we designed the custom RT2 Profiler™ microRNA Targets PCR Arrays with 84 target genes (Online Supplementary Table S2), based on target gene prediction. For each array, 300 ng total RNA was retrotranscribed using the RT2 First Strand kit (QIAGEN), according to the manufacturer’s instruction.
Further experimental procedures are described in the Online Supplementary Experimental Methods.
Results
Distinct patterns of microRNA expression in CD4+ and CD8+ T cells from patients with aplastic anemia
First, miRNA expression levels in CD4+ and CD8+ T-cell populations of AA patients were analyzed using a commercial miRNA platform. This platform allows measurement of 84 miRNA involved in lymphocyte activation. For this array, we tested 12 samples (#1 – #12) from 15 AA patients and eight samples (#26 – #33) from 11 healthy donors (Table 1). A threshold of 3-fold change (FC) in expression and a two-tailed P value cutoff of 0.01 were used to identify miRNA that were differentially expressed in the AA and control groups. Expression levels of four miRNA (miR-126-3p, miR-145-5p, miR-199a-5p, and miR-223-3p) among 84 miRNA were significantly down-regulated (>3 FC, P<0.01) in both CD4+ and CD8+ T cells of AA patients compared with those of healthy controls, as highlighted in volcano plots (Figure 1A,C and Online Supplementary Table S3). For comparison, CD19+ B cells were also examined in the same PCR array platform, showing significant down-regulation (>3 FC, P<0.01) of miR-126-3p, miR-145-5p, and miR-199a-5p (Figure 1E and Online Supplementary Table S3). We observed dysregulation of these four miRNA in the 12 AA patients relative to eight healthy donors (Figure 1B,D,F). Venn diagrams summarize expression profiles of the four miRNA in both T and B cells from AA patients compared with healthy controls: down-regulation of miR-126-3p, miR-145-5p, and miR-199a-5p in both T- and B-cell populations from AA patients and down-regulation of miR-223-3p only in T cells from AA patients (Figure 1G). Thus, three of the four miRNA expression patterns were similar across T- and B-cell lineages.
Figure 1.
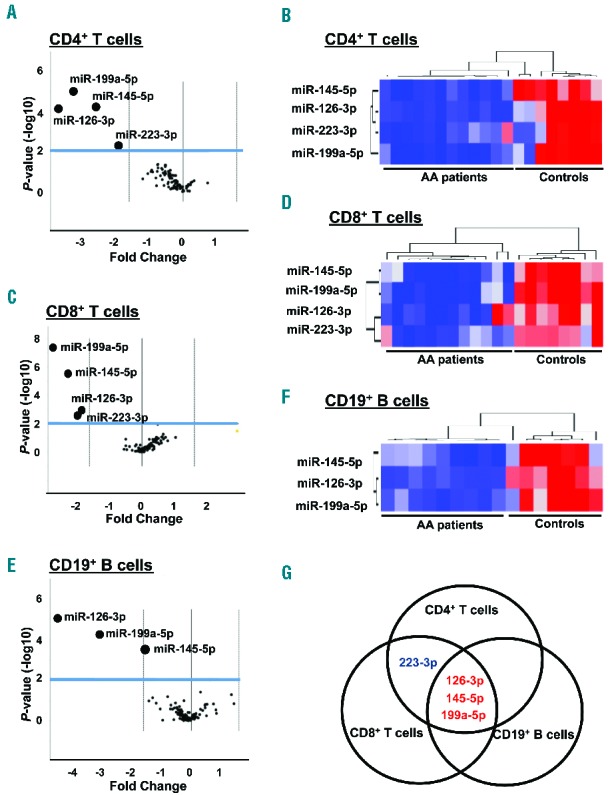
Distinct miRNA expression patterns in CD4+ and CD8+ T cells of AA patients. Volcano plots of results from 84 miRNA known to be involved in lymphocyte activation in (A) CD4+ and (C) CD8+ T cells (AA patients, n=12; healthy donors, n=8) and (E) CD19+ B cells (AA patients, n=9; healthy donors, n=7) using miRNA PCR-array. The x-axis is the estimated difference in expression measured in log2; vertical lines refer to a 3-fold difference in expression between the two groups. MiRNA highly expressed in AA or healthy donors are on the right or the left, respectively. The y-axis is the significance of the difference measured in −log10 of the P-value; the horizontal line represents our cutoff for significance at P<0.01. Hierarchical clustering of miRNA in (B) CD4+ T cells, (D) CD8+ T cells, or (F) CD19+ B cells was visualized by heatmap analysis. A red-blue color scale depicts normalized miRNA expression levels in Ct values (red: high, blue: low). (G) Venn diagram presentation of PCR array data.
Comparison of microRNA expression patterns between patients with aplastic anemia or other hematologic disorders
To determine whether the miRNA dysregulation observed in AA was distinct from miRNA expression in other hematologic diseases, miRNA levels of CD4+ and CD8+ T cells from five low-risk MDS patients and five transfusion-dependent SCD patients (Table 1) were assessed using the miRNA PCR array platform. Disease-specific miRNA expression patterns were observed (Online Supplementary Figures S2 and S3). In both CD4+ and CD8+ T cells of MDS patients, miR-182-5p was up-regulated (>3 FC, P<0.01), whereas miR-199a-5p and miR-19a-3p were down-regulated (>3 FC, P<0.01), compared with healthy controls. However, decreased miR-126-3p and miR-223-3p (>3 FC, P<0.01) expression levels were seen only in CD8+ T cells. In both CD4+ and CD8+ T cells from SCD patients, miR-34a-5p expression was enhanced (>3 FC, P<0.01) and miR-199a-5p and miR-142-5p expression was decreased (>3 FC, P<0.01) compared with levels in healthy controls.
For validation, levels of expression of four miRNA (miR-126-3p, miR-145-5p, miR-199a-5p, and miR-223-3p) in CD4+ and CD8+ T cells were measured by RT-qPCR. MiR-199a-5p expression was reduced in both CD4+ and CD8+ T cells from patients with AA, MDS, and SCD (>3 FC, P<0.05). Decreased expression of miR-223-3p (>3 FC, P<0.05) in CD4+ and CD8+ T cells was observed in patients with low-risk MDS. MiR-126-3p expression was normal in both CD4+ and CD8+ T cells from SCD patients, but it was down-regulated in CD8+ T cells from patients with MDS and CD4+ and CD8+ T cells from those with AA (>3 FC, P<0.05). In both CD4+ and CD8+ T cells from MDS patients, miR-145-5p expression was not significantly changed compared with that in healthy controls. Accordingly, concurrent down-regulation of miR-126-3p, miR-145-5p, miR-223-3p, and miR-199a-5p (>3 FC, P<0.05) in both CD4+ and CD8+ T cells was a distinguishing feature of AA (Figure 2A,B).
