Abstract
CRISPR–Cas systems silence plasmids and viruses in prokaryotes. CRISPR–Cas effector complexes contain CRISPR RNAs (crRNAs) that include sequences captured from invaders and direct CRISPR-associated (Cas) proteins to destroy corresponding invader nucleic acids. Pyrococcus furiosus (Pfu) harbors three CRISPR–Cas immune systems: a Cst (Type I-G) system with an associated Cmr (Type III-B) module at one locus, and a partial Csa (Type I-A) module (lacking known invader sequence acquisition and crRNA processing genes) at another locus. The Pfu Cmr complex cleaves complementary target RNAs, and Csa systems have been shown to target DNA, while the mechanism by which Cst complexes silence invaders is unknown. In this study, we investigated the function of the Cst as well as Csa system in Pfu strains harboring a single CRISPR–Cas system. Plasmid transformation assays revealed that the Cst and Csa systems both function by DNA silencing and utilize similar flanking sequence information (PAMs) to identify invader DNA. Silencing by each system specifically requires its associated Cas3 nuclease. crRNAs from the 7 shared CRISPR loci in Pfu are processed for use by all 3 effector complexes, and Northern analysis revealed that individual effector complexes dictate the profile of mature crRNA species that is generated.
INTRODUCTION
The CRISPR–Cas systems are a group of related, RNA-guided, adaptive immune systems that are widespread among prokaryotes (1–4). These immune systems protect host organisms from viruses, conjugative plasmids, and other potential genome invaders. Repeat sequences found in CRISPR loci alternate with ‘spacer’ sequences acquired from invaders. CRISPRs are transcribed giving rise to small CRISPR RNAs (crRNAs) that guide CRISPR-associated (Cas) protein effector complexes to silence invaders via the spacer (guide) sequence in the crRNA (5–17). Based primarily on Cas protein components and cas gene organization, CRISPR–Cas systems can be clustered into three main types (I, II and III) and at least 12 subtypes (e.g. Type I-A–Type I-G) (18,19). There are significant differences in the mechanisms of crRNA processing, target identification, and invader interference among the CRISPR–Cas systems (1–4).
The silencing of invaders occurs by an array of distinct mechanisms that increases with the characterization of each additional CRISPR–Cas system. The Type I and Type II systems utilize distinct mechanisms to directly destroy invader DNAs (15,16,20–22), while Type III systems cleave invader RNA (23–28) and also cleave DNA in a transcription-dependent manner (28). The processes underlying invader DNA cleavage are understood for several of the Type I CRISPR–Cas system subtypes, particularly the Type I-E system found in Escherichia coli (15,16,29), but essentially nothing is known regarding how Types I-C, I-D or I-G systems silence invaders.
To avoid destruction of the host genome (at the CRISPR locus where a copy of the target sequence is present), CRISPR–Cas systems employ a two-step invader recognition mechanism. Type I and Type II effector complexes require a short sequence motif adjacent to the target sequence (and not found in the CRISPR array) to activate DNA destruction (30–32). This motif, called a protospacer adjacent motif (PAM), (31,32) differs in size, sequence, and location relative to the target depending on the particular CRISPR–Cas system (6,20,30–39). Mutation of the PAM sequence in an invader disrupts DNA interference by these systems (6,20,30–33,35,36,38,39). The bioinformatically predicted PAM sequence for the CRISPR-6 repeat sequence family found in Pyrococcus furiosus (Pfu) is 5′-NGG-3′ (31,40) (Figure 1C).
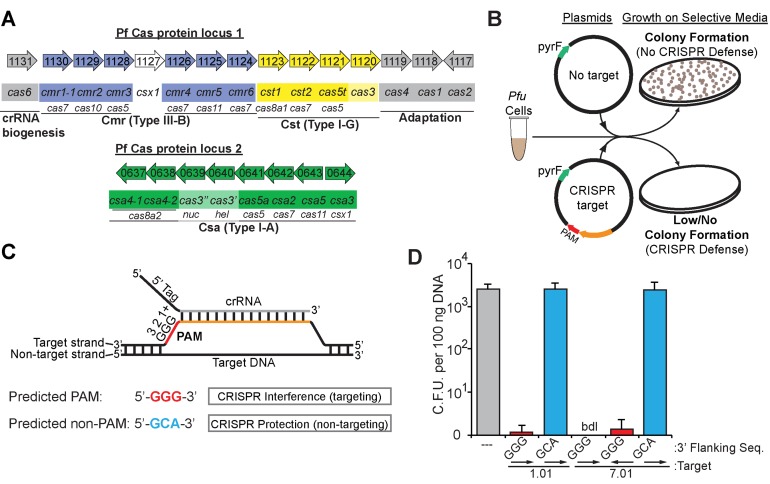
Figure 1.
P. furiosus CRISPR–Cas systems silence plasmid DNA in a PAM-dependent manner. (A) The genome organization and annotations of the predicted cas genes were adapted from the NCBI database (http://www.ncbi.nlm.nih.gov/). Cas subtype Cmr (blue), Cst (yellow), Csa (green) genes are indicated. crRNA biogenesis and adaptation genes (gray) are indicated. (B) Graphic representation of the plasmid challenge assay. CRISPR–Cas defence prevents transformation of plasmids with both a CRISPR target and a PAM from restoring uracil prototrophy in Pfu strain JFW02 (ΔpyrFΔtrpAB). (C) Diagram of crRNA base-pairing with targeted DNA molecules containing a predicted PAM. The repeat derived 5′ tag (black) and spacer derived guide (gray) crRNA sequences, invader DNA target strand (orange), and PAM (red) are displayed with complementarity indicated. (D) Plot of plasmid challenge assay results. Colony forming units in Pfu strain JFW02 are plotted on the Y-axis, with the standard deviation in 9 replicates indicated by error bars. The CRISPR target, the sequence immediately downstream (3′ flank), and target orientation on the plasmid (displayed with arrows) are indicated on the X-axis. Chart bar color additionally indicates plasmids with no target (gray), with a CRISPR target and a predicted PAM (red) or predicted non-PAM (blue). Cases in which no colonies were observed are indicated as ‘bdl’ for below detection limit of the assay.
Organisms harboring CRISPR–Cas systems may have just a single CRISPR–Cas immune system; however, many organisms contain various combinations of distinct CRISPR–Cas immune systems. For example, the model organism Streptococcus thermophilus contains modules encoding Types I, II and III CRISPR–Cas systems, and each system appears to be completely self-contained – with its own CRISPR array, CRISPR RNA processing enzyme, and set of adaptation proteins (required for invader sequence acquisition) as well as effector complex proteins (5,41–43). In other studied organisms, such as Pyrococcus furiosus, Thermococcus kodakarensis and Sulfolobus islandicus, CRISPR–Cas systems appear to be at least partially interdependent—with multiple shared CRISPR arrays, a single primary CRISPR RNA processing enzyme, and one set of adaptation proteins (6,14,44). While there are many organisms with multiple CRISPR–Cas systems encoded within their genome, there is very limited experimental information about the functionality/dormancy and any interdependence of the individual systems in these organisms.
