Abstract
The objective of the present study was to validate prognostic gene signature for estrogen receptor alpha-positive (ER03B1+) and lymph node (+) breast cancer for improved selection of patients for adjuvant therapy. In our previous study, we identified a group of seven genes (GATA3, NTN4, SLC7A8, ENPP1, MLPH, LAMB2, and PLAT) that show elevated messenger RNA (mRNA) expression levels in ERα (+) breast cancer patient samples. The prognostic values of these genes were evaluated using gene expression data from three public data sets of breast cancer patients (n = 395). Analysis of ERα (+) breast cancer cohort (n = 195) showed high expression of GATA3, NTN4, and MLPH genes significantly associated with longer relapse-free survival (RFS). Next cohort of ERα (+) and node (+) samples (n = 109) revealed high mRNA expression of GATA3, SLC7A8, and MLPH significantly associated with longer RFS. Multivariate analysis of combined three-gene signature for ERα (+) cohort, and ERα (+) and node (+) cohorts showed better hazard ratio than individual genes. The validated three-gene signature sets for ERα (+) cohort, and ERα (+) and node (+) cohort may have potential clinical utility since they demonstrated predictive and prognostic ability in three independent public data sets.
Keywords: breast cancer, gene signature, estrogen receptor, lymph node positive, relapse-free survival, prognosis, biomarkers
Introduction
Breast cancer is a heterogeneous disease,1 making it an ideal disease to study using microarrays since different expression patterns can be identified within distinct tumor groups. An increased understanding of the pathogenesis of breast cancer is imperative in the pursuit of innovative therapies for treatment and/or prognosis of patients. Gene expression studies based on microarray have been used extensively by cancer researchers to profile cancer subsets, predict patients’ outcome, and identify genes of clinical relevance.2–5
Breast cancer is no longer a single disease, but it is a heterogeneous disease consisting of different subtypes on the molecular and histopathological levels with different prognostic and therapeutic outcomes.6–9 Gene expression profiling has classified breast cancer into five biologically distinct intrinsic subtypes: luminal A, luminal B, human epidermal growth factor receptor 2 (HER2+), basal-like, and normal-like.6–9 The luminal A subtype is estrogen receptor alpha positive (ERα+), progesterone receptor positive (PR+), and HER2 (−); luminal B subtype is ERα+, PR+, and HER+, and luminal B is associated with a relatively worse outcome. Both HER2 (+) and basal-like (ER-, PR-, and HER-) breast cancers have poor outcomes. Parker et al.9 developed an efficient classifier, called PAM50, to distinguish these five intrinsic subtypes using the expression of 50 “classifier genes.” In a more recent study, a large breast cancer patient cohort (n ~ 2000) was clustered into 10 molecularly defined subgroups with apparently distinct biology and disease-specific survival characteristics.10 In addition, different breast cancer subtypes have different treatment responses.11,12
An important part of the diagnostic workup of all the breast cancer patients is the determination of the ERα status of the tumor. Clinically, an ERα (+) status is associated with improved prognosis, lower risk of relapse, and better overall survival,13 and that are key aspects for making decisions for endocrine therapy with antiestrogens. A major problem in clinical oncology is to distinguish the patients who are likely to present a relapse of the disease from those with a favorable prognosis. In recent years, it has been realized that apart from ERα, other factors are also important in deciding the therapeutic strategies of the patient. These include histological markers such as grade, tumor size, lymph node involvement, PR, and HER2 receptor status. Each of these has modest positive predictive value (30%–60%).14–17 Moreover, the current histological classifications of breast cancer do not fully represent the diverse clinical outcome of the disease. Recent approaches for patient management, which utilize histological markers in conjunction with online statistical algorithms such as “Nottingham Prognostic Index” and “Adjuvant! Online”, fail to predict the course of the disease in a significant number of breast cancer patients.18,19 Women with node (+), ERα (+), and HER2 (−) often receive adjuvant treatment with chemotherapy and hormonal therapy. Nevertheless, few patients eventually experience a recurrence. Thus, new tools are needed to allow improved definition of a risk of recurrence. If it were possible to predict cancer recurrence following standard therapy, these patients could be targeted for alternative treatment strategies.