Figure 2.
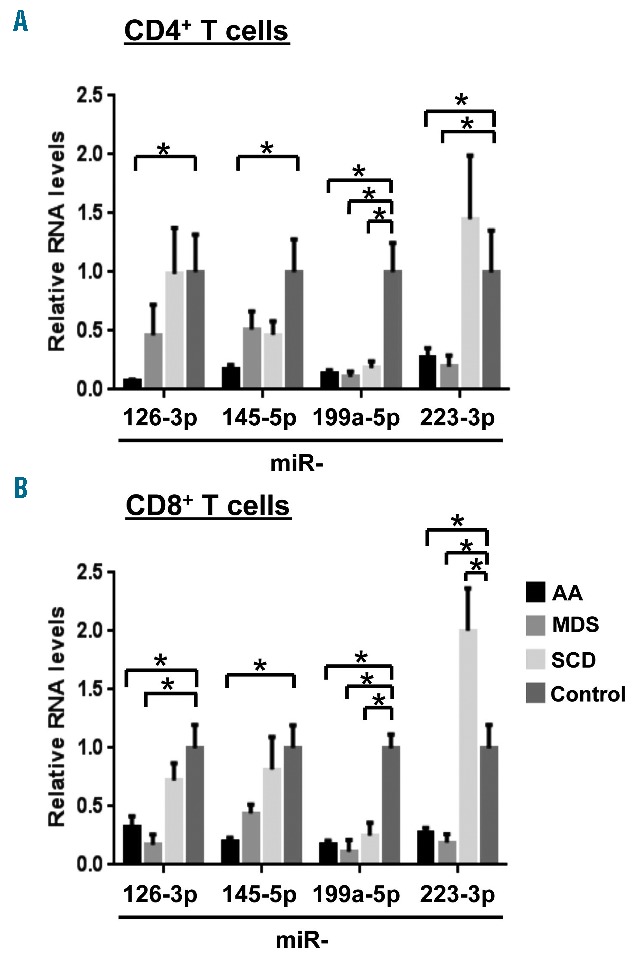
Comparison of expression levels of specific miRNA in other hematologic diseases. RT-qPCR analysis of miR-126-3p, miR-145-5p, miR-199a-5p, and miR-223-3p expression in (A) CD4+ and (B) CD8+ T cells from AA (n=12), low-risk MDS (n=5), and SCD (n=5) patients, and healthy controls (n=8). MiRNA relative expression was obtained by normalizing to RNU-2 expression. *P<0.05 [two-way analysis of variance (ANOVA)].
MicroRNA expression profiles in CD4+ and CD8+ T-cell subsets from patients with aplastic anemia
T cells from three AA patients and three healthy controls were sorted by flow cytometry: CD4+ T cells into three subsets [naïve (TN), central memory (TCM), and effector memory (TEM)] and CD8+ T cells into four subsets [TN, TCM, TEM, and T effector (TE)]. Expression of the four down-regulated miRNA in AA patients was measured in individual T-cell subsets by RT-qPCR in order to identify those cells responsible for differential expression of the four miRNA. Among CD4+ T cells, miR-126-3p and miR-223-3p were down-regulated in a TEM population in AA patients (Figure 3A, P<0.05), compared with controls. In CD8+ T cells, down-regulation of miR-126-3p, miR-145-5p, and miR-223-3p was present in both TEM and TE populations in AA (Figure 3B, P<0.05). However, no significantly different miR-199a-5p expression levels were detected in either CD4+ or CD8+ T-cell subsets. Thus there was a clear tendency of down-regulation of miR-126-3p, miR-145-5p, and miR-223-3p from TN toward TEM or TE populations in CD8+ T cells from AA patients.
Figure 3.
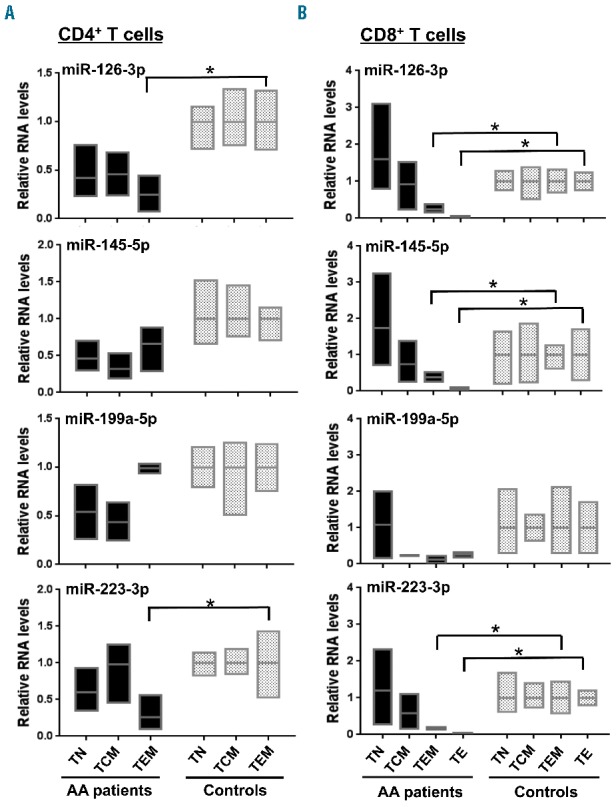
MiRNA expression in CD4+ and CD8+ T cell subsets of AA patients. RT-qPCR analysis of miR-126-3p, miR-145-5p, miR-199a-5p, and miR-223-3p expression in (A) CD4+ and (B) CD8+ T cell subsets from AA patients (n=3) and healthy controls (n=3). The TE population in CD4+ T cells was not examined, due to the low number of cells in healthy controls. Relative expression of miRNA was calculated by normalizing to RNU-2 expression. *P<0.05 [two-way analysis of variance (ANOVA)].
