Abstract
Silent information regulator 2 (Sir2), the founding member of the conserved sirtuin family of NAD+-dependent histone deacetylase, regulates several physiological processes including genome stability, gene silencing, metabolism and life span in yeast. Within the nucleus, Sir2 is associated with telomere clusters in the nuclear periphery and rDNA in the nucleolus and regulates gene silencing at these genomic sites. How distribution of Sir2 between telomere and rDNA is regulated is not known. Here we show that Sir2 is sumoylated and this modification modulates the intra-nuclear distribution of Sir2. We identify Siz2 as the key SUMO ligase and show that multiple lysines in Sir2 are subject to this sumoylation activity. Mutating K215 alone counteracts the inhibitory effect of Siz2 on telomeric silencing. SUMO modification of Sir2 impairs interaction with Sir4 but not Net1 and, furthermore, SUMO modified Sir2 shows predominant nucleolar localization. Our findings demonstrate that sumoylation of Sir2 modulates distribution between telomeres and rDNA and this is likely to have implications for Sir2 function in other loci as well.
INTRODUCTION
In the budding yeast Saccharomyces cerevisiae, transcriptionally silent heterochromatin-like regions are found at the telomeres, the silent mating type loci HMR and HML and also at the rDNA locus that encodes rRNA. Establishment and maintenance of silent chromatin involves interactions between the cis acting elements present at these loci, the trans-acting proteins and the nucleosomes. Silent Information Regulator proteins (SIR) are central to this process and are structural components of silent chromatin found at telomeres and HM loci. At the telomeres, they are assembled in a sequential manner with initial recruitment of Sir4 to these sites via interaction with Rap1 that binds to telomeric repeats and Yku70/80 heterodimer that binds to the telomere ends. Sir4 recruits Sir3 and Sir2 via protein-protein interactions (reviewed in (1,2)). Sir2 is a conserved NAD+-dependent histone deacetylase that deacetylates histone tails, primarily, H4 lysine 16 (K16) and additionally K9, K14 and H3K56 (3,4). Sir3 and Sir4 bind to the deacetylated nucleosomes and this complex recruits more Sir2 that further deacetylates adjacent nucleosomes leading to spreading of silent chromatin. This mechanism of heterochromatin establishment is conserved between HM loci and telomeres (5). The silencing of the rDNA locus does not require Sir3 and Sir4 but is dependent on Sir2, although the exact structural components of this chromatin are less well understood (6–8).
Sir2 family of histone deacetylases is conserved across eukaryotes and is involved in deacetylating several substrates. Sir2 in S. cerevisiae, apart from being an essential component of silent chromatin at all the three loci, also has roles in multiple other physiological processes, including aging, stress response, DNA repair and replication (9,10). At the rDNA, Sir2 is required to prevent recombination of the repeats of rDNA and thus maintain genome stability (11,12). Sir2 prevents premature aging through the calorie-restriction pathway. This anti-aging effect of Sir2 is conserved across species including, yeast, Drosophila, C. elegans and mammals (13–15). In yeast, Sir2 also regulates aging by promoting the retention of damaged mitochondria in mother cells rather than transporting them into daughter cells (16). In addition, it also maintains protein homeostasis (17) and functions at origins of replication (18,19). In many of these processes, the substrates of Sir2 have not been elucidated. In particular, how Sir2 is distributed in unperturbed cells, how that is altered under stress or other perturbations or during cell cycle is not known.
Sumoylation of proteins is a form of post-translational modification wherein a SUMO (small ubiquitin like protein) moiety is added to the epsilon amino groups of lysine of target proteins. SUMO, encoded by a single SMT3 locus in S. cerevisiae, is essential and is produced as a pre-protein that is proteolytically cleaved to yield the active SUMO. In a three-step process reminiscent of ubiquitination, SUMO is first activated by a heterodimer encoded by the AOS1/UBA1 loci. The activated SUMO is conjugated to a E2-SUMO ligase, Ubc9, and then transferred to target proteins by E3 SUMO ligases (20,21). As there is only one known E2 ligase, E3 ligases are proposed to confer substrate specificity. The S. cerevisiae genome encodes four SUMO ligases, namely, Siz1, Siz2, Mms21 and Zip3 (22). Siz1, Siz2 and Mms21 sumoylate proteins involved in several functions like replication, repair, recombination, transcription, RNA processing, translation, organelle transport etc. (23,24) while role of Zip3 seems to be restricted to meiosis. Several key substrates have been reported for these SUMO ligases. In addition, sumoylation is reversible by the action of two desumoylating enzymes encoded by ULP1 and ULP2 loci (25) providing a dynamic means of altering target protein activity. Sumoylation alters the property of the target protein by influencing its interaction with other proteins or its subcellular localization. Protein homeostasis is also affected by sumoylation through a specialized pathway where sumoylated proteins are recognized by specific SUMO-dependent ubiquitin ligases that target these sumoylated proteins for degradation (26).
In S. cerevisiae, sumoylation has been implicated in gene silencing. Deletion of the desumoylating enzyme Ulp2 or the SUMO-dependent ubiquitin ligase, Slx5, leads to loss in telomere position effect (TPE) (27). In both mutants, sumoylated proteins accumulate and interestingly, elevating Sir2 dosage could overcome these defects, indirectly implicating Sir2 involvement. Recently, a study showed that loss in the catalytic activity of Mms21 led to reduction in telomeric position effect (28). In our earlier work, we had shown that elevated doses of Siz2 disrupt telomeric silencing and that this phenotype is exacerbated by deletion of ESC1, encoding a protein associated with the inner nuclear membrane, and involved in localizing desumoylating enzyme, Ulp1 to the nuclear periphery (29). In this work we investigated if Sir2 was a target of sumoylation. We show here that Sir2 is sumoylated by the SUMO ligase Siz2 and the sumoylated Sir2 is compromised in its interaction with Sir4. We also identify the sumoylation sites on Sir2 and discuss the implications of the locus specific effect produced by sumoylation of Sir2.
MATERIALS AND METHODS
Yeast methods
Standard yeast manipulation methods were followed. Strains used in the study are listed in Table 1; majority of strains were derived from W303 (30). All knockouts described here are full-length open reading frame replacements unless otherwise stated. Gene silencing assays were performed by spotting 5 μl of 10-fold serially diluted cultures of strains on 5-FOA plates. Plates were incubated up to 48 h and photographed. Silencing was quantified by obtaining the ratio of colonies on test plate over total. Yeast two-hybrid assays were done in PJ69-4A (31) and Sir2 constructs were made in pGBDUC1; pGAD-SIR4 and pGAD-NET1 were obtained from David Shore. Standard protocols for recombinant DNA construction were followed. Elevated dosage (over 100-fold) of Siz2 was confirmed by RT-qPCR. Site directed mutagenesis of SIR2, carried on pRS415 and expressed from its own promoter and containing 3′ UTR, were created by KOD DNA polymerase and confirmed by sequencing. Expression of all Sir2 constructs was comparable to the chromosomally encoded Sir2. Recombination at rDNA was determined by measuring the loss of ADE2 gene inserted at the rDNA locus (32). Only half-sectored colonies that represent loss of ADE2 in the first division were considered.
Table 1. List of strains used in this study.
