Abstract
During translation, elongation factor G (EF-G) plays a catalytic role in tRNA translocation and a facilitative role in ribosome recycling. By stabilizing the rotated ribosome and interacting with ribosome recycling factor (RRF), EF-G was hypothesized to induce the domain rotations of RRF, which subsequently performs the function of splitting the major intersubunit bridges and thus separates the ribosome into subunits for recycling. Here, with systematic mutagenesis, FRET analysis and cryo-EM single particle approach, we analyzed the interplay between EF-G/RRF and post termination complex (PoTC). Our data reveal that the two conserved loops (loop I and II) at the tip region of EF-G domain IV possess distinct roles in tRNA translocation and ribosome recycling. Specifically, loop II might be directly involved in disrupting the main intersubunit bridge B2a between helix 44 (h44 from the 30S subunit) and helix 69 (H69 from the 50S subunit) in PoTC. Therefore, our data suggest a new ribosome recycling mechanism which requires an active involvement of EF-G. In addition to supporting RRF, EF-G plays an enzymatic role in destabilizing B2a via its loop II.
INTRODUCTION
Protein translation is a cyclic process which could be divided into four steps: initiation, elongation, termination and ribosome recycling (1,2). Each step is assisted by a translational GTPase (trGTPase) in combination with other translation factors. EF-G is the only trGTPase involved in two distinct steps, elongation and ribosome recycling. In the elongation step, EF-G translocates the (tRNA)2•mRNA complex by one codon in length on the ribosome, moving the peptidyl–tRNA from the A to the P site and the deacylated tRNA from the P to the E site, respectively (Figure 1A) (3). In the ribosome recycling step, EF-G together with RRF disassembles the post termination complex (PoTC) into two ribosomal subunits and releases mRNA and deacylated tRNA for the next round of translation (Figure 1B) (4–8). Subsequently, translation initiation factor 3 (IF3) binds to 30S subunit preventing reassociation of the free subunits (7,8). Both the translocation and ribosome recycling require EF-G-catalyzed GTP hydrolysis (9,10). When GTP hydrolysis is blocked by replacing GTP with its non-hydrolyzable analog GDPNP, the ribosome recycling is completely blocked (7,8), while a single round of translocation still take place, albeit slowly (9,11).
Figure 1.
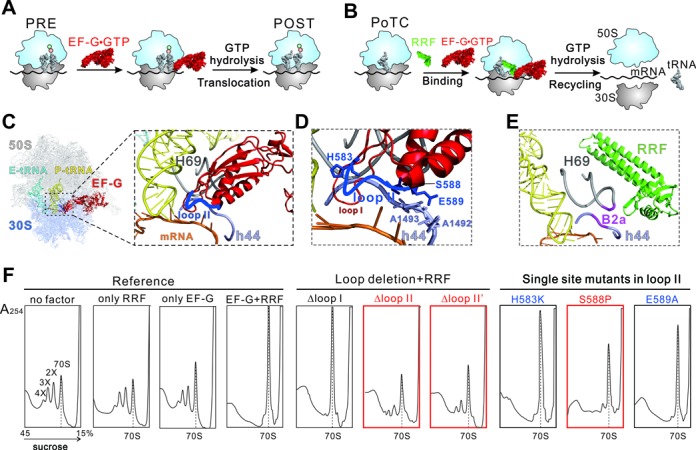
Function of EF-G during translocation and ribosome recycling. (A) Brief model of EF-G catalyzed translocation. The process from PRE to POST state is promoted by GTP hydrolysis. (B) Brief model of ribosome recycling process. EF-G and RRF function together to dissociate the PoTC into subunits. (C-E) Interactions of EF-G loop II (blue) and RRF (green) with ribosomal intersubunit bridge B2a. The B2a contains H69 (gray) of 23S rRNA and h44 (lavender) of 16S rRNA. Both EF-G and RRF interact with this bridge. The model of the POST-state ribosome(C, D) was extracted from a previous crystal structure (PDB 4V5F (19)). The model of PoTC•RRF (E) was also from a crystallography study (PDB 4V9D(26)). (F) Effect of loop II deletion and single site mutants on ribosome breakdown. The ribosome sedimentation pattern was analyzed by 15–45% sucrose density gradient ultracentrifugation. Fractions were collected from the bottom to the top of the tube and measured at A254. The peaks corresponding to the 70S ribosome, disome, trisome and tetrasome are marked as 70S, 2X, 3X and 4X, respectively. The 70S position is indicated with dashed line throughout. The red colored mutants are effective in both recycling and tRNA translocation, while the blue ones only in tRNA translocation.
EF-G is composed of five domains which could be divided into two superdomains. Superdomain I includes domains I-II and superdomain II includes domains III-V. During translocation, the two superdomains could move relatively to each other. Upon GTP hydrolysis, the distal end of superdomain II, namely domain IV tip region, has a movement of 10–15 Å and the whole factor thus transforms from the pre-translocational (PRE) to the post-translocational (POST) conformation (12). Domain IV of EF-G is a factor-specific domain (13). The structural integrity of domain IV is important for both translocation and ribosome recycling (14–18).