Altered microRNA expression patterns after immunosuppressive therapy
To address the status of miRNA expression levels after immunosuppressive therapy, we sorted CD4+ and CD8+ T cells from six AA patients after immunosuppressive therapy (#1, #7, #9, #10, #11, and #12) and measured miRNA expression by RT-qPCR. All six patients achieved partial or complete responses (Table 1). Analysis of the four miRNA (miR-126-3p, miR-145-5p, miR-199a-5p, and miR-223-3p) showed restoration of expression levels of miR-126-3p, miR-145-5p, and miR-223-3p (>2 FC, P<0.05) after successful immunosuppressive therapy as compared to the same cases tested before immunosuppressive therapy (Figure 4A,B). The percentage restoration of three miRNA (miR-126-3p, miR-145-5p, and miR-223-3p) after immunosuppressive therapy, relative to levels in healthy controls, were 54%, 96%, and 57% in CD4+ T cells, and 93%, 109% and 94% in CD8+ T cells, respectively. However, recovery of miR-199a-5p expression was not seen, suggesting that miR-199a-5p is regulated by a cellular mechanism distinct from the other three miRNA.
Figure 4.
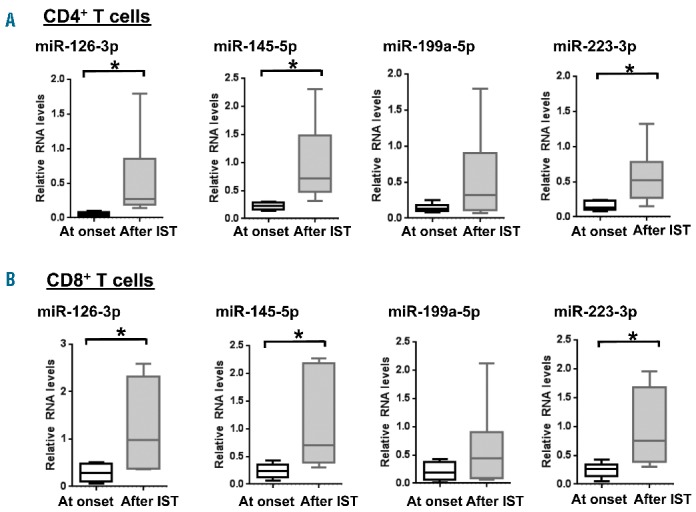
MiRNA expression changes after immunosuppressive therapy (IST). RT-qPCR analysis of miR-126-3p, miR-145-5p, miR-199a-5p, and miR-223-3p expression in (A) CD4+ and (B) CD8+ T cells from AA patients at onset (n=6) and after IST (n=6). Relative expression of miRNA was calculated with respect to RNU-2 expression. Relative expression levels were normalized to those of healthy controls. *P<0.05 (Student t-test).
MicroRNA target gene prediction and validation in CD4+ and CD8+ T cells
We generated a list of pathways likely to be specifically controlled by the four miRNA that were differentially expressed in AA and healthy controls. Using the web-based computational tool DIANA-miRPath, we performed in silico analysis of putative interactions among the four miRNAs and common signaling pathways. This computational tool estimates the impact of co-expressed miRNA in biological pathways.22 Pathway enrichment analysis in AA patients showed putative gene network interactions with the four miRNA, enriched significantly for signaling pathways of cancer, Toll-like receptor, mammalian target of rapamycin, and PI3K-Akt (Online Supplementary Table S4). Candidate target genes of the four miRNA were predicted and selected by using integrated analyses based on four computational algorithms and the Ingenuity Knowledge Base (IPAKB). From these predictions, we designed a custom PCR Array which included 84 experimentally validated or highly predicted target genes of these four miRNA (Online Supplementary Table S2).
In CD4+ T cells, the custom PCR Array analysis showed elevated expression levels of MYC and ETS1 (>1.5 FC, P<0.05), as compared with levels in healthy controls (Figure 5A), whereas increased expression levels of PIK3R2 and PAK4 (>1.5 FC, P<0.05) were observed in CD8+ T cells (Figure 5G). Up-regulation of MYC (1.66±0.25 FC, P<0.05) in CD4+ (Figure 5B) and PIK3R2 (2.26±0.21 FC, P<0.05) in CD8+ T cells (Figure 5H) was confirmed by RT-qPCR.
Figure 5.
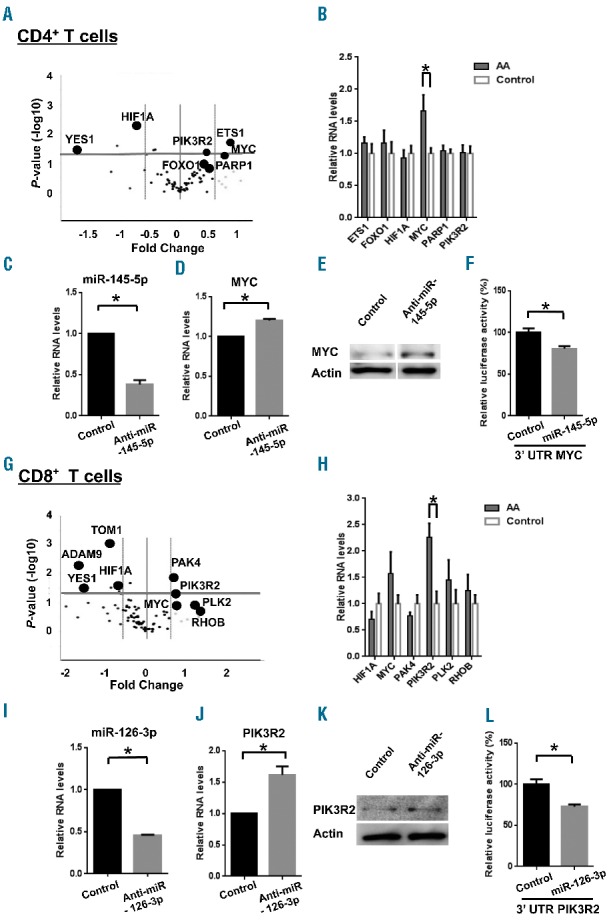
MiRNA target gene prediction and validation in CD4+ and CD8+ T cells. Volcano plots represent relative expression levels of 84 mRNA analyzed using miRNA Targets PCR Arrays. These 84 mRNA were predicted or experimentally validated to be targets of four miRNA (miR-126-3p, miR-145-5p, miR-199a-5p, and miR-223-3p) in (A) CD4+ and (G) CD8+ T cells from AA patients (n=6) and healthy donors (n=6). The x-axis is estimated difference in expression measured in log2; vertical lines refer to a 1.5-fold difference in expression between the two groups. mRNA highly expressed in AA or healthy donors are on the right or the left, respectively. The y-axis is the significance of the difference measured in −log10 of a P-value; the horizontal line indicates our cutoff for significance at P<0.05. RT-qPCR analysis of several target gene expression in (B) CD4+ or (H) CD8+ T cells from AA patients (n=12) and healthy controls (n=8) *P<0.05 [two-way analysis of variance (ANOVA)]. After treatment of CD4+ T cells with anti-miR-145-5p, relative expression of (C) miR-145-5p or (D) MYC was measured by RT-qPCR 24 h later while (E) MYC protein expression was assessed by immunoblot analysis 48 h later. After treatment of CD8+ T cells with anti-miR-126-3p, relative gene expression of (I) miR-126-3p or (J) PIK3R2 and (K) protein expression of PIK3R2 were measured in similar manners. (F, L) Dual luciferase reporter assay. The relative luciferase activity of MYC 3′UTR or PIK3R2 3′UTR construct was measured by co-transfection with miR-145-5p in CD4+ T cells or miR-126-3p in CD8+ T cells, respectively. Data are shown as percentages of controls (cells co-transfected with MYC 3′UTR or PIK3R2 3′UTR construct and control miRNA, respectively). The relative activity of firefly luciferase expression was normalized to renilla luciferase activity. Relative RNA expression of miRNA or mRNA was calculated by normalizing to RNU-2 or β-actin expression, respectively. Data are from three independent experiments (means ± SEM). *P<0.05 (Student t-test).