Pfu encodes three different immune effector complexes, including two Type I complexes—Type I-G Cst and Type I-A Csa—and the Type III-B Cmr complex (8,23,44,45) (Figure 1A). The Cst and Cmr effector complex genes are located in one locus between genes encoding the Cas6 CRISPR RNA processing endonuclease (46) and the Cas1/Cas2/Cas4 adaptation proteins, whereas the Csa effector complex genes are found at a separate locus without associated processing or adaptation genes (Figure 1A). We have previously isolated and described the RNA and protein components of complexes from each of the 3 systems in Pfu (45). Pfu contains 7 CRISPR arrays, all of which produce primary transcripts processed by the Cas6 enzyme to generate over 200 unique crRNAs (46–48). Representative species from the 7 CRISPRs are found associated with all 3 of the immunoaffinity-purified complexes (8,45). We have extensively characterized the Cmr effector complex and its homology-dependent cleavage of RNAs (8,23,49–51). However, whether the Pfu Cst and Csa system effector complexes are functional has not been determined.
In the current study, we generated strains of Pfu that encode a single CRISPR–Cas effector complex, either Cst or Csa, to allow us to assess the function of these two systems independently in vivo. Our results establish that both the Type I Cst and Csa systems silence plasmid DNA, delineate the PAM sequences able to activate each of these systems (31–33), and establish that silencing by both the Cst and Csa system requires the specifically associated Cas3 nuclease. We also show that generation of mature crRNA species depends on the effector complex proteins and that each immune system gives rise to a subset of the processed mature crRNA species found in the wild-type strain.
MATERIALS AND METHODS
Strains and growth conditions
The strains and plasmids used in this study are listed in Supplemental Table S1. Pfu strains were grown under strict anaerobic conditions at 90°C in defined medium. The medium was composed of 1× base salts, 1× trace minerals, 10× vitamin solution, 2× 19-amino-acid solution, 10 μM sodium tungstate, 0.35% cellobiose, and 1 mg/l resazurin, with added cysteine at 1 g/l, sodium sulfide at 0.5 g/l, sodium bicarbonate at 1 g/l and 1 mM sodium phosphate buffer (pH 6.5). Stock solutions were prepared as described previously (52) with the exception that vitamin solution was prepared as a 4000× solution rather than 200× and cysteine was used instead of cysteine–HCl. Medium pH was adjusted before the addition of phosphate buffer to approximately pH 6.5. Liquid cultures were inoculated with 1–2% inoculum or with a single colony and grown in anaerobic culture bottles. Anaerobic culture bottle headspace was exchanged with four cycles of vacuum and argon. Solid media preparation and culture growth was as described previously (52). Medium was supplemented with 20 μM uracil and/or 2.75 mM 5-FOA as needed for selection. E. coli strain TOP10 was used for general plasmid DNA manipulation. Cultures were grown at 37°C in Luria Broth (Millers) supplemented with apramycin sulfate (50 μg/ml).
Northern analysis
Total RNA samples were isolated from ∼50 mg of Pfu cells (JFW02, TPF07, TPF15, TPF17 or TPF20) using Trizol LS (Invitrogen). Ten microgram of total RNA samples were separated on 15% TBE–urea polyacrylamide gels beside a 32P 5′ end-labeled Decade Marker RNA (Life Technologies). RNAs were transferred to nylon Zeta-Probe membranes (Bio-Rad) using a Trans-Blot SD Semi-Dry Cell (Bio-Rad). Membranes were baked at 80°C for an hour before prehybridization in a ProBlot hydridization oven (LabNet) for an hour at 42°C. Prehybridization and hybridization were performed in hybridization buffer containing 5× SSC, 7% SDS, 20 mM NaPO4 (pH 7.0) and 1× Denhardt's solution. Deoxyribonucleotide probes (Operon) (10 pmol) were 5′ end-labeled with T4 Polynucleotide Kinase (NEB) and γ-[32P]-ATP (specific activity > 6000 Ci/mmol, Perkin Elmer) using standard protocols. Labeled probes (1 million cpm/ml) were added to prehybridization buffer, followed by hybridization at 42°C overnight. Probed membranes were washed twice in 2× SCC, 0.5% SDS for 30 min at 42°C. Membranes were reprobed for the 5S rRNA loading control following initial probing. Radioactive signals were detected by phoshorimaging (Storm 840 Scanner GE Healthcare). Probe sequences used are listed in Supplemental Table S2.
General DNA manipulation and plasmid construction
Plasmid DNAs for sequencing and routine analysis were isolated from E. coli strain Top10 using the QIAprep Spin Miniprep Kit (Qiagen). Large-scale plasmid DNA isolation from the Top10 strain was carried out for the construction of plasmids or for use in Pfu plasmid transformation assays, using the Zyppy Plasmid Maxiprep Kit (Zymo Research). Following isolation with the Maxiprep kit, plasmids were isopropanol-precipitated and resuspended in 1 mM Tris–Cl, pH 8.5. Routine PCR screening was carried out with Crimson Taq (NEB), and Splicing by Overlap Extension PCR (SOE-PCR) was carried out with Phusion polymerase (NEB). Pfu genomic DNA was isolated from 1 ml of overnight liquid cultures with the Zymo Quick gDNA Miniprep kit (Zymo Research) for routine PCR genotyping.
The expression plasmid pJE47 was constructed using the T. kodakarensis csg promoter and chiA terminator sequences found on the plasmid pLC64-ChiA (6). Primers (sequences given in Supplemental Table S2) Pcsg_F and Pcsg_NdeI_Bam_R were used to amplify the promoter region, and primers Term_NdeI_Bam_F and ChiA_Term_R were used to amplify the terminator region. The PCR products were spliced by SOE-PCR, in the process adding NdeI and BamHI restriction sites between the promoter and terminator, and cloned into the NotI/EcoRV sites of the Pfu shuttle vector plasmid pJFW18 (53).
Plasmids containing CRISPR target inserts (Supplemental Table S1) were constructed by one of three methods. Plasmids pJE18–33 were constructed by ligating annealed 5′-phosphorylated oligos (Supplemental Table S2) into the NotI site of pJFW18. Plasmid was linearized with NotI-HF (NEB) and dephosphorylated with thermosensitive alkaline phosphatase (Promega). 5′-phosphorylated CRISPR target oligo pairs were annealed in 10 mM Tris–Cl (pH 8), 1 mM EDTA, 500 mM NaCl. Annealed oligo pairs were ligated with NotI-linearized pJFW18 using T4 DNA Ligase (NEB).