The tip region of domain IV contains two highly conserved loops, i.e. loop I and loop II (19). During the process of translocation, these two loops insert into the decoding center (DC) to override the hydrogen bonds between the DC nucleotides, mainly A1492 and A1493, and A-site tRNA, hence to disturb the interactions between mRNA–tRNA2 and the 30S subunit (Figure 1C–D) (20,21). This allows the swiveling motion of the 30S head and the subsequent conformational change of the ribosome, and eventually leads to the concomitant translocation of mRNA–tRNA2 (1,21). During ribosome recycling, binding of RRF–PoTC first induces a movement of the main intersubunit bridge B2a, which locates adjacent to the DC (Figure 1E) (22–27). With subsequent loading of EF-G, RRF was hypothesized to undergo domain rotations and hence to split the B2a completely (4,25–27). In this model, EF-G plays the role of facilitator, yet the detailed mechanism remains elusive. Considering the close location of domain IV tip in the vicinity of B2a, we assume that in addition to support RRF, EF-G might play a more active role in the splitting process.
In this study, we employed in vivo and in vitro assays to investigate the effect of the conserved loops of EF-G domain IV on the tRNA translocation and PoTC disassembly. Our earlier study has revealed that the two loops of EF-G domain IV, especially loop II, is essential for triggering the process of translocation (21). Extension of this study has now revealed that the loop II, which is away from the EF-G: RRF interface, unexpectedly, is crucial for ribosome recycling. In the cryo-electron microscopy (cryo-EM) reconstitution of the PoTC•EF-G•GDPNP complex at the resolution of 4.3 Å, we observed a conformational difference of loop II compared with EF-G structures in other recycling or elongational complexes. These data suggest a catalytic role of EF-G in destabilizing the intersubunit bridges of the 70S ribosome and a common aspect of EF-G function in translation and recycling.
MATERIALS AND METHODS
Strains, plasmids, growth conditions and reagents
Bacterial strains and plasmids used in this study were prepared as described in our previous study (21). LB broth or plates were used for bacterial growth. The media was supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml). IPTG (isopropyl-β-D-thiogalactopyranoside) was added to induce protein expression. GTP and GDPNP (guanidine 5′-β, γ-imido tri-phosphate), a non-hydrolysable GTP analogue, were purchased from Sigma (USA). All other chemical were from Merck (GER) or AMRESCO (USA).
Polysome breakdown assay
Polysome was prepared from E. coli MRE600 as previously described (28). Polysome (0.2–0.6 A260 units) was incubated with puromycin, RRF, EF-G and GTP in 550 μl of RRF buffer (20 mM HEPES pH 7.6, 8.2 mM MgSO4, 80 mM NH4Cl, 4 mM β-Mercaptoethanol) at 30°C for 15 min. The sedimentation profiles of reactions were obtained using sucrose density gradient centrifugation (15–45%, w/v) sucrose in RRF buffer, Beckman SW40 rotor, 36 000 rpm, 3.5 h, 4°C. The results were analyzed by A254 measurement.
RESULTS
Conserved loop II of EF-G domain IV is required for ribosome recycling
In our previous work, we showed that both loops I and II of EF-G domain IV are essential for translocation by systematic mutagenesis of EF-G (21). Here, we further tested the capacity of these EF-G mutants in ribosome recycling. Two sets of constructs were prepared, including loop deletions and single site mutations. Three loop deletion mutants were EF-G Δloop I (507–513), Δloop II (584–586) and Δloop II’ (586–589) (Supplementary Figure S1). Single site mutants were created by replacing the residues with an amino acid (aa) of different charge or the size of side chain (Supplementary Figure S1).
The effects of EF-G mutants on PoTC dissociation were determined by measuring the reduction of naturally produced poly-PoTC complexes, which were puromycin treated polysomes with a deacylated tRNA at the P-site (4). In the presence of wild type (WT) EF-G (EF-Gwt) and RRF, almost all the polysomes were disassembled into subunits, which quickly reassociated into monosomes and resulted in a sharp increase of 70S (Figure 1F, left part). However, when EF-Gwt was substituted by the loop deletion mutants, only Δloop I was able to convert polysomes into monosomes, while the two loop II deletions abolished the activity (Figure 1F, middle part). This clearly indicates that loop II is indispensable for ribosome recycling, in contract to the negligible role of loop I.
