Abstract
Optimizing bio-production involves strain and process improvements performed as discrete steps. However, environment impacts genotype and a strain that is optimal under one set of conditions may not be under different conditions. We present a methodology to simultaneously vary genetic and process factors, so that both can be guided by design of experiments (DOE). Advances in DNA assembly and gene insulation facilitate this approach by accelerating multi-gene pathway construction and the statistical interpretation of screening data. This is applied to a 6-aminocaproic acid (6-ACA) pathway in Escherichia coli consisting of six heterologous enzymes. A 32-member fraction factorial library is designed that simultaneously perturbs expression and media composition. This is compared to a 64-member full factorial library just varying expression (0.64 Mb of DNA assembly). Statistical analysis of the screening data from these libraries leads to different predictions as to whether the expression of enzymes needs to increase or decrease. Therefore, if genotype and media were varied separately this would lead to a suboptimal combination. This is applied to the design of a strain and media composition that increases 6-ACA from 9 to 48 mg/l in a single optimization step. This work introduces a generalizable platform to co-optimize genetic and non-genetic factors.
INTRODUCTION
Industrial bioprocess development involves many optimization steps at different stages, from the genetic engineering of the initial strain to the optimization of process conditions and scale-up. Development is done iteratively in discrete steps; in other words, new strains are screened holding the environmental conditions constant, and then the growth conditions are optimized for the top strain (1). It is appropriate to separately optimize the strain and conditions if they are decoupled parameters. However, there is ample evidence to the contrary, where different genotypes are favored under different environmental conditions as changes in media nutrients, buffer pH, cultivation temperature and aeration can all influence cell physiology and metabolism (2–5). Here, we have combined approaches for the balancing of the expression levels in a metabolic pathway with those used to optimize media composition. The goal is to accelerate the search through the early identification of interdependencies between these parameters without requiring an underlying mechanistic model.
There are an enormous number of production parameters, including media components and process variables (feed rate, O2, agitation, etc.), and it is impractical to attempt all parameter combinations. As such, there has been a long history of applying design of experiments (DOE) algorithms to guide the search (6–8). The strength of DOE is that a minimum number of combinations are evaluated, each of which simultaneously varies many parameters while avoiding biases. This often takes the form of a factorial design, where each parameter is varied between two discrete levels. The design can either be ‘full’ or ‘fraction’ depending on whether all possible combinations of discrete levels are tested. There are a variety of algorithms, such as Plakett-Burman (9) and Yates (10), which guide the selection of the fraction to be tested. From these data, commercially available software can be used to determine which parameters, and combinations thereof, impact performance (6). Once the important parameters are identified, an optimization step involves experiments that move all of the parameters in favorable directions. Media optimization can be performed in very high throughput, where components are varied in 96-well and larger formats (11).
Strain engineering also involves many genetic parameters. For example, to optimize metabolic flux, it is important to balance the expression levels of enzymes to increase product production and avoid unwanted byproducts (12–18). This requires the selection of genetic parts, for example promoters or ribosome binding sites, to control the expression of each enzyme. This leads to a multi-dimensional search space, whose optimum is the set of expression levels that lead to the highest product productivities (17,19). Algorithms have been developed to aid the search of this space by guiding the generation of genetic diversity and the interpretation of screening results. For example, regression modeling has been applied to identify the optimal construct within a defined space (20). This can be further extrapolated outside of the inspected range via the incorporation of mechanistic modeling (21).
Optimizing the genetics and the media currently occur at separate stages of process development, even though it is recognized that they involve dependent parameters. In other words, strains are screened under one media composition and then the winner is evaluated under many media compositions. This has been constrained by a mismatch in the iteration times. In the past, the construction of new strains could take months, whereas various media formulations could be tested in days. In addition, the cost of new strain libraries was much greater. However, recently the cost and time of building genetic constructs has decreased such that large libraries of rationally designed multi-gene systems can be built and verified in 1–2 weeks (22–24). Automated genome engineering has also advanced, where 10 000s of strains can be built a week (25). Here, we introduce the concept that the genetic constructs and media components could be co-varied in a DOE design. Core to this idea is that each strain would not be tested in every media formulation. In fact, each construct is tested in a single media composition, as determined by a fractional factorial design.
We selected a de novo pathway for 6-aminocaproic acid (6-ACA) biosynthesis in Escherichia coli (E. coli) for proof-of-principle experiments (unpublished results) (Figure 1A). 6-ACA is the linear form of caprolactam, which is the chemical building block of nylon-6. Nylon is the most highly produced synthetic fiber globally (about 4M tons/yr). The fossil-based chemical process for producing caprolactam leads to significant greenhouse gas emission, quantified by its global warming potential (GWP). Depending on the origin of the carbon and energy sources used, the GWP for bio-based production has the potential to be 91% lower than that of the chemical caprolactam route, which is considered to be a sustainable and green process (unpublished results).
Figure 1.
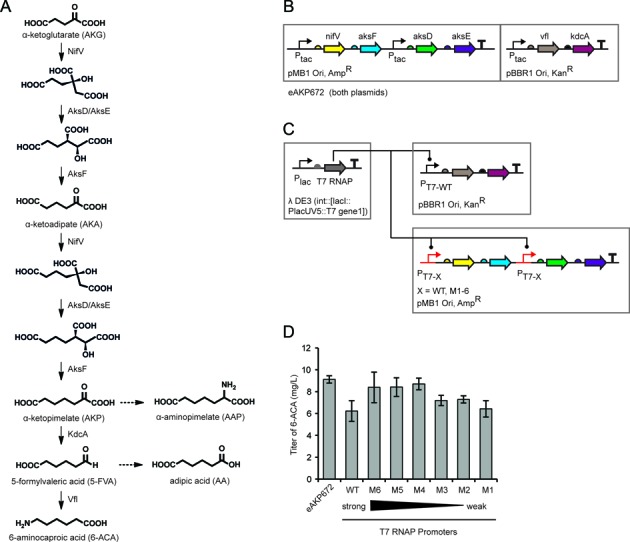
The biosynthetic pathway to 6-ACA and associated genetic designs. (A) The four-step pathway involving six enzymes is shown along with the two known byproducts (dashed arrows). (B) The organization of the initial two-plasmid system with three operons (eAKP672 includes both plasmid pAKP444 and pAKP96) is shown. The full plasmid maps are shown in Supplementary Figure S4. (C) The genome integrated T7 RNAP controller cassette is shown as the box to the left (locus shown). The operons were maintained as in part b and only the promoters were changed. (D) The 6-ACA production titers are shown for the starting construct (part a) and different T7 promoters substituted into the red positions (PT7-X) in part b. The promoters were substituted simultaneously into both positions. Error bars were calculated as the standard deviation of three independent experiments performed on different days.