Plasmids pJE186-249 were constructed by a combination of two methods. The inserts for the majority of the plasmids were constructed by primer extension of oligo 7.01_TIM_Sat_Mut_F, which contains degenerate PAM nucleotides, with primer 7.01_TIM_Sat_Mut_R. The primer extended oligo pairs were digested with NdeI (NEB) and BamHI-HF (NEB) and ligated with NdeI/BamHI-linearized pJE47. After analysis of ∼200 clones, all remaining tri-nucleotide combinations were cloned into pJE47 as described for plasmids pJE18–33, with the exception that the host vector was NdeI/BamHI-linearized pJE47. Additional 6.01 target plasmids, pJE275 & pJE299–301, were generated by ligation of annealed 5′-phosphorylated oligos with NdeI/BamHI-linearized pJE47.
Plasmids pJE252 (Cas3), 253 (Cas3”), and 269 (Cas3′+3”) were constructed using standard cloning techniques. Inserts for these plasmids were generated by PCR amplification of the genes using JFW02 genomic DNA. Each insert was generated using the following primer sets: Cas3 with PF1120_pJE47_F/R, Cas3” with PF0639_pJE47_F/R, and Cas3′+3” with PF0640_pJE47_F/PF0639_pJE47_R. Inserts were cloned into NdeI/BamHI linearized pJE47 with the GENEART Seamless Cloning and Assembly kit (Life Technologies). To generate plasmids pJE257, 258 and 270, these plasmids were linearized and dephosphorylated with NotI-HF and thermosensitive alkaline phosphatase prior to ligation with annealed PF_7.01_GGG+/- oligos.
All E. coli transformants were analyzed by PCR for inserts and sequenced to confirm insert sequence and orientation. Primer sequences used for PCR amplification of inserts and oligo pairs used for the construction of target-bearing plasmids are indicated in Supplemental Table S2, with the ‘+’ oligo being annealed with the cognate ‘-’ oligo to generate a double-stranded target for ligation.
Pyrococcus furiosus strain construction
Pfu strains were constructed to characterize individual effector complexes in vivo, using a variant of the previously described pop-in/pop-out marker replacement technique (Supplemental Figure S4) (52,54). Linear PCR products containing a counter-selectable pyrF wild type allele were transformed into Pfu host strains containing a deletion of the pyrF gene to select uracil prototrophy. Regions of homology flanking the pyrF gene were used to guide homologous recombination at desired genomic regions. Following marker replacement of the region of interest, 5-FOA, a toxic PyrF substrate, was used to select for cells that had removed the pyrF wild type allele by homologous recombination between short regions of homology, internal to the PCR fragment, flanking the pyrF gene resulting in a markerless deletion of the gene of interest. The transformed PCR products were generated by splicing four PCR products together with SOE-PCR (Supplemental Figure S4 and Supplemental Table S2). Strain TPF07 (Csa) (Supplemental Table S1) was constructed by a single-step deletion of genes PF1130 through PF1121 (Δcmr+cst). TPF15 (Cmr), TPF17 (Cst) and TPF20 (null) strains were each constructed with two consecutive deletion steps. TPF15 (Cmr) was constructed by stepwise deletions with the Δcsa (deletion of PF0637 through PF0644) and Δcst (deletion of PF1123 through PF1121) PCR constructs. TPF17 (Cst) was constructed with the Δcmr (deletion of PF1130 through PF1124) and Δcsa constructs. TPF20 (null) was constructed with the Δcmr+cst and Δcsa PCR constructs. TPF01 (ΔCas3”) and TPF02 (ΔCas3) strains were constructed by deletion of PF0639 and PF1120, respectively, from the parental strain JFW02. TPF10 (ΔCas3”, ΔCas3) was generated by deletion of PF1120 from TPF01. TPF29 (Cst ΔCas3) was generated by deletion of PF1120 from TPF17, and TPF30 (Csa ΔCas3”) was generated by deletion of PF0639 from TPF07.
Plasmid transformation assay in Pyrococcus furiosus
Plasmid transformations were accomplished by culturing Pfu strains anaerobically at 90°C to mid-to-late log phase in defined media supplemented with 20 μM uracil. 33.3 ul of the culture was mixed with plasmid DNA to a final concentration of 2.0 ng/μl (35 μl total reaction volume) and briefly incubated at room temperature for typically 5–60 min before plating on solid defined media lacking uracil. The mixture was spread on solid media using Coliroller glass beads (EMD Millipore) and incubated for ∼64 h at 90°C under anaerobic conditions. Following incubation, colonies per plate were enumerated. All transformation assays were carried out with a minimum of three technical replicates.
RESULTS
The type I-G Cst and type I-A Csa CRISPR–Cas systems in Pfu target DNA in vivo
We assayed CRISPR–Cas-mediated plasmid silencing in Pfu (strain JFW02) by challenging cells with plasmids that contain (or lack) target sequences for endogenous Pfu crRNAs 1.01 or 7.01 (arising from CRISPR1 spacer 1 or CRISPR7 spacer 1, respectively; Figure 1B). The bioinformatically-predicted PAM sequence for the CRISPR repeat sequence family found in the seven Pfu CRISPR loci is 5′-NGG-3′ (31,40). Accordingly, we included either a predicted PAM (5′-GGG-3′) or a tri-nucleotide sequence that does not conform to the predicted PAM (5′-GCA-3′) directly adjacent to the target sequence in the plasmids (Figure 1C). The plasmid also included the pyrF gene, so infected Pfu cells produce colonies on plates lacking uracil.
We found that the presence of crRNA target sequences on plasmids reduced plasmid infection by at least 3 orders of magnitude. Infection with the parent plasmid (lacking target sequences for endogenous Pfu crRNAs) produced over 103 colonies per 100 ng of plasmid DNA (Figure 1D, gray bar). The plasmids containing crRNA target sequences and the predicted PAM produced at least 3 orders of magnitude fewer colonies (Figure 1D, red bars). Disruption of the predicted PAM resulted in loss of silencing of the plasmids (Figure 1D, blue bars). Moreover, we observed that the 7.01 crRNA target sequence induces silencing in either orientation, indicative of transcription-independent silencing of DNA targets (Figure 1D, target orientation indicated by arrows). Taken together, these results indicate that Pfu contains at least one CRISPR–Cas system that is capable of crRNA homology-dependent plasmid DNA silencing in vivo.
