Abstract
Restriction-modification (R-M) systems pose a major barrier to DNA transformation and genetic engineering of bacterial species. Systematic identification of DNA methylation in R-M systems, including N6-methyladenine (6mA), 5-methylcytosine (5mC) and N4-methylcytosine (4mC), will enable strategies to make these species genetically tractable. Although single-molecule, real time (SMRT) sequencing technology is capable of detecting 4mC directly for any bacterial species regardless of whether an assembled genome exists or not, it is not as scalable to profiling hundreds to thousands of samples compared with the commonly used next-generation sequencing technologies. Here, we present 4mC-Tet-assisted bisulfite-sequencing (4mC-TAB-seq), a next-generation sequencing method that rapidly and cost efficiently reveals the genome-wide locations of 4mC for bacterial species with an available assembled reference genome. In 4mC-TAB-seq, both cytosines and 5mCs are read out as thymines, whereas only 4mCs are read out as cytosines, revealing their specific positions throughout the genome. We applied 4mC-TAB-seq to study the methylation of a member of the hyperthermophilc genus, Caldicellulosiruptor, in which 4mC-related restriction is a major barrier to DNA transformation from other species. In combination with MethylC-seq, both 4mC- and 5mC-containing motifs are identified which can assist in rapid and efficient genetic engineering of these bacteria in the future.
INTRODUCTION
Genetic engineering of bacterial species is essential to modify genetic and biochemical pathways to facilitate the large-scale production of novel drugs, valuable biofuels and bioproducts, and antibiotics among many other biomedical, bioenergy and industrial applications. However, restriction-modification (R-M) systems have proven a major barrier to DNA transformation and subsequent genetic engineering of bacterial species (1). Specifically, through the cleavage of foreign DNA sequences by either Type I or Type II restriction enzymes, bacteria protect their genomes from invasive foreign DNA (2). Recognition sequences in the host genome are protected by methylation of adenine (N6-methyladenine – 6mA) and cytosine (N4-methylcytosine – 4mC and 5-methylcytosine – 5mC) bases within the host DNA recognition motifs (3). While it took many years to identify the R-M systems in Escherichia coli (4), recent advances in genomic technologies are enabling scientists to acquire sequence level data at an unprecedented rate and resolution. For example, the precise location and sequence context of 5mC can be readily identified using whole-genome bisulfite sequencing (5,6). The combination of bisulfite-mediated conversion and next-generation sequencing technologies enable researchers to accurately identify 5mC sites in bacterial genomes at single-base resolution with relatively low cost, which make it affordable to map bacterial epigenomes in a scalable manner (hundreds to thousands of samples). This method also yields results rapidly, within days of preparing bacterial genomic DNA and sequencing libraries from fairly low inputs of total genomic DNA (<100 ng).
4mC is frequently present in thermophilic bacteria as a base modification and also exists in many bacterial mesophiles (7). DNA transformation to 4mC-containing species is extremely difficult due to the lack of systematic knowledge about the 4mC-specifc R-M systems, such as 4mC motifs and the corresponding methyltransferases and restriction endonucleases (8). Single-molecule, real time sequencing (SMRT) technology is capable of producing data useful for whole genome assemblies while at the same time being able to detect all 4mC and 6mA base modifications (3,9,10). SMRT can also be used to directly detect 5mC after conversion of 5mC to 5caC to enhance its kinetic signature (11). However, comparing with the commonly used next-generation sequencing technologies such as Illumina sequencing systems, SMRT sequencing is more costly for library preparation and sequencing, and is not a feasible solution for analysis of thousands of bacterial R-M systems for which genomes already exist in the public domain. Hence, the establishment of a 4mC specific detection method compatible with next-generation sequencing platforms will be particularly important for rapid and efficient manipulation of the genomes of industrially promising thermophiles, and will facilitate the high-throughput analysis of 4mC-involved R-M systems in many uncharacterized but potentially useful strains.
Bisulfite sequencing, which can selectively deaminate unmodified cytosine, but not 5mC, to uracil (U), which ultimately is read as thymine (T) after PCR amplification, has been widely used to resolve the location of 5mC in genomes of bacteria, plants and animals at single-base resolution (5,6,12–16). Standard bisulfite sequencing protocols may also be used to map 4mC because 4mC is partially resistant to bisulfite-mediated deamination (17). However, considering that 5mC is a prevalent base modification in many prokaryotes and eukaryotes, traditional bisulfite sequencing (MethylC-seq) is not suitable to accurately differentiate 4mC from 5mC as both will be read as C (Figure 1a). Therefore, standard MethylC-seq cannot accurately detect 5mC if 4mC and 5mC are both present in a genome.
Figure 1.