In order to experimentally validate that MYC and PIK3R2 were targets of miRNA in CD4+ and CD8+ T cells, inhibition experiments using anti-miR-145-5p and anti-miR-126-3p were performed with freshly isolated CD4+ and CD8+ T cells from healthy donors. After exposure to each specific antagomir, gene and protein expression levels of MYC in CD4+ T cells and PIK3R2 in CD8+ T cells were measured by RT-qPCR and immunoblot, respectively. Knockdown efficiencies were 62% ± 5% for miR-145-5p in CD4+ T cells and 54% ± 1% for miR-126-3p in CD8+ T cells (Figure 5C,I). After knockdown of miR-145-5p, both RNA and protein expression levels of MYC were increased in CD4+ T cells (1.20 ± 0.02 FC, P<0.05) (Figure 5D,E). Similarly, miR-126-3p knockdown up-regulated both RNA and protein expression levels of PIK3R2 in CD8+ T cells (1.62±0.14 FC, P<0.05) (Figure 5J,K). To study direct interactions between miRNA and sites on the 3′UTR, we used two commercially available plasmids containing either MYC or PIK3R2 3′UTR inserted downstream of the firefly luciferase reporter gene, and renilla luciferase gene for normalization. In CD4+ or CD8+ T cells transiently transfected with the MYC 3′UTR or PIK3R2 3′UTR construct and selected mimic miRNA or miRNA control, significant inhibition of luciferase activity was observed. Compared with levels in controls, miR-145-5p caused an 19% ± 2.9% decrease in MYC 3′UTR luciferase activity in CD4+ T cells, and miR-126-3p caused a 27% ± 2.4% decrease in PIK3R2 3′UTR luciferase activity in CD8+ T cells (P<0.05, Figure 5F,L). As confirmation, we searched miR-145-5p and miR-126-3p target sequences in MYC 3′UTR and PIK3R2 3′UTR by TargetScan. Highly preserved target sequences of the two miRNA were identified in MYC 3′UTR and PIK3R2 3′UTR, respectively (Online Supplementary Figure S4).
Functional assessment of microRNA down-regulated in T cells from patients with aplastic anemia
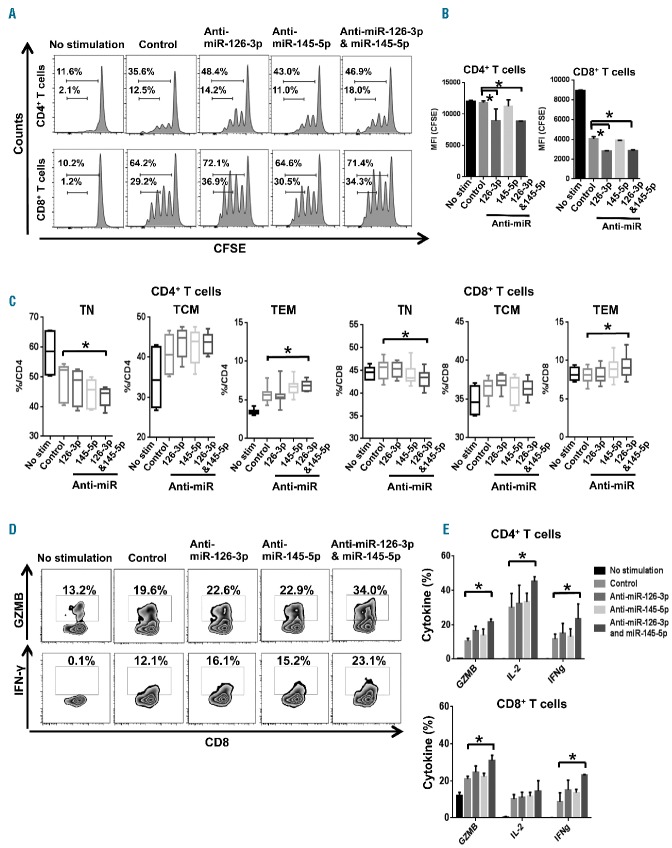
Expression levels of MYC in CD4+ T cells and PIK3R2 in CD8+ T cells were increased in AA patients, in which miR-145-5p and miR-126-3p appeared to be regulators. As miR-199a-5p and miR-223-3p were down-regulated in MDS or SCD patients, we focused on two miRNA associated with AA, miR-126-3p and miR-145-5p, in further functional experiments. Next, we tested whether miR-126-3p and miR-145-5p down-regulation affected proliferation of T cells. Purified CD4+ and CD8+ T cells from healthy controls were labeled with CFSE and transfected with anti-miR-126-3p and/or anti-miR-145-5p, followed by stimulation with anti-CD3/CD28 beads. When median fluorescence intensity of CFSE was assessed by flow cytometry on day 4, enhanced proliferation was observed in both CD4+ and CD8+ T cells co-transfected with anti-miR-126-3p and anti-miR-145-5p, as compared to those transfected with miR-negative control: 8887 ± 6 in CD4+ T cells and 2857 ± 54 in CD8+ T cells by co-transfection with anti-miR-126-3p and anti-miR-145-5p; 11842 ± 176 in CD4+ T cells and 4047 ± 157 in CD8+ T cells by transfection with negative control (P<0.05, Figure 6A,B). Knockdown of miR-126-3p and miR-145-5p decreased the numbers of TN cells and increased TEM cells among CD4+ and CD8+ T cells (P<0.05, Figure 6C, Online Supplementary Table S5). Consistently with more differentiated phenotypic features, knockdown of miR-126-3p and miR-145-5p increased production of effector molecules, granzyme B (GZMB) and IFN-γ production in CD4+ and CD8+ T cells, assessed by intracellular staining (P<0.05, Figure 6D,E, Online Supplementary Table S6). These results indicate promoted differentiation and functionality of effector cells as a result of reduced miRNA. Knockdown of miR-126-3p and miR-145-5p increased interleukin-2 (IL-2) production in CD4+ T cells (P<0.05, Figure 6D,E, Online Supplementary Table S6), but no significant difference was found in CD8+ T cells.