| Name | Description | Source/reference |
|---|---|---|
| KRY2 | W303-1A (leu2-3,112 his3-11,15 ura3-1 ade2-1 trp1-1 can1-100 rad5-535) MATa | (30) |
| KRY3 | W303-1B (leu2-3,112 his3-11,15 ura3-1 ade2-1 trp1-1 can1-100 rad5-535) MATα | (30) |
| KRY12 | KRY 3 except adh4::URA3-TelVII L MATα | (29) |
| KRY18 | KRY2 except sir2::KanMX | (50) |
| KRY19 | KRY3 except sir2::KanMX | (50) |
| KRY280 | PJ69-4A | (31) |
| KRY360 | KRY 3 except RDN1::mURA3-HIS3 hmr::TRP1 adh4::ADE2 Tel VIIL sir2::KanMX | (50) |
| KRY670 | 8XHis-SMT3::TRP1 Mat α (GA5347) | (52) |
| KRY 717 | KRY 758 except esc1::kanMX | This study |
| KRY758 | KRY670 except sir2:: kanMX Matα | This study |
| KRY788 | sir2::kanMXesc1::kanMXTel VIIL URA3 MATa | This study |
| KRY 790 | KRY670 except esc1:: kanMX Matα | This study |
| KRY793 | sir2::kanMX Tel VIIL URA3 MATa | This study |
| KRY 864 | sir2::kanMX esc1::kanMX Matα | This study |
| KRY868 | sir2::kanMX SIR4-Myc-KanMX Matα | This study |
| KRY 870 | ade-,can-,his-,leu-,ura-trp-,sir2::HIS3,NET1-myc::LEU2 | This study |
| KRY875 | -ade-,can-,his-,leu-,ura-trpNET1-myc::LEU2 | (53) |
| KRY878 | KRY 670 except siz2::HIS3 | This study |
| KRY883 | KRY 670 siz1::HIS3 MATα | This study |
| KRY1507 | KRY 670 mms21–11::KanMX MATα | This study |
| KRY1541 | MATa sir2::kanMX RDN1::ADE2 | This study |
SUMO pull down
To assess the sumoylation status of Sir2, yeast strain containing SMT3 tagged with 8X-HIS at the N-terminal under the control of the endogenous SMT3 promoter, a gift from Erica Johnson, was used (33). We confirmed that the tagged SMT3 expression levels were similar to untagged SMT3 by RT-qPCR. Pellet of 50 ml of 0.6 to 1 OD culture was flash frozen in liquid nitrogen. Cells were resuspended in 1ml of lysis buffer (1.85 N NaOH, 7.4% β-mercaptoethanol) and incubated on ice for 10 min. One millilter of 50% TCA was added to the suspension. The suspension was centrifuged at 13K rpm for 10 min at 4°C. Pellet was washed with ice-cold acetone and 1 ml of 6 M guanidinium hydrochloride, 50 mM sodium phosphate pH 8, 10 mM Tris–HCl (pH 8) was added and incubated at room temperature for 20 min. After incubation, it was centrifuged at 13K rpm at room temperature and supernatant collected and incubated with 40 μl of Ni-NTA beads overnight. Beads were pelleted down and washed with 8 M Urea, 50 mM sodium phosphate buffer pH 8, and 10 mM Tris (pH 8) thrice. Beads were boiled with 50 μl of 1× gel loading buffer containing 15 mM EDTA in boiling water bath for 5 min and supernatant was collected and 10 μl was loaded per lane for western blot.
Immunoprecipitation
Fifty milliliters of 1–1.5 OD culture was pelleted down and frozen in liquid nitrogen. One milliliter of lysis buffer (50 mM HEPES, 0.25% NP40, 500 mM sodium acetate, 5 mM magnesium acetate, 0.1 mM EDTA, 1 mM DTT, 5% glycerol) and 200 μl of 0.5 mm glass bead were added and sonicated in a bead beater with a 30 s pulse and 2 min hold (3×) followed by 13K rpm spin. To the supernatant, 0.1 μg of anti-myc antibody was added and incubated in ice for 2 h. Thirty microliters of proteinA Sepharose was then added and incubated overnight at 4°C. Beads were washed with low salt lysis buffer (50 mM HEPES, 0.25% NP40, 150 mM sodium acetate, 5 mM magnesium acetate, 5% glycerol, 1 mM DTT, 1 mM PMSF) and boiled in 60 μl of 1× Laemmli buffer at 65°C for 10 min and 10 μl of the supernatant was separated in SDS-PAGE gel for western blot. Antibodies were from AbCam (myc) and Santa Cruz (Sir2).
Immunofluorescence
Immunofluorescence was performed as described in (29,34). Briefly, sir2Δ strains carrying appropriate plasmids were fixed in formaldehyde followed by spheroplast preparation. Spheroplasts were adhered to poly-lysine coated glass slides, permeabilized with methanol and acetone and blocked with BSA. This was followed by incubation with Sir2 antibodies from Santa Cruz; anti- Nsp1or Nop1 or myc antibodies from AbCam and appropriate Alexa Flour™ conjugated secondary antibodies. Images were taken on a Zeiss Axioscope A1 microscope equipped with an AxioCam camera and processed using Zen software.
Chromatin immunoprecipitation
At least three independent chromatin immunoprecipitation (ChIP) experiments were done for each strain as described previously (29). Briefly, 1.5 to 1.0 OD600 of 50 ml culture of cells were cross-linked with 1% formaldehyde for 10 min and then quenched with 3.4 ml of 2 M glycine. The cells were then pelleted and washed with ice-cold Tris-buffered saline (TBS). Cells were lysed in 800 μl of ice-cold lysis buffer with protease inhibitor (0.1% deoxycholic acid, 1 mM EDTA, 50 mM HEPES–KOH [pH 7.5], 140 mM NaCl, 1% Triton X-100) by addition of an equal volume of glass beads and vortex at maximum speed for 20 min at 4°C. Lysate was sonicated to shear the chromatin to an average length of 200–800 bp. Fifty microliters of sample was taken in a fresh tube and used as input DNA. Samples were incubated overnight with primary antibody at 4°C with constant rotation. Fifty microliters of protein A-DynaMag beads was added to the chromatin-antibody mixture and incubated for 2 h at 4°C with constant rotation. Protein A-DynaMag beads were washed with 1 ml each of lysis buffer, lysis-500 buffer, LiCl-detergent solution and TBS buffer. Chromatin immunoprecipitate was eluted first with 100 μl of 1% SDS in Tris-EDTA and then with 150 μl of 0.67% SDS in TE buffer by incubation at 65°C for 10 min. DNA from bound and unbound chromatin (input sample) was purified by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation after RNAse and Proteinase K digestion. DNA from ChIP experiments was analyzed by real-time PCR using Sybr green master mix (Applied Biosystems) on an Applied Biosystems 7500 HT fast real-time PCR system. The primers used are located in the sub telomeric region of chromosome VI-R and described in Xu et al. (35). Relative quantification of immunoprecipitated DNA was done based on the comparative Ct value method. The fold enrichment for each sample was calculated after normalization with internal control SPS2 and then used to compute fold change.
mRNA analysis
For mRNA analysis, total RNA was extracted from 10ml of yeast culture (1OD) by hot phenol method and c-DNA was constructed using high-capacity reverse transcription kit (Applied Biosystems) using 2 μg of RNA. This was used as template for real-time PCR using SYBR green PCR master mix (Applied Biosystems) and quantified. Primer sequences are available upon request.
Computational methods
The initial structures for the Sir2 and SUMO proteins were obtained from the protein databank (PDB IDs: 4IAO (36) and 1L2N (37) respectively). MODELLER program (38) was used to model the missing residues (39 residues between S99 and L562) of Sir2. Due to the unavailability of an appropriate template structure, homology modeling for the N-terminal parts of both Sir2 (1–98) and SUMO (1–20) proteins was not attempted. However, two glycine residues (G97 and G98) were modeled in the C-terminal end of SUMO. Both the proteins were subjected to atomistic molecular dynamics (MD) simulations in explicit solvent environment for 10 ns employing the CHARMM force field (39) using the NAMD program (40).
Three individual models for sumoylated Sir2 were generated by connecting the SUMO protein to the three different lysines (K106, K132 and K215) of Sir2 via isopeptide bonds with G98 of SUMO. Initially, protein–protein docking was performed between Sir2 and SUMO using the HADDOCK program (41). From the docking calculations, those protein–protein binary complexes where G98 of SUMO was close to any of the three lysines listed above were identified. Out of several possible structures, three different orientations of SUMO with respect to Sir2 were chosen for further modeling. The position of SUMO in reference to Sir2 was changed with utmost care to avoid non-physical overlaps between the two proteins so that the formation of isopeptide bond was possible. The three structures were then subjected to energy minimization followed by 10 ns MD simulations in explicit solvent environment. Each of the three systems involved approximately 140 000 numbers of atoms and the 50 ns MD simulations reported here took about 6 weeks on an 8-processor computer. With respect to extending each of the MD simulations on the solvated sumoylated Sir2 systems for a total of 25 ns, no change in the qualitative trends originally obtained are observed, which confirmed that adequate equilibrium has been attained.
RESULTS
Sir2 is sumoylated in vivo
Several whole SUMO proteome identification studies except one, have not detected Sir2 as sumoylated in vivo (42–45). Therefore, in order to test if Sir2 is sumoylated in vivo, we enriched whole cell extracts for sumoylated proteins (46). In a strain carrying 8xHis-tagged SMT3, we enriched for sumoylated proteins by passing the total protein extract through a Ni2+ column. The enriched extract was tested for the presence of sumoylated Sir2 on a western blot using antibodies to Sir2. As shown in Figure 1A, a slower migrating Sir2 band, corresponding to the size of monosumoylated Sir2, could be clearly detected in the enriched samples (lane 4). Neither the total extract (lane 1, 2) nor the extract from untagged strain enriched on Ni2+ column (lane 3) showed this additional band, suggesting that a fraction of Sir2 is sumoylated in vivo. Our previous results had shown that elevated SIZ2 dosage induced a silencing defect that was further exacerbated by the deletion of ESC1. We, therefore, next tested if either of these conditions affected Sir2 sumoylation. Extracts enriched for sumoylated proteins were prepared from both wild type and esc1Δ strains with or without SIZ2 and tested for Sir2 sumoylation. We found that while elevated dose of SIZ2 by itself increased the population of sumoylated Sir2 slightly (2 fold), esc1Δ produced an additional slow moving population of Sir2 (Figure 1B). Furthermore, the fraction of sumoylated Sir2 was higher when SIZ2 dosage was elevated in esc1Δ (2.5fold). We could also detect sumoylated Sir2 in cells with unmodified SMT3, albeit at very low efficiency (Supplementary Figure S1). These observations establish that Sir2 is sumoylated and that Siz2 and Esc1 modulate Sir2 sumoylation.