The contribution of the individual aa residue to the disassembly activity of EF-G was next assayed. As expected from Δloop I data, all loop I single site mutants displayed no apparent defects in the polysome breakdown experiments (Supplementary Figure S2A). Similarly, most single site mutants of loop II showed no defects (Supplementary Figure S2B), except for the S588P mutant (Figure 1F, right part). In the presence of S588P, the increase of the monosome peak was significantly smaller, and residual 3X and 4X forms of polysome could be identified, indicating that the recycling activity of EF-G was considerably compromised. It is interesting to note that previous data suggested that the interactions between the upper region of EF-G domain IV and RRF is important for RRF domain rotation (29,30). Here, our data indicate that the distal loop II of domain IV, which locates far away from EF-G:RRF interface (25), is essential for ribosome recycling. Among the seven residues of loop II, S588 might be of particular importance.
Loop II single site mutants retard cell growth and protein synthesis
Considering the necessity of loop II for ribosome recycling, we next analyzed the total effect of loop II mutants on cell growth and protein synthesis in vivo. We constructed overexpression plasmids of EF-G mutants and transfected each plasmid into E. coli BL21 cells, which could overexpress the corresponding EF-G mutant upon IPTG induction. As shown in Figure 2A, the overexpression of EF-Gwt, compared to the control (pEF-28a), did not inhibit cell growth. In contrast, the single mutants H583K, S588P and E589A were all toxic. We further investigated the effect of these mutants on total protein synthesis by using 35S-methionine incorporation assay. With SDS-PAGE and subsequent radioautography, the total proteins synthesized in vivo after 35S-methionine incorporation were visualized (Figure 2B). It is clear that the three mutations had dominant negative effect on total protein synthesis and this effect could not be compensated by a prolonged incubation (Figure 2C). These results provide explanations for the observations in Figure 2A, that overexpression of EF-G loop II mutants retarded cell growth due to their inhibitive effect on translation process. However, EF-G has dual functions of translocation and recycling in translation. To specify the reason, we next performed the fluorescence measurements, biochemical analyses, and cryo-EM structural studies subsequently to determine the effect of loop II mutants in translocation and recycling.
Figure 2.
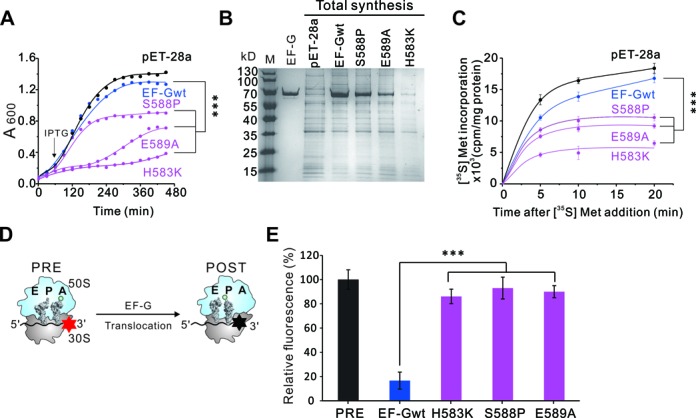
Effect of EF-G single site mutants on total protein synthesis and tRNA translocation. (A) Effect of overexpression of EF-G mutants on the growth of E.coli BL 21. Black arrow indicates the time that IPTG was added. (B and C) Effects of EF-G overexpression on total protein synthesis in vivo. Total soluble proteins from E.coli BL 21 overexpressing EF-G wt or mutants were analyzed by SDS-PAGE (B). After 35S-methionine addition, the ratio of proteins containing 35S-methionine increased over time as new proteins were continued to be synthesized in cells (C). (D) Brief illustration of the pyrene-modified mRNA translocation assay. The translocation action was monitored by tracing the fluorescence of pyrene attached to the 3′ end of the mRNA. The fluorescence emission is high (red star) before translocation and decreases (black star) upon translocation. (E) Relative fluorescence of PRE state ribosomes programmed by the labeled mRNA with incorporation of EF-Gwt or mutants. Error bars, s.e.m. (n = 3 technical replicates). ***P < 0.001.
Loop II mutants inhibit (tRNA)2•mRNA translocation
We recently identified His583, Ser588 and Glu589 as the catalytic residues for EF-G translocase activity by tRNA foot-printing studies (21). Here, we employed a steady-state fluorescence measurement to monitor the active motion of mRNA upon EF-G proteins binding onto the PRE state ribosomes (Figure 2D). In the pre-experiment of puromycin assay, we first tested the translocation efficiency of the tRNAs in the PRE complexes, which were programmed with MF-mRNAs, a deacylated tRNAfMet at the P site and an Ac[14C]Phe-tRNAPhe at the A site (21,31). After addition of EF-G proteins, a rapid increase in puromycin activity was observed in the EF-Gwt samples but not in the samples of single site mutants (Supplementary Figure S3). In the main experiment of steady-state fluorescence measurement, mRNA movement was monitored by tracing a fluorescence dye pyrene attached to the 3′ end of the mRNA (32,33). Upon mRNA translocation, the pyrene will enter the mRNA tunnel in the ribosome, thus the fluorescent signal will be quenched (Figure 2D). In this assay, PRE state ribosome complexes were prepared in a similar manner as in the puromycin assay, except that the mRNA was replaced with pyrene-labeled mRNA. When the PRE ribosomes were mixed with EF-Gwt, the pyrene signal was quenched very quickly. Contrarily, little fluorescent change was observed upon addition of EF-G mutants, demonstrating that the mRNA was not translocated (Figure 2E). Together with our previous data, these results firmly establish that the loop II mutants, even the single site mutants H583K, S588P and E589A, dramatically impaired the translocation activity of EF-G. Their pronounced toxicity to the cells as observed in Figure 2A–C is likely caused mainly by their deficiency in translocation, the main step of translation elongation. However, considering the importance of loop II, especially the residue Ser588, in recycling (Figure 1F), we next explored the impact of loop II mutants on RRF in PoTC.
