Abstract
Thermodynamic studies of ligand binding to human telomere (ht) DNA quadruplexes, as a rule, neglect the involvement of various ht-DNA conformations in the binding process. Therefore, the thermodynamic driving forces and the mechanisms of ht-DNA G-quadruplex-ligand recognition remain poorly understood. In this work we characterize thermodynamically and structurally binding of netropsin (Net), dibenzotetraaza[14]annulene derivatives (DP77, DP78), cationic porphyrin (TMPyP4) and two bisquinolinium ligands (Phen-DC3, 360A-Br) to the ht-DNA fragment (Tel22) AGGG(TTAGGG)3 using isothermal titration calorimetry, CD and fluorescence spectroscopy, gel electrophoresis and molecular modeling. By global thermodynamic analysis of experimental data we show that the driving forces characterized by contributions of specific interactions, changes in solvation and conformation differ significantly for binding of ligands with low quadruplex selectivity over duplexes (Net, DP77, DP78, TMPyP4; KTel22 ≈ <KdsDNA) and for highly selective quadruplex-specific ligands (Phen-DC3, 360A-Br; KTel22 > KdsDNA). These contributions are in accordance with the observed structural features (changes) and suggest that upon binding Net, DP77, DP78 and TMPyP4 select hybrid-1 and/or hybrid-2 conformation while Phen-DC3 and 360A-Br induce the transition of hybrid-1 and hybrid-2 to the structure with characteristics of antiparallel or hybrid-3 type conformation.
INTRODUCTION
G-quadruplexes have attracted significant attention due to their growing biological importance, mainly as possible regulators of transcription of cancer cells DNAs in gene promoter regions of oncogenes and telomeric regions (1–3). Formation of stable G-quadruplexes has been found to modulate their biological functions, therefore, the development of new anticancer G-quadruplex stability ligands seems to be a novel anticancer strategy (4,5). In this light, the extreme conformational diversity of G-quadruplexes requires the development of highly specific ligands capable to discriminate between the diverse G-quadruplex topologies (6–8) and not only between the G-quadruplexes and duplexes (9).
One typical example of such conformational diversity (10) of G-quadruplexes are various G-quadruplex structures formed from the human telomeric DNA sequence. For example, X-ray crystallography shows that the sequence 5′-AGGG(TTAGGG)3–3′ (Tel22) in the presence of K+ ions adopts the parallel structure (11) while NMR and some biophysical techniques indicate that Tel22 in K+ solutions at room temperature exists as a mixture of two energetically similar (3+1) hybrid-type G-quadruplex structures known as hybrid-1 and hybrid-2 conformations (12–14). On the other hand, it has been shown by NMR that in solutions with Na+ ions Tel22 adopts antiparallel structure (15). Furthermore, it has been shown recently that human telomeric DNA folding pathways involve multiple intermediate states (16–22).
Based on the general structure and stability of G-quadruplexes, (23) most of the ligands have planar aromatic part enabling π–π stacking interactions with G-quartets and positive charge which enables favorable electrostatic interactions with the negatively charged DNA backbone (24). Publications dealing with ligand binding to G-quadruplexes and structures of the formed complexes (RHPS4, (7) TMPyP4 (25–30), telomestatin, (31) Phen-DC3, (32) berberine, (33,34) Braco-19, (35) actinomycin D, (36) HXDV, (37) naphthalene diimide, (38) N-methyl mesoporphyrin IX, (39,40) distamycin (41) etc.) suggest that most of the ligands bind to the end quartets of G-quadruplexes with the binding constant ranging between 104 and 108 M−1.
Despite the long-standing interest in understanding the molecular recognition of human telomeric DNA by small ligands only a few studies on thermodynamics of their binding to human telomeric DNA (ht-DNA) quadruplexes have been reported (25,33,36,37,41). In spite of the fact that ht-DNA may populate various conformational states at physiological conditions (different folded and folding intermediate states) these binding studies, as a rule, neglect their involvement in the binding process. Thus, the aim of the present work is to investigate the mechanism of binding of several ligands to Tel22 through the nature of the driving forces that control these binding events and the involvement of Tel22 folding intermediates in the binding process. In our study of molecular recognition of Tel22 we compare thermodynamic and structural characteristics of binding of six different ligands (Figure 1): netropsin (Net), two dibenzotetraaza[14]annulene derivatives (DP77, DP78), cationic porphyrin (TMPyP4) and two bisquinolinium ligands (Phen-DC3, 360A-Br). Net is a typical dsDNA minor groove binder with binding constant around 108 M−1 for AATT binding sites (42–44). DP77 and DP78 are characterized as intercalators with a pronounced preference for A-T(U) sequences (binding constant higher than 106 M−1) and show moderate to high antiproliferative activity against five human tumor cell lines (45). DP77 and DP78 molecules possess large conjugated cyclic aromatic part that may, potentially, strongly interact with DNA G-quartets. TMPyP4 is one of the most studied quadruplex ligands that can induce formation of quadruplex from single-stranded DNA but does not show significant selectivity for quadruplexes over dsDNA (46). For Phen-DC3 and 360A-Br it has been shown that they both exhibit high affinity for G-quartets very likely as a result of strong π–π stacking interactions as show by the NMR structure of G-quadruplex-Phen-DC3 complex (32,47). In addition, upon binding to Tel22 in solutions with K+ ions these two ligands remove, as suggested by Marchand et al. (48) and Bončina et al., (49) one K+ ion and induce conformational switching of Tel22 quadruplex to a conformation that according to CD spectra exhibits characteristics of an antiparallel type structure. For these and for many other quadruplex targeting ligands the question on the mechanism by which ligand binding is coupled to conformational changes remains unanswered. In general, two limiting mechanisms can be considered: (i) ‘conformational selection’, in which the ligand selectively binds to a quadruplex conformation that is present in solution in small amounts eventually converting the quadruplex to the ligand-bound conformation; and (ii) ‘induced fit’, in which the ligand binds to the predominant free quadruplex conformation followed by a conformational change to give the preferred ligand-bound quadruplex conformation. Though these mechanisms can be distinguished only by kinetic measurement of rate of the conformational change the detailed thermodynamic and structural analysis may give clues whether the observed ligand–quadruplex recognition is more likely an ‘induced fit’ or a ‘conformational selection’ process.
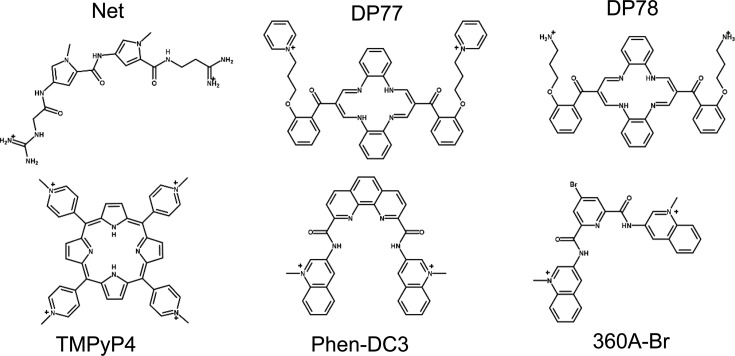
Figure 1.
Structural formulas of ligands used in this study: netropsin (Net), dibenzotetraaza[14]annulene derivatives (DP77, DP78), TMPyP4 and bisquinolinium based ligands (Phen-DC3, 360A-Br).
In this light a thorough investigation of Net, DP77, DP78, TMPyP4, Phen-DC3 and 360A-Br binding to Tel22 at various temperatures and K+ concentrations was performed using isothermal titration calorimetry (ITC), circular dichroism (CD), fluorescence spectroscopy and gel electrophoresis. Here, we present experimental results for Net, DP77, DP78 and TMPyP4 while experimental results for Phen-DC3, 360A-Br have already been presented in our previous study (49). Dissection of thermodynamic parameters, obtained from the global analysis of experimental data enabled us to estimate the dominant forces that drive binding of the mentioned ligands to Tel22. We believe that these estimates combined with CD spectroscopy and gel electrophoresis data and the structural modeling on the ligand-Tel22 complexes provide a deeper insight into the mechanisms of recognition of human telomeric G-quadruplexes by various aromatic ligands.