Figure 1.
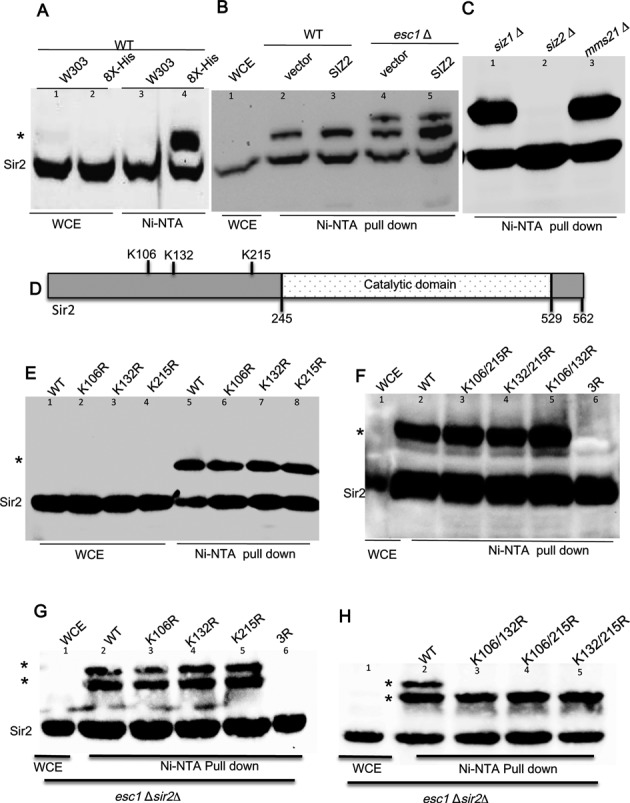
Protein extract from yeast cells expressing 8X-His tagged SUMO was pulled down with Ni-NTA beads and immunoblotted with anti-Sir2 antibody. (A) Sir2 is sumoylated by Siz2. Lanes 1 and 2 show whole cell extract (1/20) of the samples used for pull down. Lane 3 (untagged SUMO) and lane 4 show pull-down. * indicates the slow migrating sumoylated Sir2 band visible only in lane 4. (B) Hyper sumoylation of Sir2 upon esc1 deletion. NiNTA pull down and Sir2 immunoblots as in Figure 1a for wild type (KRY 670; lanes 2 and 3) and esc1Δ (KRY 790; lanes 4 and 5) with or without SIZ2. (C) Sir2 sumoylation is Siz2 dependent. Pull-down from siz1Δ, siz2Δ and mms21-11 shown in lanes 1–3 (WCE not shown). siz2 Δ abrogates sumoylation completely. (D) Sir2 is sumoylated at three different sites. A schematic representation of Sir2 showing the position of lysines that are the plausible SUMO sites. (E) Lanes 1–4 are whole cell extract (1/20) from sir2Δ (KRY758) expressing the indicated plasmids. Pull down for corresponding strains in lanes 5, 6, 7, 8. (F) Pull down for indicated double mutants and the triple mutant in sir2Δ. Slow migrating sumoylated Sir2 is absent in the triple mutant. (G and H) Pull down for esc1Δsir2Δ (KRY717) carrying indicated SIR2 mutants. Whole cell extract (1/20) from one of the samples only is shown in lane 1 in (F), (G), (H) for clarity.
S. cerevisiae genome encodes for four E3 SUMO ligases which sumoylate a differential set of substrates. In order to test which of the four E3 SUMO ligases sumoylated Sir2, we introduced deletions of SIZ1, SIZ2 or a partial deletion of MMS21 that abrogates its ligase activity (47) in the strain containing 8X His-tagged SMT3. SUMO protein enrichment and Sir2 western blots were done as described above. We found that the Sir2 was sumoylated almost exclusively by Siz2 (Figure 1C), because siz2Δ leads to complete loss of sumoylated Sir2 (lane 2). On the other hand, siz1Δ (lane 1) or the catalytically inactive mms21 (lane 3) did not alter the levels of sumoylated Sir2. These data clearly indicate that Sir2 is sumoylated primarily, if not exclusively, by Siz2 and esc1Δ leads to additional sumoylation of Sir2.
Multiple lysines in Sir2 are sumoylated
Having established that Sir2 is sumoylated in a Siz2 dependent manner, we investigated the functional consequences of this modification for Sir2. In order to address this, we first sought to identify the residues that are sumoylated in Sir2. Epsilon amino group of lysine present in a consensus sequence of ψ-K-X-E is the target for sumoylation in many cases although lysines at other sites have also been shown to be sumoylated (48). Using available online tools that predict potential sumoylation sites (49), three potential sites for Sir2 at target lysines, K106, K132 and K215 were identified in the conserved N-terminal domain (Figure 1D). Each of these lysines was replaced with arginine and three different SIR2 constructs in a CEN vector were generated. These were transformed into sir2Δ strains encoding his-tagged SMT3. After confirming that all of these mutants were stably expressed (Supplementary Figure S2A and B), we tested if these mutants were sumoylated by enrichment on Ni2+ column and western blot. As shown in Figure 1E, all three single point mutants were sumoylated. This suggests that the lysines identified may not be the targets or maybe all three could serve as targets. Because no other lysine in Sir2 appeared to be a consensus for sumoylation, we tested the second possibility by generating the double mutants, i.e. replacing any of the two lysine residues with arginine and also a triple mutant, where all three lysines were replaced with arginine. We observed that although all the double mutants were sumoylated, the triple mutant was not, suggesting that these are the three lysine residues subjected to sumoylation in vivo (Figure 1F). Based on our observation that esc1Δ induced additional sumoylation of Sir2, we asked if the mutants were also sumoylated at two sites. We found that although all the three single mutants (Figure 1G) produced the additionally sumoylated population like wild type and the triple mutant remained unsumoylated, the double mutants were only singly sumoylated in esc1Δ (Figure 1H) suggesting that additional sumoylation sites were not available. In order to further establish that the modification is sumoylation, we performed pull-downs for the double mutants in siz2Δ and found that in each case the slow moving Sir2 band was detected only when Siz2 was present (Supplementary Figure S2C). Together, these observations establish that the three residues identified in our study are sumoylated in vivo and are the sole sumoylation targets in Sir2. In addition, this analysis also confirms that esc1Δ induces multi-sumoylation, i.e. sumoylation at multiple sites rather than polysumoylation. If Sir2 were polysumoylated, then in double mutants, where only one site was available for sumoylation, it would have still produced a population of Sir2 with more than one SUMO conjugated.
K215R is refractory to inhibitory effects of perturbing sumoylation machinery
We next tested the ability of these mutants to establish silencing and also their response to elevated doses of SIZ2. All three single mutants are able to establish TPE under unperturbed conditions, suggesting that these single amino acid changes have not compromised the functions of Sir2 (Figure 2A). As expected Sir2 point mutant lacking catalytic activity (H364A) is completely deficient in silencing and the wild type establishes robust silencing (50). In contrast, in esc1Δ, where we have earlier established that there is a severe reduction in silencing with elevated doses of SIZ2, K215R could establish silencing (Figure 2B,2C). Both K106R and K132R are indistinguishable from wild type and show exacerbated loss in silencing under these conditions. To further confirm if K215R by itself was sufficient to resist the effects of Siz2, we tested the double mutants for silencing at telomeres. We first established that the double mutants complemented TPE under unperturbed conditions (Supplementary Figure S3) and then performed TPE assays in esc1Δ. The double mutant K106,132R, where the only possible sumoylation site is K215, was similar to wild type and lost silencing in the presence of elevated SIZ2 doses in esc1Δ (Figure 2D). But the two double mutants containing a K215R replacement resisted the exacerbation in silencing defect induced by Siz2. Thus, replacing lysine at 215 with arginine in all combinations protected cells from loss of silencing. Together these results confirm that lysine 215 is the key residue in Sir2 that is affected by elevated SIZ2 dosage in esc1Δ. The triple mutant is unable to complement Sir2 function at telomeres even under unperturbed conditions, although it produces stable protein (Supplementary Figures S2A and S3). This suggests that the triple mutant has defects that prevent silencing at telomeres independent of sumoylation and hence could not be used for functional analysis of Sir2 sumoylation.
Figure 2.
