Abstract
Plasmodium falciparum, the mosquito-transmitted Apicomplexan parasite, causes the most severe form of human malaria. In the asexual blood-stage, the parasite resides within erythrocytes where it proliferates, multiplies and finally spreads to new erythrocytes. Development of drugs targeting the ribosome, the site of protein synthesis, requires specific knowledge of its structure and work cycle, and, critically, the ways they differ from those in the human host. Here, we present five cryo-electron microscopy (cryo-EM) reconstructions of ribosomes purified from P. falciparum blood-stage schizonts at sub-nanometer resolution. Atomic models were built from these density maps by flexible fitting. Significantly, our study has taken advantage of new capabilities of cryo-EM, in visualizing several structures co-existing in the sample at once, at a resolution sufficient for building atomic models. We have discovered structural and dynamic features that differentiate the ribosomes of P. falciparum from those of mammalian system. Prompted by the absence of RACK1 on the ribosome in our and an earlier study we confirmed that RACK1 does not specifically co-purify with the 80S fraction in schizonts. More extensive studies, using cryo-EM methodology, of translation in the parasite will provide structural knowledge that may lead to development of novel anti-malarials.
INTRODUCTION
Plasmodium falciparum is the mosquito-transmitted Apicomplexan parasite that causes the most severe form of human malaria. All of the human malaria species require two different hosts to complete their life cycle: humans and mosquitoes. Following inoculation of the human host by an infected mosquito, the parasite travels to the liver where it differentiates into the blood-invasive form. The exponential amplification of this asexual blood-stage form of the parasite results in all of the clinical symptoms of malaria. During the asexual blood-stage, the young parasites mature from the ring to the trophozoite stage, and then mature into schizonts, which eventually rupture and release 16–32 daughter merozoites. Some of the released merozoites will invade fresh erythrocytes, continuing the asexual life cycle, and some will differentiate into sexual transmission forms, that are taken up by a female mosquito during a blood meal (1).
There is an urgent need to identify novel drug targets and develop more effective antimalarial drugs. Resistance has developed in the parasite to all anti-malarials currently in large-scale clinical use. One promising avenue of research is suggested by the success of a few antibiotics which inhibit protein synthesis in the parasites. However, our knowledge of the Plasmodium ribosome, specifically as a target for antibiotics, remains incomplete.
The ribosome, a ribonucleoprotein complex formed by two subunits, has an overall conserved core structure consisting of the decoding center, the GTPase center and the peptidyl transferase center. It has three binding sites for tRNAs, the aminoacyl (A) site, peptidyl (P) site, and exit (E) site. The ribosome actively synthesizes proteins in multiple rounds of the translation elongation cycle as dictated by the mRNA, entailing the binding of aminoacyl-tRNA to the A site (decoding), transfer of the nascent peptide chain from the tRNA at the P site to the aminoacyl group on the A-site tRNA (peptide bond formation) and movement of tRNAs and mRNA by one codon (translocation). The process of translocation is facilitated by large-scale conformational changes in the ribosome, as it equilibrates between two conformations, termed ‘rotated’ and ‘nonrotated’, distinguished by a ≈5- to ≈9-degree rotation between the two subunits (intersubunit rotation) (2–4).
The recently published cryo-EM structure of P. falciparum schizont-stage ribosome (5) identified additional P. falciparum-specific ribosomal proteins, ribosomal RNA expansion segments, and P. falciparum-specific inter-subunit bridges, as compared with ribosomes from yeast and human. However, the dynamic behavior of P. falciparum ribosomes, of critical importance for understanding the molecular mechanism of parasite translation, has remained uncharacterized. Here, we used advanced techniques of cryo-EM to image schizont-stage P. falciparum 80S ribosome complexes in several conformational states. These different states are distinguished mainly by a combination of intersubunit rotation and differences in tRNA occupancies/positions. We were also able to provide complete models of tertiary structures of helix 16 and expansion segments ES10S and ES6BS, highly flexible regions in the ribosome of P. falciparum.
These findings provide rich insights into the dynamics of the P. falciparum ribosome during the schizont-stage translation elongation process and reveal important differences from the mammalian system. In addition, our results confirm the finding of Wong et al. (5) reporting the absence of RACK1. We performed co-purification experiments which indicated that indeed RACK1 does not specifically co-purify with the 80S fraction, suggesting that RACK1's presence on the ribosome is not essential for translation.
MATERIALS AND METHODS
Isolation and purification of P. falciparum ribosomes
P. falciparum parasites (3D7 strain) were cultured in human red blood cells under standard conditions (6). Schizont-stage parasites were released from host cells by treatment with 0.15% Saponin (Sigma). Purified parasites were re-suspended in lysis buffer (50 mM Tris–HCl, pH = 7.4; 100 mM KOAc; 7 mM Mg(OAc)2; 380 mM sucrose; 6.5 mM β-mercaptoethanol; 0.14% vol/vol Triton X-100; 15 mM leupeptin and half a protease inhibitor cocktail tablet (Roche, EDTA-free)). Cells were disrupted using 0.5 mm glass beads (Sigma) and further clarified by short centrifugation. The ribosome-enriched pellets were obtained by overnight centrifugation (7) and further purified using 20K PEG precipitation methods described previously (7), with slight modifications. The final pellets were suspended in Buffer G (7) and kept at −80°C for further use.
Co-purification experiments
Ribosomes were isolated from late-stage P. falciparum according to the protocol published in Bunnik et al. (8). Immunoblot analysis of PfRACK1-HA was performed with αHA antibody (dilution 1:1000. Pierce). Immunoblot analysis of ribosome proteins were performed with αPfP0 (1E5F4) and αPfP2 (E2G12) monoclonals (dilution 1:500), generously provided by Dr Shobhona Sharma.