The pathway was previously constructed by introducing six heterologous enzymes from various pathways and organisms into E. coli (unpublished results). Beginning with the central metabolite α-ketoglutarate (AKG), the one-carbon elongation route was implemented from methanogenic archea in the biosynthesis of coenzyme B to generate key intermediate α-ketopimelate (AKP), a metabolite only naturally present in methanogens (26,27). This conversion from AKG to α-ketoadipate (AKA) and then to AKP involves four enzymes (NifV, AksD, AksE and AksF) that collectively result in the net addition of two carbon units to AKG via two iterations. NifV functions as homocitrate synthase that condenses one acetyl group to the α-keto dicarboxylic acid precursor (28). AksD and AksE form an enzyme complex containing an iron-sulfur (Fe-S) cluster for catalyzing the isomerization step via dehydration and rehydration events (29). The subsequent decarboxylation is achieved by AksF, which is homologous to NAD+-dependent isocitrate dehydrogenase (30,31). The hypothetical intermediates produced by this set of four enzymes are shown in Figure 1A. Next, AKP is decarboxylated by KdcA, a keto-acid decarboxylase to obtain the corresponding carboxylic aldehyde, 5-formylvaleric acid (5-FVA) (32). This is then converted to 6-ACA by Vfl, which performs an amino-transfer step.
There are two main side products from this pathway: α-aminopimelate (AAP) and adipic acid (AA). It is likely that AAP is converted from AKP by an E. coli endogenous amino-transferase whereas AA is derived from 5-FVA via a non-enzymatic oxidation reaction. The six enzymes were identified by database mining and grouped into three operons in two plasmids (Figure 1B). The total production of 6-ACA from the top strain (eAKP672) was 160 mg/l at 10L fermentation scale (unpublished results) and 8 mg/l under shake flask conditions. However, there was 8-fold more intermediates/side products (1.2 g/l) than 6-ACA observed in fermentation scale, indicating a misbalance in the relative enzyme activities.
In this study, we first modularized the 6-ACA pathway so that the expression levels of the genes could be independently controlled. This involves two steps. First, the genes are re-organized as monocistronic units under the control of independent T7 RNA polymerase (T7 RNAP) promoters. Second, genetic insulators are included between the cistrons so that the promoters can be changed without impacting neighboring genes (33,34). Factorial design was implemented by selecting a pair of promoters that implement a small perturbation in the expression of each gene between a low and high state. Three media components were also included in the library design (Fe3+/C, vitamin B1 and Mg2+). These were chosen because they either serve as precursors to catalytic cofactors for pathway enzymes or potentially increase cell growth. The genetic and media changes would lead to a full factorial library of 29 = 512. A fractional factorial library of 32 was chosen by DOE algorithms to efficiently sample this space. This library was constructed, screened and analyzed to determine those factors most influencing titer as well as the trends for each factor (e.g. whether the expression of an enzyme should be increased or decreased). This data set is statistically analyzed in order to simultaneously predict the optimal construct and media condition. Notably, the best construct in the optimized media is different from that in the original composition.
MATERIALS AND METHODS
Strains, plasmids and media
Escherichia coli (E. coli) DH5α was used for routine cloning and plasmid propagation (NEB, #C2987I). E. coli BL21 strain (NEB, #C2530H) was used as production strain harboring the original pathway eAKP672. The eAKP672 strain contains the pAKP444 and pAKP96 plasmids (Supplementary Figure S4A). An E. coli BL21(DE3) strain containing a genomic copy of T7 RNAP was used as production strain for 6-ACA pathway libraries (NEB, #C2527I). LB medium (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl; VWR #90003–350) with appropriate antibiotic supplementation was used for strain maintenance and plasmid construction. Terrific broth (TB) medium (1.2% tryptone, 2.4% yeast extract, 7.2 mM dipotassium phosphate, 17 mM monopotassium phosphate, 0.4% glycerol; Teknova #T3011) with appropriate antibiotics was used for 6-ACA production. Antibiotic selection was performed with kanamycin (50 mg/l; Gold Bio #K-120–5) and ampicillin (100 mg/l; Affymetrix #11259–5). Isopropyl-β-D-1-thiogalactopyranoside (IPTG; Gold Bio #I2481C25 259) was supplemented to the media at 0.2 mM unless otherwise stated. The stock solutions of ferric citrate (140 mM, Sigma F3388), thiamine hydrochloride (10 mg/ml, Alfa Aesar A19560 or L08137) and magnesium sulfate (1M, USB Corporation 18651) were added accordingly. The stock solution of L-cysteine (200 mM, Sigma C7352) was freshly prepared every time before use.
Promoter characterization
The strengths of the T7 promoters were quantified in the context of the six gene locations of the complete pathway. This was done by replacing each gene (and RBS) with the mRFP reporter (and RFP008 RBS). The background pathway was derived from library member #14 and when one position is being tested, the other five positions remain the pathway enzymes. The strains were cultured in 500 μl TB in a micro-titer plate (VWR® 96 deep-well plates, cat. No: 82007–292) at 30°C for 4.5 h with shaking at 900 rpm (Multitron HTS, Infors USA Inc., Laurel, MD, USA). When the OD600 of the cultures reached 0.8, 0.2 mM IPTG was added. After an additional growth period of 16 h, flow cytometry was performed.
Flow cytometry
All cytometry measurements were done using a BD LSR Fortessa flow cytometer with a 640-nm laser for RFP and analyzed using FlowJo v10 (TreeStar Inc., Ashland, OR, USA). Cells were diluted in PBS buffer with 2 mg/ml kanamycin and run at a rate of 0.5 μl/s. At least 50 000 events were recorded for each sample. All events were gated by forward and side scatter.