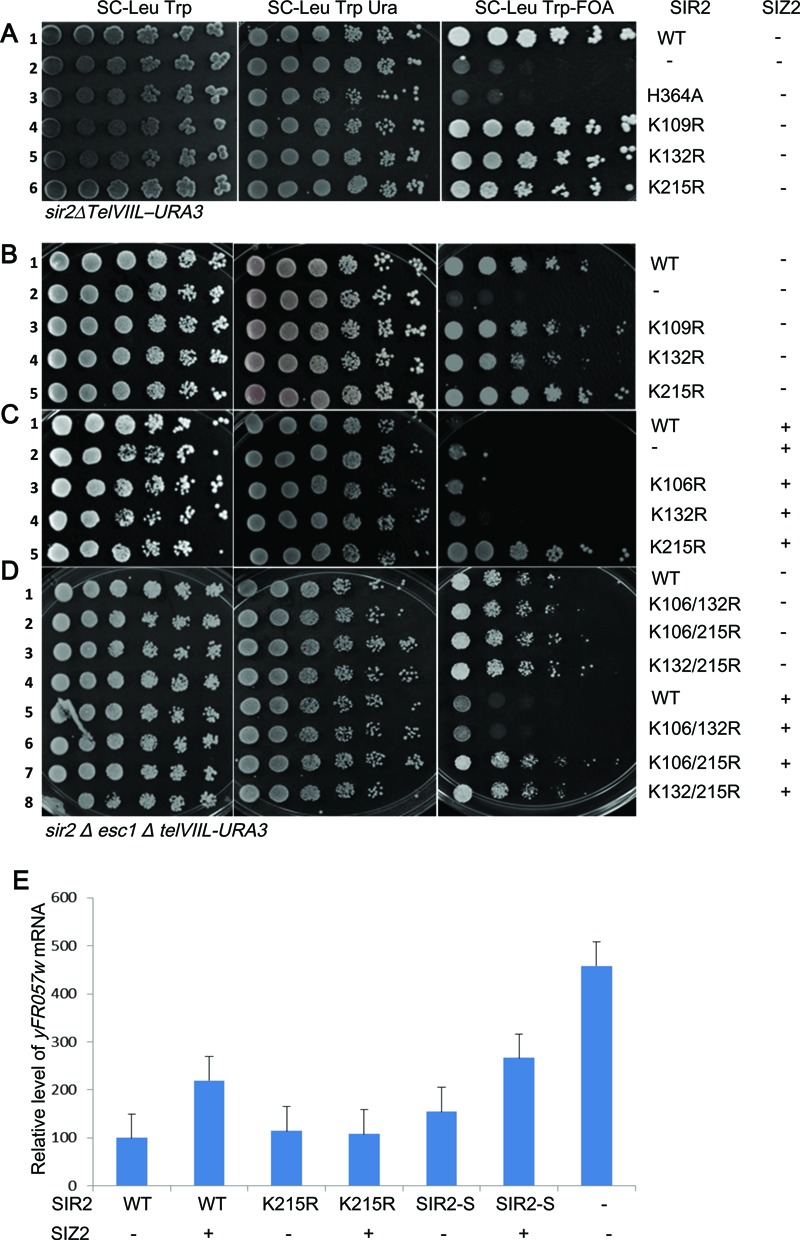
Sir2 K215R is refractory to exacerbation of silencing defect caused by elevated Siz2 dosage in esc1Δ. (A) KRY793 with either wild type or indicated mutants of Sir2 spotted on selective plates. Wild type Sir2 establishes silencing (row 1), empty vector and Sir2 catalytic mutant cannot establish silencing (rows 2 and 3). The point mutants (rows 4–6) are indistinguishable from wild type. (B) KRY 788 with either wild type or indicated plasmids along with either empty vector (B) or SIZ2 (C). Wild type and point mutants grow on 5-FOA plate in the absence of SIZ2 (rows 1, 3, 4 and 5). In (C), only K215R grows on FOA. (D) KRY788 with either wild type or indicated double mutant plasmids along with either empty vector or SIZ2. Wild type and point mutants grow on 5-FOA plate in the absence of SIZ2 (rows 1–4). Upon elevating SIZ2 dosage, WT and K106/132R are unable to establish TPE but K106/215R and K132/215R are able to. (E) Transcription of native silent loci. Transcription of yFR057w was quantified by reverse transcription and real-time PCR analysis for same strains as above. sir2Δesc1Δ with indicated 2micron plasmids are shown on X axis (1-SIR2+vector; 2-SIR2+SIZ2; 3-K215R+vector; 4-K215R+SIZ2; 5-SIR2-SMT3+vector; 6-SIR2-SMT3+SIZ2; 7-sir2Δ). Signals were normalized against actin as control. Results represent an average of at least three experiments, and error bars denote standard deviations. Expression increased both in WT and Sir2–SUMO but not in Sir2–K215R when SIZ2 dosage was elevated.
In order to further confirm that the growth phenotypes observed on 5-FOA plates were a true reflection of silencing, we also measured the expression of the endogenous locus, yFR057w, known to be silenced in a Sir protein dependent manner and was sensitive to elevated doses of SIZ2 (29,51). yFR057w (Figure 2E) was also refractive to loss of silencing induced by SIZ2 when transformed with Sir2-K215R mirroring the FOA assay. These results strongly suggest that K215 is the critical residue that influences Siz2-dependent changes in telomere position effect.
Sir2–SUMO is less effective for TPE but is competent for rDNA silencing
The results shown above establish Sir2 as a key target of Siz2 in modulating TPE. Since Siz2 has multiple substrates in the silencing complex, including Sir4, Yku70 and Yku80 and since only a fraction of the total protein is sumoylated at any given instance we decided to directly test the consequences of sumoylation of Sir2. Therefore, we made a recombinant Sir2 with SUMO fused to the C-terminus similar to the Yku80–SUMO reported (52). After confirming the robust expression of this fusion protein (Supplementary Figure S2A and B), we tested its effect on TPE. We found that this fusion protein was partially compromised for TPE under unperturbed conditions (Figure 3A, row 3) while a similar fusion with GFP was indistinguishable from wild type. This effect was further enhanced by the increased dosage of SIZ2 (row 6), probably, by additionally sumoylating the consensus lysine residues (Supplementary Figure S2A). Importantly, unlike wild type Sir2, Sir2–SUMO was less effective in silencing in the esc1Δ strain even without additional copies of SIZ2 (Figure 3B, row 3). As expected, both wild type and Sir2–SUMO were completely ineffective in establishing TPE when SIZ2 dosage was elevated in an esc1Δ mutant.
Figure 3.
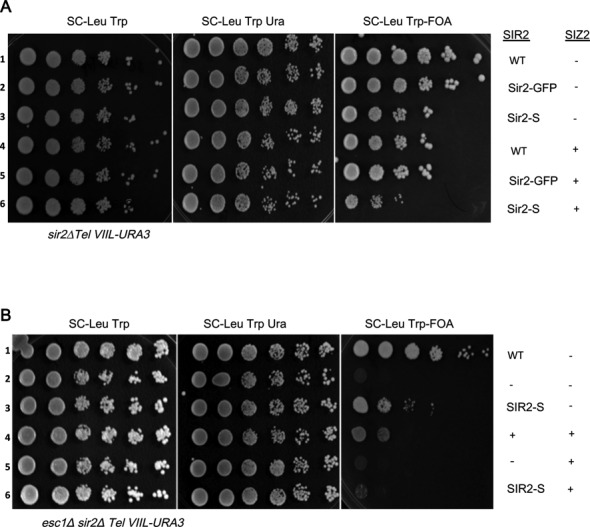
TPE in strains with Sir2–SUMO. (A) KRY793 and KRY788 (B) strains were tested for TPE with Sir2–SUMO with and without elevated doses of SIZ2. Sir2–SUMO shows reduced TPE even in WT (KRY793; compare row 1 to row 3 in panel A) and further reduction upon additional copies of SIZ2 (in panel A, row 6). In esc1Δ (KRY788) Sir2–SUMO has reduced TPE even without additional SIZ2 (panel B, row 3) and loses TPE with elevated SIZ2 dosage (panel B, row 6).
Sir2–SUMO fusion was compromised for TPE and this could be due to a reduced functionality of Sir2 fusion protein. In the rDNA locus, Sir2 is required for silencing of genes transcribed by RNA polymerase II. This function of Sir2 is not altered when SIZ2 dosage is elevated either in wild type or when esc1 is deleted (29). Therefore, we tested the Sir2–SUMO and all point mutants described above in rDNA silencing. These plasmids were introduced into a sir2Δ strain carrying URA3 at the rDNA locus. We then tested for growth on plates containing 5-FOA. All Sir2 constructs, including the point mutants and Sir2–SUMO, were competent to establish silencing at the rDNA locus (Figure 4A and B). This observation suggests that some specific property of Sir2, essential for telomeric silencing, was affected upon addition of SUMO, leading to reduced silencing at telomeres. Furthermore, Sir2 is required to repress recombination between the rDNA repeats and sir2Δ have elevated levels of recombination. We measured the recombination frequencies in a strain containing ADE2 at the rDNA by counting the half-sectored colonies, which arise due to loss of ADE2 by recombination (Figure 4B). We found that compared to wild type Sir2, Sir2–SUMO showed enhanced protection from recombination and recombination dropped to undetectable levels; and interestingly, Sir2K215R showed marginally enhanced levels of recombination. These results together, confirm that addition of SUMO is specifically detrimental to telomere position effect but not to rDNA functions of SIR2. This also suggests that the key catalytic activity of Sir2, i.e. deacetylaion of H4K16, is unlikely to be affected by the fusion as that activity is required for both telomeric and rDNA silencing. We further confirmed this by performing western blots and ChIP experiments for histone H4K16 at the telomeres (Supplementary Figure S4A and B). Total acetylated histone population is not altered and similar amounts of acetylated H4K16 is present in WT, Sir2–SUMO and Sir2K215R, whereas the sir2Δ has much higher levels of H4K16, indicating that these modifications are not detrimental to the catalytic activity of Sir2.