Electron microscopy
Four microliters of purified ribosomes were applied to holey carbon grids (carbon-coated Quantifoil R2/4 grid, Quantifoil Micro Tools, GmbH, Großlöbichau, Germany) containing an additional continuous thin layer of carbon, glow-discharged using Gatan Solarus 950 (9). Grids were blotted for 4 s at 4°C in 100% humidity and vitrified by plunging into liquid ethane cooled with liquid nitrogen, using the Mark IV Vitrobot (FEI, Hillsboro, Oregon) (10). Data were collected on a TF30 Polara electron microscope (FEI, Hillsboro, Oregon) operating at 300 kV, set up with a K2 Summit direct electron detection camera (Gatan, Warrendale, PA). Images were recorded using the automated data collection system Leginon (11) in counting mode, and taken at the nominal magnification of 23 000x, corresponding to a calibrated pixel size of 1.66 Å. The dose rate was nominally set to 8 electron counts per physical pixel per second (12) and the total exposure time was 8 s. Image stacks were collected in a defocus range of −1.5 μm to −3.5 μm, fractionated into 20 frames, each with an exposure time of 0.4 s.
Image processing
The dose-fractionated image stacks were first corrected for beam-induced motion, using the method of Li et al. (13), and averages of all 20 frames were used for image processing. A total number of 329K particles were automatically extracted from 5734 selected averaged images, using the ara-autopick and ara-crop tools in Arachnid (14), a python-encapsulated version of SPIDER. Contrast transfer function parameters were estimated using CTFFIND3 (15), and 3D classification was performed in RELION 1.2 (16), to discard defective particles and identify structurally homogeneous subsets.
Initial RELION 3D classification, with K = 10 classes and an angular sampling of 1.8°, yielded mainly three classes: rotated 80S (‘rt80S’ for brevity) with A/P and P/E tRNAs (22 793), slightly rotated 80S without tRNAs (27 158), and non-rotated 80S ribosomes (197 536). The largest class, of non-rotated ribosomes, was refined using the automated refinement procedure in RELION. This refinement resulted in a 4.4 Å (‘gold standard’ protocol with FSC = 0.143 criterion; b-factor of 176.1) density map, showing fragmented densities in the E-tRNA binding site, indicative of remaining heterogeneity. A subsequent second-stage RELION 3D classification was performed on this largest class, using K = 10 classes, a final angular sampling of 0.5° and combined local searches around the refined orientations. This classification resulted in four structurally distinct classes, non-rotated 80S (‘nrt80S’ for brevity) with E-tRNA (96 732), nrt80S with P-tRNA (14 696), nrt80S with P/P and E/E tRNAs (14 676), and nrt80S without any tRNAs (32 246). RELION auto-refinement was performed on each of these classes, as well as rt80S, yielding density maps of (i) rt80S with A/P-P/E tRNAs at average resolution of 5.8 Å (b-factor 202.4), (ii) nrt80S with E-tRNA at 4.7 Å (b-factor 157.7), (iii) nrt80S with P-tRNA at 6.7 Å (b-factor 311.1), (iv) nrt80S with P/P and E/E tRNAs at 6.7 Å (b-factor 300.4), and (v) nrt80S without tRNAs at 5.1 Å (b-factor 184.1) (Supplementary Figure S1). To further improve the resolution and correct the local beam-induced movement, the RELION statistical movie processing method (17) was applied to the class of nrt80S with E-tRNA. It was run with averages of 5 movie frames; a standard deviation of 1° for the prior on the Euler angles; and a standard deviation of 1 pixel for the translation. This step yielded the density maps of nrt80S with E-tRNA to an average resolution of 4.2 Å (b-factor 264.1) (Supplementary Figure S1).
Additionally, focused refinements were performed on the class of nrt80S with E-tRNA, where either 40S subunit or 60S subunit was masked out. This step gave slightly improved density for the 40S subunit, with an average resolution of 4.9 Å, and the 60S subunit, with an average resolution of 4.6 Å. However, in terms of structural definition at the interface of the subunits, the gain was minimal. Thus, for further analysis, we focused on the five density maps from RELION auto-refinements and used Wong et al.'s maps (5) as the reference.
Resolutions reported were based throughout on the ‘gold standard’ protocol along with the FSC = 0.143 criterion, and involved soft masking and high-resolution noise substitution (19). For the final visualization, all density maps were corrected for the effects of a soft mask in RELION post-processing (16,18), and sharpened by application of an automatically estimated negative b-factor (18). Local resolutions were all measured using the ResMap software (19).
Model building by molecular dynamic flexible fitting
An atomic model was built for the nrt80S E-tRNA complex starting from the published models for the P. falciparum 40S and 60S ribosomal subunits (5) (PDB codes: 3J79, 3J7A). Since the published models of the P. falciparum 40S and 60S subunits were refined separately, their interface contains elements that violate stereochemistry. We found a large number of spatial clashes when we attempted to put the 40S and 60S models together as a full 80S model. We first resolved the clashes manually, and then applied MDFF flexible fitting to generate the complete structure of the 80S from P. falciparum. (For details on major clashes see Supplementary Figure S6 and Table S2).
More specifically, we fitted the structures of the 40S and the 60S subunits into the segmented maps of the nrt80S-E ribosome using the molecular dynamics flexible fitting program MDFF (20). The E-site tRNA from PDB 3J7A was also fitted into the segmented map. The initial system was prepared for MDFF (20) using Visual Molecular Dynamics (VMD) (21) and run in the NAMD program (22) for 0.5 ns of simulation time. Details of the simulation are as follows: Each of these fittings was run for a length of 0.5 ns, which stabilized the structures to closely match the maps. These runs removed most of the initial clashes, and the remaining ones were removed manually. The fitted structures of the 40S, 60S and E-site tRNA were then combined for a collective fitting of the map of the nrt80S E-tRNA complex. All of the runs used the generalized-Born implicit solvent model, as implemented in NAMD (22).
The above fitted structure for the nrt80S E-tRNA complex was used as the starting model for modeling for three maps: nrt80S-P, nrt80S, and rt80S-A/P-P/E. For the nrt80S P-tRNA complex, the X-ray structure of the P-tRNA (PDB 2WRI) was fitted into the current segmented map, then the fitted structure was combined with the structure of the 80S ribosome. For the map of the empty ribosome, the E-site tRNA was simply removed from the fitted structure for the nrt80S E-tRNA complex. For the rt80S-A/P-P/E complex, the structures for the two subunits had to be fitted separately, due to the large conformational changes relative to the nrt80S E-tRNA complex. Additionally, the density for the A/P tRNA is quite weak, and thus, at the current stage, we had to leave out the A/P tRNA in the MDFF modeling of the rt80S-A/P-P/E complex. The map for the P/E-site tRNA was fitted using the existing model (PDB 3J0Z), then the fitted P/E-site tRNA structure was combined with the fitted structure for the rotated 80S ribosome to generate the final structure of the rt80S-P/E complex. The final product fitting for each map ran 0.5 ns, which was followed by 5000 steps of energy minimization.