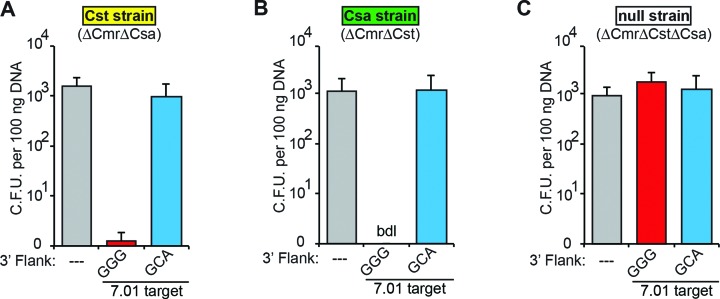
To determine whether the individual Pfu Cst and Csa CRISPR–Cas systems effect DNA silencing, genetically-engineered strains of Pfu harboring a single CRISPR–Cas effector complex module (Cst strain and Csa strain) or no effector complex modules (null strain) were generated and challenged with plasmids (Figure 2). Each strain was assayed using plasmids containing no target (—), or the 7.01 crRNA target with the predicted PAM (5′-GGG-3′) or mutated PAM (5′-GCA-3′). As expected, the null strain was infected with the plasmids irrespective of the presence of crRNA target sequences in the plasmid (Figure 2C). However, few to no colonies were observed when the plasmid with the 7.01 target and predicted PAM was transformed into either the Cst or Csa strain (Figure 2A and B). We also challenged the Cst and Csa strains with plasmids containing a 6.01 crRNA target sequence and either a predicted PAM (GGG) or one of three predicted non-PAMs (CCC, TTT, AAA; Supplemental Figure S1). Again, few to no colonies were observed when the 6.01 target plasmid with predicted PAM was transformed into either the Cst or Csa strain. These results demonstrate that both the Cst and Csa CRISPR–Cas immune systems of Pfu silence invader DNA.
Figure 2.
P. furiosus Cst and Csa CRISPR–Cas systems silence plasmid DNA in a PAM-dependent manner. Plots of plasmid challenge assay results in Pfu strains harboring the Cst (A), Csa (B) or no (C) CRISPR–Cas systems. Colony forming units are plotted on the Y-axis, with the standard deviation in six replicates indicated by error bars. The CRISPR target and the sequence immediately downstream (3′ flank) are indicated on the X-axis. Additionally, bar color indicates plasmids with no target (gray), with a CRISPR target and a predicted PAM (red) or predicted non-PAM (blue). The name and genotype of the assayed strain is indicated above each panel. Cases in which no colonies were observed are indicated as ‘bdl’ for below detection limit of the assay.
The Cst and Csa systems utilize a highly overlapping set of PAMs for DNA silencing
To comprehensively define the PAM requirements for DNA silencing by the Cst and Csa CRISPR–Cas systems, we systematically transformed the Cst and Csa strains with a set of plasmids containing the 7.01 crRNA target and the 64 possible tri-nucleotide PAM sequence combinations immediately 3′ of the target sequence (Figure 3A). Both the strain containing the Cst system and the strain containing the Csa system resisted infection by plasmids with a contiguous, discrete subset of 3′ flanking tri-nucleotide sequences (Figure 3B and C, Supplemental Figures S2 and S3). Colony formation was reduced by at least 2 orders of magnitude for these plasmids compared to the parental control plasmid (lacking the target sequence). The Cst strain efficiently silenced 14/64 targets with NGR, HAG and HCG (where H = A, C, or T) 3′ flanking sequences (Figure 3B and Supplemental Figure S2). Additionally, we observed an intermediate reduction in colony formation (between 1 and 2 orders of magnitude) for the 5/64 plasmids containing NGC or GAG 3′ flanking sequences in the Cst strain. The Csa strain efficiently silenced 13/64 plasmids with NGR, NAG, and CCG 3′ flanking sequences (Figure 3C and Supplemental Figure S3). A lesser reduction in colony formation (∼1 order of magnitude) was observed for plasmids containing ACG and TGC 3′ flanking sequences. The Cst and Csa systems utilize significantly overlapping PAM sequences, and the bioinformatically-predicted 5′-NGG-3′ PAM resulted in efficient silencing in both systems (Figure 3B and C, Supplemental Figures S2 and S3). Of note, the repeat sequence flanking the spacers in the Pfu CRISPR loci (5′-CTT-3′) is not utilized as a PAM by either the Cst or the Csa systems. Overall, these results demonstrate that both Pfu Type I CRISPR–Cas systems efficiently silence targets flanked by a specific and highly-overlapping subset of PAM sequences.
Figure 3.
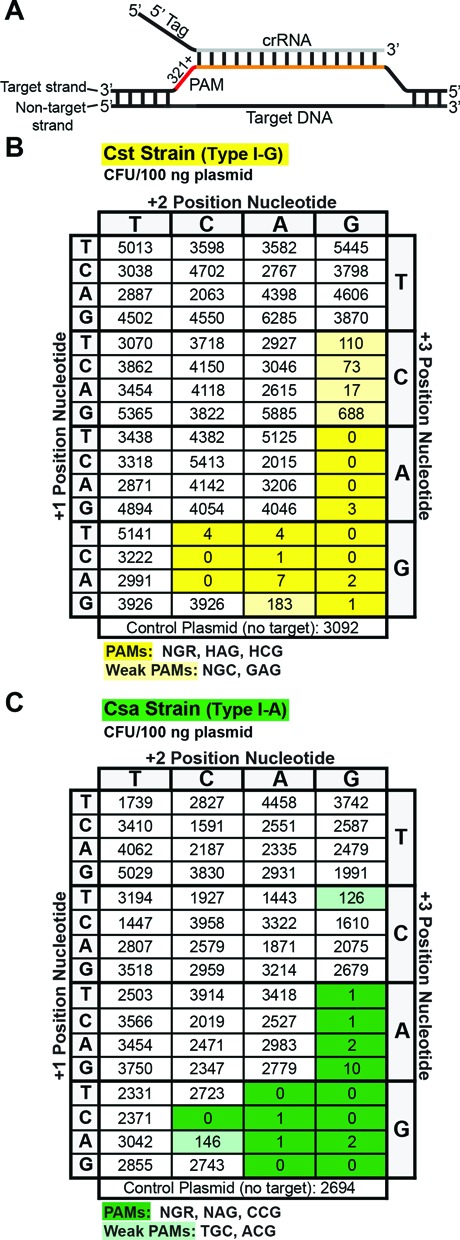
Cst and Csa strains have similar PAM requirements for plasmid interference. (A) Diagram indicating the location of the PAM sequences relative to the target sequence and an annealed crRNA. (B) and (C) show results of plasmid challenge assays for Cst and Csa strains, respectively. All target plasmids contain the 7.01 target and the indicated PAM sequence. Colony enumerations for each plasmid are the average of at least three replicates. Darkly and lightly shaded boxes indicate plasmids with PAMs considered to be effectively or intermediately silenced, respectively.
The Cst and Csa systems each require a specific Cas3 protein for plasmid interference
Both the Cst and Csa modules found in Pfu include a member of the Cas3 protein superfamily common to all Type I CRISPR–Cas systems ((18,45) and Figure 1A). Cas3 has been shown to unwind and destroy target DNA in several Type I CRISPR–Cas systems, including the Csa system in Thermoproteus tenax (15,16,20,29,55–60), so we tested the hypothesis that Cas3 was the effector nuclease in the Cst as well as Csa system in Pfu.