DNA assembly and verification
Gibson assembly was used to build the T7 RNAP expression system shown in Figure 1C (23). MoClo was used to assemble the monocistronic pathway gene expression cassettes into a single backbone plasmid (22). Supplementary Table S1 and S2 list the parts and genes used in this study. The GenBank accession No. for NifV, AksD, AksE, AksF, KdcA and Vfl are P05342 (Azotobacter vinelandii), ABR55899 (Methanococcus aeolicus Nankai-3), ABR56236 (M. aeolicus Nankai-3), ABR57060 (M. aeolicus Nankai-3), AAS49166 (Lactococcus lactis), AEA39183 (Vibrio fluvialis) respectively (optimized codon sequences are provided in Supplementary Table S2). The scheme for DNA assembly from level 0 to level 1 to the final level 2 constructs is illustrated in Supplementary Figure S3. For level 0 parts-containing plasmids, the backbone plasmid pL0 was derived from pUC19. To be golden gate compatible, the BsaI and BbsI sites were removed by introducing silent mutations. Parts including T7 promoter-ribozyme, RBS-CDSs and double terminators were ligated into pL0 by restriction ligation using SmaI and T4 ligase. 10 ng of both insert and backbone plasmid were added to a 10 μl reaction containing 0.5 μl SmaI and 1 μl T4 ligase for incubation at room temperature for 2 h. For level 0 promoter plasmids, the inserts which contain a spacer, one T7 promoter and six different ribozymes were constructed by DNA oligo annealing. The spacer sequences were designed by the Random DNA Generator using a random GC content of 50% (http://www.faculty.ucr.edu/∼mmaduro/random.htm). The different T7 promoter variants were then introduced by primers via inverse PCR. The promoter-ribozyme parts are flanked by two BsaI sites with GGAG and TTAA as four nucleotides overlaps for the subsequent type IIS reaction to build level 1 plasmids. All of the engineered ribozyme sequences end with TTAA. The RBS-CDS constructs containing the enzymes were obtained from a concurrent study (unpublished results), which we mutated to eliminate BsaI and BbsI sites. Note that the RBS for kdcA in the monocistronic design is K005 (designed using the RBS Calculator) as compared to K007, which was used for the operon-based designs in Figure 1B and C. The level 1 plasmids are based on the pL1 backbone, originating from pMJS1CD (24) with Kanamycin resistance (also free of BsaI and BbsI sites). Then, 20 fmol of each level 0 plasmid are mixed with 5 U BsaI (New England Biolabs, #R0539S) and 5 U T4 DNA Ligase (Promega, #M1794) for a total of 10 μl 1× Promega T4 DNA Ligase Buffer and incubated. Two level 1 plasmids carrying expression cassettes with high (+1) and low (−1) expression levels for each of the six pathway genes were built. In total, there are 12 level 1 plasmids for six pathway genes and 12 containing mRFP in the pathway context for part characterization. Inverse PCR (iPCR) was used to generate the level 1 plasmids for the kdcA and aksF libraries using pL1-kdcA-TU2 and pL1-aksF-TU2 as templates. To build the level 2 plasmids, the six pathway cistrons are assembled in the order shown in Figure 2A. The cistrons are PCR amplified from the level 1 plasmids to give each construct specific cohesive ends upon BsaI digestion corresponding to the assigned position. The final expression plasmid backbone pL2 is derived from pAKP444 (unpublished results), where the BsaI sites in β-lactamase gene were eliminated. The seven assembly junction regions were sequence verified by Sanger sequencing. To build the kdcA and aksF libraries shown in Figure 3B and C, only the expression cassette for either kdcA or aksF was changed, while the other gene expression cistrons are the same as the #4 construct. To achieve higher expression, an extra copy of kdcA or aksF was introduced in a separate p15a Kanr plasmid (Supplementary Figure S4C). The T7 promoter M4 is used for both kdcA and askF expression. The same plasmid for overexpressing aksF or kdcA was introduced into the strain containing construct [−1, −1, −1, +1, +1, +1] individually for Figure 3A.
Figure 2.
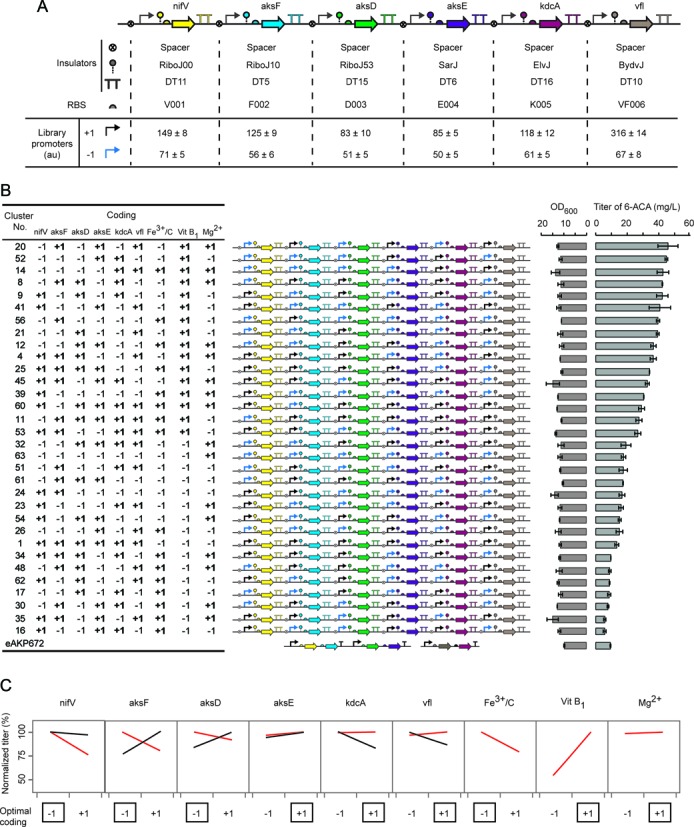
Co-variance of genetic and media factors in a DOE library. (A) The monocistronic architecture is shown, where each gene has its own promoter (arrow), ribozyme (dashed line with circle), RBS (semicircle) and strong double terminator (TT). Two promoters are chosen for each gene that generate a low (−1) and high (+1) expression level. From left to right, these promoters are: (M4/M6), (M4/M6), (M4/M6), (M4/M6), (M1/M4), (M1/M4). The strengths are in arbitrary units of fluorescence and were evaluated by creating a new construct with the promoter at each position fused to mRFP (Materials and Methods). The part sequences are provided in Supplementary Tables S1 and S2. The errors were calculated as the standard deviation of three independent experiments performed on different days. (B) The 29-4 DOE library is shown, rank ordered by the titer. From left to right, the nine factors are shown according to their high/low expression state or presence/absence in the media (+1/−1). The associated construct is shown in the center, following the genetic part coloring and format as in part a. The titer and OD600 of each construct is shown to the right. The error bars were calculated as the standard deviation of two experiments performed on different days. (C) The normalized titers (Materials and Methods) are shown for the 29-4 DOE library (red lines) and the 26 full factorial library that only varies the expression levels (black lines). The full factorial library and screening data are shown in Supplementary Figure S1. The ‘optimal coding’ represents the predicted optimal state of each factor, used to build the optimal construct from the full factorial library (evaluated in Figure 3A).
Figure 3.