Modeling of RNA expansion segments
We used the 3dRNA webserver (http://biophy.hust.edu.cn/3dRNA/3dRNA.html) (23) to predict the three-dimensional structures of RNA expansion segments, ES10S and ES6BS of 40S subunit, and helix 16 (h16) of 18S rRNA.
The identification of ES10S, ES6BS and h16 sequences were based on Wong et al.'s work, PDB 3J7A (chain A) (5). The secondary structures were predicted using the RNAfold webserver (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) with default settings (24). RNAfold returned a minimum free-energy secondary structure and an optimal thermodynamic ensemble secondary structure. These two predictions led to the same secondary structure in our target regions, and match with the structure obtained by Wong et al. (5). Thus this secondary structure was deemed quite suitable as the input for tertiary structure building. Specifically, the inputs for ES10S were (i) the RNA sequence, UAUGCUUAUAUUUGUAUCUUUGAUGCUUAUAUUUUGCAUA, and (ii) the RNA secondary structure, ((((..((((..(((((…)))))..))))…))))). The inputs for ES6BS were (i) the RNA sequence, AAAUCCCCACUUUUGCUUUUGCUUUUUUUGGGGAUUU, and (ii) the RNA secondary structure, (((((((((…..((….))……))))))))). The inputs for h16 were, (i) RNA sequence, UGCAAGGCCAAUUUUUGGUUUUGUA, and (ii) the RNA secondary structure, (((((((((((…))))))))))).
The program 3dRNA generated ten models for each expansion segment. In order to select a good model for further analysis, we rigid-body fitted each of these models into the segmented map of nrt80S-P using UCSF Chimera (25), and chose the one that had the highest cross-correlation (CC) value (details see Supplementary Figure S5 and Table S1). In addition to the rigid-body fitting, we attempted to do flexible fitting using MDFF. However, since the density in these regions is scattered, MDFF was not able to place the model into the map accurately.
For helix 16, the ten models generated by 3dRNA differed from one another only in the orientation of the hairpin tip region (for details, see Supplementary Figure S5). Rigid-body fitting the model into the map gave similar CC values, ranging from 0.82 to 0.835, and the one with highest CC value, 0.835, was selected. For ES6BS, the ten predicted models clustered into two major groups, differing in the orientation of the tip part: in one group the tip is bent outward to the 40S body, and in the other it is bent more inward (for details, see Supplementary Figure S5). The group that is closer to the 40S body has overall higher CC values, 0.76 to 0.77, as compared with 0.71 to 0.72 of the other group. The model with highest CC value, 0.77, was selected. For ES10S, due to the intrinsic flexibilities of ES10S and the 40S head region, the density is very weak and does not fully cover the predicted models. Hence the overall CC values are lower, as compared with ES6BS and h16, ranging from 0.56 to 0.66. The one with the highest CC, 0.66, was selected.
RESULTS
Cryo-EM reconstructions of schizont-stage P. falciparum 80S ribosomes
Synchronized P. falciparum parasites were harvested at the schizont-stage, the last stage of the erythrocytic cycle during which there is a proliferation of free ribosomes. To preserve the functional states as much as possible, we used a similar buffer as the one for an in vitro translation system (26), with relatively low salt concentration, and following standard ribosome purification procedures with slight modification (details described in ‘Materials and Methods’).
The purified sample was imaged using the TF30 Polara electron microscope (FEI, Hillsboro, Oregon), set up with a GATAN K2 Summit direct electron detector camera (Gatan, Warrendale, PA) in the single-electron counting mode. The dose-fractionated image stacks were first corrected for beam-induced motion, using the method of Li et al. (13). A total number of ≈329k particles were automatically extracted from 5734 selected averaged images. A two-step unsupervised 3D classification using RELION (16) was performed to discard defective particles and identify structurally homogeneous subsets, and auto-refinement was followed to further improve the resolution.
In the first step, we identified two main states of the 80S ribosomes, distinguished by intersubunit rotation and termed rotated (rt80S) and nonrotated (nrt80S). However, the reconstruction of the nrt80S still showed fragmented densities in the mRNA channel, indicative of remaining heterogeneity. A subsequent exhaustive 3D classification and ‘focused’ classification were performed to resolve this remaining heterogeneity.
In all, five major classes were identified: (i) rt80S bound with hybrid A/P and P/E tRNAs (‘rt80S-A/P-P/E’), (ii) nrt80S bound with E-tRNA (‘nrt80S-E’), (iii) nrt 80S bound with P-tRNA (‘nrt80S-P’), (iv) nrt80S bound with P/P and E/E tRNAs (‘nrt80S-P-E’), and (v) nrt80S without any tRNAs. The average resolutions of the density maps reconstructed from these classes ranged from 4.6 Å to 6.7 Å (FSC = 0.143; ‘gold-standard’ protocol) (Figure 1 and Supplementary Figure S1). Atomic models were built by flexible fitting based on the published structure of the P. falciparum ribosome (5). Specifically, for further biological analysis, we will discount complexes that contain no tRNAs (Figure 1D), as they would represent non-translating ribosomes.
Figure 1.