Loop II mutants do not retard RRF in PoTC
The binding property of recycling-defective mutants of EF-G, i.e. Δloop II, Δloop II’ and S588P, to PoTC was first determined by sucrose cushion assay. Figure 3A shows that at the molecular ratio of 10 (EF-G to 70S), the occupancy of EF-G wt on PoTC has been saturated at ∼95%. Similarly, every mutant could saturate its binding on PoTC to an extent of 95% at the ratio of 10 (Figure 3B). It indicates that the binding of EF-G loop II mutants to PoTC was not impaired by mutations. For binding condition optimization, different nucleotides (GTP, GTP+FA, GDPNP) were also tested, and similar results were obtained (Supplementary Figure S4).
Figure 3.
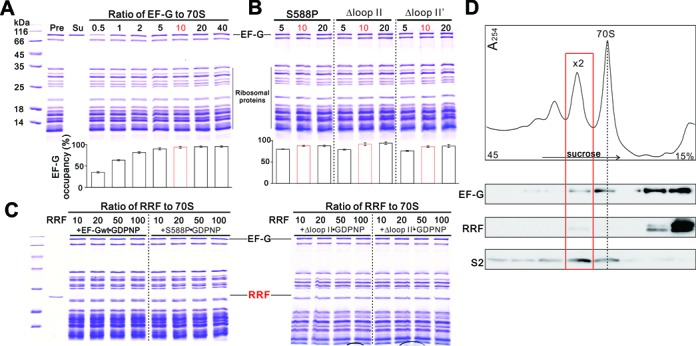
Condition optimization to achieve PoTC•EF-G•RRF complex. (A and B) Titration of EF-G wt (A) or mutants (B) to PoTC. Pre: before sucrose cushion ultracentrifugation. Su: supernatant after sucrose cushion ultracentrifugation. Quantification of EF-G occupancy on PoTC is shown beneath each gel. The optimized ratio is colored in red. Error bars, s.e.m. (n = 3 technical replicates). (C) Titration of RRF to PoTC with subsequent addition of EF-G proteins in the presence of GDPNP. (D) Polysomes fractionation and factor detection by western blotting with corresponding antibodies against EF-G, RRF and small ribosomal subunit protein S2. In the fraction of disome (red rectangle), both EF-G and RRF were detected.
In PoTC, binding of RRF will be replaced upon addition of EF-G due to the observed steric hindrance between RRF and EF-G (in GTP-like state) in the post termination state of the ribosome (30,34). Here, when the wt EF-G was exchanged into the recycling-defective mutants, the same results were obtained: upon addition of each EF-G mutant, RRF was no long detected in PoTC (Figure 3C). It suggests that all EF-G loop II mutants maintained the natural conformation of the factor, in agreement with Figure 3A and 3B, and therefore interacted normally with RRF in PoTC under the optimized condition in vitro. To determine the situation in vivo, we next analyzed the polysomes of E. coli cells overexpressing S588P. By collecting the fractions with detectable amount of both EF-G and RRF (Figure 3D), we enriched the potential PoTC with both factors. However, when we performed cryo-EM reconstitution, we did not find ribosome particles with both EF-G and RRF. Thus, all the in vitro and in vivo conditions we have tried did not support binding of RRF to PoTC in the presence of EF-G or its recycling-defective mutants. Nevertheless, these results reflect an essential aspect of recycling mechanism, that the loop II region of EF-G domain IV dose not interact with or influence RRF in PoTC, and very possibly they function independently from each other. Thus, loop II must play distinctive role in PoTC. To specify the role of EF-G loop II, we performed further molecular dynamic and cryo-EM analyses.
Loop II mutants stabilize rotated PoTC
After the release of the newly synthesized polypeptide from the ribosome, the PoTC carries the mRNA and a deacylated tRNA at the P-site, fluctuating between P/P and P/E positions. This fluctuation is coupled to the classical and the rotated states of the ribosome, respectively (25,26). The rotated state of the PoTC could be stabilized by either EF-G or RRF (20,26,35,36) and is required for the subsequent steps of disassembly. For binding condition optimization, different nucleotides (GTP, GTP+FA, GDPNP) were also tested, and similar results were obtained, we constructed PoTC•EF-G•GDPNP complexes and studied the conformational changes of the ribosome using Förster resonance energy transfer (FRET) assay.