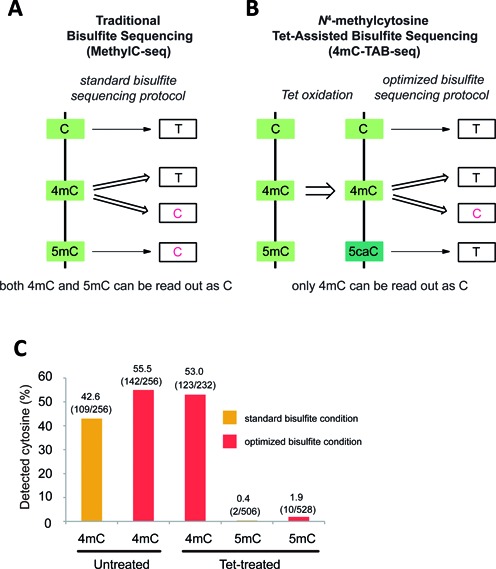
Comparison of MethylC-seq and 4mC-TAB-seq. (A) MethylC-seq converts C and a portion of 4mC to T. The remaining 4mC and almost all 5mC will be read as C. (B) 4mC-TAB-seq converts C, 5mC and a portion of 4mC to T, whereas about half of 4mC will be exclusively read as C. (c) Properties of 4mC and 5mC under different treatment conditions. Untreated or Tet-treated 4mC/5mC-containing model DNA is applied to either standard or optimized bisulfite treatment condition. Samples were PCR amplified, subcloned into TOPO vector and Sanger sequenced to quantify the number of 4mC or 5mC being read as C.
Previous studies have reported that the Tet (ten-eleven translocation) proteins are able to specifically oxidize 5mC to 5-carboxylcytosine (5caC), which is read as T in standard bisulfite sequencing protocols (18,19). In fact, Tet-mediated oxidation and bisulfite treatment to genomic DNA are two of the major chemical transformations required for Tet-Assisted Bisulfite-sequencing (TAB-seq) to detect 5-hydroxymethylcytosines (20). Here we present a novel 4mC-TAB-seq strategy that is able to accurately identify 4mC sites exclusively without interference of 5mC. In 4mC-TAB-seq, excess recombinant mouse Tet1 protein (Tet) is utilized to oxidize all 5mC to 5caC. Then, after bisulfite treatment under optimized conditions and PCR amplification, 5caC is read as T and about half of the 4mC sites are read as C (Figure 1b). This strategy results in the ability to accurately generate genome-wide, single-base resolution maps of 4mC and in the ultimate identification of 4mC-containing motifs associated with bacterial R-M systems. Because standard MethylC-seq gives the sum of 4mC+5mC, a subtraction of 4mC-TAB-seq data from MethylC-seq data would afford accurate mapping of 5mC in genomic DNA.
MATERIALS AND METHODS
Culture conditions and genomic DNA isolation
Caldicellulosiruptor kristjanssonii wild-type strain was grown anaerobically in liquid low osmolarity complex growth (LOC) medium (21) (final pH 7.0) with maltose (0.5% w/v; Sigma M5895) as the carbon source. Liquid cultures were grown from a 0.5% inoculum and incubated at 75°C in anaerobic culture bottles degassed with five cycles of vacuum and argon. Genomic DNA of C. kristjanssonii was prepared from 50 ml cultures grown to mid-log phase (∼0.1 at OD680), harvested by centrifugation at 6000 × g at 4°C for 15 min and resuspended in 800 μl of Genomic Lysis buffer (Zymo Research). Cells were lysed by a combination of three freeze-thaw cycles and sonication on ice. The following steps were performed using the Quick-gDNA™ MiniPrep (Zymo Research) according to the manufacturer's instructions. Genomic DNA concentrations were determined using the Qubit® 2.0 fluorometer (Invitrogen) and the quality of DNA was assessed by agarose gel electrophoresis.
Preparation of 304 bp model DNA with 4mC modifications
For N4-methylcytosine (4mC) containing model DNA, 0.5 ng of pUC19 vector DNA (NEB) was PCR amplified as follows in a 50 μl reaction: 2.5 U RedTaq polymerase (Sigma), 5 μl 10× reaction buffer, 1 μl N4-methyl-dCTP (4mdCTP) (Trilink)/dATP/dGTP/dTTP cocktail (10 mM each), 1 μl 10 mM forward primer (5′-GAACGAAAACTCACGTTAAGGG), 1 μl 10 mM reverse primer (5′-TGCTGATAAATCTGGAGCCG). Cycling parameters: 94°C 2 min, 30 cycles of 94°C 1 min, 37°C 2 min, 55°C 3 min, followed by 55°C 7 min. The PCR product was purified by gel electrophoresis.
Generation of spike-in controls for 4mC-TAB-seq and MethylC-seq of C. kristjanssonii
C/5mC spike-in control (CpG methylated lambda DNA) was prepared by treating unmethylated lambda cl857 DNA (Promega) with M. SssI (NEB) according to the manufacturer's instructions. The CpG methylation was confirmed by standard bisulfite sequencing. To generate the 4mC spike-in control, 0.5 ng of pUC19 vector (NEB) was PCR amplified as follows in a 50 μl reaction: 2.5 U RedTaq polymerase (Sigma), 5 μl 10× reaction buffer, 1 μl 4mdCTP (Trilink)/dATP/dGTP/dTTP cocktail (10 mM each), 1 μl 10 mM forward primer (5′-GCGGTAATACGGTTATCCAC), 1 μl 10 mM reverse primer (5′-TGCTGATAAATCTGGAGCCG). Cycling parameters: 94°C 2 min, 35 cycles of 94°C 1 min, 37°C 2 min, 55°C 5 min, followed by 55°C 7 min. The PCR product was purified by gel electrophoresis.