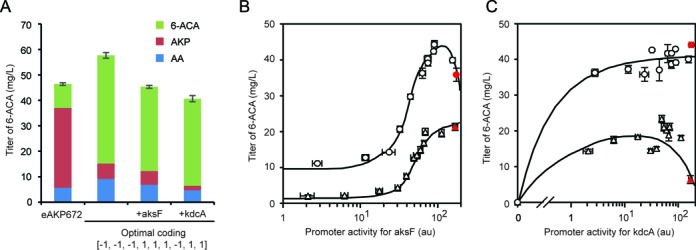
Improvement in 6-ACA production and exploration of the local fitness landscape. (A) Quantification of 6-ACA production for the starting construct (eAKP672, Figure 1B), the optimal coding one determined from the 29-4 library (Figure 2C) and the optimal coding one co-expressed with an extra copy of askF or kdcA. The eAKP672 strain was measured in TB and the optimal coding ones were in TB with the additional nutrients Vit B1 and MgSO4. In addition to the final product, the amount of an intermediate metabolite (AKP) and an undesired byproduct (AA) were quantified. The error bars are representative of three measurements performed on different days and are calculated for the 6-ACA titers only. (B) The expression level was varied for AksF by changing the promoter. Data were generated for cells grown in the optimized media (circles) and TB (triangles). The lines were drawn to guide the eye. The x-axis is the activity of the promoter, as measured using a fluorescent reporter (Supplementary Figure S4D). The red points show high expression levels that could only be achieved by including an additional plasmid carrying a second copy of the gene (see text and Materials and Methods). Error bars in both the y- and x-directions were calculated as the standard deviation of three experiments performed on different days. The promoter activities used for the x-axis were determined using a fluorescent reporter. The promoters were characterized in both media (Supplementary Figure S2). (C) The data and format are as described in part b, except the gene being controlled is kdcA.
Culture screening and LC-MS/MS quantification
A shake flask assay was used that has been previously demonstrated to correlate positively with results at the 10L fermentation scale (unpublished results). Overnight seed cultures were prepared by inoculating the strains from glycerol stock into 3 ml of LB media in 15 ml culture tubes. These were grown for 20 h at 37°C and shaken at 250 rpm. The overnight seed culture was then used to inoculate 20 ml of TB media in a 125 ml Erlenmeyer flask. In order to achieve the same initial cell densities (≈0.006) across a batch culture, the OD600 of each overnight seed culture was measured and the inoculant volume was calculated accordingly. The cells in flasks were grown for 5 h at 30°C and 250 rpm, leading to cell densities of ≈0.8. Then, IPTG was added to the culture to a final concentration of 0.2 mM. After induction, the cells were grown for another 41 h at 30°C and 120 rpm. Cells are removed from the culture by centrifugation at 4000 g for 12 min. The supernatant was further cleaned using a 0.2 μm filter (13 mm, 0.20 μm MicroLiter nylon syringe filter, Wheaton). The resulting cell broth is diluted four times and LCMS/MS analysis is used for product quantification. A XSELECTTM HSS T3 column 3.5 μm, 2.1 mm × 75 mm (Waters, part No. 186006464) and XSelect HSS T3 Sentry Guard column (Waters, 100Å, 5 μm, 2.1 mm X 10 mm, part No. 186006486) with gradient elution are used for the separation of alpha-keto acids, 6-ACA, AAP and Adipate. Solvent A consists of LCMS grade water, containing 0.1% formic acid, and solvent B consists of acetonitrile, containing 0.1% formic acid. For the HSS T3 column the flow-rate was 0.25 ml/min and the column temperature was kept constant at 30°C. The gradient started at 100% solvent A and was increased linearly to 15% solvent B over 6.5 min and then immediately increased to 80% solvent B for 3.5 min, and finally to 100% Solvent A and stabilized for 5 min. The injection volume was 10 μl for all the analyses. For metabolite quantification, the LC-MS experiments were performed on a triple-quadrupole AB Sciex 4000 QTRAP® MS/MS system (AB Sciex, Framingham, MA, USA), equipped with an Agilent 1200 LC system (Agilent Technologies Inc., Santa Clara, CA, USA), using either ESI positive or negative ionization mode using multiple reaction monitoring (MRM). m/z 116, dwell time 100 ms with the following conditions: 70 V fragmentor, 350°C drying gas temperature, 12 L N2/min drying gas, 50 psig nebuliser pressure and 3 kV capillary voltage. The ion source temperature was kept at 130°C, whereas the desolvation temperature is 350°C, at a flow-rate of 500 l/h. For 6-ACA the protonated molecule was fragmented with 13 eV, resulting in specific fragments from losses of H2O, NH3 and CO. For AKG, AKA, AKP and adipate, the deprotonated molecule was fragmented with 10–14 eV, resulting in specific fragments from losses of, e.g. H2O, CO and CO2. To determine concentrations, calibration curves of the external standards of synthetically prepared compounds spiked in blank fermentation broth was analyzed to calculate a response factor for the respective ions, and was used to calculate the concentrations in fermentation samples. The retention time for each compounds and their transition mass are listed in Supplementary Table S6.
Measurement of cell density
The cell densities were recorded in 96 well plate format (Nunc® 96-Well Optical Bottom Plates, Thermo scientific) using Synergy H1 Hybrid Microplate Reader (BioTek instruments, Inc., Winooski, USA). The culture volume was fixed at 200 μl. The final OD600 reading was obtained by subtracting the blank OD600 reading of TB media. The OD600 values reported in the screen (Figure 2B) were determined by taking a 10 μl of stationary phase cultures in flasks and diluting them into 190 μl of TB media and multiplying the resulting OD600 measurement by 20.
Statistics software and analysis
The regression and graphical analyses of the library data and the statistical analysis of variance (ANOVA) of the model were performed using JMP pro 11.0.0 software (SAS Institute Inc., Cary, NC, USA). The F-test was used as part of the ANOVA analysis to determine the statistical significance for the effect of each factor on the final output titer. This is presented in the Supplementary Information (Tables S4 and S5). In this two-level DOE library design, a larger F ratio for a factor means the variation of the two mean production titers at either the high or low level for that factor is significant, indicating that the change of this factor has a large effect on the final production titer. If there is no titer difference between the two levels, then the F ratio is close to 1. Here, the F ratio is calculated as the ratio of between-group mean square value (MSB) and within-group mean square value (MSw). For each individual factor, a group means the sub-set of the design at either high or low level. Therefore, there are two groups for each factor in the 29-4 DOE design. Each group contains 16 different designs, which also represents 16 observation counts for that factor in either the high or low level. MSB is then related to the between group sum of squared difference (SSB) divided by the between group degree of freedom (DFB). DFB is one less than the number of groups (equal to 1 for a 2-level design). MSw is accordingly related to the within group sum of squares (SSw) divided by the within group degree of freedom (DFw). DFw is the product of the levels for each factor (2) and the observation count for the factor (16) at one level minus one (DFw = 2 × (16–1) = 30). The p-value is calculated using the JMP software (with a significance of α = 0.05).