Figure 6.
Functional study of miRNA down-regulated in T cells of AA patients. Purified CD4+ and CD8+ T cells from healthy controls were labeled with CFSE and transfected with anti-miR-126-3p and/or anti-miR-145-5p, followed by stimulation with anti-CD3/CD28 beads. (A) Representative histograms of CD4+ and CD8+ T-cell proliferation. (B) Frequency of CD4+ or CD8+ T-cell proliferation was quantified as the median fluorescence intensity (MFI) of CFSE after transfection of control, anti-miR-126-3p, anti-miR-145-5p, or both miRNA. *P<0.05 [one-way analysis of variance (ANOVA)]. (C) Percentages of TN, TCM and TEM were examined in CD4+ or CD8+ T cells transfected with anti-miR-126-3p and/or anti-miR-145-5p 48 h later. *P<0.05 [one-way analysis of variance (ANOVA)]. (D) Representative plots of intracellular GZMB and IFN-γ expression following in vitro stimulation with anti-CD3/CD28 beads. (E) Percentages of T cells producing GZMB, IFN-γ, and IL-2 were examined in CD4+ or CD8+ T cells transfected with anti-miR-126-3p and/or anti-miR-145-5p 48 h later. Data are from three independent experiments (means ± SEM). *P<0.05. [two-way analysis of variance (ANOVA)].
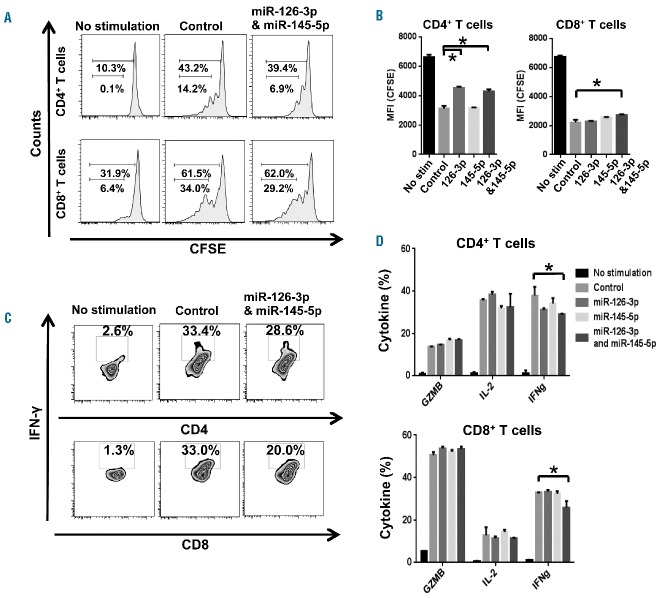
Furthermore, we tested whether overexpression of miR-126-3p and/or miR-145-5p in T cells from AA patients had opposite functional effects. Decreased proliferation was observed in both CD4+ and CD8+ T cells co-transfected with miR-126-3p and miR-145-5p, as compared to cells transfected with control miRNA (P<0.05, Figure 7A,B). Transfection of miR-126-3p and miR-145-5p decreased IFN-γ production in both CD4+ and CD8+ T cells (P<0.05, Figure 7C,D), but GZMB and IL-2 production were not significantly affected.
Figure 7.
Overexpression of miR-126-3p and miR-145-5p in T cells from AA patients decreases T-cell proliferation and inhibits IFN-γ production. Purified CD4+ and CD8+ T cells from AA patients were labeled with CFSE and transfected with miR-126-3p and/or miR-145-5p, followed by stimulation with anti-CD3/CD28 beads. (A) Representative histograms of CD4+ and CD8+ T-cell proliferation. (B) Frequency of CD4+ or CD8+ T-cell proliferation was quantified as the median fluorescence intensity (MFI) of CFSE from dividing cells, after transfection of control, miR-126-3p, miR-145-5p, or both miRNA. *P<0.05 [one-way analysis of variance (ANOVA)]. (C) Representative plots of intracellular IFN-γ expression following in vitro stimulation with anti-CD3/CD28 beads. (D) Percentages of T cells producing GZMB, IFN-γ, and IL-2 were examined in CD4+ or CD8+ T cells transfected with miR-126-3p and/or miR-145-5p 48 h later. Data are from three independent experiments (means ± SEM). *P<0.05. [two-way analysis of variance (ANOVA)].