Condition test using 4mC or 5mC-containing model DNA
The 304 bp DNA with 4mC and CpG methylated lambda DNA were prepared as described above. The recombinant catalytic domain of mouse Tet1 protein (Tet) was expressed and purified as previously described (22). Oxidation reactions were performed in a 50 μl solution with 50 mM HEPES buffer (pH 8.0), 100 μM ammonium iron (II) sulfate, 1 mM α-ketoglutarate, 2 mM ascorbic acid, 2.5 mM DTT, 100 mM NaCl, 1.2 mM ATP, 6 ng/μl sonicated mouse embryonic stem cells (mESC) genomic DNA with 0.5% (w/w) 4mC or 5mC-containing model DNA and 4.5 μM Tet. The reactions were incubated at 37°C for 1.5 h. After proteinase K treatment, the oxidized DNA was purified with Micro Bio-Spin 30 Columns (Bio-Rad) and then by QIAquick PCR Purification Kit (QIAGEN). The untreated or Tet-treated mESC genomic DNA with 4mC or 5mC-containing model DNA was applied to MethylCode Bisulfite Conversion Kit (Invitrogen) using thermal cycling program: (i) 98°C 10 min, 64°C 2.5 h or (ii) 98°C 10 min, 53°C 30 min, 8 cycles of 53°C 6 min and 37°C 30 min. After bisulfite conversion, 3 μl of purified converted DNA was PCR amplified using ZymoTaq DNA polymerase (Zymo Research) following the manufacturers’ instructions (for 4mC model, forward primer: 5′-GAATGAAAATTTATGTTAAGGG; reverse primer: 5′-ATTTAAAACTTCATTTTTAATTTAAAA; for 5mC model, forward primer: 5′-TTTGGGTTATGTAAGTTGATTTTATG; reverse primer: 5′-CACCCTACTTACTAAAATTTACACC). The PCR products were TOPO cloned using the TOPO TA cloning kit (Invitrogen), and individual clones were subjected to Sanger sequencing using the M13 Forward primer.
Quantification of 4mC and 5mC in C. kristjanssonii by LC-MS/MS
Five hundred nanograms of C. kristjanssonii genomic DNA was digested by 2 U Nuclease P1 (Wako) in 30 μl solution containing 0.01 M NH4Ac (pH 5.3) and 2 mM ZnCl2 at 42°C overnight. After adding 3.5 μl freshly prepared 1 M NH4HCO3 and 1 mU Phosphodiesterase I (Sigma P3134), the reaction was allowed to incubate at 37°C for 2 h followed by addition of 2 U Alkaline Phosphatase (Sigma) and another 2 h incubation at 37°C. The digested sample was filtered and 5 μl was subject to LC–MS/MS. The separation of nucleosides was performed using Agilent 1290 UHPLC system with a C18 reversed-phase column (2.1 × 50 mm, 1.8 μm). The mobile phase A was water with 0.9 ppm (v/v) formic acid (final pH 4.5) and mobile phase B was methanol with 0.9 ppm (v/v) formic acid. Online mass spectrometry detection was performed using an Agilent 6460 triple quadrupole mass spectrometer in positive electrospray ionization mode. Quantification was accomplished in multiple reaction monitoring (MRM) mode by monitoring the transitions of 228.2→112.1 (dC), 242.2→126.1 (4mdC/5mdC). 4mdC and 5mdC had the different retention times. dC, 4mdC and 5mdC were quantified basing on the corresponding calibration curves generated with pure standards.
4mC-TAB-seq and MethylC-seq of C. kristjanssonii genomic DNA
C/5mC and 4mC spike-in controls were added to C. kristjanssonii genomic DNA to a final concentration of 0.5% (w/w) and sonicated to average 200 bp with Covaris S220 (Peak Incident Power 175 W, Duty Factor 10%, Cycles per Burst 200, 180 s).
For MethylC-seq, sonicated C. kristjanssonii genomic DNA with spike-in controls was directly used for library preparation. For 4mC-TAB-seq, 150 ng sonicated C. kristjanssonii genomic DNA with spike-in controls was first oxidized by 4.5 μM Tet in a 50 μl solution with 50 mM HEPES buffer (pH 8.0), 100 μM ammonium iron (II) sulfate, 1 mM α-ketoglutarate, 2 mM ascorbic acid, 2.5 mM DTT, 100 mM NaCl, 1.2 mM ATP. The oxidation reaction was incubated at 37°C for 1.5 h. After proteinase K treatment, the oxidized DNA was purified with Micro Bio-Spin 30 Columns (Bio-Rad) and then by QIAquick PCR Purification Kit (QIAGEN).
For both MethylC-seq and 4mC-TAB-seq, libraries were constructed as previously described (16) with the bisulfite conversion thermocycling adapted slightly as follows: 98°C 10 min, 53°C 30 min, 8 cycles of 53°C 6 min and 37°C 30 min.
4mC-TAB-seq data analysis and motif detection
MethylC-seq and 4mC-TAB-seq analyses were performed as previously described (12). Two control DNA sequences were added as quality controls. For the 4mC control, an ∼1 kb region of the pUC19 vector was constructed by PCR amplification with 4mdCTP. This approach achieves >98.5% of cytosines present as 4mC. Unmethylated lambda DNA was treated by M. SssI to methylate all cytosines in CpG context to 5mC. The 5mCs in CpG context were used to calculate the 5mC conversion rate and non-CpG sites were used to compute the sodium bisulfite reaction non-conversion rate of unmodified cytosines. Only cytosine sites with a minimum coverage (set as 3) were allowed for subsequent analysis. Binomial test coupled with Benjamini-Hochberg correction was adopted to determine the methylation status of each cytosine.