RESULTS
Design of a 6-ACA pathway with modular genetics
The first step of DOE involves a ‘screening phase,’ where the factors are identified that most contribute to performance. This guides how the factors are tuned in a subsequent ‘optimization phase.’ In essence, the idea is to map the local search landscape surrounding the starting construct and then use this information to guide the direction of the search. Ideally, the mapping would be done via small perturbations in the factors as small changes are more likely to be additive. Larger changes can result in complicating effects; for example, the overexpression of an enzyme can have a dominating impact on performance that obscures the effects caused by changing the expression of the other pathway enzymes. An obstacle in implementing this approach is that the precision of genetic parts is limited and impacted by the local genetic context, making it difficult to reliably obtain small changes. Another problem is that the organization of the genetic system can contribute to more coupling between factors. For example, operons and transcriptional readthrough can make it impossible to vary one factor without also impacting others simply because of the way in which parts are organized. All of these challenges limit the ability to perturb each enzyme between two expression states as part of a factorial DOE library without impacting the expression of other enzymes in the pathway.
To address these challenges, we sought to redesign the genetic architecture of the 6-ACA pathway, so that the expression levels of the six enzymes could be independently controlled. The original design (eAKP672) divided the six genes into three two-gene operons across two plasmids (unpublished results) (Figure 1B). One plasmid contains two operons that are not separated by a terminator, leading to transcriptional readthrough. Three changes were made to modularize the architecture. First, genes were organized into monocistronic expression units on a single plasmid. This allows each gene to be controlled independently. Second, strong double terminators were placed after each gene to turn off transcriptional readthrough (35,36). Third, genetic insulators were added to allow promoters to be changed without invoking context effects. Specifically, we used a set of ribozyme insulators (RiboJ variants) that allow the promoters to be varied without impacting RBS strength or mRNA stability (33). Upstream spacers (25 bp) were also included to further insulate the promoters and these contained the sequences required for Golden Gate assembly (Materials and Methods). To avoid recombination, the sets of double terminators, spacers and ribozyme insulators all had to be chosen to have sufficiently diverse sequences. Supplementary Table S1 and S2 provide the part and gene sequences used in the designs.
All of the operons in the initial construct (eAKP672) were controlled with the IPTG-inducible Ptac promoter. These were replaced by T7 promoters, which are small (12 bp) and easy to swap to change expression levels. A separate ‘controller’ was integrated into the genome from which wild-type T7 RNAP is expressed under IPTG control (Figure 1C) (Materials and Methods). We decided to first substitute the T7 promoters into the Ptac locations in eAKP672 before fully breaking up the system into individual insulated cistrons. To do this, the same strong promoter (PT7-WT) was substituted for the three Ptac promoters. The promoters controlling nifVaksF and aksDE were then varied and an optimum expression level was observed (Figure 1D). These substitutions yielded a construct that produced 6-ACA titers comparable to eAKP672, as quantified by LC-MS (Figure 1D) (Materials and Methods).
The complete monocistronic pathway design is shown in Figure 2A. For each gene, two promoters were selected based on the results in Figure 1D to reflect high (+1) and low (−1) expression levels. To map the local space, these were chosen to minimize the change in expression while still maintaining statistically significant differences between the levels. T7 promoter variants M4/M6 were selected for the expression of nifV, aksF, aksD and aksE genes and M4/M1 were used for the expression of kdcA and vfl. The strength of these promoters was measured in the context of the pathway by replacing each gene individually with red fluorescent protein (mRFP) under the control of each promoter, leading to a set of 12 constructs (Materials and Methods and Supplementary Figure S4B). These were assayed to measure fluorescence using flow cytometry (Materials and Methods) and their strengths are shown in Figure 2A. The ratio between the high (+1) and low (−1) promoters is approximately 2-fold, with absolute levels determined by the identity of the RiboJ insulator. Notably, there is no left-to-right increase in expression, which demonstrates the efficient insulation provided by the double terminators. Because all of the genes are oriented in the same direction, readthrough from upstream genes would lead to a systematic increase in expression across the construct.
To ensure that the fully monocistronic design maintained activity, two constructs were constructed and tested that varied the identity of the promoter for each gene (Figure 2B). One construct (#25) consisted of a pattern of high and low promoters as [+1 +1 +1 +1 −1 −1] and the other (#14) has the opposite pattern [−1 −1 −1 −1 +1 +1]. These variants yielded 6-ACA titers of 11.2 mg/l and 18.3 mg/l, respectively, both of which are higher than the initial (eAKP672) and operon-based constructs. This indicated that the insulated monocistronic designs did not disrupt activity and that the promoter substitutions would explore the local space without leading to non-functional variants.
Integration of genetic and media permutation in a 29-4 factorial library
For DOE factorial library design, each factor is varied between two discrete states. In terms of the six genes in the pathway, this involves the selection of low (−1) and high (+1) promoters (Figure 2A). Note, however, that factors do not have to relate to expression levels or even genetic changes. To demonstrate this, we also included factors that capture the media composition. The baseline media is terrific broth (TB) (1.2% tryptone, 2.4% yeast extract, 7.2 mM dipotassium phosphate, 17 mM monopotassium phosphate, 0.4% glycerol) to which we evaluated three additional components. The first is ferric citrate and cysteine (Fe3+/C), which is reported to be beneficial for the expression and activity of iron-sulfur cluster containing enzymes (AksD and AksE) (27,37,38). The second is vitamin B1 (Vit B1), which provides the cofactor precursor for thiamine pyrophosphate (TPP) dependent decarboxylase (KdcA) (32,39). Finally, the magnesium ion (Mg2+) concentration often affects high-density bacterial growth (40). These three components were included as factors, where −1 indicates the absence of the supplement and +1 indicates its presence (1 mM Fe3+/C, 0.17 mg/ml Vit B1 or 2 mM Mg2+). Together, the genetic and media factors lead to 29 = 512 possible permutations of −1/+1 states. When generating the fractional factorial library, the algorithm does not distinguish the genetic and media factors. This makes it trivial to include other aspects of process development into the search as additional factors, such as agitation, feed rate or temperature.