Recently, we published gene expression profile of breast tumors and identified seven genes (GATA3, NTN4, SLC7A8, MLPH, ENPP1, LAMB2, and PLAT) that showed high messenger RNA (mRNA) expression levels in ERα (+) compared to ERα (−) breast tumors.20 In the present study, we have delineated the association between mRNA expression of these aforementioned seven genes with clinicopathological parameters such as PR, HER2, tumor grade, and lymph node status. To demonstrate that these genes have significantly better prediction for RFS than standard prognostic factors, we investigated the clinical utility of the aforementioned seven genes on 340 patient samples from three public data sets as validation cohort and used Kaplan–Meier survival curve using univariate and multivariate analysis.
Materials and Methods
Patients and breast cancer tissues
Human cells and specimens were obtained for previous research,20 deidentified, and reused for this study. This research was therefore exempt from the requirement for ethics committee approval under US regulations §46.101(b)(4). The research was conducted in accordance with the standards in the Declaration of Helsinki. Breast tumor samples were obtained from patients undergoing surgery after informed written consent (Apollo Hospital). The excised tumor specimens were immediately preserved in RNA-later (Life Technologies) and stored at 4 °C until shipment. All tumor samples utilized in this study were invasive ductal carcinoma (Supplementary Table 1). All the histopathological information used in the analysis was directly documented from the original pathology reports. Grading, tumor type, ERα status, PR status, and HER2 status had been routinely recorded at the Apollo Hospital.
Public microarray data sets
Due to limited availability of clinical information in our data set, we used independent data sets to assess the predictive ability of our gene signatures, which will give us additional confidence in clinical validity. The data sets with Gene Expression Omnibus (GEO) accession numbers GSE2740, GSE1992, and GSE2607 generated using Agilent microarray platform have been previously described.21–23 A total of 141 patient samples in GSE2740, 152 patient samples in GSE1992, and 102 patient samples in GSE2607 were used. A total of 395 patient samples with histopathological information are available in Supplementary Table 2. Patient samples with missing survival information were omitted; hence, a total of 340 samples were used for further analysis (Table 1). The clinical data were extracted from the gene expression data files downloaded from GEO. The ERα status, nodal status, survival data, and gene expression data were used for Kaplan–Meier survival curve and Cox multivariate analysis.
Table 1.
The range of expression for seven genes and a median value used to stratify patient samples from public data sets into the groups of high expression and low expression.
| GATA3 | NTN4 | SLC7A8 | MLPH | ENPP1 | LAMB2 | PLAT | |||
|---|---|---|---|---|---|---|---|---|---|
| Cohort I: 195 estrogen receptor positive (ER +) patient samples | |||||||||
| mRNA expression range | 1.7 to −4.1 | 2.7 to −3.9 | 1.5 to −4.0 | 2.2 to −2.1 | 2.5 to −2.5 | 1.1 to −1.2 | 2.8 to −3.2 | ||
| Cut-off value | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| Number of high expression samples | 95 | 96 | 97 | 91 | 97 | 96 | 95 | ||
| Number of low expression samples | 100 | 99 | 98 | 104 | 98 | 99 | 100 | ||
| Cohort II: 109 ER (+) and node (+) patient samples | |||||||||
| mRNA expression range | 1.4 to −3.0 | 1.0 to −3.3 | 2.3 to −1.4 | 2.8 to −2.7 | 1.6 to −2.0 | 1.2 to −1.1 | 2.8 to −3.2 | ||
| Cut-off value | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| Number of high expression samples | 22 | 22 | 22 | 22 | 22 | 22 | 22 | ||
| Number of low expression samples | 87 | 87 | 87 | 87 | 87 | 87 | 87 | ||
| Cohort III: 67 ER (+) samples with 3-gene combined expression | Cohort IV: 43 ER (+) Node (+) samples with 3-gene combined expression | ||||||||
| GATA3/NTN4/MLPH | GATA3/SLC7A8/MLPH | ||||||||
| Cut-off value | 0.0 | 0.0 | |||||||
| Number of high expression samples | 30 | 20 | |||||||
| Number of low expression samples | 37 | 23 | |||||||
Statistical analysis
To visualize gene expression values using heat maps, the values for each probe were centered by subtracting the mean expression value across patients. No gene-specific scaling (standardization) was performed, and, thus, information about the relative signal strength between probes was retained. The color tone in the heat maps was calibrated so that saturated red and saturated green were reached at values equal to 3.5-fold the standard deviation of the expression values of the entire matrix. Red and green reflect high- and low-expression levels (log 2-transformed scale), respectively.