MATERIALS AND METHODS
Sample preparation
HPLC pure DNA oligonucleotide 5′-AGGG(TTAGGG)3–3′ (Tel22) was obtained from Midland Co., U.S.A. The buffer solutions used in our experiments consisted of 20 mM cacodylic acid, 1 mM EDTA and various concentrations of K+ ions. KOH was added to cacodylic acid to reach pH 6.9. Then, KCl was added to obtain the desired concentration of K+ ions (25 or 200 mM K+ in cacodylic buffer). DNA was first dissolved in water and then extensively dialyzed against the buffer using a dialysis tube Float-A-Lyser (Spectrum Laboratories, USA, Mw cutoff 500–1000 Da). The starting oligonucleotide solution was first heated up to 95 °C in an outer thermostat for 5 min to make sure that all DNA transforms into the unfolded form and then cooled down to 5 °C at the cooling rate of 0.05 °C min−1 to allow DNA to adopt G-quadruplex structure(s). Concentration of Tel22 in the buffer solution was determined at 25 °C spectrophotometrically using Cary 100 BIO UV/Visible Spectrophotometer (Varian Inc.) equipped with a thermoelectric temperature controller. Accurate concentrations at 25 °C were obtained from the melting curves monitored at 260 nm. Absorbance of the unfolded form (high temperatures) was extrapolated to 25 °C. For the extinction coefficient of the unfolded form at 25 °C we used the value ϵ260 = 228 500 M−1cm−1 estimated from the nearest-neighbor data of Cantor et al. (50).
Ligands netropsin (Net) (purchased from Sigma-Aldrich), dibenzotetraaza[14]annulene derivatives DP77, DP78 (synthesized at Ruđer Bošković Institute, Croatia and Jagiellonian University, Poland) and TMPyP4 (purchased from Sigma-Aldrich) were first dissolved in the same buffer as DNA. Their concentrations were determined spectrophotometrically at 25 °C using extinction coefficients 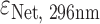 = 21 500 M−1 cm−1,
= 21 500 M−1 cm−1, 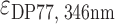 = 47 730 M−1 cm−1 and
= 47 730 M−1 cm−1 and 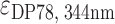 = 47 610 M−1 cm−1,
= 47 610 M−1 cm−1, 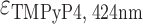 = 226 000 M−1 cm−1.
= 226 000 M−1 cm−1.
CD spectroscopy
CD spectroscopic titrations were conducted at 25 °C by titrating ligand solution (cL ≈ 300 μM) into a DNA solution (VDNA = 600 μl, cDNA ≈ 10 μM) in the case of Net, DP77 and DP78 while in the case of TMPyP4 DNA solution (cDNA ≈ 70 μM) was titrated into a ligand solution (VL = 600 μl, cL ≈ 10 μM) to avoid aggregation of TMPyP4-quadruplex complexes. Ellipticity, Θ, was measured in the wavelength range between 240 and 350 nm in a 1.0 cm cuvette with signal averaging time of 3 s and 5 nm bandwidth. Experiments were performed using AVIV CD Spectrophotometer 62A DS (Aviv Biomedical, Lakewood, NJ, USA) equipped with a thermoelectric temperature controller.
Fluorimetry
Fluorescence titrations were conducted at 25 °C by titrating DNA solution (cDNA ≈ 30 μM) into a 1000 μl TMPyP4 ligand solution (cL ≈ 2.5 μM). Emission spectra were recorded between 600 and 750 nm (excitation wavelength λex = 445 nm, ex. slit 5 nm, em. slit 10 nm, absorbance < 0.1) in a 1.0 cm cuvette with scanning speed of 50 nm min−1. Experiments were performed using Perkin Elmer LS 55 luminescence spectrometer (Perkin Elmer, Waltham, MA, USA) equipped with a thermally controlled cell holder.
Isothermal titration calorimetry (ITC)
In the case of Net, DP77 and DP78 ITC experiments were performed at 10 and 25 °C by titrating a solution of ligand (cL ≈ 700 μM) into a DNA solution (cDNA ≈ 20 μM, V = 1.386 ml) while in the case of TMPyP4 at 25, 30 and 35 °C by titrating a solution of DNA (cDNA ≈ 70 μM) into a ligand solution (cL ≈ 10 μM) (reverse titration was used to avoid aggregation of TMPyP4-quadruplex complexes detected by absorbance jump (Supplementary Figure S2B) when TMPyP4 solution was titrated into DNA solution at r = [L]t / [DNA]t > 4) using a VP-ITC isothermal titration calorimeter from Microcal Inc. (Northampton, MA, USA). The area under the peak following each injection of titrant solution was obtained by integration of the raw signal, corrected for the corresponding heat of dilution of the added titrant (blank titration) and expressed per mole of added titrant per injection, to give the enthalpy of interaction (ΔHT). The experimental ΔHT data were modeled as described in SI.
PAGE electrophoresis
DNA dissolved in the buffer solution containing 200 mM K+ ions was mixed with an appropriate amount of ligand in the same buffer solution to achieve the desired ligand/DNA molar ratio. Ten microliters of these samples were mixed with 3 μl of 40% (w/v) sucrose solution and then 10 μl of such modified sample solutions were loaded onto a 22 (w/v) polyacrylamide gel containing 200 mM K+ ions and subjected to a constant voltage of 110 V for 3 h. The running TBE buffer, pH = 8.2, contained 0.09 M Tris, 0.09 M boric acid and 1 mM EDTA. Electrophoresis cell was placed in a water bath at 25 °C. After the electrophoresis, the gels were photographed under short and long UV light using G-box (Syngene, Cambridge, UK) or directly with a camera detecting the color of a ligand (TMPyP4).
Molecular modeling
Yasara Structure (51) has been used for molecular docking of a ligand to various Tel22 conformations and the subsequent refinement of the resulting complexes. Docking calculations were performed on four human quadruplex conformations: the hybrid-1 (PDB: 2HY9 and 2JSM), hybrid-2 (PDB: 2JPZ and 2JSK), hybrid-3 (PDB: 2KF) and the antiparallel (PDB: 143D). Non-Tel22 sequences were modified to Tel22 and structurally optimized. Analysis of Phen-DC3 and 360A-Br binding by CD spectroscopy suggests that their binding induces conformational change of the hybrid-1 and/or hybrid-2 Tel22 conformation into the structure with characteristics of the antiparallel and/or hybrid-3 conformation (Figure 5). Therefore, Phen-DC3 and 360A-Br docking was performed with the Tel22 antiparallel and hybrid-3 conformation. In the first step, coronene was used as a ‘pore making ligand’ and placed on the top of guanine quartet (dG4, dG8, dG16, dG20) of the antiparallel and (dG3, dG8, dG16, dG20) of the hybrid-3 conformation. Restrained minimization using AMBER03 force field (52) has been run for the initial complex. In this first minimization all three quartets were fixed. The resulting coronene–quadruplex complex was used as an entry for Phen-DC3 and 360A-Br docking experiment. In the second step, Autodock–Vina (53) (integrated in YASARA) docking experiment has been performed to obtain the structural model of the complex between the ligand (Phen-DC3 or 360A-Br) and the antiparallel or hybrid-3 Tel22 quadruplex. In addition, docking of Phen-DC3 and 360A-Br with hybrid-1 and hybrid-2 conformation was performed, for comparative purposes. We run 50 independent docking experiments. The pose with the best score was minimized in explicit water (TIP3P) (54) using AMBER03 force field.