A three-nucleotide (nt) seed method was used for motif searching and identification. First, all methylated cytosines and flanking sequences (12 nt in each direction) were extracted from the reference genome. Next, these methylated regions were separated by NCN (C = methylated C, N = A, T, C, and G) context around methylated cytosines to 16 subsets. The number of methylated regions combined with single cytosine methylation level distributions in each subset was applied to determine methylated seeds. These methylated three-base NCN seeds were then extended from both directions to identify potential motif combinations. The bidirectional extension of every single potential motif was terminated after the percentage of the number of methylated motifs fell below a threshold of 80%. Sequence conservation analysis was performed using program WebLogo 3.4 (23). Finally, single cytosine methylation level distributions of each motif were drawn to determine the efficiency to which each cytosine was methylated in the original genomic DNA. Weighted methylation levels were performed as previously described (24).
SMRT sequencing and bioinformatic analysis of SMRT sequencing data
Genomic DNA samples were submitted to the University of California-Irvine Sequencing Core for sequencing on a Pacific Biosciences RS II instrument. Libraries of 5 kb were constructed and sequenced using P6-C4 chemistry. C. kristjanssonii was sequenced on two SMRT Cells yielding a coverage of ∼700. Data were analyzed using the SMRT Analysis version_2.3.0 and Motif Finder 1.3.1.
De novo assembly of C. kristjanssonii was generated using RS_HGAP_Assembly.3 Protocol and polished by Quiver consensus calling algorithm. This assembly served as the reference input for the base modification analysis using PacBio RS_Modification_and_Motif_Analysis Protocol as previously described at http://www.pacb.com/pdf/TN_Detecting_DNA_Base_Modifications.pdf. Modification.gff file was used as input for PacBio's Motif-Finder software 1.0.0.21 (https://github.com/PacificBiosciences/DevNet/wiki/Motiffinder) to cluster the sequence motifs. Minimum modification detection score of 50 was chosen based on the kinetic distribution observed in the data.
RESULTS
Coupling Tet oxidation with a modified bisulfite treatment procedure for 4mC-specific detection
In a previous study, treatment of model DNA with M. MvaI, a 4mC methyltransferase, resulted in roughly 55% modified cytosines displaying as C under standard bisulfite treatment conditions (17). In order to accurately and sensitively detect 4mC using a genome-wide approach it is critical to have both highly active Tet and an improved bisulfite treatment condition under which most 4mCs are resistant to deamination. The former can ensure almost complete conversion of 5mC to 5caC to eliminate interference from 5mC, whereas the latter can maximize the percentage of 4mC read as C after bisulfite treatment. In the previous application, we demonstrated that purified recombinant Tet is highly active and converts over 97% of 5mC to 5caC in mammalian genomic DNA under appropriate reaction conditions (20). As the readouts of both 5caC and 4mC in bisulfite sequencing are subject to change upon the adjustment of bisulfite treatment parameters, the ideal bisulfite treatment condition should meet the following two requirements to achieve optimal detection of 4mC: 1) mild enough to retain as much 4mC as C after treatment and amplification; 2) strong enough to convert most 5caC (generated from 5mC oxidation) to final readout of T after treatment and amplification to reduce false positives.
To quantify how much 4mC is still read as C after bisulfite treatment, we generated a 304 base-pair (bp) DNA sequence containing multiple 4mC sites by PCR amplification with N4-methyl-dCTP (4mdCTP) and used it as a model DNA applying to different conditions. In this case, every cytosine on the generated model DNA is nearly 100% of 4mC (except for the cytosines of PCR primers). As shown in Figure 1c, 43% of 4mC sites were identified as C after standard bisulfite treatment. To further reduce the deamination rate of 4mC, we changed the thermal cycling program in a standard bisulfite treatment from 98°C 10 min, 64°C 2.5 h to 98°C 10 min, 53°C 30 min, 8 cycles of 53°C 6 min and 37°C 30 min, whereas the concentration and the pH of bisulfite reagent remained unchanged. Under the optimized condition, the percentage of 4mC displayed as C increased from 43% to 55%. Introduction of the Tet oxidation step only slightly affected the ability to detect 4mC, which may be due to a weak demethylation activity of Tet towards 4mC (Supplementary Figure S1).
To test whether this optimized bisulfite treatment condition is also compatible with 5caC deamination and ascertain the capability of Tet converting 5mC to 5caC, we generated a model DNA with sequence identical to the lambda genome that contained all CpG sites as 5mC. Treatment of the 5-methylated CpG lambda DNA with Tet and measurement of the 5mC conversion rate to a final readout of T under standard bisulfite treatment conditions reveals highly efficient oxidation of 5mC as only ∼0.4% of 5mCs were read as C. Next, the same Tet-treated 5-methylated CpG lambda DNA substrate was subjected to the optimized bisulfite treatment condition and an ∼2% 5mC non-conversion rate was observed, revealing that the optimized bisulfite treatment condition did not significantly reduce the deamination rate of 5caC (Figure 1c).
Application of 4mC-TAB-seq and MethylC-seq to the Caldicellulosiruptor kristjanssonii genome
We next applied both 4mC-TAB-seq (Tet-treated, using the optimized bisulfite treatment condition) and MethylC-seq (untreated with Tet, using the optimized bisulfite treatment condition) to genomic DNA from C. kristjanssonii, a prokaryote that contains both 4mC and 5mC modifications at comparable levels according to LC–MS/MS results (Figure 2d). Caldicellulosiruptor is a genus of extremely thermophilic bacteria which has the potential to directly convert biomass to biofuel and bioproducts (25). However, restriction of DNA from E. coli has been shown to be an absolute barrier to transformation of members of this genus to further improve their ability in generating biofuels with higher efficiency (25).