We applied a FRET strategy described earlier (37,38), where L9 and S6 were modified with the donor and acceptor dyes AF555 and AF647, respectively. A counter-clockwise rotation of the 30S subunit relative to the 50S subunit will cause a longer separation distance between the two fluorophores and a consequent lower FRET efficiency (Figure 4A). In the experiments, binding of EF-Gwt to the PoTC in the presence of GDPNP induced a decrease in energy transfer (Figure 4B). Similar results were also observed in Δloop II, Δloop II’ or S588P samples, indicating that all these recycling-defective mutants were able to stabilize the rotated state of the PoTC (Figure 4C). Hence, loop II should not contribute to the hybrid state stabilizer role of EF-G in recycling phase.
Figure 4.
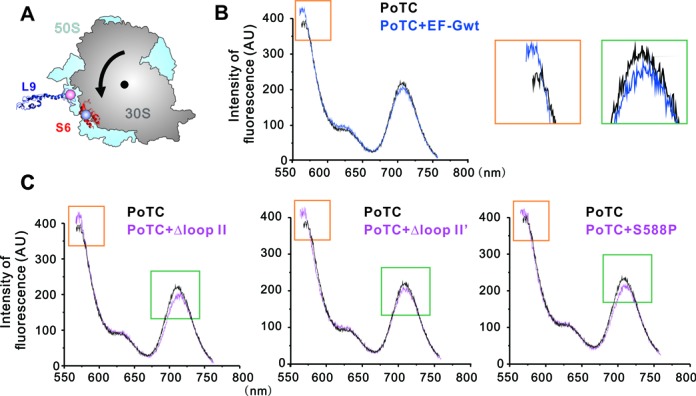
FRET analyses of ribosomal rotation. (A) Locations of fluorescence labeled residues in 30S and 50S subunits. Small ribosomal subunit protein S6 was labeled with AF555 (donor) and large ribosomal subunit protein L9 was labeled with AF647 (acceptor). The black arrow indicates the direction of rotation of PoTC induced by EF-G binding. (B) Fluorescence emission of the S6/L9 ribosome constructs. Black curve is the spectrum of vacant ribosome; blue curve is the PoTC complex in the presence of EF-G·GDPNP. The excitation wavelength was 550 nm. Addition of EF-G wt led to a significant decrease in FRET efficiency, indicated by orange and green rectangles, and enlarged in the right panels. (C) Fluorescence emission of the PoTC in the presence of EF-G mutants Δloop II, Δloop II’ and, S588P.
Loop II neighbors the groove of H69/h44 close to B2a
To obtain the structural insights into the interaction between loop II and the PoTC, we performed cryo-EM structural analyses of the PoTC•EF-G (S588P)•GDPNP complex. Thirty thousand raw particles were collected and were analyzed by RELION (39) and the final structure was solved at the resolution of 4.3 Å (Supplementary Figure S5). The PoTC•EF-G (S588P)•GDPNP complex was featured with a rotated ribosome and a hybrid P/E-site tRNA (Figure 5A). Compared to the previous structures (40), the S588P-induced ribosomal conformation exhibited a very similar 30S rotation as observed in ribosome•EF-Gwt complexes (Figure 5C). When the structure was further zoomed in, we observed a distinct conformation of loop II, which locates next to the groove of H69/h44, i.e. the major intersubunit bridge B2a of PoTC (Figure 5B). When we compared our result with a recent cryo-EM structure of 70S•EF-G (41), whose EF-G was GTPase center mutated (H91A) but with an intact loop II (Figure 5D), we found the S588P mutation caused a much less extended conformation of loop II (Figure 5E), which is also true when a recent crystal structure of 70S•EF-G (35) (Figure 5F) was used for comparison.
Figure 5.