The gene expression data from 340 patient samples were dichotomized according to the median (cutoff value 0.0) of the complete cohort. The expression data higher than the median were grouped into the “high-expression” group, and the expression values lesser than the median were grouped into the “low-expression” group (Table 1). Mann–Whitney t-test was used to evaluate the difference between mRNA levels of genes and clinicopathological parameters. Survival distributions were estimated using the Kaplan–Meier method (univariate), and the significance of differences between survival rates was ascertained by the log-rank test using the GraphPad software. Candidate prognostic factors for RFS with a 0.05 significance level in univariate analysis were entered in a multivariate Cox model.24 Multivariate analysis was evaluated by step-wise forward Cox’s regression analysis. The Cox proportional hazard model was used to calculate the hazard ratio (HR) and 95% confidence interval (CI) in the analysis of relapse-free survival (RFS). RFS was measured from the date of diagnosis to relapse or censored at the last follow-up. A P-value less than 0.05 were considered statistically significant.
Results
Relationship between mRNA levels of seven genes and clinicopathological parameters
Initially, we sought to corroborate the findings of our recent study.20 We correlated the mRNA expression of these seven genes with clinicopathological parameters. In our previous study,20 we had performed reverse transcription quantitative polymerase chain reaction (RT-qPCR) analyses of 76 tumor specimens and showed that the expression of aforementioned seven genes was significantly associated with ERα (+) tumors (P < 0.01). Herein, we utilized the same RT-qPCR data and classified it based on PR, HER2, tumor grade, and lymph node status (Supplementary Table 1). We observed that increased expression of the aforementioned seven genes was significantly associated with PR (+) breast tumors (P < 0.05). In contrast, no such association was found between mRNA expressions of these genes with HER2 receptor status and lymph node status (Table 2). Interestingly, six out of the seven genes did not show any association with regard to tumor grade. The only mRNA expression level of MLPH (P = 0.013) was significantly higher in grade I than grade III tumor. Given that the increased expression of each of the seven genes was associated with not only ERα (+) status but also PR (+) status, we ascertained if there was any correlation between the expression of these genes with ERα (+) and PR (+) breast tumors. We observed that this latter group of patients expresses statistically significant higher mRNA levels of GATA3, NTN4, SLC7A8, MLPH, ENPP1, LAMB2, and PLAT compared to triple negative [ERα (−)/PR (−)/HER2 (−)] breast tumors (Table 2). Interestingly among endocrine-treated patients, the previous report25 also showed that the presence of both ERα and PR was a stronger marker for the benefit of adjuvant endocrine therapy than ER alone. Accordingly, high expression of our gene signature could be used as a prediction marker for endocrine therapy.
Table 2.
The relation between mRNA levels and clinicopathological parameters.
| ESTROGEN RECEPTOR (ER) | PROGESTERONE RECEPTOR (PR) | HER2 | HISTOLOGICAL GRADE | NUMBER OF LYMPH NODES POSITIVE | PHENOTYPE | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POSITIVE (40) | NEGATIVE (36) | POSITIVE (42) | NEGATIVE (34) | POSITIVE (22) | NEGATIVE (54) | I (8) | II (36) | III (26) | 0 (25) | 1–3 (20) | >3 (22) | ER(+) | LUMINAL A SUBTYPE | LUMINAL B SUBTYPE | TRIPLE NEGATIVE (BASAL) | ||