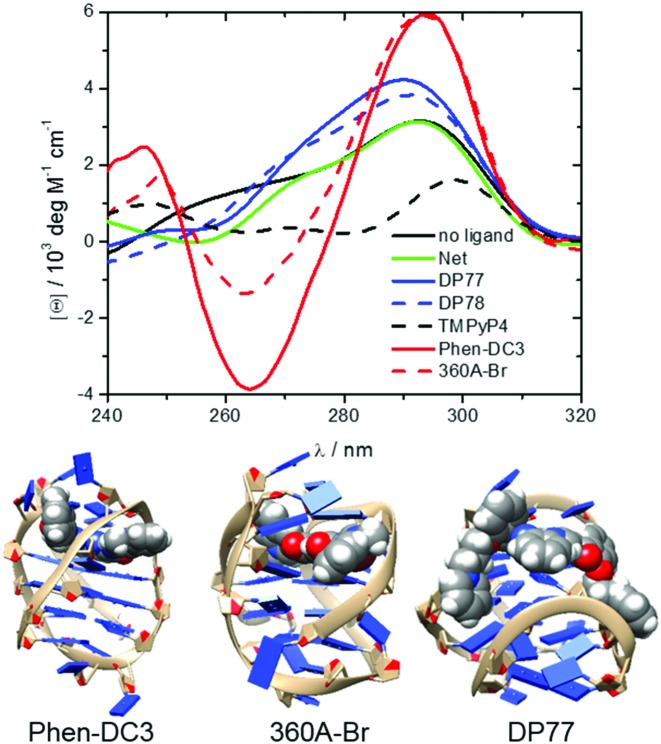
Figure 5.
CD spectra of fully bound G-quadruplexes at 25 °C and in the presence of 200 mM K+ ions (top). The measured CD signal, [θ], can be presented as a linear combination (sum of the contributions: [θ] = ∑i[θ]iαi) of characteristic signals of each species i, [θ]i, weighted by its fraction in the solution, αi = [ci] / [DNA]t, (i = ligand free G-quadruplex or any ligand-Tel22 complex). Therefore, CD spectra ([θ]i versus λ) of the ligand-Tel22 complexes were calculated by deconvolution of the measured spectra based on the model-predicted αi values and the corresponding spectrum of the ligand-free G-quadruplex. Structural models of some of the studied ligand-Tel22 complexes: Phen-DC3-antiparallel, 360A-Br-hybrid-3 and DP77-hybrid-1 (bottom).
On the other hand, analysis of DP77 and DP78 binding to Tel22 by CD spectroscopy suggests that DP77 and DP78 form complexes with hybrid-1 and/or hybrid-2 (Figure 5). Thus, structural models of the complexes between the DP77 or DP78 and hybrid-1 and hybrid-2 quadruplexes were obtained through the Autodock–Vina docking experiment.
Structure based thermodynamic calculations
Changes of solvent accessible polar, ΔAP, and non-polar, ΔAN, surface area (SASA) accompanying the formation of ligand–quadruplex complexes were calculated as the differences between SASA of the complex and the summed SASA of the ligand-free Tel22 and the free (unbound) ligand. SASA of the ligand-free Tel22 was calculated as the average of two hybrid-1 and two hybrid-2 SASAs. SASAs were calculated using the NACCESS (55) program. In these calculations we used a probe radius of 1.4 Å and the program default sets of atomic radii and polarity. The corresponding ΔC°P values were calculated using a relation ΔC°P = aΔAN + bΔAP where a = (0.45 ± 0.02) cal mol−1 K−1 Å−2 and b = (−0.26 ± 0.02) cal mol−1 K−1 Å−2, (56) which appears to be appropriate for describing ΔC°P of ligand–DNA association (42,34).
RESULTS AND DISCUSSION
Model analysis of ligand binding
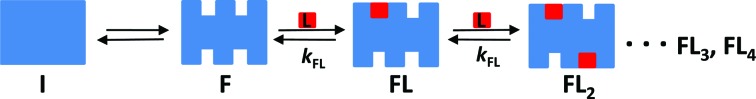
Binding isotherms characterizing the sequential binding of Phen-DC3 and 360A-Br to Tel22 were analyzed elsewhere (49). Net, DP77, DP78 and TMPyP4 binding characteristics were obtained from binding experiments performed at conditions at which at the beginning of the experiment only the folded G-quadruplex, F, and the folding intermediate, I, can be present in the solution (16,49). In this light the proposed model mechanism of Net, DP77, DP78 and TMPyP4 binding (Figure 2) assumes equilibrium between I, F and various ligand-G-quadruplex complexes, FLi (i = 1, 2,…N).
Figure 2.
Schematic presentation of ligand binding to human telomeric DNA fragment Tel22. The model mechanism describes Net, DP77, DP78 and TMPyP4 binding at various salt concentrations and temperatures where no unfolded Tel22 is present. It assumes the conversion between the folding intermediate, I, that contains no binding sites, and the folded intramolecular G-quadruplex structure, F, which contains a set of independent equivalent binding sites, N, characterized with the binding constant kFL.
The model mechanism is translated into the model functions that describe the experimental data obtained by spectroscopic (CD, fluorescence) and calorimetric (ITC) titrations (see Supplementary Material). Global fitting of these model functions to the experimental spectroscopic (CD, fluorescence) and ITC data measured at different salt concentrations and temperatures was based on the non-linear Levenberg–Marquardt χ2 regression procedure. Only the standard thermodynamic parameters of ligand binding to the G-quadruplex binding site (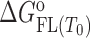 = Gibbs free energy and
= Gibbs free energy and 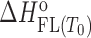 = enthalpy at the reference temperature T0 and
= enthalpy at the reference temperature T0 and 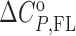 = heat capacity, N = no. of binding sites, nFL = no. of ions released or uptaken, all three assumed to be temperature independent) were adjusted in the global fitting procedure (Supplementary Table S1) while the parameters describing the I → F conversion were obtained from the recently presented global model analysis of folding/unfolding process of ligand-free Tel22 in the presence of K+ ions, (16,49) and were used in the fitting procedure as fixed values. In case that due to the aggregation of the ligand–quadruplex complexes the binding of a given ligand can be studied only in a narrow salt concentration range nFL cannot be determined (eqn. S10) and the described fitting procedure can lead only to apparent binding parameters specific for the salt concentration at which the binding process can be investigated (Supplementary Table S1-DP77, DP78, TMPyP4).
= heat capacity, N = no. of binding sites, nFL = no. of ions released or uptaken, all three assumed to be temperature independent) were adjusted in the global fitting procedure (Supplementary Table S1) while the parameters describing the I → F conversion were obtained from the recently presented global model analysis of folding/unfolding process of ligand-free Tel22 in the presence of K+ ions, (16,49) and were used in the fitting procedure as fixed values. In case that due to the aggregation of the ligand–quadruplex complexes the binding of a given ligand can be studied only in a narrow salt concentration range nFL cannot be determined (eqn. S10) and the described fitting procedure can lead only to apparent binding parameters specific for the salt concentration at which the binding process can be investigated (Supplementary Table S1-DP77, DP78, TMPyP4).
Figure 3 shows that all the calorimetric (ITC) and spectroscopic (CD) data measured for a given ligand-Tel22 system are successfully described by the model functions translated from the mechanism presented in Figure 2. It should be mentioned that in the absence of ligand at physiological temperatures and ion concentrations the folded Tel22 forms are dominant (Figure 3B,3D), (16,49) the population of intermediates is small but significant and increases up to about 40% at 25 mM concentration of K+ ions (Figure 3D) (16,17,49). Though our model analysis is based on rather simplistic mechanism (Figure 2) that does not distinguish between various folding intermediates (17) and folded forms (hybrid-1, hybrid-2) present in the solution at given conditions it shows that the description of ligand binding to Tel22 without considering intermediate(s) (neglection of the I → F step) leads to unsatisfactory agreement of the binding model with experimental data (Supplementary Figure S3). In addition, an assumption that also the folding intermediate contains a set of independent binding sites does not lead to better description of experimental data by the model. Moreover, so modified model has to be discarded since it involves a large number of highly correlated adjustable parameters whose values (especially ΔH° and ΔC°P) cannot be determined with sufficient accuracy. In other words, the proposed model (Figure 2) is the simplest model that gives a satisfactory agreement between experimental data and the corresponding model functions (eqs S3, S4 and S7) and is characterized by sufficiently reliable and accurate thermodynamic parameters.