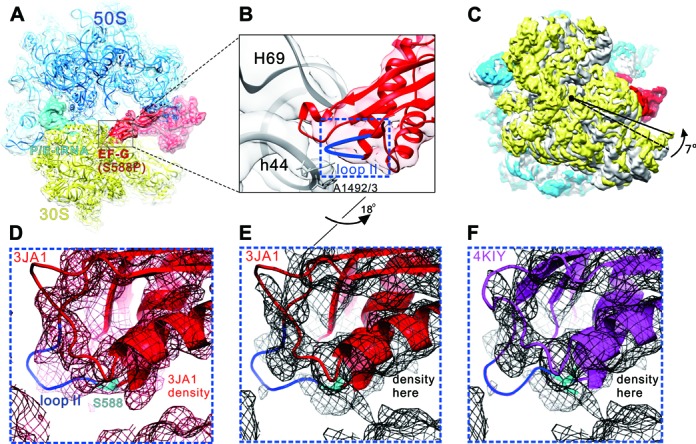
Cryo-EM structure of PoTC•EF-G (S588P)•GDPNP. (A) Overview of the PoTC with EF-G (S588P) (red) and a deacylated tRNA (cyan) in the P/E state.To model this complex, the 70S ribosome crystal structure (PDB 4KIX and 4KIY (35)) were used. (B) Zoom-in view of B2a region composed of H69 and h44 (gray), and loop II (blue) of EF-G domain IV. (C) Rotation of the 30S subunit (yellow) relative to the 50S subunit (transparent blue) caused by EF-G (S588P) binding. Compared to the classical state (gray, PDB 4V51 (50)), 30S shows a counter-clockwise rotation by 7°(viewed from the solvent side of 30S subunit). The structures are aligned using the 23S rRNA as reference. (D) Superimposition of the density map of a cryo-EM structure of 70S•EF-G (H91A) with the derived atomic model (PDB 3JA1 (41)). (E) Superimposition of our density map of the PoTC•EF-G (S588P)•GDPNP complex with the atomic model from 3JA1. (F) Superimposition of our density map of the PoTC•EF-G (S588P)• GDPNP complex with the atomic model from 4KIY (35). Compared to (B), the orientation in (D–F) has been turned ∼18° along the long axis of EF-G domain IV.
DISCUSSION
The translocation and recycling are essential steps for protein synthesis in both prokaryotes and eukaryotes. In these two steps, the ribosome undergoes similar conformational changes (25,29,36) but applies different mechanisms. In the translocation step, EF-G stabilizes the rotated state of the PRE ribosomes and catalyzes the (tRNA)2•mRNA translocation by disrupting the interaction between the ribosomal DC and the codon-anticodon duplex at the A site (1,21,42). In the recycling step, EF-G stabilizes the rotated PoTC similarly as in PRE state, and facilitates the domain rotation of RRF, which consequently catalyzes the dissociation of the ribosome into subunits and the release of mRNA and deacylated tRNA for a new round of translation (43,44).
The interplay between RRF and PoTC has been studied by foot-printing (45) and various structural approaches (22–24,26,27,29,30,45–48). However, the exact role of RRF in splitting the PoTC remains unclear. For example, in a POST•RRF complex, the inclusion of RRF did not induce any changes in bridges connecting the two subunits, suggesting that RRF cannot take full responsibility for breaking the intersubunit bridges (22). More importantly, it is difficult to visualize the functional state of PoTC with both RRF and EF-G loaded concurrently, because the whole 70S ribosome will easily be split into subunits in the presence of both factors (7,8,49). A recent cryo-EM structural study suggested that EF-G and RRF from different species could coexist in PoTC (25). In this structure, EF-G domains III, IV and V contacted RRF interdomain hinges, head domain and C-terminus, respectively, and hence induced a series of domain rotations of RRF in the splitting process. With reoriented head domain and C-terminus, RRF destabilized the main intersubunit bridge B2a (26,32). But, the final step of B2a separation remains elusive.
The distal end of EF-G domain IV has been reported to interact with the DC and the neighbor B2a region in h44 of 16S rRNA and undergo flexible conformations (Supplementary Figure S6) during translocation (19,20,36). Here, in the structure of PoTC•EF-G (S588P)•GDPNP complex, we observed the deformed EF-G loop II (S588P) neighboring the groove of H69/h44, which locates adjacent to the B2a but far from the interface between EF-G and RRF. In native conformation, the loop II of EF-Gwt might be directly involved in splitting the B2a bridge in PoTC (Figure 5E and F). In polysome breakdown study, WT RRF with EF-G loop II deletion or S588P mutants abolished largely the disassembly activity. Taken together, these results demonstrate that EF-G is involved in the final splitting process by playing an active role in addition to facilitating RRF orientation. Through the highly conserved loop II, EF-G together with RRF disassembles the PoTC into the subunits. These findings shed new light on the molecular mechanism of EF-G/RRF catalyzed ribosome recycling: upon binding of RRF to PoTC state ribosomes, it promotes the rotated conformation of the ribosome and destabilizes the interactions between H69 and h44. Subsequently, loading of EF-G•GTP facilitates RRF reorientation and actively splits the B2a bridge. Finally, upon GTP hydrolysis, EF-G transforms into a GDP conformation and thus separates the 70S ribosome into two subunits completely (Figure 6). In this process, loop II plays a mechanistic role to disrupt the interactions in the vicinity of DC, in a similar manner as during translocation. Thus, the interactions of loop II with the DC region constitute a threshold event in both the translocation and recycling processes. It reflects a common aspect of EF-G function in these two translational phases.
Figure 6.