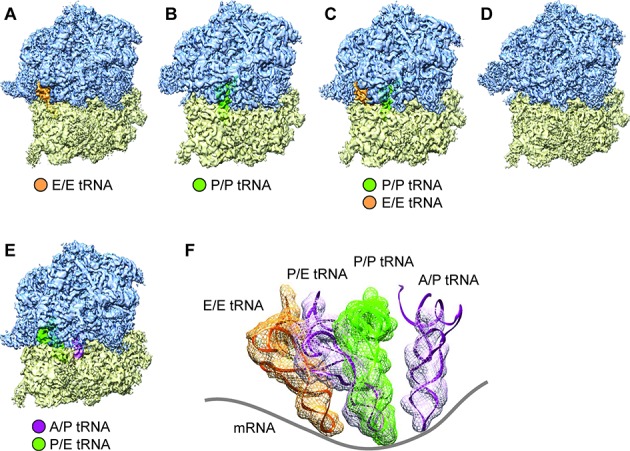
Cryo-EM reconstructions of P. falciparum 80S ribosomes obtained by classification. (A–D) Density maps of the P. falciparum 80S ribosome in non-rotated states (A) bound with E-tRNA (96 732 particles; 53.4%) at an average resolution of 4.7 Å; (B) bound with P-tRNA (14 696 particles; 8.1%) at 6.7 Å; (C) bound with P/P- and E/E-tRNAs (14 676 particles; 8.1%) at 6.7 Å; and (D) without tRNAs (32 246 particles; 17.8%) 5.1 Å. (E) rotated state (22 793 particles; 12.6%) at 5.8 Å resolution. 60S subunits are colored in blue and 40S subunits are in yellow. (F) Positions of tRNAs for all 80S complexes in (A)–(D). The structures of E-tRNA, P-tRNA and P/E-tRNA were obtained by MDFF fitting, and the structure of A/P-tRNA is from the existing model, PDB 3J0Z, rigid body-fitted into the segmented map in UCSF Chimera (25). Contours of cryo-EM densities are displayed in mesh; structures of tRNA are displayed as ribbons; and mRNA path has been added as cartoon.
As for the rotated P. falciparum 80S ribosome bound with two hybrid-state tRNAs, the 40S subunit displays a ≈9° counterclockwise rotation (when viewed from the solvent side of the 40S subunit) of the body relative to the 60S subunit (Figure 2C and D), and a moderate back-swiveling of the head (i.e. counterclockwise rotation when viewed from the top) by about 2 degrees, compared with the nrt80S-P state. Concurrent with the 40S subunits movements, we also observe that the L1 stalk, a mobile region of 60S subunit, moves inward by about 42° as it goes from nrt80S-P to rt80S-A/P-P/E (for details, see Supplementary Figure S2).
Figure 2.
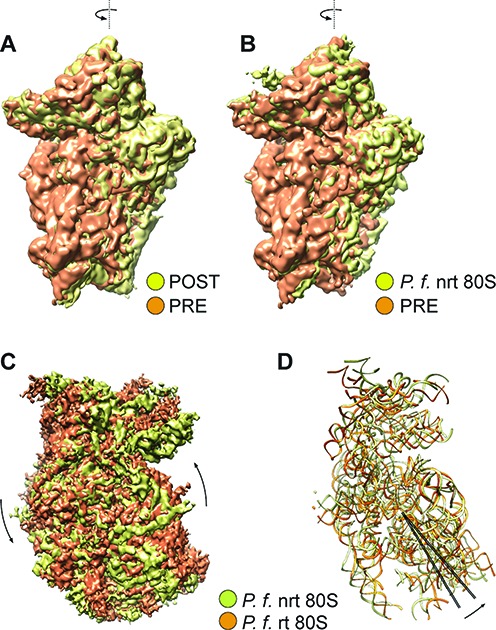
Global movements of P. falciparum 40S ribosomal subunits. (A, B) ‘Subunit rolling’ in POST states P. falciparum 80S. Comparison of the 40S subunit positions in POST state (yellow) (EMD 2621) with the PRE states (orange): (A) mammalian POST complex (data adapted from (27); EMD 2620), (B) P. falciparum P-tRNA bound POST 80S. (C, D) Inter-subunit rotation of 40S subunit in rotated PRE P. falciparum 80S ribosomes. (C) Cryo-EM maps of 40S subunit and (D) atomic models of 18S rRNA of 40S, in nrt80S-P (yellow) and rt80S-A/P-P/E (orange) states. The axes (black lines) were calculated in UCSF Chimera based on residues 1955–2033 of 18S rRNA (approximately h44 region). Crossing angle of the axes is ≈9°. Proteins are omitted for clarity. All comparisons were obtained by structural alignment on the 60S subunits of the 80S ribosomes using UCSF Chimera (25).
We then analyzed this rotated state by comparing it with mammalian rotated PRE-state ribosomes in earlier studies by Budkevich et al. (4,27). This comparison shows a good overall agreement, particularly with respect to the locations of the tRNA binding sites. Thus, the rt80S-A/P-P/E complex can be assumed to represent the pretranslocational (PRE) state of P. falciparum, a state after peptidyl-transfer but before translocation.
Of the three tRNA-containing non-rotated states, the nrt80S-P (Figure 1B), and the nrt80S-P-E (Figure 1C), complexes are candidates for POST states of the P. falciparum ribosome. Comparison of these maps with those of published mammalian POST 80S complexes containing classically configured P- and E-tRNAs (27,28), indeed reveals good overall structural agreement. In particular, the P-tRNAs in our nrt80S-P and nrt80S-P-E complexes have the same position and configuration as the one in mammalian systems (27,28). In addition, we can characterize the spatial relationship between the 40S subunit and the 80S ribosome by the former's ‘rolling’ position, in reference to a motion discovered by Budkevich et al. (27) in which the small subunit rotates around its long axis. In the P. falciparum P-tRNA-containing nrt80S complexes the 40S subunit has the same ‘unrolled’ configuration (Figure 2B) as the mammalian POST-state complex (27), when compared with the mammalian PRE-classical state complexes (27).
In addition to the nrt80S-P and nrt80S-P-E complexes, we have a third well-populated non-rotated class, the nrt80S-E state (Figure 1A). It has overall good structural similarities with P. falciparum POST-like nrt80S-P state; however, with several exceptions. Specifically, comparison with the nrt80S-P complex shows that, in the nrt80S-E complex, the 40S subunit head displays a ≈6 degree back-swiveling rotation, and the L1 stalk has rotated toward the E site on the large ribosomal subunit by ≈30 degrees.
Dynamics of P. falciparum small 40S subunits
The recent study of the P. falciparum ribosome, in a state comparable to our nrt80S-E, provided a detailed analysis of the P. falciparum- specific elements, including the large rRNA expansion segments (ESs) (5). Structurally, the rRNA ESs can be divided into two types. Expansion segments of one type are tightly associated with r-proteins or other rRNA ESs, while the second type comprises long rRNA helices that are attached to the ribosome only at their bases, and the extended part can adopt different conformations, as in the case of ES10S and ES6S (for positions of the ESs on the P. falciparum 80S ribosome, see Supplementary Figure S3.).