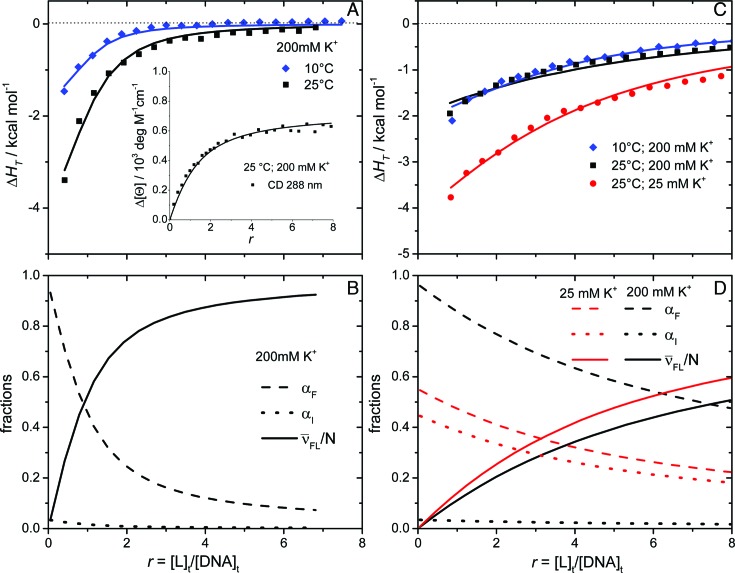
Figure 3.
Model analysis of calorimetric (ITC) and spectroscopic (CD) data describing the binding of DP78 (A) and Net (C) to Tel22. The best-fit model functions (eqs S3 and S7; presented by lines) show reasonably good agreement with ITC and CD spectroscopy (inset) experimental data (points). The corresponding fractions of species F, I and FL (C, D) at 25 °C and different concentrations of K+ ions as a function of ligand/Tel22 molar ratio r were calculated using the ‘best fit’ parameters reported in Supplementary Table S1. 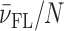 represents the average fraction of occupied binding sites on the G-quadruplex molecule. Results obtained for DP77 and TMPyP4 binding to Tel22 are presented in Supplementary Figures S4 and S5, respectively.
represents the average fraction of occupied binding sites on the G-quadruplex molecule. Results obtained for DP77 and TMPyP4 binding to Tel22 are presented in Supplementary Figures S4 and S5, respectively.
Additionally, the appropriateness of the proposed binding model (Figure 2) for the description of ligand binding to Tel22 was checked by reverse titrations (titration of Tel22 solution into the ligand solution). We observed that in the case of Net, DP77 and DP78 the Tel22-to-ligand titration data may be successfully described by the same values of thermodynamic parameters than the corresponding ligand-to-Tel22 titration data (Supplementary Figure S1). Based on this observation we believe that Tel22-to-ligand titrations give reliable information on the binding thermodynamics also in cases (TMPyP4, Phen-DC3 and 360A-Br) in which aggregation of the ligand-Tel22 complexes prevents reliable thermodynamic analysis of the observed ITC curves (Supplementary Figure S2A). In this light the thermodynamic analysis of TMPyP4 binding to Tel22, as well as of the recently studied Phen-DC3 and 360A-Br binding to Tel22, (49) was based on Tel22-to-ligand titration data.
Thermodynamics of binding
Thermodynamic analysis reveals that the studied ligands differ significantly in their binding affinity for Tel22 G-quadruplexes and selectivity for Tel22 G-quadruplexes over DNA duplexes. The binding constants follow the order Net < DP77 < DP78 < TMPyP4 < 360A-Br < Phen-DC3 and range from 104 M−1 to 107 M−1 (Figure 4, Supplementary Table S1). The studied ligands differ also in the maximum no. of binding sites they can occupy on Tel22. Thus, Tel22 can accommodate one (DP77, DP78), two (360A-Br, Phen-DC3; sequential binding) (49), three (Net) and four (TMPyP4) ligand molecules. The significant difference in binding thermodynamics between DP77 and DP78 indicates that in the binding process besides the conjugated cyclic aromatic part also the side chains play an important role. Since the binding constants for Net, DP77, DP78 and TMPyP4 association with Tel22 are lower or close to those reported for their binding to dsDNA (45,46,57), they may be considered as ligands with low selectivity for ht-G-quadruplex over duplexes. By contrast, the quadruplex specific ligands Phen-DC3 and 360A-Br bind to Tel22 with significantly higher affinity (k > 106 M−1) than to dsDNA (58). To verify this information we checked the binding selectivity for the 360A-Br ligand and found that the corresponding kquadruplex / kdsDNA value is higher than 50 (see Supplementary Table S4).
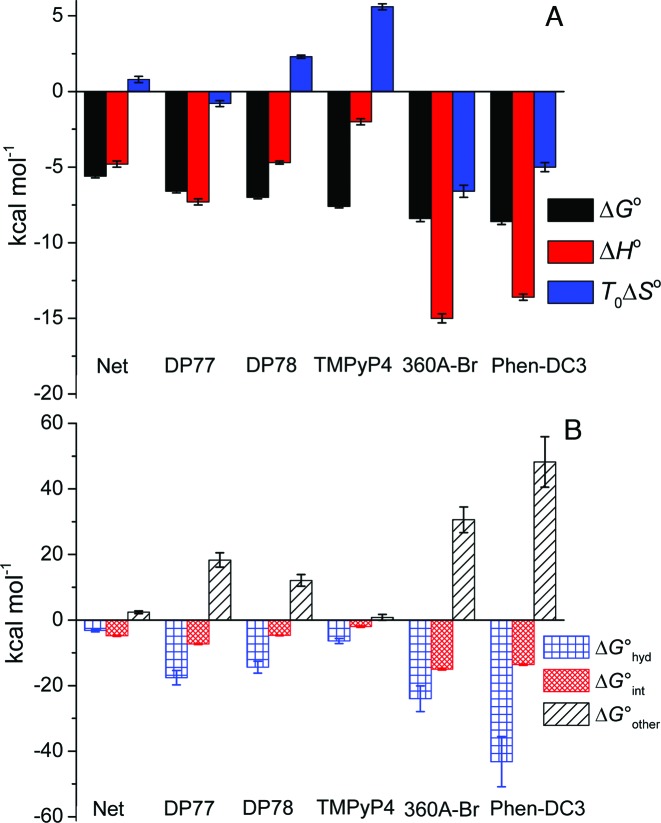
Figure 4.
Thermodynamic profiles (A) and Gibbs free energy contributions (driving forces) (B) of ligand binding to Tel22 at 200 mM concentration of K+ ions and 25 °C. Thermodynamic binding profiles of Phen-DC3 and 360A-Br are presented only for their binding to the high affinity Tel22 binding site (49). Error bars in ΔG° and ΔH° represent 2x standard deviations obtained as square roots of diagonal elements of variance-covariance matrixes obtained from global model analysis of experimental data. Errors in TΔS° were calculated from the errors of ΔG° and ΔH°. The errors in ΔG° contributions were calculated combining the errors of experimental quantities (ΔG°, ΔH°, ΔC°P; global model analysis) and errors reported in the literature (ΔG°hyd, ΔG°rt) (60–62).