Figure 2.
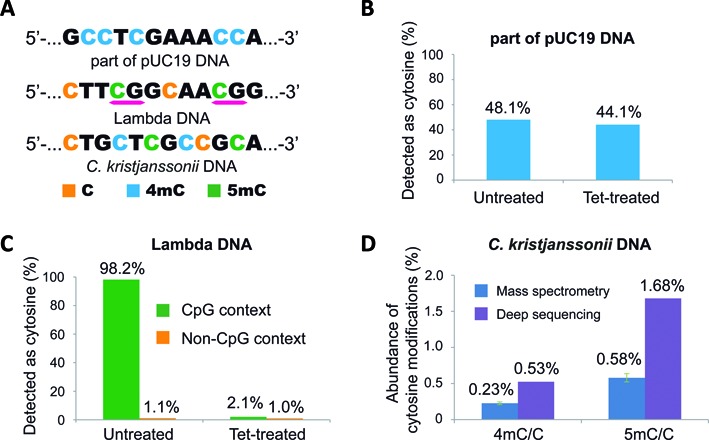
Data analysis of spike-in controls from MethylC-seq and 4mC-TAB-seq in the context of C. kristjanssonii genomic DNA. (A) Composition of pUC19 DNA, lambda DNA, and C. kristjanssonii genomic DNA. (B) The percentage of detected as cytosine reads on 4mC sites in untreated and Tet-treated samples. (C) The detected as cytosine reads percentage on unmodified cytosine sites (non-CpG context) and 5mC sites (CpG context) in untreated and Tet-treated samples. (D) Quantification of 4mC and 5mC in C. kristjanssonii genomic DNA, determined by LC-MS/MS and deep-sequencing respectively. Error bars indicate mean ± SD, n = 4.
The accurate detection of 4mC sites relies on three parameters: (i) the conversion rate of unmodified C to T in final sequencing reads; (ii) the conversion rate of 5mC to T in final sequencing reads; and (iii) the percentage of 4mC resistant to deamination. To assess all three parameters, spike-in control DNA samples bearing C/4mC/5mC were added to C. kristjanssonii genomic DNA prior to treatment. The 4mC control is generated by PCR amplification with 4mdCTP using part of the pUC19 vector (∼1 kb) as a template, whereas the C/5mC control is generated by treating unmethylated lambda DNA with the CpG methyltransferase (M. SssI) (Figure 2a). The lambda DNA therefore contains methylated 5mC in the CpG context and unmethylated non-CpG, which is used to measure the efficiency of the bisulfite-mediated conversion and Tet oxidation. In accordance with the model test results, over 40% of 4mC sites on the pUC19 vector were read as C in both untreated and Tet-treated samples (Figure 2b). In all CpG sites of lambda DNA, 98.2% of them were read as C in the untreated sample, whereas only 2.1% of them remained as C after being treated with Tet, indicating high fidelity of both the M. SssI and Tet enzymatic reactions (Figure 2c). Furthermore, unmodified cytosines as a result of non-conversion by bisulfite treatment were observed in both untreated (1.1%) and Tet-treated (1.0%) samples at very low levels, indicating the optimized bisulfite treatment condition is strong enough to convert most unmodified cytosines to uracils and our modification of bisulfite treatment condition will not introduce extra difficulty in data analysis. The analysis of C/5mC/4mC spike-in controls all together confirmed the capability of the 4mC-TAB-seq method to differentiate 4mC from 5mC in genomic DNA. The summary mapping statistics for the C. kristjanssonii are described in Supplementary Table S1. The abundance of 4mC and 5mC in the C. kristjanssonii genome revealed by 4mC-TAB-seq also correlated well with the quantification results from LC–MS/MS, which further supported the reliability of the 4mC-TAB-seq method (Figure 2d).
Because a consistent percentage of 4mC is read as T in 4mC-TAB-seq, more accurate information regarding the methylation level at each 4mC site can also be obtained. To achieve this goal, we proceeded to generate a calibration curve using 76-mer probes that contain known fractions of 4mC (Supplementary Figure S2). This curve provides a solution to accurately calculate methylation levels at each 4mC site in genomic DNA once 4mC-containing motifs are identified.
Identification of 4mC- and 5mC-containing motifs in Caldicellulosiruptor kristjanssonii
To identify 4mC- and 5mC-containing motifs respectively from the data generated by MethylC-seq and 4mC-TAB-seq, a three-nucleotide (nt) seed method was used for both untreated and Tet-treated samples (see ‘Material and Methods’ section). First, nine distinct methylated motifs in total were identified in the untreated sample (MethylC-seq dataset) (Figures 3a and 4a). To evaluate the accuracy of each revealed motif, sequence conservation analysis was performed for the flanking two bases in both directions by calculating entropy dynamics at each position. The result indicated that for each individual motif, there is not an additional base in either flanking or more distant regions (up to 30 bases – Supplementary Figure S3) either upstream or downstream (Figures 3a and 4a). To further assess the accurate identification of each base modification within each motif, we traced back each motif distribution in the reference genome and the untreated sample. After removing low coverage and unmappable motifs, all nine motifs revealed sufficient genome-wide detection (>93%) (Figures 3b and 4b). Additionally, for each individual motif, over 91% of the detected motif sites were methylated. The high methylation rate of each motif confidently supports the accuracy of each revealed motif. If any additional bases were required for any of the detected motifs, either upstream or downstream, the methylation detection rate of the motif would drastically decrease from >90% depending on the base composition of the flanking regions of any given motif. Furthermore, in the untreated sample, a total of 22 200 cytosines were identified as methylated cytosines and 96.9% of these were associated with the nine detected methylated motifs. No conserved motif(s) was found within the remaining methylated cytosines, which likely indicates that these methylated cytosines are unconverted cytosines from the sodium bisulfite reaction, nucleotide polymorphisms between sequenced DNA and the reference assembly, or off-target sites of methyltransferases.