Figure 4.
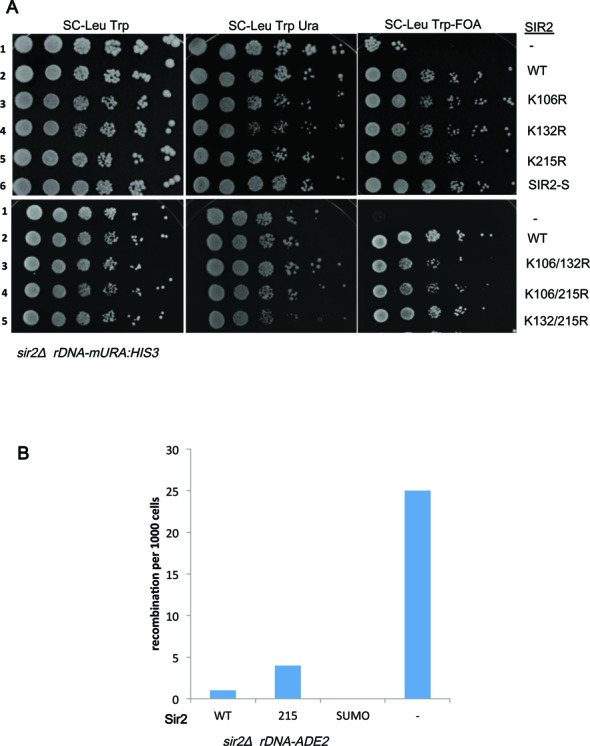
Sir2 mutants are functional in rDNA silencing and recombination. (A) KRY360 was transformed with either vector or Sir2 wild type or Sir2 mutant plasmids along with SIZ2 and tested for growth on 5-FOA to measure silencing of rDNA locus. Wild type Sir2 and all point mutants of Sir2 restore silencing at the rDNA. (B) KRY 1541 was transformed with plasmid encoding either WT Sir2 or Sir2K215R or Sir2–SUMO and recombination frequencies were estimated. Only red/white half sectors were counted.
Sir2–SUMO does not interact with Sir4 and localizes predominantly to the nucleolus
One key difference between silencing at the telomeres and rDNA is the requirement for different proteins that target Sir2 to these loci. Sir4 recruits Sir2 to telomeres and Net1 recruits Sir2 to rDNA. Therefore, we asked if the interaction of Sir2 with these proteins is influenced by sumoylation status of Sir2. To do these experiments we generated sir2Δ in strains carrying a myc epitope tagged Sir4 or Net1 (53). After introducing different Sir2 plasmids, namely wild type, K215R or Sir2–SUMO, protein extracts were made and immunoprecipitated using antibodies to the myc epitope. The immunoprecipitates were tested for the co-precipitation of Sir2. As shown in Figure 5(A and B), all constructs stably expressed Sir2 (lanes 1–3) and Sir2–SUMO appears clearly as a higher molecular weight protein compared to native Sir2. When immunoprecipitation was done in extracts from strains expressing Sir4-myc, we found that Sir2 was co-immunoprecipitated as expected. However, in case of Sir2–SUMO, we found only the lower molecular weight form, that corresponds to the non-sumoylated form, appearing (Figure 5A, lane 6). The flow-through samples has the Sir2–SUMO (lane 9) ruling out the possibility of complete cleavage of Sir2–SUMO. This is particularly significant because the sole Sir2 in these cells is the Sir2–SUMO. It appears that the Sir2–SUMO undergoes some cleavage and the cleaved, free Sir2 is able to interact with Sir4. This cleavage is evident in western blots from whole cell extracts in Sir2–SUMO lanes (Supplementary Figure S2A). This is probably why we see some silencing even in Sir2–SUMO strains. The Net1-myc, in contrast, was able to pull down sumoylated Sir2 from the Sir2–SUMO cells efficiently (Figure 5B). These results suggest that in the same cells, Net1 was able to stably interact with Sir2–SUMO but Sir4 was not. Taken together, these results provide strong evidence that Sir4 interacts with the non-sumoylated Sir2 population while Net1 interacts with both free and sumoylated Sir2.
Figure 5.
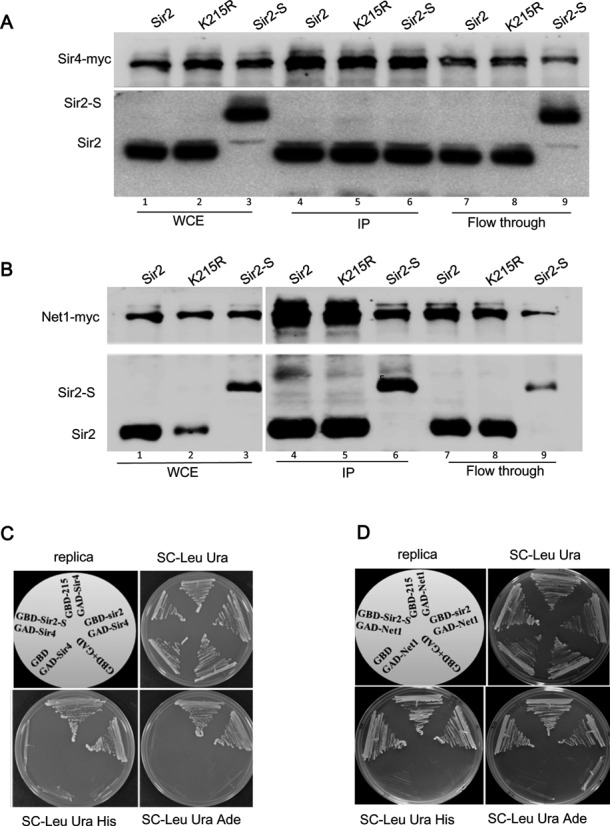
Interaction of Sir2 with Sir4 and Net1. (A) Protein extracts from KRY868 containing myc-tagged Sir4 or KRY875 containing myc-tagged Net1 (B) with indicated plasmids were immunoprecipitated with anti-myc antibodies and immunoblotted with myc or Sir2 antibody to test co-immunoprecipitation of Sir2 with Sir4 and Net1. Whole cell extracts and flow though are shown for comparison. (C) Interaction of Sir4 with Sir2 in yeast 2-hybrid assay. Gbd fusion of Sir2, Sir2K215R and Sir2–SUMO were cotransformed with either empty GAD vector or GAD vector encoding SIR4 (C) or Net1 (D). Growth on SC-Leu Ura indicates that both plasmids are present. Growth on SC-His and SC-Ade indicates interaction between the Gbd and Gad fusion plasmids.
Our observations discussed above indicated that sumoylated Sir2 is probably unable to interact with Sir4. Sir2 interaction with Sir4 and Net1 has been demonstrated by yeast two-hybrid previously (50). In order to test if Sir2–SUMO interacted with Sir4 and Net1 directly, we made Gbd fusions of Sir2K215R and Sir2–SUMO and asked if they could interact with Sir4 in a yeast two-hybrid assay. All constructs expressed stably and we could demonstrate that both wild type and Sir2-K215R interacted with both Sir4 and Net1, seen as robust growth on plates lacking histidine and adenine. In contrast, the Sir2–SUMO, although interacted robustly with Net1, did not interact with Sir4 (Figure 5C and D). These results support our immunoprecipitation data and confirm that Sir2–SUMO is defective in interaction with Sir4.
Based on the interaction data obtained above, we hypothesized that sumoylated Sir2 is non-functional in TPE possibly due to defective recruitment to telomeres. We therefore, performed sub-cellular localization of Sir2 in sir2Δ strains expressing either wild type or sumoylated Sir2. Costaining with Sir4 was done to demarcate the telomeres and with Nop1 to indicate the nucleolus (Figure 6A and B). Unmodified Sir2 was found to localize to the nucleolus and telomeres as shown previously and seen here as multiple spots that colocalize either with Sir4 or Nop1 (29,54). Strikingly, we found that Sir2–SUMO was localized almost exclusively to the nucleolus as seen by spots colocalizing with Nop1, with very few cells showing additional telomere spots (<10%). These data show that Sir2–SUMO is not recruited to the telomeres.