Because of the intrinsic flexibilities of ES10S and ES6BS, their models were only partially built in the published structure (5) using model building tools adopted from X-ray crystallography. In our work we used a combination of different strategies to build complete models for ES10S and ES6BS. We identified the region of ES10S and ES6BS based on Wong et al.'s work (5) and used the 3dRNA software (23), an automated RNA tertiary structure-building program, to complete the tertiary structure of these components (for details, see Figure 3, Supplementary Figure S5 and ‘Materials and Methods’). Both ES10S and ES6BS fulfill the requirements for application of the 3dRNA software as the length of these expansion segments is within the benchmarked length, and the secondary structures are known (5). Considering the good quality of cryo-EM map and the biologically functional meaning of the state, we chose to use the map of the nrt80S-P complex for further detailed analysis and model building.
Figure 3.
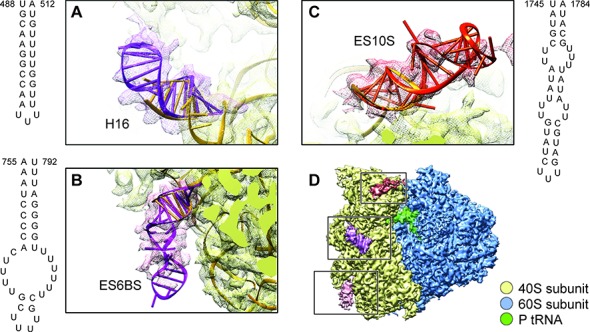
Predicted models of ES10S, ES6BS and helix 16 of P. falciparum 40S subunits using 3dRNA software. (A–C) Cryo-EM maps (meshed) and atomic models of (A) helix 16, (B) ES6BS and (C) ES10S. The secondary structures shown in (A-C) were predicted using RNAfold (24). The optimal tertiary structures shown in (A-C) were predicted using 3dRNA (23) and rigid-body fitted into density maps in UCSF Chimera (25). (D) Cryo-EM map of nrt80S-P. ES10S, ES6BS and h16 are highlighted in colors. 40S subunit is colored in yellow and 60S subunit is in blue.
ES10S
The most prominent P. falciparum-specific ES on 40S subunit is ES10S, which includes about 50 nucleotides and is located at the top of the 40S subunit head. (For details of modeling, see Figure 3C and Supplementary Figure S5 and ‘Materials and Methods’). The optimal model was selected on account of its highest CC value against the segmented density. We attribute the fact that the tip part of the model is not fully covered by the density to the high flexibility of ES10S. A comparison between our nrt80S-P and rt80S-A/P-P/E density maps shows overall good agreement in the ES10S region (structural alignment on 40S subunits head region), indicating absence of significant conformational changes attributable to inter-subunit rotation.
Compared with some other well-known eukaryotes, S. cerevisiae, T. brucei, T. cruzi, D. melanogaster and H. sapiens, (7,29–31), the ES10S in P. falciparum is the largest one, both in terms of secondary structure and size of density visible in the cryo-EM density map. It is important to note that in P. falciparum, ES10S is expressed differently—in terms of length and secondary structures—at different life stages of the parasite, in mosquito and human hosts. This variation of ES10S in the parasite's different life stages and in different eukaryotes could indicate that ES10S possesses an extra P. falciparum-specific role in translation; for instance, it may recruit necessary translation factors via its long expanded helices. It is therefore quite likely ES10S has extra functions that are associated with the transition between two life stages in the development of the parasite.
ES6S
Another striking example of a large ES in P. falciparum is the ≈280 nucleotide long ES6S, which is located at the solvent side of the 40S subunit platform. According to the secondary-structure prediction (5), P. falciparum ES6S is formed by five helices, namely ES6AS, ES6BS, ES6CS, ES6DS and ES6ES. Particularly, ES6BS is located at the lower part of the 40S subunit body, close to left foot region. In this case, only the base region was modeled in Wong et al. (5), leaving a ≈40 nucleotides-long helix un-modeled. We completed the tertiary-structure model of ES6BS by using 3dRNA (23) (For details of modeling see Figure 3, Supplementary Figure S5, and ‘Materials and Methods’). We rigid body-fitted the resulting models into the density map of nrt80S-P, and selected the one with highest cross-correlation value (for details, see Supplementary Figure S5). Still, the density is averaged out at the tip and does not fully cover the model, as a consequence of the flexibility of ES6BS. When going from the nrt80S-P to the rt80S-A/P-P/E state, ES6BS does not show conformational changes accompanying the inter-subunit rotation. In other words, ES6BS moves along rigidly with the 40S body region. For the other helices of ES6S, ES6AS, ES6CS, ES6DS and ES6ES, we used the Wong et al. model (5) to analyze the dynamics going from the nrt80S-P to the rt80S-A/P-P/E state, but we did not find significant local conformational changes in addition to the inter-subunit rotation.
It is tempting to speculate that the structural variation of ES6S reflects functional differences of eukaryotic translation among different species, and that the conformationally flexible regions may enable ES6S to recruit translation factors to the ribosome. In support of this idea, we note that the binding sites of eIF3 and eIF4G (31–33) to the 40S subunit are situated very close to the ES6S region.
Helix 16
The helix16 (h16) of 18S rRNA is located between the 40S subunit's head and shoulder, and involved in the formation of the mRNA entry channel. Inspection of the sequences and structures of ribosomes from S. cerevisiae (PDB 4V88; ref 7), P. falciparum (PDB 3J79; ref 5), D. melanogaster (PDB 4V6W; ref 29) and H. sapiens (PDB 4V6X; ref 29) makes it highly likely that this helix is universally conserved among all eukaryotes. However, due to the intrinsic conformational flexibility, about 7 base pairs of h16 were not modeled in (5). Here, we have completed the modeling of h16 using 3dRNA (23) and rigid-body fitted the best model into our density map. Comparison among nrt80-P, nrt80S-E nrt80S-P-E and rt80S-A/P-P/E states shows that h16 in all these structures is bent toward protein uS3 in a similar manner without displaying additional conformation changes. Moreover, h16 adopts a similar conformation in Wong et al.'s study (5).