T cells from a murine model of bone marrow failure show distinct microRNA profiles
MiRNA profiling was performed in T-cell populations from a murine model of minor histocompatibility antigen-mismatched bone marrow failure (Online Supplementary Figure S5). Fatal bone marrow failure was induced by infusion of LN cells from B6 mice into C.B10 recipients which were mismatched at multiple minor histocompatibility antigens, including the immunodominant antigen H60. In comparison to mice that received total body irradiation only without LN cell infusion, four recipient mice that received B6 LN cells were alive but had severe neutropenia (0.21 ± 0.02 109/L versus 0.05 ± 0.03 109/L, P<0.05), thrombocytopenia (126.0 ± 13.4 109/L versus 73.8 ± 11.8 109/L, P<0.05), and reduction in total bone marrow cells (23.4 ± 0.7× 106 cells/mL versus 7.0 ± 0.3 × 106 cells/mL, P<0.05) (Online Supplementary Figure S5). Bone marrow cells from the four surviving mice were sorted into CD4+ and CD8+ T cells by flow cytometry, followed by miRNA expression profiling of individual T-cell populations. Unexpectedly, 22 miRNA, including miR-126a-3p and miR-145a-5p, were increased in CD4+ and CD8+ T cells in the bone marrow failure model mice (>2 FC, P<0.05). MiR-155-5p expression, which is known to be up-regulated in the major antigen MHC-mismatched GVHD model mice,23 was increased (>2 FC, P<0.05) both in CD4+ and CD8+ T cells, whereas 32 miRNA and 29 miRNA were decreased in CD4+ or CD8+ T cells, respectively (Online Supplementary Figure S5). Both CD4+ and CD8+ T cells had reduced levels of expression (>2 FC, P<0.05) of miR-142-3p, miR-142-5p, and miR-29a-3p which have been reported to be miRNA down-regulated in allo-reactive T cells using AGO-CLIP-ChIP procedures and mRNA CLIP-ChIP profiles.24
Discussion
Rossi et al. performed miRNA profiling of 17 highly purified human lymphocyte subsets and identified miRNA signatures that were distinct among various subsets and different from those of mouse lymphocytes.25 In our study, we used flow cytometry-sorted CD4+ and CD8+ T cells to identify novel genes involved in the regulation of immune responses of T cells in AA. To characterize miRNA signatures in T-cell populations purified from AA patients, we employed the miScript miRNA PCR Array platform, because it is considered appropriate in terms of reproducibility, sensitivity, accuracy, specificity, and concordance of differential expression.26 Applying these strategies and qPCR validation, we found that miR-145-5p and miR-126-3p were specifically down-regulated in CD4+ and CD8+ T cells in AA patients, appeared to regulate key cellular functions that contribute to aberrant T-cell activation, and identified MYC/PIK3R2 as targets of these miRNA. Knockdown of miR-126-3p and miR-145-5p promoted proliferation and differentiation from TN toward the TEM phenotype in both CD4+ and CD8+ T cells. Moreover, increased GZMB and IFN-γ production was also observed in CD4+ and CD8+ T cells, indicating enhanced effector differentiation and functionality of CD4+ and CD8+ T cells as a result of reduced activity of the two miRNA. These results are consistent with previous reports describing increased GZMB and IFN-γ expression in CD8+ T cells of AA patients.2,27
We speculated that transfusion might affect the miRNA signatures in T cells as a result of allogenic antigen exposure in AA patients.28 We, therefore, also examined miRNA profiles of T cells from age-matched healthy donors and patients with transfusion-dependent SCD and low-risk MDS for comparison with AA. In addition, we compared the miRNA signatures in CD4+ and CD8+ T cells in SCD patients before and 1 week after red blood cell transfusion, but miRNA signatures did not change after transfusion (data not shown). Low-risk MDS patients are characterized by progressive pancytopenia and may need transfusion of red blood cells and platelets, which are also required in AA. Distinguishing between AA and MDS is often difficult.29 We found differences in miRNA signatures in T cells between AA and MDS. Previous studies have described the association of miRNA expression with MDS subtypes and disease outcome, clinical implications of miRNA in MDS, and miRNA deregulation in mouse MDS model systems.30 Reduced expression levels of miR-378a-3p, miR-143-3p, miR-143-5p, miR-145-5p, and miR-146a-5p have been reported in MDS with del(5q).31,32 Knockdown of miR-145 and miR-146a together or enforced expression of TRAF6 in mouse hematopoietic stem and progenitor cells resulted in thrombocytosis, mild neutropenia, and megakaryocytic dysplasia.31 However, these reports mostly focused on miRNA expression in hematopoietic stem and progenitor cells and not in immune cells. Similarly, some studies of miRNA profiles in SCD patients used erythrocytes33 or platelets,34 but not T-cell populations. Thus, we have clarified the miRNA signatures in T cells in low-risk MDS and SCD patients. In addition, we also found reduced miR-199a-5p expression in CD4+ and CD8+ T cells from patients not only with AA but also with MDS and SCD, and decreased miR-223-3p expression in MDS. It seems likely that miR-199a-5p and miR-223-3p do not reflect specific immunological features of AA and that reduced levels of these two miRNA may be non-specific.
The three miRNA (miR-126-3p, miR-145-5p, and miR-223-3p) which were seen to be down-regulated in CD4+ T, CD8+ T, and CD19+ B cells in AA patients, were shown to be regulators of hematopoiesis in previous studies. MiR-145-5p is abundant in hematopoietic stem and progenitor cells and participates in megakaryopoiesis by activating the innate immunity target, TIRAP (Toll-interleukin-1 receptor domain-containing adaptor protein).31 MiR-145-5p regulates multiple gene targets, such as MYC,35 which regulates various processes involved in cell proliferation, survival, differentiation, and metabolic changes in T cells.36 MiR-126-3p is expressed in CD34+ human cord blood and mobilized peripheral blood cells37 and modulates primitive erythropoiesis by non-hematopoietic Vcam-1+ cells.38 Reduction in miR-126-3p induces expansion without hematopoietic stem cell exhaustion.39 MiR-126-3p regulates multiple gene targets, including PIK3R2 and other immune response genes.40 In T-cell biology, silencing miR-126-3p increased expression of its target PIK3R2 and altered activation of the PI3K/Akt pathway, responsible for reduced induction and suppressive function of regulatory T cells.41 In AA patients, regulatory T cells are decreased in number and functionally abnormal.42 Down-regulation of miR-126-3p in CD4+ T cells might partly explain the reduced induction and suppressive function of regulatory T cells in AA. MiR-223-3p reduces the commitment of erythroid progenitors and has a crucial role in granulocyte progenitor proliferation and function, and its expression is activated in neutrophils.43 Thus, miR-126-3p, miR-145-5p, and miR-223-3p are involved in various stages of hematopoietic cell regulation.
Individual autoimmune diseases display unique miRNA signatures; miR-155 and miR-146a dysregulation is commonly observed.44 Unlike miR-155, which acts to potentiate the immune response, miR-146a is a negative regulator of the immune response, especially through inhibition of Toll-like receptor signaling.45 There were no significant changes in these two miRNA in T cells from AA patients, suggesting that miRNA signatures in AA are distinct from those in other immune-mediated diseases. However, there is some commonality in miRNA dysregulation in other autoimmune diseases. For example, decreased expression of miR-145-5p was detected in T cells from patients with myasthenia gravis46 and systemic lupus erythematosus.47 MiR-223-3p is overexpressed in T cells from patients with rheumatoid arthritis,48 indicating the similarity and difference of miRNA expression in different type of diseases.