Figure 3.
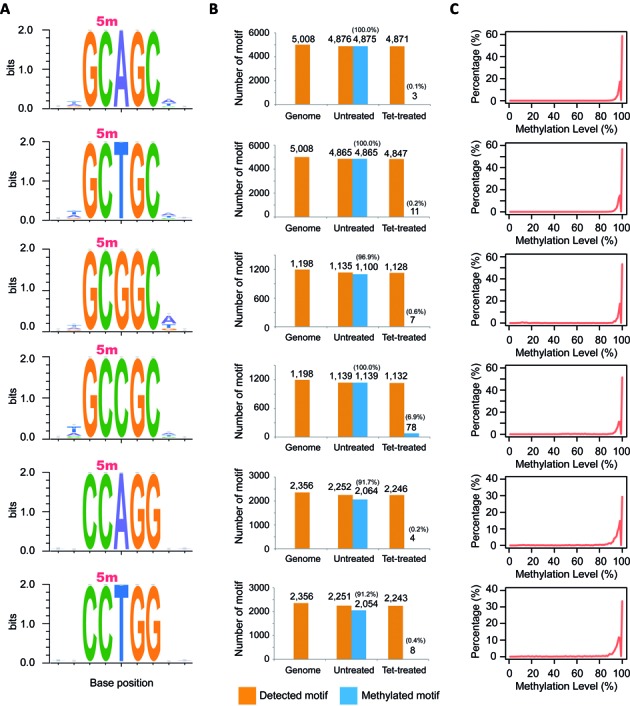
5mC-containing motif characterizations and distributions in C. kristjanssonii. (A) Motif sequence profile and sequence conservation analysis. Methylated cytosine is indicated with 5m. (B) Motif distributions in the reference genome, untreated- and Tet-treated samples. (C) Single cytosine methylation level distributions of each motif in the untreated sample.
Figure 4.
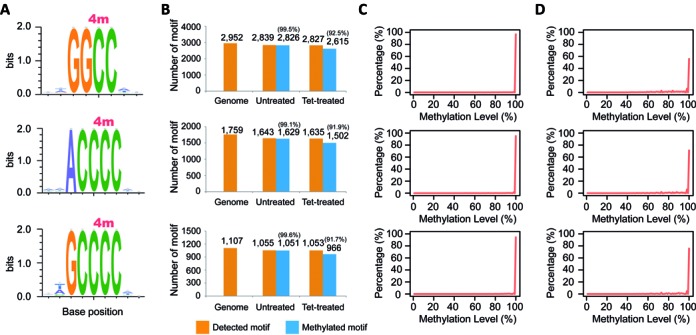
4mC-containing motif characterizations and distributions in C. kristjanssonii. (A) Motif sequence profile and sequence conservation analysis. Methylated cytosine is indicated with 4m. (B) Motif distributions in reference genome, untreated- and Tet-treated samples. (C) Single cytosine methylation level distributions of each motif in the untreated sample. (D) Single cytosine methylation level distributions of each motif in the Tet-treated sample. The levels in both samples were scaled by 4mC conversion rate generated from 76-mer probes (Supplementary Figure S2).
To determine whether these detected methylated sites are 4mC or 5mC, we further analyzed the Tet-treated sample (4mC-TAB-seq dataset) and observed that the number of methylated motif sites of the first six motifs, including GC*AGC, GC*TGC, GC*GGC, GC*CGC, CC*AGG, and CC*TGG (* indicates previous base is methylated), was substantially reduced to almost zero (Figure 3b), implying they are 5mC-containing motifs. In contrast, even in the presence of Tet treatment, the methylation status of the last three motifs (GGC*C, ACCC*C and GCCC*C) remained methylated (Figure 4b), implying that they are 4mC-containing motifs and are not affected by Tet oxidation. Again, we attribute the slightly reduced number of 4mC-containing motif sites upon Tet treatment to a weak demethylation activity of Tet towards 4mC. Lastly, methylation levels of 4mC-containing motifs were scaled by 4mC conversion rate according to the calibration curve generated with 76-mer probes. By plotting the methylation level distributions of each motif, we observed that all of the methylated cytosines in each motif exhibit strong methylation signals (Figures 3c and 4c, and d), implying the high efficiency of corresponding methyltransferase and also confirming the integrity of each motif.
To validate this novel method we performed SMRT sequencing on C. kristjanssonii genomic DNA, as SMRT is a completely orthogonal assay that does not rely on bisulfite treatment. Using this method the exact same three 4mC-containing motifs and modified positions (GGC*C, ACCC*C and GCCC*C) were identified with high confidence (>97%), which further supported the reliability of using the 4mC-TAB-seq method to identify 4mC sites in the genome (Supplementary Table S2). However, one suspected cytosine methylation motif, GC*NGCVGC, was reported, although with lower confidence (∼82%). Based on our sequencing results discussed above using 4mC-TAB-seq, this suspected motif should be a subset of 5mC-containing motifs (GC*NGC), which were almost fully oxidized in Tet-treated sample (Figure 3). Lastly, three 6mA-containing motifs (GA*TC, CGDA*G, and CCA*TTY) are also revealed by SMRT sequencing (Supplementary Table S2).