In addition, in the h16 area, we do not observe the stress-related protein stm1, which is often co-purified together with 80S ribosomes (7,29) and can inhibit translation. Stm1 binds to the head domain of the 40S subunit and prevents mRNA access by inserting an α-helix through the mRNA entry channel (7). The absence of stm1 in our present work supports the notion that the P. falciparum 80S complexes represent actively translating ribosomes.
The absence of RACK1 on P. falciparum 40S subunits
Another striking structural difference between the eukaryotic and parasite ribosome is the absence of RACK1 (receptor for activated kinase 1). RACK1, a key player in multiple signaling pathways (34), and a well-established component of the small ribosomal subunit in most eukaryotic ribosomes, is completely missing from its conserved binding sites. (For details see Supplementary Figure S4)
From yeast to mammalian ribosomes, RACK1 has a highly conserved binding site, interacting with ribosomal proteins eS17, uS3, and h39 and h40 of 18S rRNA (7,35–38). Notably, both in our work and in Wong et al.'s results (5), RACK1 is completely missing in all maps, and its interacting partners in other species, proteins eS17 and uS3, exhibit weak densities, indicative of flexibility.
To further investigate the unusual behavior of RACK1 (PF3D7_0826700) in P. falciparum, we performed co-purification experiments to detect any physiological interaction between RACK1 and ribosomes. Our results show that RACK1 is expressed throughout the asexual life cycle, including the schizont stage. However RACK1 did not specifically co-purify in the fractions of isolated ribosomes from late-stage parasites and the majority of RACK1 was found in the unbound fractions (Figure 4). These results suggest that RACK1 mainly functions in a ribosome-unbound, free state in P. falciparum during the blood-stage, which would point to differences in translational regulation between the human host and the evolutionary divergent parasite (see Discussion for more details).
Figure 4.
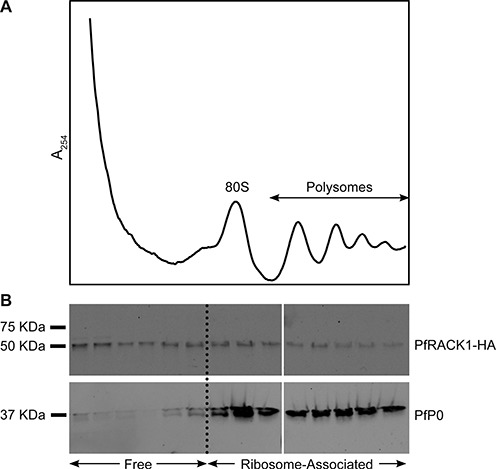
P. falciparum RACK1 does not specifically co-purify with isolated ribosomes from late-stage P. falciparum parasites. (A) Ribosome isolation profile, with monosome (80S) and polysome peaks indicated. (B) Western blots against PfRACK1-HA (50 kDa) and the ribosomal protein PfP0 (37 kDa) for the different fractions.
Structural rearrangements in the inter-subunit interface
The two ribosomal subunits are held together by a number of inter-subunit bridges, and the flexibility of these bridges facilitates the transition of the ribosome between the two main conformational states, rotated and nonrotated. As the dynamic behavior of the bridges is of great functional importance (3), we analyzed the way they are reconfigured during intersubunit rotation, in going from nrt80S-E (chosen for highest resolution) to rt80S-A/P-P/E in P. falciparum.
We found two distinct local movements which are effected by inter-subunit rearrangements. One involves the eukaryotic-specific inter-subunit bridge eB12, which is mainly formed by eL19 and ES6S (28,38), and the other is the P. falciparum-specific inter-subunit bridge which involves contacts between the C-terminal helix extension of protein eL8 and the C-terminal helix of protein eS1 (5) (for details see Figure 5).
Figure 5.
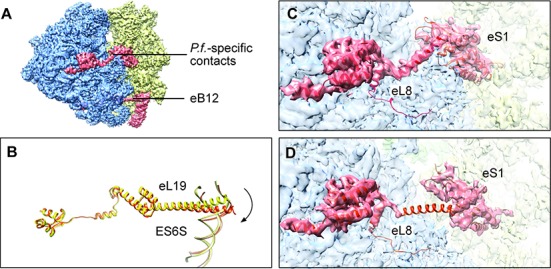
Structural re-arrangements of inter-subunit bridges. (A) Eukaryotic-specific bridge eB12 and P. falciparum-specific small bridge between eL8 and eS1 are shown in red. (B) The remarkably extended α-helix of eL19, in bridge eB12, is rotated by ≈17° in concert with the inter-subunit rotation (nonrotated state colored in yellow and rotated state in orange). Measurement was completed in UCSF Chimera (25). (C, D) The P. falciparum-specific small bridge is intact in nonrotated state (C), while it is broken in the rotated state (D). (In panels (C, D) density contours are displayed as transparency, and atomic models generated from MDFF are colored in red.)
Going from the P. falciparum nrt80S state to the rt80S-A/P-P/E state, the remarkably extended α-helix of eL19 (Figure 5A; B) rotates by ≈17° as it retains the integrity of the eB12 bridge. This persistency of eB12 in the dynamic ribosome was also observed in the human ribosome (28,38), indicating the evolutionary conservation of this feature between ribosomes from human malaria parasite and human host. Structural information on eB12 for any other eukaryotic ribosomes is not available at this time. The P. falciparum-specific bridge is found to be intact in all nrt80S complexes (nrt80S-E, nrt80S-P and nrt80S-P-E), but broken in the rt80S-A/P-P/E complex (for details, see Figure 5).
Besides the changes in the two inter-subunit bridges, we also noticed a lateral movement of h69 of 28S rRNA, which forms the core contacts in the interface area. Going from the POST nrt80S-P to the PRE rt80S-A/P-P/E state, h69 shifts as far as 6 Å toward to the E site, coupled with the inter-subunit rotation, such that bridge B2a stays intact. H69 has direct contacts with P-tRNA in the nrt80S complexes, but it loses these contacts when tRNA binds in the hybrid P/E site in the rt80S complexes. These movements are consistent with the one observed in the rotated 80S ribosome from yeast (2), but different from the one in the mammalian ribosome (27).