We examined miRNA signatures in T cells from mice used as a model of minor histocompatibility antigen-mismatched bone marrow failure. The discrepancy in T-cell miRNA signatures between human AA and the murine bone marrow failure models may be due to the difference of species and/or mouse bone marrow failure models based on minor-histocompatibility antigen-mismatched graft-versus-host disease reactions, given the fact that the dysregulations of miRNA in our model overlapped with those in previous reports.23,24 We speculate that the mouse models based on MHC mismatching at least in part share miRNA signatures associated with allo-reactivity.
Our study had some limitations, such as the small number of patients’ samples and limited number of analyzed miRNA and target genes. We may have missed important miRNA dysregulation and target gene changes that might have been identified using global miRNA and mRNA arrays. MiR-126-3p and miR-145-4p are known to be down-regulated in B-cell malignancies, such as chronic lymphocytic leukemia and diffuse large B-cell lymphoma, respectively.49,50 Moreover, autoantibodies are frequently detected in patients with AA.51,52 Perhaps dysregulations of B cells is related to aberrant B-cell activation. Previous research in patients with systemic lupus erythematosus demonstrated overlapping signatures in T- and B-cell-expressed miRNA which are differentially expressed.53 Our data showed rather modest miRNA expression change. However, miRNA profiling studies often show that subtle changes in miRNA expression, such as a 1.5-fold difference, can have a significant impact on the biology of the cell.54 The exact mechanisms by which these four miRNA were down-regulated in AA are unclear although in silico analysis suggested possible mechanisms by which transcription factors might affect the expression of the four miRNA (Online Supplementary Figure S6). We could not show the dysregulation of these miRNA was related to antigen specificity, as the autoantigens remain elusive in AA. We could not examine the miRNA expression in self-reactive T cells because of limited cell numbers. However, this miRNA expression pattern was more likely to represent an early molecular event rather than a signature for auto-reactive T cells, as auto-reactive T cells are usually very infrequent.55,56 Our data strongly suggest that aberrant miRNA expression is predominantly disease-driven, especially as restoration of the expression levels of miRNA was observed after successful immunosuppressive therapy.
Indeed, normalization of expression of miRNA after immunosuppressive therapy suggests that down-regulation of miRNA may contribute to the immunopathophysiology of AA. Additional studies in large cohorts of patients would be required to define whether miRNA are useful as disease biomarkers and as potential targets for therapeutics. In conclusion, we have provided evidence of altered expression of miRNA in CD4+ and CD8+ T cells in AA, underscoring the importance of this novel class of genes in the regulation of immune responses and pathogenesis of autoimmunity. Our work describes previously unknown potential regulatory roles of the miRNA 145-5p and 126-3p in T-cell activation in AA, in which MYC and PIK3R2 are the respective targets of these miRNA. We suggest that dysregulated miR-145-5p and miR-126-3p promote T-cell proliferation and increase GZMB and IFN-γ production. More importantly, overexpression of these miRNA inhibited the proliferation and cytokine production of CD4+ and CD8+ T cells of AA patients. Targeting or employing miRNA mimics might be novel molecular therapeutic approaches in AA. Understanding these novel pathways may provide new therapeutic strategies and novel mechanistic insights into the epigenetic control of human T-cell effector responses.
Acknowledgments
We thank Marie Desierto, Susan Wong, and Pilar Fernandez for technical assistance; Olga Rios and Kinneret Broder for assistance in obtaining samples from AA patients and healthy volunteers; Barbara Weinstein for obtaining patients’ clinical information; Delong Liu for assistance in conducting the statistical analysis; and Heather Fox-Brashears for designing the custom plate.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
This research was supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Young NS, Bacigalupo A, Marsh JC. Aplastic anemia: pathophysiology and treatment. Biol Blood Marrow Transplant. 2010;16(1 Suppl):S119–S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sloand E, Kim S, Maciejewski JP, Tisdale J, Follmann D, Young NS. Intracellular interferon-gamma in circulating and marrow T cells detected by flow cytometry and the response to immunosuppressive therapy in patients with aplastic anemia. Blood. 2002;100(4):1185–1191. [DOI] [PubMed] [Google Scholar]
- 3.Risitano AM, Kook H, Zeng W, Chen G, Young NS, Maciejewski JP. Oligoclonal and polyclonal CD4 and CD8 lymphocytes in aplastic anemia and paroxysmal nocturnal hemoglobinuria measured by V beta CDR3 spectratyping and flow cytometry. Blood. 2002;100(1):178–183. [DOI] [PubMed] [Google Scholar]
- 4.Dufour C, Capasso M, Svahn J, et al. Homozygosis for (12) CA repeats in the first intron of the human IFN-gamma gene is significantly associated with the risk of aplastic anaemia in Caucasian population. Br J Haematol. 2004;126(5):682–685. [DOI] [PubMed] [Google Scholar]
- 5.Solomou EE, Keyvanfar K, Young NS. T-bet, a Th1 transcription factor, is up-regulated in T cells from patients with aplastic anemia. Blood. 2006;107(10):3983–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng W, Kajigaya S, Chen G, Risitano AM, Nunez O, Young NS. Transcript profile of CD4+ and CD8+ T cells from the bone marrow of acquired aplastic anemia patients. Exp Hematol. 2004;32(9):806–814. [DOI] [PubMed] [Google Scholar]
- 7.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of RNA-mediated gene silencing. Cell. 2008;132(1):9–14. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. [DOI] [PubMed] [Google Scholar]
- 9.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–349. [DOI] [PubMed] [Google Scholar]
- 10.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microrRNAs in acute myeloid leukemia. Blood. 2011;117(4):1121–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123(1):11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32(4): 199–206. [DOI] [PubMed] [Google Scholar]
- 15.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. [DOI] [PubMed] [Google Scholar]
- 16.Nakasa T, Miyaki S, Okubo A, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58(5):1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du C, Liu C, Kang J, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10(12):1252–1259. [DOI] [PubMed] [Google Scholar]
- 18.Iborra M, Bernuzzi F, Invernizzi P, Danese S. MicroRNAs in autoimmunity and inflammatory bowel disease: crucial regulators in immune response. Autoimmun Rev. 2012; 11(5):305–314. [DOI] [PubMed] [Google Scholar]