| GATA3 mRNA | Median | 5.7 | 0.33 | 5.0 | 0.79 | 2.2 | 1.43 | 11.2 | 2.3 | 0.42 | 0.76 | 0.4 | 2.7 | 6.5 | 8.26 | 4.49 | 0.22 |
| P-value | <0.0001 | <0.0001 | 0.785 | 0.069 | 0.219 | 0.0039 | |||||||||||
| NTN4 mRNA | Median | 3.8 | 0.52 | 3.9 | 0.72 | 2.2 | 1.28 | 1.6 | 1.3 | 0.9 | 1.1 | 0.66 | 2.5 | 1.3 | 8 | 2.5 | 0.41 |
| P-value | <0.0001 | 0.0003 | 0.93 | 0.305 | 0.348 | 0.0127 | |||||||||||
| SLC7A8 mRNA | Median | 15.3 | 1.9 | 15.75 | 3.25 | 6.48 | 9.48 | 13.6 | 10.04 | 4.245 | 9.46 | 5.52 | 9.48 | 9.82 | 17 | 12.2 | 0.67 |
| P-value | <0.0001 | <0.0001 | 0.428 | 0.54 | 0.454 | 0.014 | |||||||||||
| MLPH mRNA | Median | 6.85 | 0.3 | 6.75 | 0.405 | 4.385 | 2.66 | 21.7 | 3.3 | 1.36 | 2.02 5 | 1.15 | 5.46 | 6.15 | 6.88 | 5.16 | 0.13 |
| P-value | <0.0001 | <0.0001 | 0.271 | 0.013 | 0.212 | 0.02 | |||||||||||
| ENPP1 mRNA | Median | 4.55 | 0.79 | 3.81 | 0.95 | 1.92 | 2.72 | 2.8 | 3.25 | 1.18 | 1.95 | 0.95 | 4.1 | 4.55 | 5.93 | 2.35 | 0.38 |
| P-value | <0.0001 | <0.0002 | 0.838 | 0.253 | 0.527 | 0.0003 | |||||||||||
| LAMB2 mRNA | Median | 5.87 | 0.94 | 6.1 | 1.05 | 2.81 | 3.16 | 7.53 | 3 | 1.01 | 3 | 1.09 | 3.69 | 2.74 | 7.63 | 3.68 | 0.7 |
| P-value | 0.0006 | <0.0001 | 0.628 | 0.698 | 0.179 | 0.0014 | |||||||||||
| PLAT mRNA | Median | 1.62 | 0.59 | 1.62 | 0.6 | 1.28 | 1.05 | 2.41 | 1.7 | 0.67 | 1.15 | 0.78 | 2.26 | 0.69 | 2.96 | 1.04 | 0.21 |
| P-value | 0.03 | 0.027 | 0.975 | 0.544 | 0.101 | 0.6759 | |||||||||||
Note: Significant P-values are in italics.
Meta-analysis of seven genes deregulated in ERα (+) breast tumors
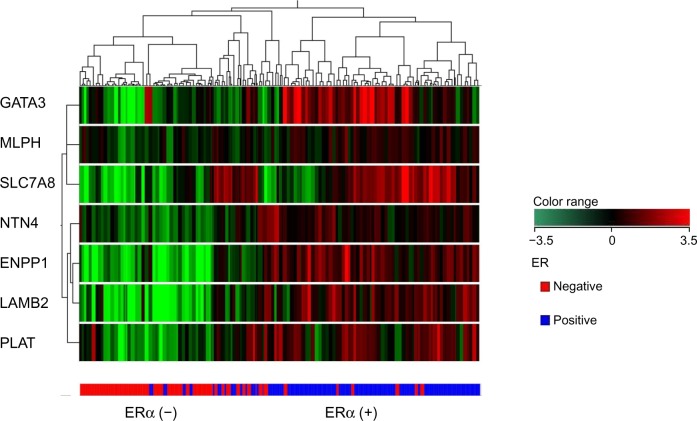
Next, we sought to correlate the mRNA expression of these seven genes with long-term survival data. Since we did not have an adequate number of clinical samples, we utilized published data sets to validate our predictive gene set. Further use of independent data sets to assess the predictive ability of our gene signatures will give us additional confidence in clinical validity. Accordingly, gene expression data were obtained from three independent public data sets (n = 395).21–23 The 340 patient samples from public data sets were considered for further analysis based on the available survival information (Table 1; Supplementary Table 2). Out of 340 samples, there were 195 ERα (+) and 145 ERα (−) samples. The seven genes dysregulated in ERα (+) breast tumors (GATA3, NTN4, SLC7A8, MLPH, ENPP1, LAMB2, and PLAT) were utilized to cluster all the tumors and, subsequently, the data were visualized in the form of heat map (Fig. 1). Out of 195 ERα (+) and 145 ERα (−) samples, our seven genes could classify 181 ERα (+) samples and 135 ERα (−) samples correctly with 93% positive prediction. These proved prediction value of mRNA levels of these seven genes to classify ERα (+) and ERα (−) samples.
Figure 1.
Dendrogram of 340 breast cancer samples from public data sets. Unsupervised, hierarchical, uncentered Pearson distance (co-relation) clustering was performed to classify the seven genes into homogeneous clusters.
Note: The columns in the dendrogram represent the patient’s tumor samples, while the rows represent the genes classified into clusters based on similar expression patterns. The expression color bar demonstrates the limits of regulation on either direction. The ERα (+) tumor samples are colored blue and ERα (−) samples colored red.