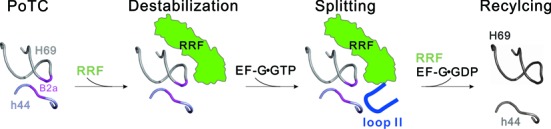
Schematic representation of the catalytic role of EF-G loop II in ribosome recycling. At the end of translation, RRF binds to PoTC inducing the conformational change of H69 and h44 to destabilize B2a. Upon EF-G•GTP binding, loop II (blue) locates in the vicinity of B2a. Together with RRF, loop II disassembles the ribosome into the subunits.
ACCESSION NUMBERS
The cryo-EM map has been deposited at EMDB under accession code EMD-6474.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Profs. Akira Kaji (University of Pennsylvania) and Hideko Kaji (Thomas Jefferson University) for help and discussions. We also thank the China National Center for Protein Sciences (Beijing) and Tsinghua National Laboratory for Information Science and Technology (“Explorer 100” cluster system) for providing computation resource.
FUNDING
Major State Basic Research of China 973 [2012CB911001 and 2013CB531200 to Y.Q.]; National Natural Science Foundation of China [31170756, 31270847 and 31322015 to Y.Q., 31170677 and 31422016 to N.G.]. Funding for open access charge: Major State Basic Research of China 973 [2012CB911001 to Y.Q].
Conflict of interest statement. None declared.
REFERENCES
- 1.Yamamoto H., Qin Y., Achenbach J., Li C., Kijek J., Spahn C.M., Nierhaus K.H. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat. Rev. Microbiol. 2014;12:89–100. doi: 10.1038/nrmicro3176. [DOI] [PubMed] [Google Scholar]
- 2.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 3.Agirrezabala X., Frank J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 2009;42:159–200. doi: 10.1017/S0033583509990060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirokawa G., Demeshkina N., Iwakura N., Kaji H., Kaji A. The ribosome-recycling step: consensus or controversy? Trends Biochem. Sci. 2006;31:143–149. doi: 10.1016/j.tibs.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Frank J., Gao H., Sengupta J., Gao N., Taylor D.J. The process of mRNA-tRNA translocation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirashima A., Kaji A. Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J. Biol. Chem. 1973;248:7580–7587. [PubMed] [Google Scholar]
- 7.Zavialov A.V., Hauryliuk V.V., Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Peske F., Rodnina M.V., Wintermeyer W. Sequence of Steps in Ribosome Recycling as Defined by Kinetic Analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Hirokawa G., Iwakura N., Kaji A., Kaji H. The role of GTP in transient splitting of 70S ribosomes by RRF (ribosome recycling factor) and EF-G (elongation factor G) Nucleic Acids Res. 2008;36:6676–6687. doi: 10.1093/nar/gkn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal R.K., Heagle A.B., Penczek P., Grassucci R.A., Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 11.Rodnina M.V., Savelsbergh A., Katunin V.I., Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 12.Brilot A.F., Korostelev A.A., Ermolenko D.N., Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang D., Qin Y. The paradox of elongation factor 4: highly conserved, yet of no physiological significance? Biochem. J. 2013;452:173–181. doi: 10.1042/BJ20121792. [DOI] [PubMed] [Google Scholar]
- 14.Ito K., Fujiwara T., Toyoda T., Nakamura Y. Elongation factor G participates in ribosome disassembly by interacting with ribosome recycling factor at their tRNA-mimicry domains. Mol. Cell. 2002;9:1263–1272. doi: 10.1016/s1097-2765(02)00547-6. [DOI] [PubMed] [Google Scholar]
- 15.Savelsbergh A., Matassova N.B., Rodnina M.V., Wintermeyer W. Role of Domains 4 and 5 in Elongation Factor G Functions on the Ribosome. J. Mol. Biol. 2000;300:951–961. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
- 16.Martemyanov K.A., Gudkov A.T. Domain IV of elongation factor G from Thermus thermophilus is strictly required for translocation. FEBS Lett. 1999;452:155–159. doi: 10.1016/s0014-5793(99)00635-3. [DOI] [PubMed] [Google Scholar]
- 17.Martemyanov K.A., Yarunin A.S., Liljas A., Gudkov A.T. An intact conformation at the tip of elongation factor G domain IV is functionally important. FEBS Lett. 1998;434:205–208. doi: 10.1016/s0014-5793(98)00982-x. [DOI] [PubMed] [Google Scholar]
- 18.Guo X., Peisker K., Backbro K., Chen Y., Koripella R.K., Mandava C.S., Sanyal S., Selmer M. Structure and function of FusB: an elongation factor G-binding fusidic acid resistance protein active in ribosomal translocation and recycling. Open Biol. 2012;2:120016. doi: 10.1098/rsob.120016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y.G., Selmer M., Dunham C.M., Weixlbaumer A., Kelley A.C., Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J., Lancaster L., Donohue J.P., Noller H.F. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G.Q., Song G.T., Zhang D.Y., Zhang D.J., Li Z.K., Lyu Z.X., Dong J.S., Achenbach J., Gong W.M., Zhao X.S., et al. EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon-anticodon duplex. Nat. Struct. Mol. Biol. 2014;21:817–824. doi: 10.1038/nsmb.2869. [DOI] [PubMed] [Google Scholar]