The fractional factorial library was generated using the Yates algorithm (41,42), which allows orthogonal and balanced fractional sets chosen from the full combination for effective estimation of all the main factor effect and some two-factor interactions. A resolution IV library was generated, which is sufficient to estimate the main effects of factors (10). This leads to a 29-4 = 32-member library, where each factor is evenly distributed between the −1 and +1 states. In this design, 21 two-factor interactions were clear of confounding. Thus their parameters can be evaluated individually. While the rest of 15 two-factor interactions were aliased with others that their parameters were convoluted. Each library member is a unique genetic construct and each construct is tested in one media formulation that varies across the library. The library was screened twice in a 20 ml shake flask culture assay and the average titer computed (Materials and Methods). The titers in the library spanned ≈10-fold from 5.7 mg/l to 46.8 mg/l (Figure 2B). The titer by eAKP672 in this screen is 9.3 ± 0.3 mg/l. The OD600 of the strains is shown in Figure 2B. There is no correlation between titer and growth rate and there is little variation across the library, including due to media changes. For statistical analysis, the titer was normalized by the OD600 to reduce day-to-day variation.
A regression analysis was performed on the data to determine the factors that most influence the titer. The JMP software package was used to perform the analysis, which is commonly applied to bioprocess optimization (7,43). The model used for the 29-4 fractional factorial to estimate the response curve is the quadratic polynomial
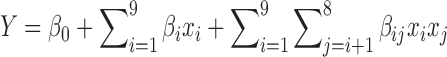 |
(1) |
where Y is the predicted titer and xi is the state [1, −1] of factor i. The β are fit parameters that are obtained via a least squares regression method and are listed in Supplementary Table S3. An ANOVA analysis was performed on the quadratic model, which determined it to be statistically significant (α = 0.05, P < 0.0001, F value = 35) (Materials and Methods and Supplementary Table S4).
From this analysis, the presence of Vit B1 (X8) has the strongest positive effect on titer (β = 1.06). This media component relates to KdcA activity by serving as precursor to its catalytic cofactor TPP. This points to KdcA activity as the bottleneck of the pathway. Using the parameterized Equation (1), the predicted titer can be calculated for each factor to estimate its impact across the library as that factor is changed (red lines in Figure 2C). This calculation is performed for each factor by changing it between the high and low states while holding the values of the remaining factors to their states predicted to achieve the maximal titer. This is normalized by dividing by the maximum predicted titer. In essence, this is looking at the shape of the search landscape around the predicted optimum within the library. This analysis shows that the strongest negative correlations with titer are nifV, askF and Fe3+/C.
The regression model predicts a genetic system and media composition that is optimal for 6-ACA production amongst the 512 binary possibilities. The predicted pattern of promoter activities is [−1, −1, −1, +1, +1, +1] and the media is supplemented with Vit B1 and Mg2+. We built the associated construct and evaluated it in this media composition. This strain and media combination yielded 48.0 mg/l of 6-ACA, which is ≈5-fold higher than that of eAKP672 and is close to that predicted by Equation (1) (46.6 mg/l) (Figure 3A). Further, we measured the presence of a precursor that accumulates in the eAKP672 strain (AKP) and this decreased from 31 to 6 mg/l, which is consistent with increased KdcA activity. Note that this construct is not intended to be the final optimized system; this would need to be obtained by fine-tuning expression using the analysis as a guide. Rather, this validates the approach and predictive power of the model built with incomplete information. Starting with the optimal construct and media formulation, we then sought to determine how changes in the expression of KdcA and AksF further impact the accumulation of intermediates. To do this, we overexpressed these genes and measured the accumulation of AKP and AA (Figure 3A). The overexpression of KdcA reduces AKP (1.8 versus 6 mg/l). This further supports the conclusion that KdcA is the bottleneck of the pathway.
Comparison with full factorial analysis under one media condition
We compared the above predictions with those that are obtained by only varying the genetic factors and screening in a single media composition. Considering only the six genetic factors, the full factorial library consists of 26 = 64 constructs. The complete library was built (Supplementary Figure S1) and tested in TB media. The resulting titers ranged from 9.7 to 19.4 mg/l. The library was analyzed identically as before and fit to a version of Equation (1) containing parameters for the six factors (Supplementary Tables S3 and S5). The normalized titers were calculated for each of the factors and compared to those determined from the 29-4 library (black lines in Figure 2C). For some factors, opposite slopes are predicted by the analysis of the two data sets. This leads to different predictions in the directions that the expression levels should change to further optimize the system. For example, when media changes are not considered, AksF should be decreased, but if media can change then it should increase. Thus, co-varying both media and genetic changes is critical to accurately guide the next constructs to build.
The opposite signs for some model parameters also infer different curvatures in the expression search space (Supplementary Table S3). For example, when considering AksF, β3 = –0.08 when media is varied and β3 = 0.08 when it is not. To explore this effect, we expanded the expression space in one dimension for AksF and the enzyme participating in the rate-limiting step (KdcA). Starting with cluster #4, which ranked high in both of the libraries, two new libraries were made where promoters that span a large range of expression were used to control these two genes (Supplementary Table S1 and Figure S2). Because the starting construct is close to the strongest promoter in the library, higher expression levels were obtained by introducing another plasmid copy of either aksF or kdcA (Supplementary Figure S4C). The library was characterized both in TB and the +Vit B1/+Mg2+ media. For AksF, an optimum level of expression appears in the +Vit B1/+Mg2+ media (Figure 3B). In contrast, there is no optimum when measured in TB and higher expression outside of the range studied is predicted to lead to higher titers. Similarly, the titer is flat for a wide range of KdcA expression in the +Vit B1/+Mg2+ media, but in TB the titer decreases at high levels of expression (Figure 3C). The flatness of this expression profile could have implied endogenous background activity in E. coli. However, when kdcA is not included at all in the construct, no activity is observed. The decrease in titer when KdcA is overexpressed without Vit B1 could be due to substrate sequestration by the presence of inactive apo-enzymes.
DISCUSSION
Until recently, strain engineering—including genome modifications and DNA construction/integration—has been a slow process. Thus, the development time for strains was much larger than the time required to implement a screen (20,24). This mismatch has led to the paradigm in industry of separating these steps such that a library of strains is built and screened under a single set of conditions and the top variants progress to media and process development (1,4). Because of investment in strain engineering technologies, it is increasingly common for genetic engineers to have access to pipelines that can build 104+ per week. Further, these modification have become more rationally guided with improvements in computer aided design (44), DNA assembly (45,46) and a decline in DNA sequencing cost. Previously, libraries of similar size relied on random methods and many of the genetics of variants that did not survive the screen were unknown. These changes bring the time scale of strain construction closer to the time scale for screening and this challenges the current development paradigm (25,47). Some process variables can be tested in high-throughput assays, such as media composition, temperature, pH, aeration and feed (e.g., glucose/O2) rates. Others are lower throughput, especially with regards to scale-up, and will continue to be performed as a separate step.