Figure 6.
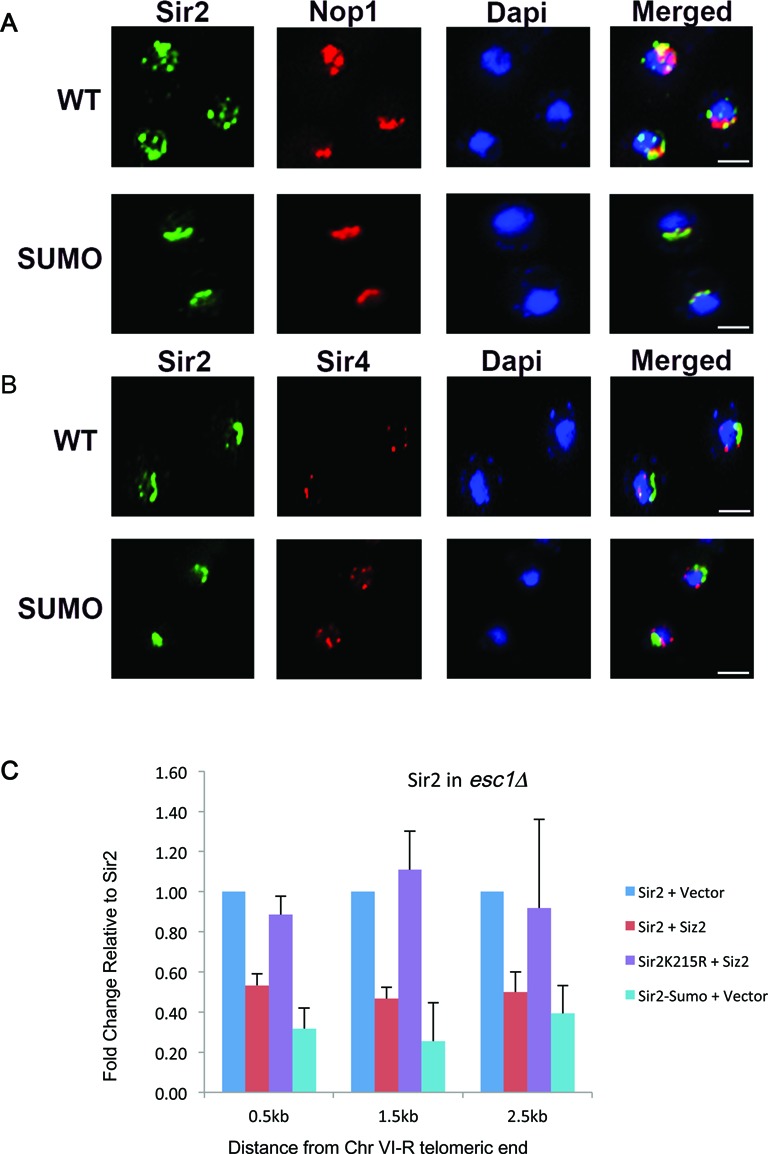
Sir2–SUMO is localized predominantly to the nucleolus. sir2Δ expressing wild type Sir2 or Sir2–SUMO was stained with antibodies to Sir2 along with nucleolar protein Nop1 (A) or telomeric protein Sir4 (B). WT Sir2 is localized to nucleolus and telomeres (multiple spots) but Sir2–SUMO is mainly localized to nucleolus. Dapi, in blue, indicates nucleus. Scale bar is 2 μm. (C) Sir2K215R remains associated with telomeres. Chromatin from the indicated transformants in sir2Δ esc1Δ was precipitated with Sir2 antibody. The fold change values for Sir2 + Siz2 relative to Sir2 + vector and Sir2K215R + Siz2 relative to Sir2K215R + vector are plotted. Sir2–SUMO relative to Sir2 is plotted. Error bars represent standard deviation. At all sites, Sir2K215R occupancy does not reduce upon elevated Siz2 dosage.
In our previous work, we had shown that upon elevation of Siz2 dosage, the levels of Sir2 diminished at the telomeres concomitant with loss in TPE in esc1Δ. Since Sir2K215R did not lose TPE upon elevation of Siz2 dosage, we asked if Sir2 levels at the telomeres were restored. We performed ChIP experiments as before and compared the occupancy of Sir2 and Sir2K215R when Siz2 dosage was elevated in esc1Δ (Figure 6C) at the sub-telomeric region of chromosome VIR. We confirmed that Sir2 levels diminished by half but importantly found that Sir2K215R levels were not altered under these conditions. We also tested the localization of Sir2–SUMO by ChIP and found that the levels of Sir2–SUMO at the telomeres in esc1Δ even without additional Siz2 was comparable to wild type Sir2 when Siz2 dosage was elevated in esc1Δ, reflecting the reduced TPE observed in these cells. These observations, taken together, suggest strongly that sumoylated Sir2 is not recruited to the telomeres leading to defective silencing at the telomeres.
Sumoylation of Sir2 at K215 may interfere with Sir4–Sir2 interactions
To obtain some insight into the possible differences in Sir4 interaction with unmodified Sir2 and sumoylated Sir2, as these SUMO sites fall within the N-terminal domain of Sir2 that interacts with Sir4, we modeled these interactions. The equilibrated structures from the last 2 ns of all the MD simulations (see Materials and Methods) were subjected to structural and energetic analyses. Firstly, the effect of sumoylation on the structure of Sir2 protein was examined by calculating the pairwise distances between the centers of masses of all residues (Supplementary Figure S5). Overall signatures of the four contact maps remain very similar to each other indicating that the structure of Sir2 remains unaltered in the presence and absence of SUMO. Similarly, the contact maps of the SUMO protein were calculated in the three sumoylated Sir2 models and were found to be very similar albeit marginal variations (Supplementary Figure S5).
The effect of sumoylation on Sir2–Sir4 binding was assessed by examining the interfacial residues between Sir2 and SUMO. The interaction energies between SUMO and Sir2 indicate significant non-bonded interactions (−96.9, −120.5 and −106.2 kcal/mol for sumoylations at K106, K132 and K215, respectively) between them in addition to both being covalently bound. These interfacial residues on Sir2 in turn were compared with the residues that have been shown to take part in Sir4 binding (36). The contact maps corresponding to the Sir2–SUMO interaction for the three sumoylated proteins are given in Figure 7A. The stretches of Sir2 residues that interact with Sir4 are marked by pink highlights in these plots. Existence of contact between the highlighted residues and the residues from SUMO indicate that these residues may not be available for Sir2–Sir4 interaction. The contact maps indicate that there are overlapping residues in all of them. However, the number of such overlapping residues is higher in case of sumoylation at K215 compared to the other two. To further quantify the availability of these residues for Sir4 interaction after sumoylation, the solvent accessible surface areas (SASA) were calculated. Since the approaching molecule for binding is the Sir4 protein, the usual probe radius of 1.4 Å (corresponding to the radius of water molecule) for the SASA calculation may not be appropriate. Hence, the SASA values were calculated using probes with several values of radii (2.57, 3.00, 3.12, 3.43 and 3.84 corresponding to the radii of the amino acids GLY, ASP, THR, LYS and TRP, respectively (55), and 5–10 Å with 1 Å increment) in addition to 1.4 Å. The average values of these SASA values of the Sir2 residues that have been shown to interact with Sir4 in unsumoylated and sumoylated Sir2 models are given in Figure 7B. Compared to the unsumoylated Sir2, these residues are less accessible in the sumoylated Sir2 proteins in general, consistent with the contact maps. Decrease in the accessibility of these residues is marginal in the Sir2 proteins sumoylated at K106 and K132 irrespective of the probe size. However, with respect to increase in the probe size, the accessibility of the Sir2–Sir4 interfacial residues drastically decreases in the Sir2 protein sumoylated at K215. This suggests possible adverse effect that sumoylation at K215 may have on Sir2-Sir4 binding. The structures of the sumoylated Sir2 proteins were subjected to further examination by modeling the Sir2-Sir4 interactions. The experimental structure of Sir2-Sir4 (incomplete; PDB ID: 4IAO (36)) binary complex was aligned on to the Sir2 part of the sumoylated proteins, and the resultant complexes (sumoylated Sir2–Sir4) were visually inspected for any possible non-physical contact between SUMO and Sir4. Consistent with the discussion above, marginal contact between SUMO and Sir4 are observed in Sir2 protein sumoylated at K106 and K132 (Figure 7C). Minimal rearrangement of few residues from Sir4 and/or SUMO is expected to facilitate sumoylated Sir2–Sir4 binding in both the cases. On the other hand, a large number of residues from both SUMO and Sir4 seem to interact with the same set of residues of Sir2 (Figure 7D) consistent with SASA calculations. Additionally, the Sir4 protein binds to Sir2 very close to the sumoylation site (K215). Hence, even a major rearrangement of the positions of the overlapping residues on SUMO and/or Sir4 is not expected to facilitate successful binding between Sir4 and Sir2 sumoylated at K215. Thus, the computational modeling and MD simulations strongly suggest that sumoylation of K215 interferes with Sir4 binding to Sir2.