In the Pfu Cst system (and most Type I systems), the Cas3 protein includes both an HD nuclease domain and an ATP-dependent DExH helicase domain (60–62), though in the Pfu Csa system, the Cas3 nuclease (Cas3”) and helicase (Cas3′) domains are encoded by two separate genes (18,61). In wild-type Pfu, deletion of either the Cst-associated cas3 gene (ΔCas3; PF1120) or the Csa-associated cas3” nuclease gene (ΔCas3”; PF0639), is insufficient to eliminate crRNA-targeted eradication of the plasmid, but deletion of both genes disrupts defense (Figure 4A).
Figure 4.
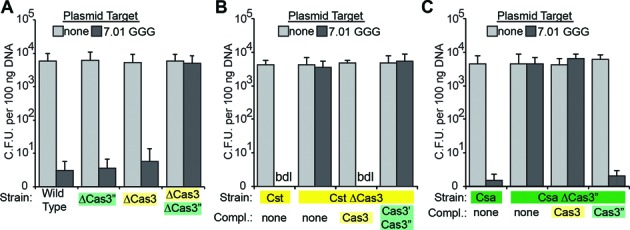
The cognate cas3 is required for Cst and Csa plasmid DNA silencing. Plasmid challenge assay results plotted for single or double cas3 gene deletion strains (A), Cst derivative strains (B), and Csa derivative strains (C). Colony forming units are plotted on the Y-axis, with error bars indicating the standard deviation in at least three replicates. Transformed plasmids contained no target (light gray bars) or the 7.01 target with GGG PAM (dark gray bars). Strain transformed and proteins(s) expressed from plasmids are indicated on the X-axis. Cases in which no colonies were observed are indicated as ‘bdl’ for below detection limit of the assay.
To assay the individual requirements of the Cst and Csa systems and to determine if a given Cas3 protein functions selectively with its affiliated system (or can function redundantly between the systems), we performed plasmid transformation assays and cas3 gene deletion/complementation analysis in Pfu strains containing isolated Cst and Csa systems (Figure 4B and C, respectively). In strains containing only the Cst or Csa system, deletion of the affiliated cas3 gene disrupted crRNA-targeted plasmid defense (Cst ΔCas3, Figure 4B and Csa ΔCas3”, Figure 4C). Furthermore, in each case, plasmid defense was restored by exogenous expression of the affiliated cas3 but not of the other cas3 (Figure 4B and C). These results demonstrate that Cas3 is required for plasmid defense by the Cst as well as the Csa CRISPR–Cas system, and that these Cas3 proteins function specifically with the associated CRISPR–Cas system.
CRISPR RNA profiles in Pfu strains with single CRISPR–Cas systems
Our previous analysis demonstrated that crRNAs derived from all 7 Pfu CRISPR loci are constitutively expressed and processed into several different size forms of mature crRNA species that differ at their 3′ ends (8,45,48). Subsets of the mature species are associated with each of the immunopurified Cst, Csa and Cmr complexes (45). Cas6 cleaves CRISPR locus transcripts within each repeat to release individual crRNA precursors (46,47), but the mechanism by which the various 3′ end-processed mature size species are generated is not known. To test the importance of effector complex proteins in the biogenesis of the specific crRNA species, we examined crRNA profiles in the wild-type strain (All), the individual effector complex strains (Csa, Cmr, Cst), and a strain with no effector complexes (null) by northern analysis (Figure 5). All strains contain Cas6, which cleaves eight nts upstream of the spacer within each repeat of the CRISPR transcript (46,47). A probe for the CRISPR repeat sequence (common to all seven CRISPR loci) detected two prominent RNAs in all of the strains: ∼67 nt and ∼135 nt RNAs typical for the previously described 1× Cas6 CRISPR transcript cleavage product containing a single crRNA unit and the 2× intermediate product containing two crRNA units (46,47) (Figure 5A).
Figure 5.
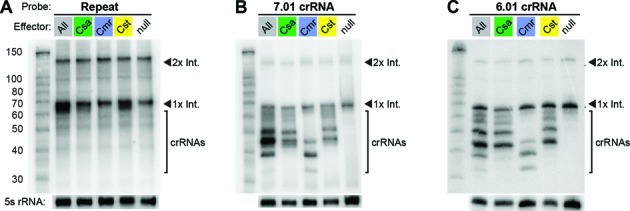
Northern analysis of crRNA expression and maturation in P. furiosus strains with a single or no effector complex. Blots were probed for the consensus CRISPR repeat sequence (A) or two representative crRNAs: 7.01 (B) and 6.01 (C). The left lane in each panel contains the Decade Marker RNA, and all other lanes contain Pfu total RNA from the strain indicated above the panel. Probing for 5S rRNA served as a loading control. RNAs corresponding to the Cas6 cleavage products, 1× intermediate and 2× intermediate, are indicated. Processed mature crRNAs are indicated with brackets.
We found that 3′ end processing and accumulation of mature crRNAs depends on the effector complex proteins. Multiple mature species can be seen in the wild-type strain (with all 3 CRISPR–Cas systems) in Northern blots probed for the first crRNAs encoded by CRISPR loci 7 and 6 (Figure 5B and C, ‘All’ lanes). A subset of these mature crRNAs is found in each single effector strain. In the Cmr strain, crRNAs are processed into two primary size forms of 45 and 39 nt in length, and a third minor size form of 33 nt (Figure 5B and C, Cmr) (8,48). The mature crRNA profiles in the Cst and Csa strains are very similar (and distinct from the Cmr crRNA profile). The 7.01 and 6.01 crRNAs are found in two prominent size forms of ∼45 and 50 nt in the Cst and Csa strains (Figure 5B and C). The sizes of the 7.01 crRNAs found in each of the single effector complex strains (Figure 5B) correspond well with the 7.01 crRNA species that co-immunopurify with the individual complexes from the wild-type strain (45). Note that most of the mature crRNAs observed in the wild-type strain can be accounted for by the sum of mature crRNAs in the individual effector complex strains (Figure 5B and C). The crRNA profile in wild-type Pfu may be the cumulative result of three independent crRNA maturation pathways, in which the effector complexes themselves contribute to the generation of the distinct functional species of crRNAs.