DISCUSSION
Following the discovery of 6mA and 5mC, 4mC was found to be the third DNA methylation base modification present in bacterial genomic DNA in 1980s (26). Similar to 5mC and 6mA, 4mC is also part of bacterial R-M systems and several 4mC methyltransferases and 4mC-sensitive restriction endonucleases have been identified in different strains (8,25–29). 4mC can exist in a variety of bacteria but is mostly prevalent in thermophiles (7,30). Although some of these thermophiles may be used for generating biofuels and bioproducts in high efficiency and at lower costs, the presence of 4mC serves as a major barrier to engineer their genomes. Attempts to transform either DNA in vitro methylated by purified methyltransferases or DNA in vivo methylated from E. coli to thermophiles fails due to the limited number of known 4mC methyltransferases and the lack of 4mC modifications in E. coli. Accurate identification of 4mC in bacterial genomes is critical to efficiently manipulate genomes containing this base modification as well as to identify novel 4mC methyltransferases. Restriction enzyme-based methods can only detect 4mC at specific motifs with prerequisite knowledge of 4mC-sensitive restriction enzymes required. Moreover, many 4mC-sensitive restriction enzymes are also sensitive to the presence of 5mC, which make it more difficult to reach conclusive assignments of target motifs (29). The recent developed SMRT technology is capable of detecting 4mC directly and is a suitable method for studying base modifications in bacteria that do not have a publicly available reference genome assembly.
Here, we report a rapid and accurate next-generation sequencing method to identify genome-wide locations of 4mC in any bacterial genome containing this base modification by modifying the widely used bisulfite sequencing method for 5mC. By coupling Tet-mediated oxidation of 5mC to 5caC with the optimized bisulfite treatment condition, only 4mC can be read as C. The significant difference between the readout of 4mC and 5mC after the treatment ensures the reliability and reproducibility of 4mC-TAB-seq in detecting 4mC and differentiating it from 5mC in genome. Moreover, the low cost of 4mC-TAB-seq makes it possible to rapidly map epigenomes of the thousands of bacterial genomes that have already been assembled in a scalable manner. For example, only ∼4 million 150 nt-length reads (represents >200X genome coverage of C. kristjanssonii) were required to map both 4mC and 5mC positions in the C. kristjanssonii genome, which is ∼$33 in sequencing reagents at the University of Georgia Genomics Facility. At this cost, thousands of bacterial epigenomes can be mapped for a significantly lower cost than could be performed with SMRT. Furthermore, only ∼100ng of total genomic DNA are required for library preparation which can be executed in a 96-well format as opposed to the >10 ug of genomic DNA requested by many core facilities preparing SMRT libraries.
Systematic application of 4mC-TAB-seq in combination with MethylC-seq can reveal accurate genomic locations of both 4mC and 5mC. The application of this technique to C. kristjanssonii has revealed three 4mC-containing motifs and six 5mC-containing motifs. Some 5mC-containing and 4mC-containing motifs are likely methylated by the same DNA methyltransferase (GC*AGC, GC*TGC, GC*GGC and GC*CGC; CC*AGG and CC*TGG; ACCC*C and GCCC*C), which will require further genetic experiments to confirm. Among the three 4mC-containing motifs, a 4mC methyltransferase (M. CbeI) responsible for GGC*C motif has been identified in C. bescii, another member of the same genus (8), and the presence of 4mC-containing GGC*C motif is also indicated in C. kristjanssonii and a few other members of Caldicellulosiruptor genus (31). Although in vitro modified DNA by purified M. CbeI can be transformed into C. bescii, the apparent transformation frequency is still very low, indicating the existence of additional active R-M systems in the genome. In accordance with these observations, 4mC-TAB-seq on C. kristjanssonii not only confirms the existence of 4mC-containing GGC*C motif, but also reveals two previously unknown 4mC-containing motifs (ACCC*C and GCCC*C), which are of high confidence, as both are nearly fully methylated (>99%) in detected sites across the whole genome. Their identification will assist in genetic engineering of this organism, which will advance the ability to use this species to convert biomass into biofuels. This new 4mC-specific sequencing strategy will also uncover and facilitate the investigations of 4mC in bacterial R-M systems, and potential roles of 4mC in the genomes of different organisms.
ACCESSION NUMBERS
The data generated for this study have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through accession number GSE63371.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Georgia [to R.J.S.]; National Institutes of Health [R01 HG006827 to C.H.]; The BioEnergy Science Center, a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science [to JW]; and M.Y. is a Howard Hughes Medical Institute predoctoral fellow; C.H. is an investigator of the Howard Hughes Medical Institute. Funding for open access charge: University of Georgia [to R.J.S.]
Conflict of interest statement. The TAB-seq technology has been patented by the University of Chicago Technology Transfer Office previously.