High mRNA expressions of GATA3, NTN4, and MLPH are associated with longer RFS in ERα (+) breast tumors
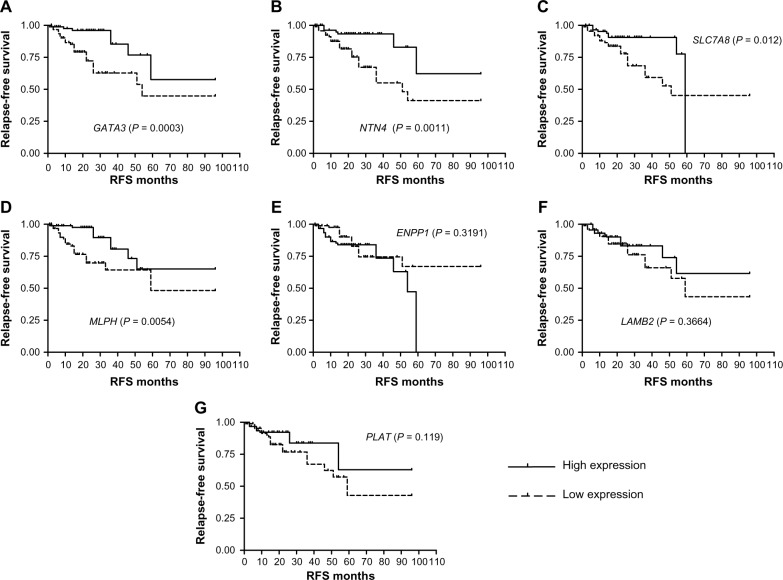
The gene expression values from the public data sets were dichotomized according to the median of the complete cohort, and expression data higher than the median were grouped into the high-expression group, and the expression values lesser than the median were grouped into the low-expression group (Table 1). Univariate analysis on ERα (+) test data sets (n = 195; Supplementary Table 3) revealed that high mRNA expression levels of GATA3 (P = 0.0003), NTN4 (P = 0.0011), SLC7A8 (P = 0.012), and MLPH (P = 0.0054) were significantly associated with longer RFS. Cox multivariate analysis revealed that GATA3 (P = 0.0167), NTN4 (P = 0.0044), and MLPH (P = 0.0321) were independent prognostic markers and significantly associated with RFS (Fig. 2; Table 3).
Figure 2.
Kaplan–Meier survival curve using high and low mRNA expression among ERα (+) breast tumors from public data sets (n = 195). Univariate analysis (n = 195) revealed that high mRNA expression levels of (A) GATA3 (P = 0.0003), (B) NTN4 (P = 0.0011), (C) SLC7A8 (P = 0.012), (D) MLPH (P < 0.0054), (E) ENPP1 (P = 0.3191), (F) LAMB2 (P = 0.3664), and (G) PLAT (P = 0.119) were significantly associated with longer relapse-free survival. A significance of difference between survival rates was ascertained by the log-rank test and P-value less than 0.05 was considered statistically significant.
Table 3.
The univariate and multivariate analysis in relation to RFS among 195 ERα (+) breast cancer patient samples from public data sets.
| GENES | UNIVARIATE | MULTIVARIATE | |||||
|---|---|---|---|---|---|---|---|
| HAZARD RATIO | 95% CI | P-VALUE | HAZARD RATIO | 95% CI | P-VALUE | LIKELIHOOD RATIO | |
| GATA3 | 0.248 | 0.1360 to 0.5570 | 0.0003 | 0.338 | 0.139 to 0.822 | 0.0167 | 271.19 |
| NTN4 | 0.274 | 0.1542 to 0.6257 | 0.0011 | 0.275 | 0.113 to | 0.0044 | 261.97 |
| SLC7A8 | 0.392 | 0.1998 to 0.8195 | 0.012 | N/A | N/A | NS | N/A |
| MLPH | 0.367 | 0.1796 to 0.7425 | 0.0054 | 0.418 | 0.188 to | 0.0321 | 256.55 |
| ENPP1 | 1.417 | 0.7054 to 2.918 | 0.3191 | N/A | N/A | N/A | N/A |
| LAMB2 | 0.725 | 0.3599 to 1.458 | 0.3664 | N/A | N/A | N/A | N/A |
| PLAT | 0.570 | 0.2805 to 1.156 | 0.119 | N/A | N/A | N/A | N/A |
Note: Significant P-values are in italics.
Abbreviations: NS, non-significant; N/A, not available; CI, confidence interval; ER, estrogen receptor; RFS, relapse-free survival.