DISCUSSION
Our previous biochemical analysis demonstrated that three compositionally distinct CRISPR–Cas effector complexes co-exist in Pyrococus furiosus (Pfu): the Type I-G Cst system, the Type I-A Csa system, and the Type III-B Cmr system (8,45,48). The functionality of the Pfu Cmr system both in vivo and in vitro against RNA targets has been demonstrated (8,23,49–51). In this study, we obtained the first evidence that the recently identified Pfu Cst and Csa effector complexes (45) are also functional (Figure 2). These two Pfu Type I CRISPR–Cas systems exhibit many commonalities: they display highly similar crRNA association profiles (Figure 5 and (45)), they each utilize a largely overlapping and specific subset of PAM sequences (Figure 3), and they silence the same DNA targets (Figures 2 and 3, and Supplementary Figure S1). Despite these similarities, the Cst and Csa effector complexes each require a dedicated Cas3 effector nuclease for target DNA destruction (Figure 4). Our data indicate that Pfu has an impressive arsenal of three active CRISPR–Cas immune systems that presumably provides robust protection from the diverse viruses and other mobile genetic elements that it naturally encounters. Furthermore, this work provides the first experimental evidence that Cst CRISPR–Cas immune systems function by invader DNA silencing.
The Cst and Csa CRISPR–Cas systems eliminate plasmids by a DNA targeting mechanism that requires the Cas3 nuclease and the presence of a PAM
Our findings show that both Type I systems of Pfu, Cst and Csa, are active and utilize a Cas3- and PAM-dependent mechanism to silence invader DNA (Figures 2–4). DNA interference by the Cst CRISPR–Cas system was not reported prior to this study. Cst complexes appear to be the most minimal Type I CRISPR–Cas invader surveillance complexes, being comprised of a crRNA and just three stably associated Cas proteins (Cst1 (large subunit), Cas5t and Cst2 (Cas7) (45,63). In addition we show that a fourth protein, the Cas3 nuclease/helicase protein, is required for silencing (Figures 1A and 4).
Csa systems are the only CRISPR–Cas systems thought to be specific to archaea (not found in bacteria) (18,61). Csa complexes or subcomplexes from Pyrococcus furiosus (45), Sulfolobus solfataricus (10) and Thermoproteus tenax (20,64) have been isolated and/or reconstituted in vitro. The protein components of these three complexes are similar, each containing Cas5a, Cas8 (large subunit), Cas7, and Csa5 (small subunit). The complexes from P. furiosus and T. tenax further contained the Cas3′ (helicase) and Cas3” (nuclease). Consistent with our findings (Figure 4), Cas3” is required for DNA cleavage by the T. tenax Csa complex (20). Our results demonstrate that Csa CRISPR–Cas systems utilize a Cas3- and PAM-dependent mechanism to silence invader DNA.
Cas3 protein diversity in Type I CRISPR–Cas systems
Cas3 superfamily proteins are very likely the effector nucleases for all Type I CRISPR–Cas systems. Recombinant Cas3 proteins from multiple Type I systems (including the Cas3” protein from the Csa system in Pfu (56)) exhibit sequence-independent nuclease activity against single-stranded DNA substrates in vitro (56,58–60). Moreover, genetic analyses indicate that the cas3 genes are essential for DNA interference activity in vivo (Figure 4 and (14,65,66)). However, there are significant differences among Cas3 proteins. For example, evidence indicates that Cas3 is a stably associated and integral functional component of some Type I complexes, such as Csa (20,45), but is recruited to other crRNP complexes, including likely the Cst complex, only as the result of PAM recognition and target DNA binding (29,60). Accordingly, our work demonstrates that the Cas3 protein components of the Cst and Csa systems in Pfu (Cas3 and Cas3”) are not functionally interchangeable (Figure 4). As a general rule, we predict that following invader DNA recognition, Type I crRNP complexes will license specific Cas3 proteins for DNA cleavage through subtype-specific protein-protein interactions.
CRISPR RNA 3′ end maturation and accumulation depends on effector complexes
For the majority of Type I and Type III CRISPR–Cas systems, the CRISPR repeat-specific endoribonuclease, Cas6, is responsible for the initial processing of the primary CRISPR transcript, resulting in formation of 1x intermediate crRNAs containing an 8 nt 5′ tag sequence, a spacer-derived guide sequence, and 3′ repeat sequence (46,65,67,68). In some systems, such as the Type I-E (Cse) and I-F (Csy) CRISPR–Cas systems, this initial cleavage product serves as the mature crRNA species associated with the effector complex and Cas6 remains bound to the 3′ repeat (17,69–72). In many other systems, including the Cst, Csa and Cmr systems in Pfu (45), the crRNA is further processed into smaller forms that retain the 5′ repeat tag sequence but typically lack 3′ repeat sequences (6,8,14,23–26,45,64,68). In this study, we observed that an effector complex is required for crRNA 3′ end maturation and generation of the complex-specific profiles of Csa, Cst and Cmr crRNA species (Figure 5). Effector complexes are not required for production and accumulation of the Cas6-generated 1× crRNA intermediates, which accumulate in a Pfu strain lacking all three CRISPR–Cas effector complexes (null; Figure 5). The profiles of crRNAs accumulating in strains containing the individual CRISPR–Cas systems (Cst, Csa or Cmr; Figure 5) are highly similar to those present in the biochemically-isolated crRNPs (45). This finding suggests that the structure of the effector complex defines (perhaps indirectly) the ultimate 3′ ends of mature crRNAs, which may be processed by non-Cas (3′-5′ exo)ribonucleases (68) to produce the distinct mature crRNA forms that function with each effector complex.
Cst and Csa CRISPR–Cas systems recognize largely overlapping PAMs
All previously characterized Type I systems utilize a short ∼2–5 nt protospacer adjacent motif (PAM) immediately flanking the target sequence to discriminate invader (protospacer with adjacent PAM) from host CRISPR (spacer lacking PAM) DNA (6,20,31,33–37,39). DNAs containing a perfectly complementary target sequence but lacking a PAM are not targeted by Type I CRISPR–Cas systems. Bioinformatic analysis has revealed a relationship between CRISPR repeat sequence classes and associated PAM sequences, which has yet to be understood at the molecular level (31,40,73). Indeed, the 5′-NGG-3′ PAM sequence predicted for the Pfu CRISPR repeat sequence (31,40,73) functions effectively with both the Cst and Csa systems: plasmids with crRNA targets flanked by 5′-NGG-3′ are silenced in our in vivo assays (Figures 1–3). However, comprehensive analysis revealed a distinct set of 13–14 additional sequence combinations that also activate DNA silencing by the Cst and Csa systems (see strong and weak PAMs shown in Figure 3, Supplementary Figure S2 and S3). It will be interesting to determine the molecular basis for the CRISPR repeat/PAM sequence relationships that are detectable by bioinformatic analysis (31,40,73) versus the additional functional PAMs detected by biological assay in this work.