High-throughput strain construction also enables more exploratory experiments, rather than every construct being an attempt to improve titer. Such exploration is an essential principle of DOE and is a common approach in bioprocess development (7,48), but this has not been applied to genetic engineering. The co-variation of genetic and process factors allow for interdependencies to be discerned early. For the 6-ACA pathway, our initial screening data would have led to the construction of a strain that would have turned out to be suboptimal after optimizing the media. There is essentially no correlation in the titers produced by the strains in the two data sets. This would have misguided the engineer as to the direction that the expression levels should be balanced in designing the next generation of constructs.
Advances in DNA construction have made it possible to build far more constructs than can be effectively screened and as the price declines, screening becomes increasingly infeasible. Here, we present an approach to aid the search of this space by guiding the construction of a subset of designs that maximize the information that can be gleaned from a screen. Part of the approach is in the organization of the genetic system itself. This involves building as modular of a system as possible, where insulating parts are selected to decouple the expression levels between enzymes. The co-development of search algorithms and genetic systems designed to be searched will be a powerful tool in strain development.
Supplementary Material
Acknowledgments
The authors thank Clary Clish and Kerry Pierce at Broad institute for their help in LCMS quantification. The authors thank Lauren Woodruff and D. Benjamin Gordon at Broad institute-MIT living foundry for their help in library statistical analysis. B.V., J.A.R. and R.A.L.B. are supported by Royal DSM, The Netherlands.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The US Defense Advanced Research Projects Agency (DARPA) Living Foundries award [HR0011-12-C-0067, HR0011-15-C-0084 to C.A.V.]; H.Z. and C.A.V. are part of the US National Science Foundation Synthetic Biology Engineering Research Center [SynBERC EEC0540879]. Funding for open access charge: The US Defense Advanced Research Projects Agency (DARPA) Living Foundries award [HR0011-12-C-0067, HR0011-15-C-0084 to C.A.V.]
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhang C.Q., Chen X.X., Zou R.Y., Zhou K., Stephanopoulos G., Too H.P. Combining genotype improvement and statistical media optimization for isoprenoid production in E. coli. PLoS One. 2013;8:e75164. doi: 10.1371/journal.pone.0075164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardinale S., Joachimiak M.P., Arkin A.P. Effects of genetic variation on the E. coli host-circuit interface. Cell reports. 2013;4:231–237. doi: 10.1016/j.celrep.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Deutschbauer A., Price M.N., Wetmore K.M., Tarjan D.R., Xu Z., Shao W., Leon D., Arkin A.P., Skerker J.M. Towards an informative mutant phenotype for every bacterial gene. J. Bacteriol. 2014;196:3643–3655. doi: 10.1128/JB.01836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S.L., Ye B.C., Chu J., Zhuang Y.P., Guo M.J. From multi-scale methodology to systems biology: to integrate strain improvement and fermentation optimization. J. Chem. Technol. Biotechnol. 2006;81:734–745. [Google Scholar]
- 5.Sagt C.M. Systems metabolic engineering in an industrial setting. Appl. Microbiol. Biotechnol. 2013;97:2319–2326. doi: 10.1007/s00253-013-4738-8. [DOI] [PubMed] [Google Scholar]
- 6.Mandenius C.F., Brundin A. Bioprocess optimization using design-of-experiments methodology. Biotechnol. Prog. 2008;24:1191–1203. doi: 10.1002/btpr.67. [DOI] [PubMed] [Google Scholar]
- 7.Kumar V., Bhalla A., Rathore A.S. Design of experiments applications in bioprocessing: concepts and approach. Biotechnol. Prog. 2014;30:86–99. doi: 10.1002/btpr.1821. [DOI] [PubMed] [Google Scholar]
- 8.Franceschini G., Macchietto S. Model-based design of experiments for parameter precision: State of the art. Chem. Eng. Sci. 2008;63:4846–4872. [Google Scholar]
- 9.PLACKETT R.L., BURMAN J.P. The design of optimum multifactorial experiments. Biometrika. 1946;33:305–325. [Google Scholar]
- 10.Box G., Hunter W., Hunter S. Statistics for experimenters: an introduction to design, data analysis, and model building. New York: John Wiley & Sons; 1978. [Google Scholar]
- 11.Olsson I.-M., Johansson E., Berntsson M., Eriksson L., Gottfries J., Wold S. Rational DOE protocols for 96-well plates. Chemometrics and Intelligent Laboratory Systems. 2006;83:66–74. [Google Scholar]
- 12.Pfleger B.F., Pitera D.J., Smolke C.D., Keasling J.D. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 13.Du J., Yuan Y., Si T., Lian J., Zhao H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012;40:e142. doi: 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zelcbuch L., Antonovsky N., Bar-Even A., Levin-Karp A., Barenholz U., Dayagi M., Liebermeister W., Flamholz A., Noor E., Amram S., et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 2013;41:e98. doi: 10.1093/nar/gkt151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliver J.W., Machado I.M., Yoneda H., Atsumi S. Combinatorial optimization of cyanobacterial 2,3-butanediol production. Metab. Eng. 2014;22:76–82. doi: 10.1016/j.ymben.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Nowroozi F.F., Baidoo E.E., Ermakov S., Redding-Johanson A.M., Batth T.S., Petzold C.J., Keasling J.D. Metabolic pathway optimization using ribosome binding site variants and combinatorial gene assembly. Appl. Microbiol. Biotechnol. 2014;98:1567–1581. doi: 10.1007/s00253-013-5361-4. [DOI] [PubMed] [Google Scholar]