High mRNA expressions of GATA3, SLC7A8, and MLPH are associated with longer RFS in ERα (+) and node (+) breast tumors
Having studied the prognostic significance of the seven dysregulated genes in ERα (+) patients, we next sought to perform our analysis with the clinically important issue of the metastatic spread of the tumor. The determination of the extent of lymph node involvement in primary breast cancer is the single most important risk factor in disease outcome. Accordingly, we next investigated the correlation of these seven dysregulated genes with ERα (+) and node (+) cohort (n = 109; Supplementary Table 4). In univariate analysis, high mRNA expression of GATA3 (P = 0.003), SLC7A8 (P = 0.0045), MLPH (P = 0.0021), and PLAT (P = 0.0311) showed significantly longer RFS. In Cox multivariate analysis, high mRNA expression of GATA3 (P = 0.0009), SLC7A8 (P = 0.0431), and MLPH (P = 0.0170) showed significantly longer RFS. In contrast, high mRNA expression of ENPP1 (P = 0.0001) was significantly associated with the worst RFS (Table 4). Interestingly, in a subset analysis of ERα (+) and node (−) cohort of patients (n = 84), none of the genes persisted in multivariate analysis (Table 5).
Table 4.
Univariate and multivariate analysis in relation to RFS among 109 ERα (+) and node (+) breast cancer patient samples from public data sets.
| GENES | UNIVARIATE | MULTIVARIATE | |||||
|---|---|---|---|---|---|---|---|
| HAZARD RATIO | 95% CI | P-VALUE | HAZARD RATIO | 95% CI | P-VALUE | LIKELIHOOD RATIO | |
| GATA3 | 0.2861 | 0.1490 to | 0.003 | 0.148 | 0.047 to | 0.0009 | 191.67 |
| NTN4 | 0.5296 | 0.2480 to 1.134 | 0.1018 | N/A | N/A | N/A | N/A |
| SLC7A8 | 0.3033 | 0.1522 to | 0.0045 | 0.372 | 0.140 to | 0.0431 | 180.58 |
| MLPH | 0.2785 | 0.1392 to | 0.0021 | 0.293 | 0.106 to | 0.0170 | 210.32 |
| ENPP1 | 2.27 | 1.099 to 5.103 | 0.0278 | 5.853 | 2.318 to | 0.0001 | 200.47 |
| LAMB2 | 0.584 | 0.2566 to 1.209 | 0.1389 | N/A | N/A | N/A | N/A |
| PLAT | 0.4233 | 0.2055 to | 0.0311 | N/A | N/A | NS | N/A |
Note: Significant P-values are in italics.
Abbreviations: NS, non-significant; N/A, not available; CI, confidence interval; ER, estrogen receptor; RFS, relapse-free survival.
Table 5.
Univariate and multivariate analysis in relation to RFS among 84 ERα (+) and node (−) breast cancer patient samples from public data sets.
| GENES | UNIVARIATE | MULTIVARIATE | |||||
|---|---|---|---|---|---|---|---|
| HAZARD RATIO | 95% CI | P-VALUE | HAZARD RATIO | 95% CI | P-VALUE | LIKELIHOOD RATIO | |
| GATA3 | 0.451 | 0.06059 to 3.606 | 0.4656 | N/A | |||
| NTN4 | 0.2637 | 0.02665 to 1.949 | 0.1769 | N/A | |||
| SLC7A8 | 0 | 0.007922 to 0.5021 | 0.009 | NS | |||
| MLPH | N/A | N/A | 0.172 | N/A | |||
| ENPP1 | 0.2637 | 0.02665 to 1.949 | 0.1769 | N/A | |||
| LAMB2 | 0 | 0.01232 to 0.7328 | 0.0239 | NS | |||
| PLAT | 4.5 | 0.7140 to 78.62 | 0.0931 | N/A | |||
Note: Significant P-values are in italics.
Abbreviations: NS, non-significant; N/A, not available; CI, confidence interval; RFS, relapse-free survival; ER, estrogen receptor.