It is clear that the Pfu Cst and Csa systems utilize a largely overlapping set of PAMs; 12 of the 14 strong Cst PAMs and 13 strong Csa PAMs activate silencing in both systems, and just one strong PAM (the Cst PAM 5′-TCG-3‘) is not at least partially recognized by the other system (Figure 3, Supplemental Figures S2 and S3). Our findings predict similar mechanisms of PAM recognition in the Pfu Cst and Csa defense complexes. It is thought that invader target sequences with adjacent PAMs are actively selected for incorporation into CRISPRs by the adaptation machinery (32,74). While the utilization of multiple PAMs is not unique among CRISPR–Cas systems (6,20,32–37,39,75), this work provides the first example of multiple co-existing CRISPR–Cas systems utilizing a highly similar set of PAMs, and likely indicates co-evolution of the two systems to function with shared CRISPR loci as well as a single common set of adaptation machinery (see Figure 1).
Co-function of similar CRISPR–Cas systems
Based on the current analysis, the functionalities of the two Type I CRISPR–Cas systems of Pfu appear to be largely redundant, which raises the question of why might it be advantageous for an organism to have two such CRISPR–Cas systems. One possibility is that the performance of each system may be differentially regulated to respond to different stimuli and invader signalling pathways. Indeed, there is evidence that csa gene expression is significantly up-regulated in Pfu (as well as T. tenax (64)) upon exposure to oxidative stress or ionizing radiation, while cst gene expression is not (76,77). In addition, the recent discovery that viruses can encode subtype-specific anti-CRISPR–Cas proteins (78,79) provides an additional rationale for the maintenance of otherwise functionally redundant CRISPR–Cas systems. Multiple operational CRISPR–Cas systems may provide robust responses to diverse types of intruding mobile genetic elements. Further delineation of the mechanisms by which the Cst and Csa silencing complexes seek and destroy invaders will provide insight into unique and shared functional principles of Type I CRISPR–Cas systems.
Supplementary Material
Acknowledgments
We thank Claiborne V.C. Glover, III for critical review of the manuscript and helpful discussions throughout the course of this work. We also thank members of the Terns lab: Caryn Hale, Anisha Hedge, Erica Figueroa, Janet Figueroa, Hyowon Ahn and Chang-Hao Wu, for help with strain construction. We thank UGA colleagues, Barney Whitman and Felipe Sarmiento, for technical advice with anaerobic culturing, Farris Poole for assistance in setting up anaerobic culturing equipment, and Joel Farkas for helpful advice regarding working with P. furiosus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 GM54682 to M.P.T. and R.M.T.]. Funding for open access charge: National Institutes of Health [R01 GM54682].
Conflict of interest statement. None declared.
REFERENCES
- 1.Terns M.P., Terns R.M. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsui T.K., Li H. Structure Principles of CRISPR–Cas Surveillance and Effector Complexes. Annu. Rev. Biophys. 2015;44:229–255. doi: 10.1146/annurev-biophys-060414-033939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson R.N., Wiedenheft B. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol. Cell. 2015;58:722–728. doi: 10.1016/j.molcel.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 6.Elmore J.R., Yokooji Y., Sato T., Olson S., Glover C.V., 3rd, Graveley B.R., Atomi H., Terns R.M., Terns M.P. Programmable plasmid interference by the CRISPR–Cas system in Thermococcus kodakarensis. RNA Biol. 2013;10:828–840. doi: 10.4161/rna.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadan A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 8.Hale C.R., Majumdar S., Elmore J., Pfister N., Compton M., Olson S., Resch A.M., Glover C.V., 3rd, Graveley B.R., Terns R.M. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul U., Wurm R., Wagner R., et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 10.Lintner N.G., Kerou M., Brumfield S.K., Graham S., Liu H., Naismith J.H., Sdano M., Peng N., She Q., Copie V., et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J. Biol. Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier L.K., Lange S.J., Stoll B., Haas K.A., Fischer S., Fischer E., Duchardt-Ferner E., Wohnert J., Backofen R., Marchfelder A. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B. RNA Biol. 2013;10:865–874. doi: 10.4161/rna.24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulepati S., Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng W., Li H., Hallstrom S., Peng N., Liang Y.X., She Q. Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol. 2013;10:738–748. doi: 10.4161/rna.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinkunas T., Gasiunas G., Waghmare S.P., Dickman M.J., Barrangou R., Horvath P., Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westra E.R., van Erp P.B., Kunne T., Wong S.P., Staals R.H., Seegers C.L., Bollen S., Jore M.M., Semenova E., Severinov K., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiedenheft B., Lander G.C., Zhou K., Jore M.M., Brouns S.J., van der Oost J., Doudna J.A., Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vestergaard G., Garrett R.A., Shah S.A. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014;11:156–167. doi: 10.4161/rna.27990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plagens A., Tripp V., Daume M., Sharma K., Klingl A., Hrle A., Conti E., Urlaub H., Randau L. In vitro assembly and activity of an archaeal CRISPR–Cas type I-A Cascade interference complex. Nucleic Acids Res. 2014;42:5125–5138. doi: 10.1093/nar/gku120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karvelis T., Gasiunas G., Siksnys V. Programmable DNA cleavage in vitro by Cas9. Biochem. Soc. Trans. 2013;41:1401–1406. doi: 10.1042/BST20130164. [DOI] [PubMed] [Google Scholar]
- 23.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Rouillon C., Kerou M., Reeks J., Brugger K., Graham S., Reimann J., Cannone G., Liu H., Albers S.V., et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staals R.H., Agari Y., Maki-Yonekura S., Zhu Y., Taylor D.W., van Duijn E., Barendregt A., Vlot M., Koehorst J.J., Sakamoto K., et al. Structure and activity of the RNA-targeting Type III-B CRISPR–Cas complex of Thermus thermophilus. Mol. Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staals R.H., Zhu Y., Taylor D.W., Kornfeld J.E., Sharma K., Barendregt A., Koehorst J.J., Vlot M., Neupane N., Varossieau K., et al. RNA targeting by the type III-A CRISPR–Cas Csm complex of Thermus thermophilus. Mol. Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamulaitis G., Kazlauskiene M., Manakova E., Venclovas C., Nwokeoji A.O., Dickman M.J., Horvath P., Siksnys V. Programmable RNA shredding by the type III-A CRISPR–Cas system of Streptococcus thermophilus. Mol. Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 28.Samai P., Pyenson N., Jiang W., Goldberg G.W., Hatoum-Aslan A., Marraffini L.A. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR–Cas Immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochstrasser M.L., Taylor D.W., Bhat P., Guegler C.K., Sternberg S.H., Nogales E., Doudna J.A. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deveau H., Barrangou R., Garneau J.E., Labonte J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 32.Shah S.A., Erdmann S., Mojica F.J., Garrett R.A. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almendros C., Guzman N.M., Diez-Villasenor C., Garcia-Martinez J., Mojica F.J. Target motifs affecting natural immunity by a constitutive CRISPR–Cas system in Escherichia coli. PLoS One. 2012;7:e50797. doi: 10.1371/journal.pone.0050797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cady K.C., Bondy-Denomy J., Heussler G.E., Davidson A.R., O'Toole G.A. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 2012;194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer S., Maier L.K., Stoll B., Brendel J., Fischer E., Pfeiffer F., Dyall-Smith M., Marchfelder A. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J. Biol. Chem. 2012;287:33351–33363. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gudbergsdottir S., Deng L., Chen Z., Jensen J.V., Jensen L.R., She Q., Garrett R.A. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manica A., Zebec Z., Teichmann D., Schleper C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol. Microbiol. 2011;80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 38.Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westra E.R., Semenova E., Datsenko K.A., Jackson R.N., Wiedenheft B., Severinov K., Brouns S.J. Type I-E CRISPR–Cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westra E.R., Swarts D.C., Staals R.H., Jore M.M., Brouns S.J., van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 41.Carte J., Christopher R.T., Smith J.T., Olson S., Barrangou R., Moineau S., Glover C.V., 3rd, Graveley B.R., Terns R.M., Terns M.P. The three major types of CRISPR–Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol. Microbiol. 2014;93:98–112. doi: 10.1111/mmi.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei Y., Terns R.M., Terns M.P. Cas9 function and host genome sampling in Type II-A CRISPR–Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terns R.M., Terns M.P. The RNA- and DNA-targeting CRISPR–Cas immune systems of Pyrococcus furiosus. Biochem. Soc. Trans. 2013;41:1416–1421. doi: 10.1042/BST20130056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majumdar S., Zhao P., Pfister N.T., Compton M., Olson S., Glover C.V., 3rd, Wells L., Graveley B.R., Terns R.M., Terns M.P. Three CRISPR–Cas immune effector complexes coexist in Pyrococcus furiosus. RNA. 2015 doi: 10.1261/rna.049130.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carte J., Wang R., Li H., Terns R.M., Terns M.P. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carte J., Pfister N.T., Compton M.M., Terns R.M., Terns M.P. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hale C., Kleppe K., Terns R.M., Terns M.P. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramia N.F., Spilman M., Tang L., Shao Y., Elmore J., Hale C., Cocozaki A., Bhattacharya N., Terns R.M., Terns M.P., et al. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR–Cas complex. Cell Rep. 2014;9:1610–1617. doi: 10.1016/j.celrep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spilman M., Cocozaki A., Hale C., Shao Y., Ramia N., Terns R., Terns M., Li H., Stagg S. Structure of an RNA silencing complex of the CRISPR–Cas immune system. Mol. Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hale C.R., Cocozaki A., Li H., Terns R.M., Terns M.P. Target RNA capture and cleavage by the Cmr type III-B CRISPR–Cas effector complex. Genes Dev. 2014;28:2432–2443. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lipscomb G.L., Stirrett K., Schut G.J., Yang F., Jenney F.E., Jr, Scott R.A., Adams M.W., Westpheling J. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 2011;77:2232–2238. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farkas J., Chung D., DeBarry M., Adams M.W., Westpheling J. Defining components of the chromosomal origin of replication of the hyperthermophilic archaeon Pyrococcus furiosus needed for construction of a stable replicating shuttle vector. Appl. Environ. Microbiol. 2011;77:6343–6349. doi: 10.1128/AEM.05057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farkas J., Stirrett K., Lipscomb G.L., Nixon W., Scott R.A., Adams M.W., Westpheling J. Recombinogenic properties of Pyrococcus furiosus strain COM1 enable rapid selection of targeted mutants. Appl. Environ. Microbiol. 2012;78:4669–4676. doi: 10.1128/AEM.00936-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han D., Krauss G. Characterization of the endonuclease SSO2001 from Sulfolobus solfataricus P2. FEBS Lett. 2009;583:771–776. doi: 10.1016/j.febslet.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 56.Beloglazova N., Petit P., Flick R., Brown G., Savchenko A., Yakunin A.F. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–4627. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Howard J.A., Delmas S., Ivancic-Bace I., Bolt E.L. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem. J. 2011;439:85–95. doi: 10.1042/BJ20110901. [DOI] [PubMed] [Google Scholar]
- 58.Mulepati S., Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J. Biol. Chem. 2011;286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinkunas T., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huo Y., Nam K.H., Ding F., Lee H., Wu L., Xiao Y., Farchione M.D., Jr, Zhou S., Rajashankar K., Kurinov I., et al. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat. Struct. Mol. Biol. 2014;21:771–777. doi: 10.1038/nsmb.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haft D.H., Selengut J., Mongodin E.F., Nelson K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson R.N., Lavin M., Carter J., Wiedenheft B. Fitting CRISPR-associated Cas3 into the Helicase Family Tree. Curr. Opin. Struct. Biol. 2014;24:106–114. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menon A.L., Poole F.L., 2nd, Cvetkovic A., Trauger S.A., Kalisiak E., Scott J.W., Shanmukh S., Praissman J., Jenney F.E., Jr, Wikoff W.R., et al. Novel multiprotein complexes identified in the hyperthermophilic archaeon Pyrococcus furiosus by non-denaturing fractionation of the native proteome. Mol. Cell. Proteomics : MCP. 2009;8:735–751. doi: 10.1074/mcp.M800246-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plagens A., Tjaden B., Hagemann A., Randau L., Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 67.Haurwitz R.E., Jinek M., Wiedenheft B., Zhou K., Doudna J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hatoum-Aslan A., Maniv I., Marraffini L.A. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc. Natl. Acad. Sci. U.S.A. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiedenheft B., van Duijn E., Bultema J.B., Waghmare S.P., Zhou K., Barendregt A., Westphal W., Heck A.J., Boekema E.J., Dickman M.J., et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jackson R.N., Golden S.M., van Erp P.B., Carter J., Westra E.R., Brouns S.J., van der Oost J., Terwilliger T.C., Read R.J., Wiedenheft B. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mulepati S., Heroux A., Bailey S. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao H., Sheng G., Wang J., Wang M., Bunkoczi G., Gong W., Wei Z., Wang Y. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014 doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- 73.Kunin V., Sorek R., Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yosef I., Goren M.G., Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J., Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams E., Lowe T.M., Savas J., DiRuggiero J. Microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus exposed to gamma irradiation. Extremophiles. 2007;11:19–29. doi: 10.1007/s00792-006-0002-9. [DOI] [PubMed] [Google Scholar]
- 77.Strand K.R., Sun C., Li T., Jenney F.E., Jr, Schut G.J., Adams M.W. Oxidative stress protection and the repair response to hydrogen peroxide in the hyperthermophilic archaeon Pyrococcus furiosus and in related species. Arch. Microbiol. 2010;192:447–459. doi: 10.1007/s00203-010-0570-z. [DOI] [PubMed] [Google Scholar]
- 78.Pawluk A., Bondy-Denomy J., Cheung V.H., Maxwell K.L., Davidson A.R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR–Cas system of Pseudomonas aeruginosa. mBio. 2014;5:e00896. doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bondy-Denomy J., Pawluk A., Maxwell K.L., Davidson A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.