DISCUSSION
Structural models of the P. falciparum ribosome in different states
In our study of ribosomes purified from Plasmodium falciparum in the schizont stage, we found five subpopulations distinguished both by conformation and composition. In terms of conformation, we observe two main degrees of freedom, namely inter-subunit rotation and 40S head swivel movement. In terms of composition, we see tRNA in several combinations and binding positions. The resolution of the density maps reconstructed from these subpopulations was sufficient to obtain detailed atomic models of the ribosomal complexes based on recently published atomic models of the two ribosomal subunits. Importantly, we generated a full, stereochemically sound 80S ribosome model for each different state, by reconciling the special clashes of at subunit interface area and fitting the model into the map using MDFF.
At this point a general comment is in order. In cryo-EM of multi-component complexes such as the ribosome, which are subject to local conformational heterogeneity, the highest resolution can be often be gained by separate refinement of the individual components. It is inevitable that the individual structures obtained from these refinements are inconsistent along their interfaces. If the stated objective of these studies is the determination of the full structure, which will be of use for a large community of scholars who are not necessarily structural biologists, publication of separate structures of the individual components without reconciliation would seem to fall short of this objective, and ‘repairs’ of the kind we have undertaken here should not be necessary.
We also complemented the published atomic model by furnishing portions of the rRNA expansion segments ES6S and ES10S as well as h16 that were left incomplete. In the following discussion we first focus on the tRNA binding and the ribosomal movement. We then move on to a discussion of RACK1 and the possible significance of its absence on the P. falciparum ribosome.
Propensity for 40S head swiveling is dependent on the presence of P-site tRNA
We found a head swiveling movement that is dependent on the presence or absence of P-tRNA, but independent of intersubunit rotation. Compared with the nrt80S-P complex, the 40S subunit head domain in the nrt80S-E complex is in the back-swiveled position. A similar movement was also observed by comparing nrt80S-P-E and nrt80S-E complexes. We analyzed the interaction between P-tRNA and the ribosome in detail. In the nrt80S-P complex, the P-tRNA has a direct interaction with uL5 P-site loop in its T-stem loop region, and has an additional tentative contact with protein eL44 of the 60S subunit. Also, there is a direct interaction of the P-tRNA acceptor stem and the PTC regions, especially around A3178, A3179, G2958 and G2959 regions (PDB 5AJ0, POST-state human ribosome (28), numbering A4358, A4359, G4158 and G4159 respectively). Thus, it is very likely that it is the extensive interactions between P-tRNA and ribosomal subunits which helps stabilizing the 40S subunit head region. Hence, while the presence of P-tRNA stabilizes the subunit head, its absence apparently enables the spontaneous (i.e. thermally driven) change of the head's position, independent of the intersubunit rotation.
The absence of RACK1
Many studies have established RACK1 as a key player in multiple signaling pathways, some of which connect signaling with the translation machinery (34,35,39–42). Surprisingly, RACK1 displays a dramatically different behavior in P. falciparum than other species, such as yeast and mammalian cells. From yeast (7,35,43) to mammalian cells (29,37), RACK1 or its counterpart cpc2 is a well-established component of the small ribosomal subunits. In contrast, it is absent from its conserved binding site on the small 40S subunit head in cryo-EM reconstructions both in our work and previous work (5), even at low density threshold settings (data not shown).
Early studies pointed out that RACK1 has both ribosome localization-dependent and -independent functions in vivo (34). Indeed, in P. falciparum, RACK1 is constitutively expressed throughout the asexual stage (44). Our co-purification experiments show that RACK1 is not specifically co-purified with the isolated ribosomes from late-stage parasites. The majority of RACK1 was detected in the unbound fractions, but some amount was also detected in the polysome fractions. One possibility is that it is the presence of RACK1, but not its strong and direct interaction with the ribosome, that is important for RACK1-mediated translation regulation in late blood-stage P. falciparum. Another possibility is that, on the contrary, its localization on the ribosome is necessary for translation regulation, but that at this life-stage of the organism the ratio of ribosome-bound to ribosome-unbound RACK1 could be modulated in response to the environmental changes, as reported for yeast (34). In this context it is also interesting that no typical Protein Kinase C ortholog, an important substrate of RACK1 in other systems, has been identified in P. falciparum (45).
In conclusion, we have confirmed that RACK1's absence on the ribosome is not a consequence of EM specimen preparation, but a reflection of weak binding on the 40S subunit in the late blood-stage of the organism. The implications of this finding for translation control at this stage warrant further exploration.
ACCESSION NUMBERS
The cryo-EM maps for rt80S-A/P-P/E, nrt80S-P and nrt80S-E have been deposited with the accession numbers EMD-6452, EMD-6456 and EMD-6454, respectively. The MDFF-fitted atomic models of rt80S-P/E, nrt80S-P and nrt80S-E have been deposited with the accession numbers 3JBO, 3JBN and 3JBP, respectively.
Supplementary Material
Acknowledgments
We thank Robert A. Grassucci for assistance with the data collection and Melissa Thomas-Baum for assistance with the preparation of the illustrations. We would like to thank Sjors Scheres and colleagues for making a preprint and coordinates of their structure available prior to publication of their paper (5). We thank Shobhona Sharma, Department of Biological Sciences, TIFR, Mumbai, India for sharing the anti-PfP0 and anti-PfP2 antibodies. We are also indebted to other members of the Frank lab for help and discussions, in particular Amedee des Georges, Harry Kao, Robert Langlois, and Danny Nam Ho.