- 22.Weixlbaumer A., Petry S., Dunham C.M., Selmer M., Kelley A.C., Ramakrishnan V. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat. Struct. Mol. Biol. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- 23.Barat C., Datta P.P., Raj V.S., Sharma M.R., Kaji H., Kaji A., Agrawal R.K. Progression of the Ribosome Recycling Factor through the Ribosome Dissociates the Two Ribosomal Subunits. Mol. Cell. 2007;27:250–261. doi: 10.1016/j.molcel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Agrawal R.K., Sharma M.R., Kiel M.C., Hirokawa G., Booth T.M., Spahn C.M., Grassucci R.A., Kaji A., Frank J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoyama T., Shaikh T.R., Iwakura N., Kaji H., Kaji A., Agrawal R.K. Structural insights into initial and intermediate steps of the ribosome-recycling process. EMBO J. 2012;31:1836–1846. doi: 10.1038/emboj.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunkle J.A., Wang L., Feldman M.B., Pulk A., Chen V.B., Kapral G.J., Noeske J., Richardson J.S., Blanchard S.C., Cate J.H. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pai R.D., Zhang W., Schuwirth B.S., Hirokawa G., Kaji H., Kaji A., Cate J.H.D. Structural Insights into Ribosome Recycling Factor Interactions with the 70S Ribosome. J. Mol. Biol. 2008;376:1334–1347. doi: 10.1016/j.jmb.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujiwara T., Ito K., Nakamura Y. Functional mapping of ribosome-contact sites in the ribosome recycling factor: a structural view from a tRNA mimic. RNA. 2001;7:64–70. doi: 10.1017/s1355838201001704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao N., Zavialov A.V., Ehrenberg M., Frank J. Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits. J. Mol. Biol. 2007;374:1345–1358. doi: 10.1016/j.jmb.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao N., Zavialov A.V., Li W., Sengupta J., Valle M., Gursky R.P., Ehrenberg M., Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Qin Y., Polacek N., Vesper O., Staub E., Einfeldt E., Wilson D.N., Nierhaus K.H. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 32.Ermolenko D.N., Noller H.F. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat. Struct. Mol. Biol. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studer S.M., Feinberg J.S., Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J. Mol. Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 34.Zavialov A.V., Hauryliuk V.V., Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 35.Pulk A., Cate J.H. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tourigny D.S., Fernandez I.S., Kelley A.C., Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hickerson R., Majumdar Z.K., Baucom A., Clegg R.M., Noller H.F. Measurement of internal movements within the 30 S ribosomal subunit using Förster resonance energy transfer. J. Mol. Biol. 2005;354:459–472. doi: 10.1016/j.jmb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Ermolenko D.N., Majumdar Z.K., Hickerson R.P., Spiegel P.C., Clegg R.M., Noller H.F. Observation of intersubunit movement of the ribosome in solution using FRET. J. Mol. Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 39.Scheres S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valle M., Zavialov A., Li W., Stagg S.M., Sengupta J., Nielsen R.C., Nissen P., Harvey S.C., Ehrenberg M., Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 41.Li W., Liu Z., Koripella R.K., Langlois R., Sanyal S., Frank J. Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G. Sci. Adv. 2015;1:e1500169. doi: 10.1126/sciadv.1500169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 43.Karimi R., Pavlov M.Y., Buckingham R.H., Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- 44.Kaji A., Kiel M.C., Hirokawa G., Muto A.R., Inokuchi Y., Kaji H. The fourth step of protein synthesis: disassembly of the posttermination complex is catalyzed by elongation factor G and ribosome recycling factor, a near-perfect mimic of tRNA. Cold Spring Harb. Symp. Quant. Biol. 2001;66:515–529. doi: 10.1101/sqb.2001.66.515. [DOI] [PubMed] [Google Scholar]
- 45.Lancaster L., Kiel M.C., Kaji A., Noller H.F. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- 46.Wilson D.N., Schluenzen F., Harms J.M., Yoshida T., Ohkubo T., Albrecht R., Buerger J., Kobayashi Y., Fucini P. X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borovinskaya M.A., Pai R.D., Zhang W., Schuwirth B.S., Holton J.M., Hirokawa G., Kaji H., Kaji A., Cate J.H. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 48.Dunkle J.A., Cate J.H. Ribosome structure and dynamics during translocation and termination. Annu. Rev. Biophys. 2010;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- 49.Hirokawa G., Nijman R.M., Raj V.S., Kaji H., Igarashi K., Kaji A. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selmer M., Dunham C.M., Murphy F.V.t., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