Figure 7.
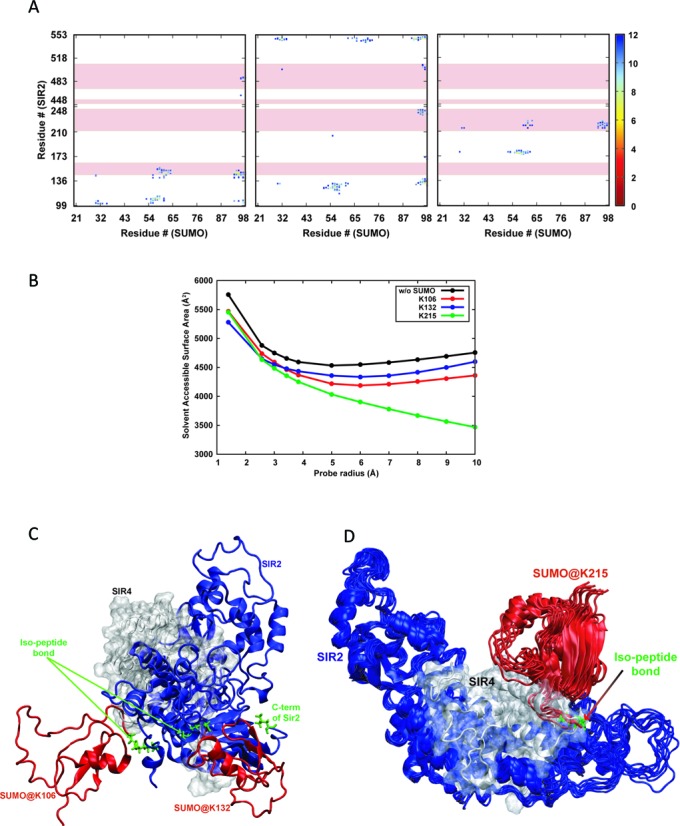
Molecular modeling of Sir2–Sir4 interaction. (A) Contact maps corresponding to Sir2–Sir4 interactions where the Sir2 protein is sumoylated at K106, K132 and K216. Those distances above 12 Å are not plotted for clarity. The pink highlights correspond to the residues on Sir2 that interact with Sir4. (B) Average solvent accessible surface areas of the Sir2 residues that take part in Sir4 binding in unsumoylated and sumoylated Sir2 with respect to the probe radius (see text). (C) Representation of the structures of sumoylated Sir2 at K106 and K132, and (D) a small ensemble of structures of sumoylated Sir2 at K215 and the experimental structure of Sir2–Sir4 binary complex aligned based on the Sir2 protein. Sir2, Sir4 and SUMO are depicted in blue, gray and red colors, respectively, and the isopeptide bonds along with the two residues are in green stick representation.
DISCUSSION
We have investigated the molecular basis for the regulation of silencing by sumoylation in yeast. Our earlier studies had established that elevated dosage of the SUMO ligase Siz2 disrupts silencing at the telomeres and HM loci but had no effect on rDNA silencing. We had also shown that Sir2 association with telomeric DNA was reduced in these cells. Therefore, we directly tested if Sir2 was the target of Siz2. For the first time we show that Sir2 is sumoylated in vivo by Siz2. Other SUMO ligases tested, Siz1 and Mms21, do not sumoylate Sir2. We also demonstrate that Sir2 interacts differentially with Sir4 and Net1 in a sumoylation-dependent manner and sumoylated Sir2 preferentially localizes to the nucleolus.
Although three sites, K106, K132, K215, can be sumoylated, apparently with equal efficiency, only one site is sumoylated at any given time as we detect mainly a singly sumoylated population. We cannot rule out the possibility that one of them is the preferred site and other sites are sumoylated only when that site is unavailable. Our mutant analyses show that K215R is able to robustly protect telomeric silencing from elevated doses of SIZ2 suggesting that this residue has a key role in this regulation. In support of this, Sir2K215R, in contrast to Sir2, is bound to telomeres even when Siz2 dosage is elevated in a esc1Δ. As under these conditions we see that a larger fraction of Sir2 is sumoylated at two sites, it reinforces the idea that protecting from sumoylation at K215R is key to TPE. Sir2–SUMO is defective in silencing at the telomeres, but competent for silencing at the rDNA and suppressing recombination, reinforcing the functionality of this molecule at the nucleolus. In addition, increasing the dose of SIZ2 leads to further reduction in telomeric silencing by Sir2–SUMO but does not impact rDNA silencing. This suggests that these amino acid changes and SUMO modification are unlikely to affect the deacetylase activity of Sir2 which is essential for all silencing activities of Sir2. We find that even when Sir2–SUMO is the sole Sir2 present in the cell, a fraction gets desumoylated and that it is this fraction that seems to interact with Sir4. Our modeling studies further support the notion that sumoylation of Sir2 at K215 interferes with Sir4 interaction. The loss in telomeric silencing, although evident at elevated dose of SIZ2 in wild type cells, is much more prominent in esc1Δ cells. Under these conditions we find that Sir2 is sumoylated at multiple sites. Based on these findings, we propose a model for sumoylation regulated telomeric silencing (Figure 8). Under unperturbed conditions, a fraction of Sir2 is sumoylated at one of the three positions identified and the fraction that is sumoylated at K215 does not interact with Sir4 and does not participate in telomeric silencing. When sumoylation is increased or when more Sir2 gets sumoylated, the population of Sir2 that is sumoylated at K215 increases considerably and is unable to interact with Sir4 and therefore, unable to establish silencing at the telomeres. Once Sir2 is sumoylated at multiple sites or when the fraction of sumoylated Sir2 increases, the desumoylating machinery is unable to cleave off all the SUMO residues to promote Sir4 interaction. In esc1Δ with increased SIZ2 dose, which produces such conditions, telomeric silencing is completely lost. In this context, ulp1Δ mutant could not be tested, as it is inviable.
Figure 8.
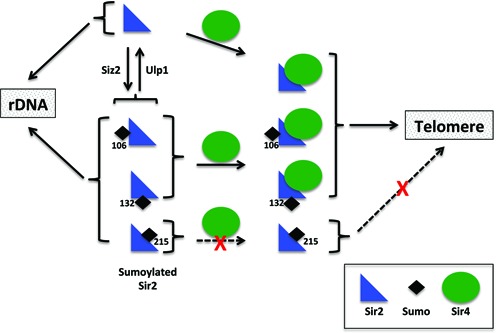
Proposed model for regulation of telomeric silencing by sumoylation. Sir2 is sumoylated by Siz2 and in unperturbed cells both unmodified and sumoylated Sir2 coexist. The fraction that is sumoylated at K215 is unable to interact with Sir4 and hence is not recruited to telomeres. Conditions that promote sumoylation, especially at K215, would shift the equilibrium towards decreased telomeric silencing.
Sir2 is a key histone deacetylase in yeast and regulates silencing at the telomeres, silent mating loci and rDNA. Apart from silencing, Sir2 has been implicated in multiple other processes including initiation of replication, repression of specific genes that are down-regulated during diauxic shift, protein homeostasis and transport of organelles to daughter cells. Many of these latter functions of Sir2 do not require Sir4 and it is not known how Sir2 is targeted to these loci. In the case of relatively well-studied functions of Sir2, namely telomeric and rDNA silencing, it has been demonstrated that there is an underlying competition for Sir2 between these loci and reduced recruitment to one enhances the recruitment to the other locus (56). Our study opens up a new avenue that post-translational modification, especially sumoylation, may lead to differential interaction of Sir2 with its partners leading to preferential recruitment to selected loci. This possibility is particularly relevant given that Ulp1, a SUMO protease, is associated with the nuclear membrane where telomeres are located in yeast and its localization is regulated by Esc1 and certain other nuclear pore complex proteins (57). We suggest that the desumoylation at the nuclear periphery by Ulp1 retains the ability of Sir2 to interact with Sir4 and remain in complex with Sir proteins. Sumoylation by Siz2 would release Sir2–SUMO from the telomere compartment, which could then be recruited to other targets including the nucleolus. In addition, others and we have shown that several proteins involved in telomere position effect, including, Yku70, Yku80, Sir4 and Sir3 are also sumoylated (29,52). We found that Yku80–SUMO and Yku70–SUMO fusions, shown to function in anchoring telomeres, had no effect on TPE with or without Siz2 (unpublished observation). However, how sumoylation of Sir4 would affect TPE and particularly its ability to recruit Sir2 and Sir3 remains to be investigated. It would be interesting to test how these multiple modifications influence one another and if there is any evidence for modification of several proteins in the pathway acting synergistically to confer both tunability and specificity (58). Given that Ulp1 and possibly other SUMO modification enzymes have dynamic localizations in stress conditions (59,60), the possibility of modifying Sir2 and other such critical proteins under these situations could be a quick and robust mechanism to respond to stress.