Elevated expression of three-gene signature improves RFS
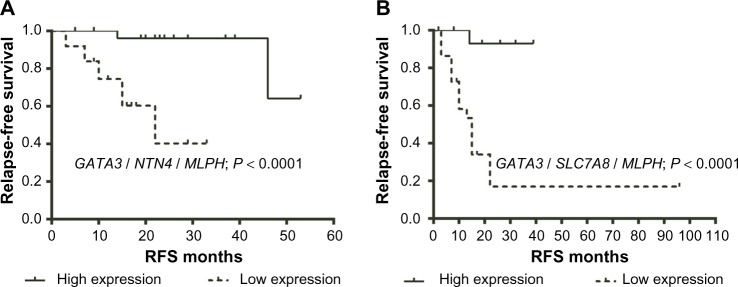
In our previous analyses, we had ascertained the predictive power of “individual” genes. We next sought to determine the “combined” predictive power of the three-gene signature. Accordingly, patients who expressed high mRNA levels of “all” three genes were studied against those who expressed low mRNA levels of all three genes with the cutoff value of zero. Initially, we looked into the 195 samples of ERα (+) cohort. In multivariate analysis, the three genes GATA3, NTN4, and MLPH showed independent prognostic value and were significantly associated with RFS. Hence, to determine the combined prediction value of all three genes, the 67 patient samples expressing similar expression of all three genes were selected (Supplementary Table 5). Patients having the high mRNA expression of all three genes GATA3, NTN4, and MLPH (n = 30) were compared with patients having the low mRNA expression of all three genes (n = 37). Multivariate analyses revealed that the three-gene signature showed better HR (0.056) and longer RFS than individual gene signature (Table 6; Fig. 3). Next, we looked into the 109 samples of ERα (+) and node (+) cohort. To determine the combined prediction value of all three genes GATA3, SLC7A8, and MLPH in this cohort, the 43 patient samples expressing similar expression of all three genes were selected (Supplementary Table 6). Patient samples showing high mRNA expression of all three genes GATA3, SLC7A8, and MLPH (n = 20) were compared with patients having low mRNA expression (n = 23) and observed that the combined three-gene signature showed better HR (0.057) than individual genes (Table 6; Fig. 3).
Table 6.
Univariate and multivariate analysis of three-gene signature in relation to RFS among ERα cohort (n = 67), and ERα (+) and node (+) samples (n = 43) from public data sets.
| SAMPLE CLASSIFICATION | UNIVARIATE | MULTIVARIATE | ||||||
|---|---|---|---|---|---|---|---|---|
| HIGH OR LOW EXPRESSION OF ALL 3-GENES | HAZARD RATIO | 95% CI | P-VALUE | HAZARD RATIO | 95% CI | P-VALUE | LIKELIHOOD RATIO | |
| ERα (+) cohort | GATA3/NTN4/MLPH | 0.1058 | 0.052 to 0.376 | <0.0001 | 0.056 | 0.0074 to 0.4268 | 0.0053 | 115.19 |
| ERα (+) and node (+) | GATA3/SLC7A8/MLPH | 0.0581 | 0.052 to 0.367 | <0.0001 | 0.057 | 0.0076 to 0.4320 | 0.0055 | 99.93 |
Note: Significant P-values are in italics.
Abbreviations: CI, confidence interval; ER, estrogen receptor.
Figure 3.
(A) Kaplan–Meier survival curve using high mRNA expression of all three genes GATA3, NTN4, and MLPH (n = 30) was compared with patients having the low mRNA expression of all three genes (n = 37) among ERα (+) breast cancer samples from public data sets (n = 67); (B) Kaplan–Meier survival curve using high mRNA expression of all three genes GATA3, SLC7A8, and MLPH (n = 20) was compared with patients having the low mRNA expression (n = 23) among ERα (+) and node (+) breast tumors from public data sets (n = 43). A significance of difference between survival rates was ascertained by the log-rank test and P-value less than 0.05 was considered statistically significant.
Discussion
The current method of determination of ERα status by immunohistochemistry under clinical setup provides information about the expression pattern of ERα with no information on possibly disabled downstream ER pathway.14,17 Thus, it is plausible that the status of the ER pathway is also clinically relevant and may explain variable response to endocrine therapy in ERα (+) patients. Hence, measurements of gene expression profiles that reflect the activity of ER pathway could provide an important insight in understanding the behavior of breast cancers. We reported the expression pattern for seven genes that were regulated by ERα and demonstrated the differential expression in various phenotypes of breast cancer pathology. We observed the highest expression of five genes in ERα (+) and PR (+) cohort of patients (luminal A subtype). Interestingly, among endocrine-treated patients, previous report25 also showed that the presence of both ERα and PR was a stronger marker for the benefit of adjuvant endocrine therapy than ER alone.