- 17.Xu P., Gu Q., Wang W., Wong L., Bower A.G., Collins C.H., Koffas M.A. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat. Commun. 2013;4:1409. doi: 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- 18.Ajikumar P.K., Xiao W.H., Tyo K.E., Wang Y., Simeon F., Leonard E., Mucha O., Phon T.H., Pfeifer B., Stephanopoulos G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biggs B.W., De Paepe B., Santos C.N., De Mey M., Kumaran Ajikumar P. Multivariate modular metabolic engineering for pathway and strain optimization. Curr. Opin. Biotechnol. 2014;29:156–162. doi: 10.1016/j.copbio.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Lee M.E., Aswani A., Han A.S., Tomlin C.J., Dueber J.E. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013;41:10668–10678. doi: 10.1093/nar/gkt809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farasat I., Kushwaha M., Collens J., Easterbrook M., Guido M., Salis H.M. Efficient search, mapping, and optimization of multi-protein genetic systems in diverse bacteria. Mol. Syst. Biol. 2014;10:731. doi: 10.15252/msb.20134955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R.Y., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M., et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 24.Smanski M.J., Bhatia S., Zhao D., Park Y., L B.A.W., Giannoukos G., Ciulla D., Busby M., Calderon J., Nicol R., et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 2014;32:1241–1249. doi: 10.1038/nbt.3063. [DOI] [PubMed] [Google Scholar]
- 25.Horwitz A.A., Walter J.M., Schubert M.G., Kung S.H., Hawkins K., Platt D.M., Hernday A.D., Mahatdejkul-Meadows T., Szeto W., Chandran S.S. Efficient Multiplexed Integration of Synergistic Alleles and Metabolic Pathways in Yeasts via CRISPR-Cas. Cell Syst. 2015;1:88–96. doi: 10.1016/j.cels.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Howell D.M., Harich K., Xu H., White R.H. Alpha-keto acid chain elongation reactions involved in the biosynthesis of coenzyme B (7-mercaptoheptanoyl threonine phosphate) in methanogenic Archaea. Biochemistry. 1998;37:10108–10117. doi: 10.1021/bi980662p. [DOI] [PubMed] [Google Scholar]
- 27.Drevland R.M., Waheed A., Graham D.E. Enzymology and evolution of the pyruvate pathway to 2-oxobutyrate in Methanocaldococcus jannaschii. J. Bacteriol. 2007;189:4391–4400. doi: 10.1128/JB.00166-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng L., White R.H., Dean D.R. Purification of the Azotobacter vinelandii nifV-encoded homocitrate synthase. J. Bacteriol. 1997;179:5963–5966. doi: 10.1128/jb.179.18.5963-5966.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drevland R.M., Jia Y., Palmer D.R., Graham D.E. Methanogen homoaconitase catalyzes both hydrolyase reactions in coenzyme B biosynthesis. J. Biol. Chem. 2008;283:28888–28896. doi: 10.1074/jbc.M802159200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bult C.J., White O., Olsen G.J., Zhou L., Fleischmann R.D., Sutton G.G., Blake J.A., FitzGerald L.M., Clayton R.A., Gocayne J.D., et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 31.Howell D.M., Graupner M., Xu H., White R.H. Identification of enzymes homologous to isocitrate dehydrogenase that are involved in coenzyme B and leucine biosynthesis in methanoarchaea. J. Bacteriol. 2000;182:5013–5016. doi: 10.1128/jb.182.17.5013-5016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smit B.A., van Hylckama Vlieg J.E., Engels W.J., Meijer L., Wouters J.T., Smit G. Identification, cloning, and characterization of a Lactococcus lactis branched-chain alpha-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 2005;71:303–311. doi: 10.1128/AEM.71.1.303-311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lou C., Stanton B., Chen Y.J., Munsky B., Voigt C.A. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat. Biotechnol. 2012;30:1137–1142. doi: 10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen A.A., Segall-Shapiro T.H., Voigt C.A. Advances in genetic circuit design: novel biochemistries, deep part mining, and precision gene expression. Curr. Opin. Chem. Biol. 2013;17:878–892. doi: 10.1016/j.cbpa.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y.J., Liu P., Nielsen A.A., Brophy J.A., Clancy K., Peterson T., Voigt C.A. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods. 2013;10:659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- 36.Cambray G., Guimaraes J.C., Mutalik V.K., Lam C., Mai Q.A., Thimmaiah T., Carothers J.M., Arkin A.P., Endy D. Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res. 2013;41:5139–5148. doi: 10.1093/nar/gkt163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuchenreuther J.M., Grady-Smith C.S., Bingham A.S., George S.J., Cramer S.P., Swartz J.R. High-yield expression of heterologous [FeFe] hydrogenases in Escherichia coli. PLoS One. 2010;5:e15491. doi: 10.1371/journal.pone.0015491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yacoby I., Tegler L.T., Pochekailov S., Zhang S., King P.W. Optimized expression and purification for high-activity preparations of algal [FeFe]-hydrogenase. PLoS One. 2012;7:e35886. doi: 10.1371/journal.pone.0035886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed G.H., Ragsdale S.W., Mansoorabadi S.O. Radical reactions of thiamin pyrophosphate in 2-oxoacid oxidoreductases. Biochim Biophys. Acta. 2012;1824:1291–1298. doi: 10.1016/j.bbapap.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lusk J.E., Williams R.J., Kennedy E.P. Magnesium and the growth of Escherichia coli. J. Biol. Chem. 1968;243:2618–2624. [PubMed] [Google Scholar]
- 41.Tulumello A., Tulumello J.D. Yates method analysis of 2n factorial design of experiments using the Ti-59, for N = 3,4,5,6. Comput. Chem. 1981;5:55–66. [Google Scholar]
- 42.Riedwyl H. Modifying and using Yates’ algorithm. Stat. Pap. 1998;39:41–60. [Google Scholar]
- 43.Harms J., Wang X., Kim T., Yang X., Rathore A.S. Defining process design space for biotech products: case study of Pichia pastoris fermentation. Biotechnol. Prog. 2008;24:655–662. doi: 10.1021/bp070338y. [DOI] [PubMed] [Google Scholar]
- 44.Appleton E., Tao J., Haddock T., Densmore D. Interactive assembly algorithms for molecular cloning. Nat. Methods. 2014;11:657–662. doi: 10.1038/nmeth.2939. [DOI] [PubMed] [Google Scholar]
- 45.Ellis T., Adie T., Baldwin G.S. DNA assembly for synthetic biology: from parts to pathways and beyond. Integrative biology: quantitative biosciences from nano to macro. 2011;3:109–118. doi: 10.1039/c0ib00070a. [DOI] [PubMed] [Google Scholar]
- 46.Ma S., Tang N., Tian J. DNA synthesis, assembly and applications in synthetic biology. Curr. Opin. Chem. Biol. 2012;16:260–267. doi: 10.1016/j.cbpa.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096-1–1258096-9. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 48.Formenti L.R., Norregaard A., Bolic A., Hernandez D.Q., Hagemann T., Heins A.L., Larsson H., Mears L., Mauricio-Iglesias M., Kruhne U., et al. Challenges in industrial fermentation technology research. Biotechnol. J. 2014;9:727–738. doi: 10.1002/biot.201300236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