Next, we would like to show how the thermodynamic driving forces reflect the observed differences in binding affinity and binding specificity for a certain quadruplex conformation. Since the binding of Phen-DC3 and 360A-Br to Tel22 is sequential (49) the corresponding thermodynamic properties are compared with those of non-selective ligands (equivalent independent binding sites) only for binding of the (first) Phen-DC3 or 360A-Br molecule that binds to Tel22 quadruplexes with the highest affinity. The observed Gibbs free energy change, ΔG°, was dissected into the enthalpy, ΔH°, and entropy, TΔS°, contribution (ΔG° = ΔH° − TΔS°; Figure 4A). In addition, we attempted to analyze ΔG° in terms of various more fundamental driving forces using the additivity approach (23,42,43,59) in which ΔG° is considered as a sum of three main contributions: ΔG° = ΔG°hyd + ΔG°int + ΔG°other (Figure 4B). The ΔG°hyd contribution refers to the solvation/desolvation effects. At 25 °C, it is considered to reflect mainly the entropy of dehydration of hydrophobic groups upon ligand binding and may be estimated as ΔG°hyd = 80(±10)K·ΔC°P (60,61). The ΔG°int contribution is ascribed to the specific inter- and intra-molecular interactions (π–π stacking, H-bonds, electrostatic). It can be considered mainly as an enthalpic contribution (49) and therefore approximated by the measured enthalpy change ΔG°int ≈ ΔH°. The ΔG°other term is considered as a sum of two main contributions, i.e. the negative entropic contribution, ΔG°rt, due to the changes of rotational and translational degrees of freedom lost upon ligand binding and the entropic contribution, ΔG°conf, that may be ascribed predominantly to the bound ligand induced conformational changes of Tel22. Since ΔG°other = ΔG°rt + ΔG°conf and ΔG°other = ΔG° − ΔG°hyd − ΔG°int one may estimate ΔG°conf as: ΔG°conf = ΔG° − ΔG°hyd − G°int − ΔG°rt.
Dissection of energetics reveals some general features of Net, DP77, DP78, TMPyP4, 360A-Br and Phen-DC3 binding to Tel22 (Figure 4). The binding of all the ligands is accompanied with a negative enthalpy and heat capacity change (Figure 4A, Supplementary Table S1) suggesting that the specific interactions occurring at the ligand–Tel22 binding interface and the removal of water from this (predominantly hydrophobic) interface play a major role in the recognition of ht-DNA by small aromatic ligands. This is consistent with the observation (Figure 4B) that the corresponding ΔG°hyd and ΔG°int contributions overcompensate the unfavorable rotational-translational and conformational contributions; (ΔG°hyd + ΔG°int) < −(ΔG°rt + ΔG°conf). The dissection further suggests that the dominant forces that drive ligand binding to Tel22 are not the specific intra- and inter-molecular interactions but removal of water from the interacting surfaces (33) (ΔG°hyd < ΔG°int), except in the case of Net where these two driving forces appear to be of about equal importance (ΔG°int ≈ ΔG°hyd). It also shows that for the G-quadruplex specific ligands (360A-Br, Phen-DC3) specific short-range interactions are essential for successful binding (if ΔG°int = 0 ⇒ ΔG°hyd + ΔG°int > −ΔG°other ⇒ ΔG° > 0; binding not favorable) while for the ligands with low selectivity for Tel22 quadruplexes (Net, DP77, DP78, TMPyP4) this is not the case (if ΔG°int = 0 ⇒ ΔG°hyd + ΔG°int < −ΔG°other ⇒ ΔG° < 0; binding favorable). Finally, the estimation of the thermodynamic signature for the binding-induced conformational change, ΔG°conf, based on the above mentioned dissection is also possible, however, it requires reasonable estimation of ΔG°rt. According to the extensive debate in the literature over the acceptable approximation of ΔG°rt contribution its value depends on the strength and the nature of ligand binding and may vary from several kcal mol−1 up to 15 kcal mol−1 (62,63). Combining ΔG°rt within these estimates with the corresponding ΔG°other leads for Net, DP77, DP78, TMPyP4 to small ΔG°conf values that may result also from the loss of conformational freedom of the ligand. This suggests that the binding of the ligands with low selectivity for Tel22 quadruplexes is coupled with relatively small Tel22 quadruplex conformational changes. The result is significantly different from ΔG°conf > 20 kcal mol−1 (estimated from ΔG°other (Figure 4B) and ΔG°rt (0 < ΔG°rt / kcal mol−1 < 15)) which is indicative for large binding-induced conformational changes of Tel22 by G-quadruplex specific ligands 360A-Br and Phen-DC3.
At this point we would like to mention that in our thermodynamic analysis of experimental data the parameter, nFL, that accounts for a number of ions released or uptaken upon binding (eqn. S10) does not distinguish between the binding-coupled release of the specifically and non-specifically bound K+ ions. Thus the value of nFL between 1.5 and 1.2 observed for Phen-DC3 and 360A-Br binding to Tel22, respectively (Supplementary Table S1), may be due to a release of one interquartet K+ ion, as suggested by Marchand et al. (48) and/or due to a release of non-specifically bound K+ ions. According to the polyelectrolyte theory (64) the corresponding electrostatic contribution to the standard free energy of binding is relatively small (its absolute value is smaller than the error of the least accurate contribution in eqn. ΔG° = ΔG°hyd + ΔG°int + ΔG°other) and was neglected in our dissection of energetic contributions. Moreover, due to the extensive burial of Phen-DC3 and 360A-Br within the quadruplex structure ((48) and our section Thermodynamics, Structural Features and Mechanism of Recognition) we assumed that the unfavorable contribution of desolvation of the ligand ionic groups is compensated by the favorable contribution of K+ ion solvation.
Thermodynamics, structural features and mechanism of recognition
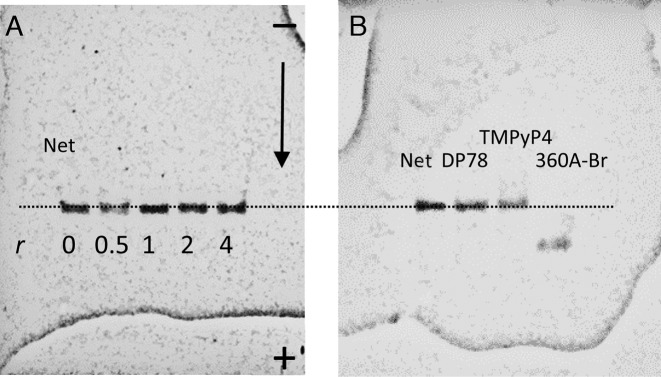
Gel electrophoresis experiments (Figure 6) suggest that gel-mobility of Tel22 complexed with the studied quadruplex non-selective ligands is the same as that of the ligand-free Tel22. Since the non-selective ligands, Net, DP77, DP78 and TMPyP4 are positively charged and bind relatively weakly to Tel22 (∼104 to ∼105 M−1) they may dissociate from Tel22 during the electrophoresis experiment and this could explain the same mobility of ligand-free Tel22 and Tel22 saturated with the ligand at the beginning of the experiment. The possibility of extensive ligand dissociation was checked for TMPyP4 and DP78. TMPyP4 was followed on gel directly under the visible light (Supplementary Figure S7) while DP78 was detected under the long UV light (exhibits lower but still detectable intensity, not shown). So detected band positions are the same as those detected under the short UV light (detection of Tel22) which confirms that the bands belong to TMPyP4–Tel22 and DP78–Tel22 complexes and not to the ligand-free Tel22.
Figure 6.
PAGE electrophoresis performed at 25 °C. Panel A: bands correspond to ligand-free Tel22 and/or Tel22-Net complexes prepared at different Net/Tel22 molar ratios, r. Panel B: Typical mobility shifts of Tel22 saturated by the quadruplex specific ligand (360A-Br) or by the quadruplex non-selective ligands (Net, DP78, TMPyP4).