REFERENCES
- 1.Schweizer H. Bacterial genetics: past achievements, present state of the field, and future challenges. BioTechniques. 2008;44:633–634. doi: 10.2144/000112807. [DOI] [PubMed] [Google Scholar]
- 2.Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 3.Davis B.M., Chao M.C., Waldor M.K. Entering the era of bacterial epigenomics with single molecule real time DNA sequencing. Curr. Opin. Microbiol. 2013;16:192–198. doi: 10.1016/j.mib.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loenen W.A., Dryden D.T., Raleigh E.A., Wilson G.G., Murray N.E. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 2014;42:3–19. doi: 10.1093/nar/gkt990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lister R., O'Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrlich M., Wilson G.G., Kuo K.C., Gehrke C.W. N4-methylcytosine as a minor base in bacterial DNA. J. Bacteriol. 1987;169:939–943. doi: 10.1128/jb.169.3.939-943.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung D., Farkas J., Huddleston J.R., Olivar E., Westpheling J. Methylation by a unique alpha-class N4-cytosine methyltransferase is required for DNA transformation of Caldicellulosiruptor bescii DSM6725. PLoS One. 2012;7:e43844. doi: 10.1371/journal.pone.0043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C., Clark T.A., Korlach J., Turner S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ecker J.R. Zeroing in on DNA methylomes with no BS. Nat. Methods. 2010;7:435–437. doi: 10.1038/nmeth0610-435. [DOI] [PubMed] [Google Scholar]
- 11.Clark T.A., Lu X., Luong K., Dai Q., Boitano M., Turner S.W., He C., Korlach J. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 2013;11:4. doi: 10.1186/1741-7007-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz R.J., He Y., Valdes-Lopez O., Khan S.M., Joshi T., Urich M.A., Nery J.R., Diers B., Xu D., Stacey G., et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013;23:1663–1674. doi: 10.1101/gr.152538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahramanoglou C., Prieto A.I., Khedkar S., Haase B., Gupta A., Benes V., Fraser G.M., Luscombe N.M., Seshasayee A.S. Genomics of DNA cytosine methylation in Escherichia coli reveals its role in stationary phase transcription. Nat. Commun. 2012;3:886. doi: 10.1038/ncomms1878. [DOI] [PubMed] [Google Scholar]
- 15.Schmitz R.J., Schultz M.D., Lewsey M.G., O'Malley R.C., Urich M.A., Libiger O., Schork N.J., Ecker J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urich M.A., Nery J.R., Lister R., Schmitz R.J., Ecker J.R. MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat. Protoc. 2015;10:475–483. doi: 10.1038/nprot.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilkaitis G., Klimasauskas S. Bisulfite sequencing protocol displays both 5-methylcytosine and N4-methylcytosine. Anal. Biochem. 1999;271:116–119. doi: 10.1006/abio.1999.4116. [DOI] [PubMed] [Google Scholar]
- 18.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B., et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farkas J., Chung D., Cha M., Copeland J., Grayeski P., Westpheling J. Improved growth media and culture techniques for genetic analysis and assessment of biomass utilization by Caldicellulosiruptor bescii. J. Ind. Mcrobiol. Biotechnol. 2013;40:41–49. doi: 10.1007/s10295-012-1202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu M., Hon G.C., Szulwach K.E., Song C.X., Jin P., Ren B., He C. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat. Protoc. 2012;7:2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz M.D., Schmitz R.J., Ecker J.R. ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 2012;28:583–585. doi: 10.1016/j.tig.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung D., Farkas J., Westpheling J. Overcoming restriction as a barrier to DNA transformation in Caldicellulosiruptor species results in efficient marker replacement. Biotechnol. Biofuels. 2013;6:82. doi: 10.1186/1754-6834-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janulaitis A., Klimasauskas S., Petrusyte M., Butkus V. Cytosine modification in DNA by BcnI methylase yields N4-methylcytosine. FEBS Lett. 1983;161:131–134. doi: 10.1016/0014-5793(83)80745-5. [DOI] [PubMed] [Google Scholar]
- 27.Klimasauskas S., Steponaviciene D., Maneliene Z., Petrusyte M., Butkus V., Janulaitis A. M.Smal is an N4-methylcytosine specific DNA-methylase. Nucleic Acids Res. 1990;18:6607–6609. doi: 10.1093/nar/18.22.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindstrom W.M. Jr, Malygin E.G., Ovechkina L.G., Zinoviev V.V., Reich N.O. Functional analysis of BamHI DNA cytosine-N4 methyltransferase. J Mol. Biol. 2003;325:711–720. doi: 10.1016/s0022-2836(02)01282-2. [DOI] [PubMed] [Google Scholar]
- 29.Butkus V., Petrauskiene L., Maneliene Z., Klimasauskas S., Laucys V., Janulaitis A. Cleavage of methylated CCCGGG sequences containing either N4-methylcytosine or 5-methylcytosine with MspI, HpaII, SmaI, XmaI and Cfr9I restriction endonucleases. Nucleic Acids Res. 1987;15:7091–7102. doi: 10.1093/nar/15.17.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrlich M., Gama-Sosa M.A., Carreira L.H., Ljungdahl L.G., Kuo K.C., Gehrke C.W. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985;13:1399–1412. doi: 10.1093/nar/13.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung D.H., Huddleston J.R., Farkas J., Westpheling J. Identification and characterization of CbeI, a novel thermostable restriction enzyme from Caldicellulosiruptor bescii DSM 6725 and a member of a new subfamily of HaeIII-like enzymes. J. Ind. Microbiol. Biotechnol. 2011;38:1867–1877. doi: 10.1007/s10295-011-0976-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