- 19.Li YT, Chen SY, Wang CR, et al. Brief report: amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis Rheum. 2012;64(10):3240–3245. [DOI] [PubMed] [Google Scholar]
- 20.Camitta BM, Thomas ED, Nathan DG, et al. Severe aplastic anemia: a prospective study of the effect of early marrow transplantation on acute mortality. Blood. 1976;48(1):63–70. [PubMed] [Google Scholar]
- 21.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vlachos IS, Kostoulas N, Vergoulis T, et al. DIANA miRpath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40(Web Server issue):W498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranganathan P, Heaphy CE, Costinean S, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood. 2012;119(20):4786–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Tawara I, Zhao M, et al. Allogeneic T cell responses are regulated by a specific miRNA-mRNA network. J Clin Invest. 2013;123(11):4739–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi RL, Rossetti G, Wenandy L, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. 2011;12(8): 796–803. [DOI] [PubMed] [Google Scholar]
- 26.Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11(8):809–815. [DOI] [PubMed] [Google Scholar]
- 27.Xing L, Liu C, Fu R, et al. CD8+HLA-DR+ T cells are increased in patients with severe aplastic anemia. Mol Med Rep. 2014;10(3): 1252–1258. [DOI] [PubMed] [Google Scholar]
- 28.Brand A. Immunological aspects of blood transfusions. Transpl Immunol. 2002;10(2–3):183–190. [DOI] [PubMed] [Google Scholar]
- 29.Afable MG, 2nd, Wlodarski M, Makishima H, et al. SNP array-based karyotyping: differences and similarities between aplastic anemia and hypocellular myelodysplastic syndromes. Blood. 2011;117(25):6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhyasen GW, Starczynowski DT. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia. 2012;26(1):13–22. [DOI] [PubMed] [Google Scholar]
- 31.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q-syndrome phenotype. Nat Med. 2010;16(1): 49–58. [DOI] [PubMed] [Google Scholar]
- 32.Votavova H, Grmanova M, Dostalova Merkerova M, et al. Differential expression of microRNAs in CD34+ cells of 5q- syndrome. J Hematol Oncol. 2011;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen SY, Wang Y, Telen MJ, Chi JT. The genomic analysis of erythrocyte microRNA expression in sickle cell diseases. PLoS One. 2008;3(6):e2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain S, Kapetanaki MG, Raghavachari N, et al. Expression of regulatory platelet microRNAs in patients with sickle cell disease. PLoS One. 2013;8(4):e60932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sachdeva M, Zhu S, Wu F, et al. P53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106(9):3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang R, Dillon CP, Shi LZ, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donahue RE, Jin P, Bonifacino AC, et al. Plerixafor (AMD3100) and granulocyte colony-stimulating factor (G-CSF) mobilize different CD34+ cell populations based on global gene and microRNA expression signatures. Blood. 2009;114(12):2530–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sturgeon CM, Chicha L, Ditadi A, et al. Primitive erythropoiesis is regulated by miR-126 via nonhematopoietic Vcam-1+ cells. Dev Cell. 2012;23(1):45–57. [DOI] [PubMed] [Google Scholar]
- 39.Lechman ER, Gentner B, van Galen P, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11(6):799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim EH, Suresh M. Role of PI3K/Akt signaling in memory CD8 T cell differentiation. Front Immunol. 2013;4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin A, Wen Z, Zhou Y, et al. MicroRNA-126 regulates the induction and function of CD4(+) Foxp3(+) regulatory T cells through PI3K/AKT pathway. J Cell Mol Med. 2013;17(2):252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi J, Ge M, Lu S, et al. Intrinsic impairment of CD4(+)CD25(+) regulatory T cells in acquired aplastic anemia. Blood. 2012;120(8):1624–1632. [DOI] [PubMed] [Google Scholar]
- 43.Felli N, Pedini F, Romania P, et al. MicroRNA 223-dependent expression of LMO2 regulates normal erythropoiesis. Haematologica. 2009;94(4):479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connell RM, Rao DS, Baltimore D. Microrna regulation of inflammatory responses. Annu Rev Immunol. 2012;(30): 295–312. [DOI] [PubMed] [Google Scholar]
- 45.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappab-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006; 103(33):12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Zheng S, Xin N, et al. Identification of novel microRNA signatures linked to experimental autoimmune myasthenia gravis pathogenesis: down-regulated mir-145 promotes pathogenetic Th17 cell response. J Neuroimmune Pharmacol. 2013;8(5):1287–1302. [DOI] [PubMed] [Google Scholar]
- 47.Lu MC, Lai NS, Chen HC, et al. Decreased microRNA(mir)-145 and increased miR-224 expression in T cells from patients with systemic lupus erythematosus involved in lupus immunopathogenesis. Clin Exp Immunol. 2013;171(1):91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu MC, Yu CL, Chen HC, Yu HC, Huang HB, Lai NS. Increased miR-223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin-like growth factor-1-mediated interleukin-10 production. Clin Exp Immunol. 2014;177(3): 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Lisio L, Sanchez-Beato M, Gomez-Lopez G, et al. MicroRNA signatures in B-cell lymphomas. Blood Cancer J. 2012;2(2):e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caramuta S, Lee L, Ozata DM, et al. Role of microRNAs and microrna machinery in the pathogenesis of diffuse large B-cell lymphoma. Blood Cancer J. 2013;3:e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirano N, Butler MO, Von Bergwelt-Baildon MS, et al. Autoantibodies frequently detected in patients with aplastic anemia. Blood. 2003;102(13):4567–4575. [DOI] [PubMed] [Google Scholar]
- 52.Takamatsu H, Feng X, Chuhjo T, et al. Specific antibodies to moesin, a membrane-cytoskeleton linker protein, are frequently detected in patients with acquired aplastic anemia. Blood. 2007;109(6):2514–2520. [DOI] [PubMed] [Google Scholar]
- 53.Stagakis E, Bertsias G, Verginis P, et al. Identification of novel microrna signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis. 2011;70(8): 1496–1506. [DOI] [PubMed] [Google Scholar]
- 54.Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gargiulo L, Papaioannou M, Sica M, et al. Glycosylphosphatidylinositol-specific, CD1d-restricted T cells in paroxysmal nocturnal hemoglobinuria. Blood. 2013;121(14): 2753–2761. [DOI] [PubMed] [Google Scholar]
- 56.Inaguma Y, Akatsuka Y, Hosokawa K, et al. Induction of HLA-B*40:02-restricted T cells possessing cytotoxic and suppressive functions against haematopoietic progenitor cells from a patient with severe aplastic anaemia. Br J Haematol. 2015. April 30 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]