CD spectroscopy is often used to obtain qualitative information on the G-quadruplex conformations in the solution. It has been shown that the CD spectra of Tel22 G-quadruplex in the presence of K+ ions are characterized by a maximum at ∼293 nm and a plateau at ∼265 nm which is typical of hybrid-1 and hybrid-2 conformations. Additions of Net, DP77 and DP78 have small effect on the shape of CD spectra which is in accordance with gel electrophoresis results and with our thermodynamics based suggestion that the corresponding ligand binding-coupled conformational changes are small (Figure 5). In the case of TMPyP4 the CD-spectrum of TMPyP4-Tel22 complex is significantly different from the one observed for the ligand free Tel22. Since the gel-electrophoretic mobility of the TMPyP4-Tel22 complex and ligand free Tel22 is the same (Figure 6) the observed difference in CD spectra is unlikely a consequence of Tel22 conformational changes. This explanation is consistent with the relatively small ΔG°conf estimated for the TMPyP4–Tel22 complex formation. By contrast, the difference between similar CD spectra of 360A-Br-Tel22 and Phen-DC3-Tel22 complexes (Figure 5) and CD spectrum of the ligand-free Tel22 may be ascribed to ligand-induced Tel22 conformational transitions into similar ligand bound structures (49). This suggestion is in accordance with the gel electrophoresis experiments (Figure 6, (49)) and with the relatively large ΔG°conf > 20 kcal mol−1 estimated for these transitions. The shape of the CD spectra with characteristics of the antiparallel conformation of the ligand-free quadruplex observed with 360A-Br-Tel22 and Phen-DC3-Tel22 complexes (Figure 5) appears to be a common feature of various G-quadruplex complexes with specific ligands such as RHPS4, (7) berberine, (34) telomestatin, (31) etc. Moreover, similar ‘antiparallel type’ CD spectrum has been observed with the ligand-free 22GT (5′-(GGGTTAA)3GGGT-3′) sequence that forms the two quartet hybrid-3 structure that incorporates only one specifically bound K+ ion (48). To summarize, the measured CD spectra qualitatively support the conclusions drawn from the suggested dissection of the corresponding binding thermodynamics and suggest that the Net, DP77, DP78 and TMPyP4 form complexes with Tel22 hybrid-1 and/or hybrid-2 conformations while Phen-DC3 and 360A-Br may form complexes with antiparallel or/and hybrid-3 type conformations.
To obtain additional information on Tel22 conformation in the ligand-Tel22 complexes we performed molecular modeling of Tel22 in the presence of ligands. According to the binding characteristics of Net and TMPyP4 (non-specific binding of three or four molecules) it is not possible to define Net and TMPyP4 characteristic binding sites on Tel22. Thus, no modeling was carried out for these two systems. On the other hand, molecular modeling was successfully performed with DP77, DP78, Phen-DC3 and 360A-Br ligands in the 1:1 complexes with different Tel22 topologies that were selected on the basis of information obtained from CD spectroscopy (Figure 5). In this light the modeling with DP77 and DP78 suggests that the ligand molecules interact with the exposed G-quarters and groves of the Tel22 hybrid-1 and hybrid-2 conformation (Figure 5). Such binding is not accompanied by significant conformational changes which is consistent with the estimated relatively small ΔG°conf. To support this consistency of the structural models of the ligand-Tel22 complexes (Figure 5) with the binding thermodynamics we performed structure-based calculations of the corresponding ΔC°P. It turns out that ΔC°P values estimated from changes of solvent accessible surface areas (SASA) accompanying binding of DP77 or DP78 to hybrid-1 and hybrid-2 conformations mixed in 1:1 ratio agree well with the corresponding ΔC°P values obtained from the model analysis of experimental data (Supplementary Table S3).
The modeling of the Phen-DC3 and 360A-Br-Tel22 1:1 complexes with Tel22 in the antiparallel conformation suggests that ligand molecules may bind to its terminal G-quartet (Figure 5) which is not exposed in the ligand-free antiparallel structure (similar structural model has been proposed for telomestatin-Tel22 complex) (65). On the other hand, the modeling of the Phen-DC3 and 360A-Br-Tel22 1:1 complexes with Tel22 in the hybrid-3 conformation suggests that ligand molecules may bind between the G-triplet and G-quartet. The formation of antiparallel or hybrid-3 type conformations upon binding is accompanied by significant conformational changes which is consistent with the estimated relatively large ΔG°conf. The formation of the ligand-Tel22 (antiparallel type) or ligand-Tel22 (hybrid-3 type) complexes from ligand (Phen-DC3 or 360A-Br) and Tel22 (hybrid-1:hybrid-2 = 1:1) is accompanied by changes in SASA which result in ΔC°P values that agree reasonably well with the corresponding experimentally determined ΔC°P values (Supplementary Table S3). Though ΔC°P estimations from SASAs suggest (Supplementary Table S3) that Phen-DC3 and 360A-Br likely bind antiparallel-type and/or hybrid-3 type conformation they are not sufficiently reliable to independently predict the preferential Tel22 conformation in the bound state. It appears that the structural modeling of Phen-DC3-Tel22 and 360A-Br-Tel22 1:1 complexes is consistent with our dissection of energetics and SASA calculations only if the two ligands are extensively buried inside the quadruplex structure (dehydration of the upper and the bottom surface of Phen-DC3 and 360A-Br). This is possible with Tel22 in an antiparallel or hybrid-3 type conformation, but less likely with hybrid-1, hybrid-2 or the all parallel structure where one side of the ligand would remain largely exposed to the solvent.
According to Marchand et al., (48) the observed quadruplex conformational changes accompanying Phen-DC3 and 360A-Br binding may be due to disruption of the three quartet hybrid-1 and hybrid-2 structures (induced fit binding) or due to conformational selection of the two quartet hybrid-3 structure. The appropriateness of the conformational selection hypothesis may be discussed on the basis of the observed large difference in the Gibbs free energy (ΔG°conf > 20 kcal mol−1) between the ligand-free Tel22 (mixture of hybrid-1 and hybrid-2) and the antiparallel or hybrid-3 type conformation adopted by Tel22 in the bound state. Large ΔG°conf suggests that the concentration of the molecules in the antiparallel or hybrid-3 type conformation in the studied Tel22 solutions in the absence of the ligand would be so low that the probability of selecting such conformation by the ligand is practically negligible. In this light our thermodynamic data are more consistent with the induced fit binding mechanism in which the ligand disrupts the hybrid-1 and hybrid-2 structure to form the antiparallel or/and hybrid-3 type conformation. The high potency of Phen-DC3 and 360A-Br to disrupt Tel22 quadruplex structure is also in accordance with our previous observations of Phen-DC3 and 360A-Br binding to Tel22 in the presence of Na+ ions where the ligand-bound quadruplex structure appears to be very similar to the one observed in the presence of K+ ions (49).
Taken together, the obtained thermodynamic information, mobility of ligand–quadruplex complexes in the gel, the shape of the CD spectra and the results of molecular modeling suggest that in the process of binding Net, DP77, DP78 and TMPyP4 select hybrid-1 and/or hybrid-2 conformation, as the most populated conformations of Tel22 in solutions with K+ ions, while binding of Phen-DC3 and 360A-Br more likely results in the binding induced formation of the ligand-Tel22 complexes with Tel22 in the antiparallel or/and hybrid-3 conformation. Evidently, due to the lack of high-resolution structural information on exact folds that Tel22 adopts in its ligand-free and ligand-bound states, our structural interpretation of contributions to binding thermodynamics has to take into account several possible ligand-Tel22 binding modes. This suggests that to improve the structural interpretation of ligand–quadruplex binding thermodynamics it would be necessary in the future to study ligand binding to those quadruplex forming sequences that adopt single folds (verified by NMR) in their ligand-free and ligand-bound states. In this case a more rigorous correlation of the thermodynamic contributions obtained by dissection of the measured binding energetic with the corresponding high-resolution structural information would be possible.
CONCLUSION
In our study, we compared binding characteristics of six ligands (Net, DP77, DP78, TMPyP4, 360A-Br, Phen-DC3) to ht-DNA fragment Tel22 in the presence of K+ ions. To the best of our knowledge, it represents one of the first experimental studies that take into account the folding intermediates in the analysis of ligand binding to ht-DNA. We found that the simplest model mechanism successfully describing all experimental data measured at physiological temperatures and salt concentrations must include conversion between the intermediate and folded G-quadruplex. The global thermodynamic analysis applied to obtain simultaneously the driving forces of binding and conversion involves a large number of adjustable parameters and does not provide the sufficient accuracy of their values. It is therefore necessary, to estimate the driving forces for the conversion between intermediate and folded quadruplex separately (16,49).