Author contributions: M.S., Y.H., J.F. and J.D. designed the experiments. K.B. and J.D. prepared the biological samples and the co-purification experiments. M.S. and S.D. purified ribosomes. M.S. performed cryo-EM experiments, classification, 3D reconstructions, and refinement. W.L. and M.S. performed modeling and fitting. M.S., W.L., K.B., Y.H., J.F. and J.D. interpreted the results. M.S., J.D. and J.F. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Howard Hughes Medical Institute and National Institutes of Health [R01 GM29169 to J.F.]; National Institutes of Health [R01 AI102907, DP2 AI112219 to J.D.]; Swedish Research Council: dnr 2013-367 [to K.B.]; LABEX: ANR-10-LABX-0036_NETRNA and the ANR grant @RAction program ‘ANR CryoEM80S’ [to Y.H.]. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Haldar K., Murphy S.C., Milner D.A., Taylor T.E. Malaria: Mechanisms of erythrocytic infection and pathological correlates of severe disease. Annu. Rev. Pathol.-Mech. 2007;2:217–249. doi: 10.1146/annurev.pathol.2.010506.091913. [DOI] [PubMed] [Google Scholar]
- 2.Svidritskiy E., Brilot A.F., Koh C.S., Grigorieff N., Korostelev A.A. Structures of yeast 80S ribosome-tRNA complexes in the rotated and nonrotated conformations. Structure. 2014;22:1210–1218. doi: 10.1016/j.str.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spahn C.M., Gomez-Lorenzo M.G., Grassucci R.A., Jorgensen R., Andersen G.R., Beckmann R., Penczek P.A., Ballesta J.P., Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budkevich T., Giesebrecht J., Altman R.B., Munro J.B., Mielke T., Nierhaus K.H., Blanchard S.C., Spahn C.M. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong W., Bai X.C., Brown A., Fernandez I.S., Hanssen E., Condron M., Tan Y.H., Baum J., Scheres S.H. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. eLife. 2014;3:e03080. doi: 10.7554/eLife.03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trager W., Jensen J.B. Human malaria parasites in continuous culture. J. Parasitol. 2005;91:484–486. doi: 10.1645/0022-3395(2005)091[0484:HMPICC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 8.Bunnik E.M., Chung D.W.D., Hamilton M., Ponts N., Saraf A., Prudhomme J., Florens L., Le Roch K.G. Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum. Genome Biol. 2013;14:R128. doi: 10.1186/gb-2013-14-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grassucci R.A., Taylor D.J., Frank J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubochet J., Adrian M., Chang J.J., Homo J.C., Lepault J., Mcdowall A.W., Schultz P. Cryo-Electron Microscopy of Vitrified Specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 11.Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Liao M.F., Cao E.H., Julius D., Cheng Y.F. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X.M., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., Cheng Y.F. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langlois R., Pallesen J., Ash J.T., Ho D.N., Rubinstein J.L., Frank J. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. J. Struct. Biol. 2014;186:1–7. doi: 10.1016/j.jsb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mindell J.A., Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 16.Scheres S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai X.C., Fernandez I.S., McMullan G., Scheres S.H.W. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S.X., McMullan G., Faruqi A.R., Murshudov G.N., Short J.M., Scheres S.H.W., Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trabuco L.G., Villa E., Mitra K., Frank J., Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 22.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y.J., Huang Y.Y., Gong Z., Wang Y.J., Man J.F., Xiao Y. Automated and fast building of three-dimensional RNA structures. Sci Rep-Uk. 2012;2:734. doi: 10.1038/srep00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Ferreras A., Triana L., Correia H., Sanchez E., Herrera F. An in vitro system from Plasmodium falciparum active in endogenous mRNA translation. Mem. Inst Oswaldo Cruz. 2000;95:231–235. doi: 10.1590/S0074-02762000000200017. [DOI] [PubMed] [Google Scholar]
- 27.Budkevich T.V., Giesebrecht J., Behrmann E., Loerke J., Ramrath D.J., Mielke T., Ismer J., Hildebrand P.W., Tung C.S., Nierhaus K.H., et al. Regulation of the mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell. 2014;158:121–131. doi: 10.1016/j.cell.2014.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behrmann E., Loerke J., Budkevich T.V., Yamamoto K., Schmidt A., Penczek P.A., Vos M.R., Burger J., Mielke T., Scheerer P., et al. Structural snapshots of actively translating human ribosomes. Cell. 2015;161:845–857. doi: 10.1016/j.cell.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 30.Gao H.X., Ayub M.J., Levin M.J., Frank J. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10206–10211. doi: 10.1073/pnas.0500926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashem Y., des Georges A., Fu J., Buss S.N., Jossinet F., Jobe A., Zhang Q., Liao H.Y., Grassucci R.A., Bajaj C., et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature. 2013;494:385–389. doi: 10.1038/nature11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Pestova T.V., Hellen C.U.T., Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siridechadilok B., Fraser C.S., Hall R.J., Doudna J.A., Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 34.Coyle S.M., Gilbert W.V., Doudna J.A. Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol. 2009;29:1626–1634. doi: 10.1128/MCB.01718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson J., Sengupta J., Frank J., Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams D.R., Ron D., Kiely P.A. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun. Signal. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandramouli P., Topf M., Menetret J.F., Eswar N., Cannone J.J., Gutell R.R., Sali A., Akey C.W. Structure of the mammalian 80S ribosome at 8.7 angstrom resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 39.McLeod M., Shor B., Caporaso A., Wang W., Chen H., Hu L. Cpc2, a fission yeast homologue of mammalian RACK1 protein, interacts with Ran1 (Pat1) kinase to regulate cell cycle progression and meiotic development. Mol. Cell. Biol. 2000;20:4016–4027. doi: 10.1128/mcb.20.11.4016-4027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu H.C., Swindell E.C., Sierralta W.D., Eichele G., Thaller C. Evidence for a role of protein kinase C in FGF signal transduction in the developing chick limb bud. Development. 2001;128:2451–2460. doi: 10.1242/dev.128.13.2451. [DOI] [PubMed] [Google Scholar]
- 41.McCahill A., Warwicker J., Bolger G.B., Houslay M.D., Yarwood S.J. The RACK1 scaffold protein: A dynamic cog in cell response mechanisms. Mol. Pharmacol. 2002;62:1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 42.Ron D., Mochlyrosen D. Agonists and antagonists of protein-kinase-C function, derived from its binding-proteins. J. Biol. Chem. 1994;269:21395–21398. [PubMed] [Google Scholar]
- 43.Yusupova G., Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu. Rev. Biochem. 2014;83:467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- 44.Madeira L., DeMarco R., Gazarini M.L., Verjovski-Almeida S., Garcia C.R.S. Human malaria parasites display a receptor for activated C kinase ortholog. Biochem. Biophys. Res. Commun. 2003;306:995–1001. doi: 10.1016/s0006-291x(03)01074-x. [DOI] [PubMed] [Google Scholar]
- 45.Sartorello R., Amaya M.J., Nathanson M.H., Garcia C.R.S. The plasmodium receptor for activated C kinase protein inhibits Ca2+ signaling in mammalian cells. Biochem. Biophys. Res. Commun. 2009;389:586–592. doi: 10.1016/j.bbrc.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