Supplementary Material
Acknowledgments
We thank D. Shore, E. Johnson, L. Pillus, X. Zhao, A. Amon and S. Gasser for strains and plasmids. We acknowledge Geetika Gupta for initiating work on modeling, R.K. Mishra for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Council for Scientific and Industrial Research (CSIR), Department of Biotechnology and Department of Science and Technology-FIST, India (to K.M.); University Grants Commission and CSIR, respectively (to A.H. and N.M.A.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kueng S., Oppikofer M., Gasser S.M. SIR proteins and the assembly of silent chromatin in budding yeast. Annu. Rev. Genet. 2013;47:275–306. doi: 10.1146/annurev-genet-021313-173730. [DOI] [PubMed] [Google Scholar]
- 2.Young T.J., Kirchmaier A.L. Cell cycle regulation of silent chromatin formation. Biochim. Biophys. Acta. 2013;1819:303–312. doi: 10.1016/j.bbagrm.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Shore D. The Sir2 protein family: a novel deacetylase for gene silencing and more. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14030–14032. doi: 10.1073/pnas.011506198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanny J.C., Dowd G.J., Huang J., Hilz H., Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 5.Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 6.Smith J.S., Boeke J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 7.Straight A.F., Shou W., Dowd G.J., Turck C.W., Deshaies R.J., Johnson A.D., Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 8.Bryk M., Banerjee M., Murphy M., Knudsen K.E., Garfinkel D.J., Curcio M.J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 9.Blander G., Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 10.North B.J., Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb S., Esposito R.E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 12.Benguria A., Hernandez P., Krimer D.B., Schvartzman J.B. Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:893–898. doi: 10.1093/nar/gkg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee K.K., Ayyub C., Sengupta S., Kolthur-Seetharam U. dSir2 deficiency in the fatbody, but not muscles, affects systemic insulin signaling, fat mobilization and starvation survival in flies. Aging (Albany, N.Y.). 2012;4:206–223. doi: 10.18632/aging.100435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarente L. Sirtuins in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 15.Kim E.J., Um S.J. SIRT1: roles in aging and cancer. BMB Rep. 2008;41:751–756. doi: 10.5483/bmbrep.2008.41.11.751. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi R., Vevea J.D., Swayne T.C., Chojnowski R., Hill V., Boldogh I.R., Pon L.A. Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr. Biol. 2013;23:2417–2422. doi: 10.1016/j.cub.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orlandi I., Bettiga M., Alberghina L., Nystrom T., Vai M. Sir2-dependent asymmetric segregation of damaged proteins in ubp10 null mutants is independent of genomic silencing. Biochim. Biophys. Acta. 2010;1803:630–638. doi: 10.1016/j.bbamcr.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Pappas D.L. Jr, Frisch R., Weinreich M. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 2004;18:769–781. doi: 10.1101/gad.1173204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida K., Bacal J., Desmarais D., Padioleau I., Tsaponina O., Chabes A., Pantesco V., Dubois E., Parrinello H., Skrzypczak M., et al. The histone deacetylases sir2 and rpd3 act on ribosomal DNA to control the replication program in budding yeast. Mol. Cell. 2014;54:691–697. doi: 10.1016/j.molcel.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 20.Barry J., Lock R.B. Small ubiquitin-related modifier-1: wrestling with protein regulation. Int. J. Biochem. Cell. Biol. 2011;43:37–40. doi: 10.1016/j.biocel.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson K.A., Henley J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reindle A., Belichenko I., Bylebyl G.R., Chen X.L., Gandhi N., Johnson E.S. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 2006;119:4749–4757. doi: 10.1242/jcs.03243. [DOI] [PubMed] [Google Scholar]
- 23.Cremona C.A., Sarangi P., Zhao X. Sumoylation and the DNA damage response. Biomolecules. 2012;2:376–388. doi: 10.3390/biom2030376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cubenas-Potts C., Matunis M.J. SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell. 2013;24:1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickey C.M., Wilson N.R., Hochstrasser M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell. Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garza R., Pillus L. STUbLs in chromatin and genome stability. Biopolymers. 2013;99:146–154. doi: 10.1002/bip.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darst R.P., Garcia S.N., Koch M.R., Pillus L. Slx5 promotes transcriptional silencing and is required for robust growth in the absence of Sir2. Mol. Cell. Biol. 2008;28:1361–1372. doi: 10.1128/MCB.01291-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan Y., Zuo X., Zhuo Y., Zhu M., Danziger S.A., Zhou Z. The functional role of SUMO E3 ligase Mms21p in the maintenance of subtelomeric silencing in budding yeast. Biochem. Biophys. Res. Commun. 2013;438:746–752. doi: 10.1016/j.bbrc.2013.07.096. [DOI] [PubMed] [Google Scholar]
- 29.Pasupala N., Easwaran S., Hannan A., Shore D., Mishra K. The SUMO E3 ligase Siz2 exerts a locus-dependent effect on gene silencing in Saccharomyces cerevisiae. Eukaryot. Cell. 2012;11:452–462. doi: 10.1128/EC.05243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 31.James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang J., Brito I.L., Villen J., Gygi S.P., Amon A., Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wohlschlegel J.A., Johnson E.S., Reed S.I., Yates J.R. 3rd. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45662–45670. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 34.Tirupataiah S., Jamir I., Srividya I., Mishra K. Yeast Nkp2 is required for accurate chromosome segregation and interacts with several components of the central kinetochore. Mol. Biol. Rep. 2014;41:787–797. doi: 10.1007/s11033-013-2918-3. [DOI] [PubMed] [Google Scholar]
- 35.Xu F., Zhang Q., Zhang K., Xie W., Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu H.C., Wang C.L., Wang M., Yang N., Chen Z., Sternglanz R., Xu R.M. Structural basis for allosteric stimulation of Sir2 activity by Sir4 binding. Genes Dev. 2013;27:64–73. doi: 10.1101/gad.208140.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheng W., Liao X. Solution structure of a yeast ubiquitin-like protein Smt3: the role of structurally less defined sequences in protein-protein recognitions. Protein Sci. 2002;11:1482–1491. doi: 10.1110/ps.0201602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiser A., Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 39.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dominguez C., Boelens R., Bonvin A.M. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 42.Hannich J.T., Lewis A., Kroetz M.B., Li S.J., Heide H., Emili A., Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 43.Panse V.G., Hardeland U., Werner T., Kuster B., Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- 44.Wohlschlegel J.A., Johnson E.S., Reed S.I., Yates J.R. 3rd. Improved identification of SUMO attachment sites using C-terminal SUMO mutants and tailored protease digestion strategies. J. Proteome Res. 2006;5:761–770. doi: 10.1021/pr050451o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wykoff D.D., O'Shea E.K. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell. Proteomics. 2005;4:73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Johnson E.S., Gupta A.A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 47.Zhao X., Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flotho A., Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., Ren J. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42:W325–W330. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuperus G., Shafaatian R., Shore D. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 2000;19:2641–2651. doi: 10.1093/emboj/19.11.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang B., Kirchmaier A.L. Bypassing the catalytic activity of SIR2 for SIR protein spreading in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:5287–5297. doi: 10.1091/mbc.E06-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferreira H.C., Luke B., Schober H., Kalck V., Lingner J., Gasser S.M. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat. Cell. Biol. 2011;13:867–874. doi: 10.1038/ncb2263. [DOI] [PubMed] [Google Scholar]
- 53.Garcia S.N., Pillus L. A unique class of conditional sir2 mutants displays distinct silencing defects in Saccharomyces cerevisiae. Genetics. 2002;162:721–736. doi: 10.1093/genetics/162.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gotta M., Strahl-Bolsinger S., Renauld H., Laroche T., Kennedy B.K., Grunstein M., Gasser S.M. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harpaz Y., Gerstein M., Chothia C. Volume changes on protein folding. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 56.Smith J.S., Brachmann C.B., Pillus L., Boeke J.D. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis A., Felberbaum R., Hochstrasser M. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 2007;178:813–827. doi: 10.1083/jcb.200702154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Psakhye I., Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 59.Srikumar T., Lewicki M.C., Raught B. A global S. cerevisiae small ubiquitin-related modifier (SUMO) system interactome. Mol. Syst. Biol. 2013;9:668. doi: 10.1038/msb.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sydorskyy Y., Srikumar T., Jeram S.M., Wheaton S., Vizeacoumar F.J., Makhnevych T., Chong Y.T., Gingras A.C., Raught B. A novel mechanism for SUMO system control: regulated Ulp1 nucleolar sequestration. Mol. Cell Biol. 2010;30:4452–4462. doi: 10.1128/MCB.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