In contrast to 360A-Br and Phen-DC3, the ligands Net, DP77, DP78 and TMPyP4 interact with G-quadruplex less or equally strongly than with dsDNA and may therefore be classified as G-quadruplex non-selective ligands. Our dissection of energetics, gel electrophoresis experiments, CD and fluorescence spectroscopy measurements and molecular modeling result in the following thermodynamic and structural features that appear to be characteristic for binding of here studied G-quadruplex non-selective and quadruplex specific ligands to Tel22: (i) Binding of the non-selective ligands is accompanied by less extensive compensation of various Gibbs free energy contributions (magnitudes of ΔG° contributions are significantly lower) than the binding of the specific ligands; (ii) For the non-selective ligands dehydration of hydrophobic surfaces appears to be sufficient for their recognition of the quadruplex, while successful binding of the specific ones (360A-Br, Phen-DC3) that are extensively buried within the quadruplex structure has to be driven also by specific interactions taking place at the binding interface; (iii) Binding of the non-selective ligands with relatively low affinity does not induce significant conformational change on the G-quadruplex. In this light upon binding Net, DP77, DP78 and TMPyP4 select the predominant hybrid-1 and/or hybrid-2 quadruplex conformation. By contrast, binding of the G-quadruplex specific ligands 360A-Br and Phen-DC3 induces structural changes in the quadruplex conformation which may be described as a transition from the hybrid-1 and/or hybrid-2 to an antiparallel or hybrid-3 type conformation.
To summarize, our global analysis provides thermodynamic fingerprints of ligand binding to ht-DNA quadruplexes. Significantly, these independent thermodynamic predictions are consistent with the observed structural features. In other words, our study shows how to explore the predictive power of thermodynamics for better understanding of molecular recognition of G-quadruplexes by aromatic ligands.
Supplementary Material
Acknowledgments
The authors thank Dr Anton Granzhan for carefully reading the manuscript and giving detailed comments and suggestions that have been helpful to improve the quality of manuscript. Authors also thank Mrs Majda Pavlin, Mrs Katja Vovčko, Mr Domen Oblak and Mr Uroš Mavsar for performing some experiments. This work would not have been done without financial support of the Slovenian Research Agency through the Grants P1–0201 and J1-5448 and of the EU FP7 project InnoMol (RBI).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Slovenian Research Agency [P1–0201, J1-5448]. Funding for open access charge: Slovenian Research Agency [P1–0201, J1-5448] and EU FP7 project InnoMol (RBI).
Conflict of interest statement. None declared.
REFERENCES
- 1.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Commun. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Sullivan R.J., Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamon F., Largy E., Guèdin-Beaurepaire A., Rouchon-Dagois M., Sidibe A., Monchaud D., Mergny J.-L., Riou J.-F., Nguyen C.-H., Teulade-Fichou M.-P. An acyclic oligoheteroaryle that discriminates strongly between diverse G-quadruplex topologies. Angew. Chem. Int. Ed. Engl. 2011;50:8745–8740. doi: 10.1002/anie.201103422. [DOI] [PubMed] [Google Scholar]
- 7.Garner T.P., Williams H.E.L., Gluszyk K.I., Roe S., Oldham N.J., Stevens M.F.G., Moses J.E., Searle M.S. Selectivity of small molecule ligands for parallel and anti-parallel DNA G-quadruplex structures. Org. Biomol. Chem. 2009;7:4194–4200. doi: 10.1039/b910505k. [DOI] [PubMed] [Google Scholar]
- 8.Dhamodharan V., Harikrishna S., Bhasikuttan A.C., Pradeepkumar P.I. Topology specific stabilization of promoter over telomeric. ACS Chem. Biol. 2015;10:821–833. doi: 10.1021/cb5008597. [DOI] [PubMed] [Google Scholar]
- 9.Dhamodharan V., Harikrishna S., Jagadeeswaran C., Halder K., Pradeepkumar P.I. Selective G-quadruplex DNA stabilizing agents based on bisquinolinium and bispyridinium derivatives of 1,8-naphthyridine. J. Org. Chem. 2012;77:229–242. doi: 10.1021/jo201816g. [DOI] [PubMed] [Google Scholar]
- 10.Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chem. Eur. J. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- 11.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 12.Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D. Human telomeric sequence forms a hybrid-typeintramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phan A.T., Kuryavyi V., Luu K.N., Patel D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K +solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray R.D., Petraccone L., Trent J.O., Chaires J.B. Characterization of a K+-induced conformational switch in a human telomeric DNA oligonucleotide using 2-Aminopurine fluorescence. Biochemistry. 2010;49:179–194. doi: 10.1021/bi901357r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 16.Bončina M., Lah J., Prislan I., Vesnaver G. Energetic basis of human telomeric DNA folding into G-quadruplex structures. J. Am. Chem. Soc. 2012;134:9657–9663. doi: 10.1021/ja300605n. [DOI] [PubMed] [Google Scholar]
- 17.Gray R.D., Buscaglia R., Chaires J.B. Populated intermediates in the thermal unfolding of the human telomerc quadruplex. J. Am. Chem. Soc. 2012;134:16834–16844. doi: 10.1021/ja307543z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buscaglia R., Gray R.D., Chaires J.B. Thermodynamic characterization of human telomere quadruplex unfolding. Biopolymers. 2013;99:1006–1018. doi: 10.1002/bip.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mashimo T., Yagi H., Sannohe Y., Rajendran A., Sugiyama H. Folding Pathways of Human Telomeric Type-1 and Type-2 G-Quadruplex Structures. J. Am. Chem. Soc. 2010;132:14910–14918. doi: 10.1021/ja105806u. [DOI] [PubMed] [Google Scholar]
- 20.Koirala D., Mashimo T., Sannohe Y., Yu Z., Mao H., Sugiyama H. Intramolecular folding in three tandem guanine repeats of human telomeric DNA. Chem. Commun. 2012;48:2006–2008. doi: 10.1039/c2cc16752b. [DOI] [PubMed] [Google Scholar]
- 21.Zhang A.Y.Q., Balasubramanian S. The Kinetics and Folding Pathways of Intramolecular G-Quadruplex Nucleic Acids. J. Am. Chem. Soc. 2012;134:19297–19308. doi: 10.1021/ja309851t. [DOI] [PubMed] [Google Scholar]
- 22.Stadlbauer P., Trantírek L., Cheatham T.E., Koča J., Sponer J. Triplex intermediates in folding of human telomeric quadruplexes probed by microsecond-scale molecular dynamics simulations. Biochimie. 2014;105:22–35. doi: 10.1016/j.biochi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Lane A.N., Chaires J.B., Gray R.D., Trent J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monchaud D., Teulade-Fichou M.-P. A hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 25.Martino L., Pagano B., Fotticchia I., Neidle S., Giancola C. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B. 2009;113:14779–14786. doi: 10.1021/jp9066394. [DOI] [PubMed] [Google Scholar]
- 26.Parkinson G.N., Ghosh R., Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46:2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 27.Freyer M.W., Buscaglia R., Kaplan K., Cashman D., Hurley L.H., Lewis E.A. Biophysical studies of the c-MYC NHE III1 promoter: model quadruplex interactions with a cationic porphyrin. Biophys. J. 2007;92:2007–2015. doi: 10.1529/biophysj.106.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cummaro A., Fotticchia I., Franceschin M., Giancola C., Petraccone L. Binding properties of human telomeric quadruplex multimers: a new route for drug design. Biochimie. 2011;93:1392–1400. doi: 10.1016/j.biochi.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Phan A.T., Kuryavyi V., Yan H., Patel D.J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yakua H., Murashimab T., Tateishi-Karimatac H., Nakanob S., Miyoshib D., Sugimoto N. Study on effects of molecular crowding on G-quadruplex-ligand binding and ligand-mediated telomerase inhibition. Methods. 2013;64:19–22. doi: 10.1016/j.ymeth.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 31.Rezler E.M., Seenisamy J., Bashyam S., Kim M.-Y., White E., Wilson W.D., Hurley L.H. Telomestatin and Diseleno Sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 32.Chung W.J., Heddi B., Hamon F., Teulade-Fichou M.P., Phan A.T. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC3. Angew. Chem. Int. Ed. Engl. 2014;53:999–1002. doi: 10.1002/anie.201308063. [DOI] [PubMed] [Google Scholar]
- 33.Arora A., Balasubramanian C., Kumar N., Agrawal S., Ojha R.P., Maiti S. Binding of berberine to human telomeric quadruplex – spectroscopic, calorimetric and molecular modeling studies. FEBS J. 2008;275:3971–3983. doi: 10.1111/j.1742-4658.2008.06541.x. [DOI] [PubMed] [Google Scholar]
- 34.Bessi I., Bazzicalupi C., Richter C., Jonker H.R.A., Saxena K., Sissi C., Chioccioli M., Bianco S., Bilia A.R., Schwalbe H., et al. Spectroscopic, molecular modeling, and NMR-spectroscopic investigation of the binding mode of the natural alkaloids berberine and sanguinarine to human telomeric G-quadruplex DNA. ACS Chem. Biol. 2012;7:1109–1119. doi: 10.1021/cb300096g. [DOI] [PubMed] [Google Scholar]
- 35.Campbell N.H., Parkinson G.H., Reszka A.P., Neidle S. Structural basis of DNA quadruplex recognition by an Acridine drug. J. Am. Chem. Soc. 2008;130:6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 36.Hudson J.S., Brooks S.C., Graves D.E. Interactions of Actinomycin D with human telomeric G-quadruplex DNA. Biochemistry. 2009;48:4440–4447. doi: 10.1021/bi900203z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbieri C.M., Srinivasan A.R., Rzuczek S.G., Rice J.E., LaVoie E.J., Pilch D.S. Defining the mode, energetics and specificity with which a macrocyclic hexaoxazole binds to human telomeric G-quadruplex DNA. Nucleic Acids Res. 2007;35:3272–3286. doi: 10.1093/nar/gkm188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collie G.W., Promontorio R., Hampel S.M., Micco M., Neidle S., Parkinson G.N. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012;134:2723–2731. doi: 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- 39.Nicoludis J.M., Barrett S.P., Mergny J.L., Yatsunyk L.A. Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012;40:5432–5447. doi: 10.1093/nar/gks152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicoludis J.M., Miller S.T., Jeffrey P.D., Barrett S.P., Rablen P.R., Lawton T.J., Yatsunyk L.A. Optimized end-stacking provides specificity of N-Methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012;134:20446–20456. doi: 10.1021/ja3088746. [DOI] [PubMed] [Google Scholar]
- 41.Pagano B., Virno A., Mattia C.A., Mayol L., Randazzo A., Giancola C. Targeting DNA quadruplexes with distamycin A and its derivatives: an ITC and NMR study. Biochimie. 2008;90:1224–1232. doi: 10.1016/j.biochi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 42.Lah J., Drobnak I., Dolinar M., Vesnaver G. What drives the binding of minor groove-directed ligands to DNA hairpins? Nucleic Acids Res. 2008;30:897–904. doi: 10.1093/nar/gkm1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lah J., Vesnaver G. Energetic diversity of DNA minor-groove recognition by small molecules displayed through some model ligand-DNA systems. J. Mol. Biol. 2004;342:73–89. doi: 10.1016/j.jmb.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Freyer M.W., Buscaglia R., Cashman D., Hyslop S., Wilson W.D., Chaires J.B., Lewis E.A. Binding of netropsin to several DNA constructs: Evidence for at least two different 1:1 complexes formed from an –AATT-containing ds-DNA construct and a single minor groove binding ligand. Biophys. Chem. 2007;126:186–196. doi: 10.1016/j.bpc.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Stojković M.R., Marjanović M., Pawlica D., Dudek D., Eilmes J., Kralj M., Piantanida I. Cationic side-chains control DNA/RNA binding properties and antiproliferative activity of dicationic dibenzotetraaza[14]annulene derivatives. New J. Chem. 2010;34:500–507. [Google Scholar]
- 46.Wang P., Ren L., He H., Liang F., Zhou X., Tan Z. A phenol quaternary ammonium porphyrin as a potent telomerase inhibitor by selective interaction with quadruplex DNA. Chembiochem. 2006;7:1155–1159. doi: 10.1002/cbic.200600036. [DOI] [PubMed] [Google Scholar]
- 47.De Cian A., DeLemos E., Mergny J.-L., Teulade-Fichou M.-P., Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 48.Marchand A., Granzhan A., Iida K., Tsushima Y., Ma Y., Nagasawa K., Teulade-Fichou M.P., Gabelica V. Ligand-induced conformational changes with cation ejection upon binding to human telomeric DNA G-quadruplex. J. Am. Chem. Soc. 2015;137:750–756. doi: 10.1021/ja5099403. [DOI] [PubMed] [Google Scholar]
- 49.Bončina M., Hamon F., Islam B., Teulade-Fichou M.P., Vesnaver G., Haider S., Lah J. Dominant driving forces in human telomere quadruplex binding-induced structural alterations. Biophys. J. 2015;108:2903–2911. doi: 10.1016/j.bpj.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cantor C.R., Warshaw M.M., Shapiro H. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 51.Krieger E., Koraimann G., Vriend G. Increasing the precision of comparative models with YASARA NOVA - a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 52.Duan Y., Wu C., Chowdhury S., Lee M.C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T. A point-charge force field for molecular mechanics simulations of proteins. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 53.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comp. Chem. 1992;13:952–962. [Google Scholar]
- 55.Hubbard S.J., Thornton J.M. NACCESS computer program. London: Department of Biochemistry and Molecular Biology, University College London; 1993. [Google Scholar]
- 56.Murphy K.P., Freire E. Thermodynamics of structural stability and cooperative folding behaviour in proteins. Adv. Protein Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. [DOI] [PubMed] [Google Scholar]
- 57.Lah J., Vesnaver G. Binding of Distamycin A and Netropsin to the 12mer DNA duplexes containing mixed AT•GC sequences with at most five or three successive AT base pairs. Biochemistry. 2000;39:9317–9326. doi: 10.1021/bi000748u. [DOI] [PubMed] [Google Scholar]
- 58.Pillet F., Romera C., Trevisiol E., Bellon S., Teulade-Fichou M.P., Francois J.M., Pratviel G., Leberre V.A. Surface plasmon resonance imaging (SPRi) as an alternative technique for rapid and quantitative screening of small molecules, useful in drug discovery. Sensor. Actuat. B-Chem. 2011;157:304–309. [Google Scholar]
- 59.Haq I., Ladbury J.E., Chowdhry B.Z., Jenkins T.C., Chaires J.B. Specific Binding of Hoechst 33258 to the d(CGCAAATTTGCG)2 Duplex: Calorimetric and Spectroscopic Studies. J. Mol. Biol. 1997;271:244–257. doi: 10.1006/jmbi.1997.1170. [DOI] [PubMed] [Google Scholar]
- 60.Baldwin R.L. Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. U.S.A. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spolar R.S., Record M.T. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 62.Finkelstein A.V., Janin J. The price of lost freedom - entropy of bimolecular complex-formation. Protein Eng. 1989;3:1–3. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- 63.Williams D.H., Westwell M.S. Aspects of weak interactions. Chem. Soc. Rev. 1998;27:57–63. [Google Scholar]
- 64.Record M.T., Jr, Anderson C.F., Lohman T.M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 65.Kim M-Y., Vankayalapati H., Shin-ya K., Wierzba K., Hurley L.H. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.