Due to limited availability of clinical information in our data set, we used independent data sets21–23 to assess the predictive ability of our gene signatures, which will give us additional confidence in clinical validity. Meta-analysis of seven genes showed that a gene expression is reliable and robust to determine ER expression in three independent data sets comprising 340 tumor samples. These analyses strengthen our recent findings and suggest that a seven-gene signature can stratify/classify ERα (+) and ERα (−) tumors in an independent data set. Of the seven genes analyzed in the ERα (+) tumors, the high mRNA expression of GATA3, NTN4, and MLPH emerged as an independent prognostic factor in multivariate analysis. In ERα (+) and node (+) tumors, the high mRNA levels of GATA3, SLC7A8, and MLPH emerged as an independent prognostic factor in multivariate analysis. In our findings, we did not find any literature describing the importance of SLC7A8 and MLPH in prognosis and overall disease-free survival of breast cancer patients.
Classic parameters, such as ER, PR, HER2 status, number of lymph nodes positive, and tumor size, have been integrated into software applications such as Adjuvant! Online19 to help doctors in calculating a risk of relapse and benefit from adjuvant therapy. However, uncertainty remains in many cases even with the use of this software. The well-established prognostic and/or therapeutic breast cancer markers are hormone receptors (ER and PR),26 HER2,27 Ki-67 antigen,28 tumor protein p53,29 carbohydrate 15–3 and carcinoembryonic antigens (CA 15–3 and CEA),30,31 and breast cancer susceptibility genes (BRCA1 and BRCA2).32 Gene signatures can complement classic prognostic factors to obtain more accurate prognostic information. The 70-gene signature (MammaPrint; Agendia) and the 21-gene signature (OncoType; Genomic Health) are being used in selected patients with early ERα (+) disease to identify those women who will be cured even if they do not receive adjuvant chemotherapy.4,33 These signatures have been extensively studied and are widely used in Europe and USA.34–36 The National Cancer Comprehensive Network guidelines indicate that the 21-gene signature can be considered in women with tumors >0.5 cm, HER2-negative disease, and node-negative disease.37 Limitations of the 21-gene and 70-gene signatures are intended to be used by women with node-negative breast cancer diagnosis. Many other gene signatures have been developed and have undergone validation. One of them is the breast cancer gene expression ratio test, which only measures the ratio of HOXB13 to IL17BR.38,39 A high mRNA expression ratio was associated with a high risk of recurrence in tamoxifen-treated patients. Recently, the accuracy of this test could be improved by including proliferation-associated genes of the molecular grade index,40 which is an RT-qPCR assay consisting of five genes that are able to identify a subgroup of ER (+) patients with a worse outcome despite endocrine therapy. The Rotterdam 76-gene signature was created on the basis of predicting the development of metastatic disease within 5 years using an unselected patient cohort regarding age, tumor size, grade, and hormone receptor status. 41,42
The 5-year survival for patients with the node-negative disease is 82.8% compared with 73% for 1–3 positive nodes, 45.7% for 4–12 positive nodes, and 28.4% for ≥13 positive nodes.43 These data demonstrate that the risk of recurrence is significant enough with lymph node-positive disease to warrant adjuvant systemic therapy since, generally, a future risk of distant recurrence of 20% or greater is regarded significant enough to consider the risks of therapy. Hence, it is important to stratify ERα (+) and node (+) patients into a low- and high-risk group for RFS. In this study’s sample population, out of 109 ER (+) and node (+) samples, 67 samples showed either high or low expression of all GATA3/NTN4/MLPH genes, whereas 43 samples showed either high or low expression of one or two GATA3/SLC7A8/MLPH genes. So the limitation of this gene signature is that not all patient samples will exhibit either low or high gene expression of all three genes and has to be excluded from the prediction.
The GATA3 transcription factor has been studied intensively in the immune system but has most recently been shown to be important in the context of breast cancer and the ERα pathway. Recently, it was demonstrated that GATA3 is required for estradiol stimulation of cell-cycle progression of breast cancer cells.44 Meta-analysis of four breast cancer microarray data sets revealed GATA3 as a promising novel prognostic biomarker in breast cancer with HR of 0.12 and P-value of 0.05.45 As reported in the present study, combined three-gene signature showed better HR and improved RFS. The netrins are a family of secreted proteins that are highly conserved through evolution. Three netrins, netrin1 (NTN1), netrin 3 (NTN3), and netrin 4 (NTN4), have been identified. Both the netrins and their receptors are widely expressed and have been implicated in a wide range of development processes including axon guidance, angiogenesis, and mammary gland development.46,47 In an earlier study, NTN4 expression was demonstrated to be an independent predictor of improved outcome with a HR of 0.2544 and P-value of 0.015.48 Our study reports three-gene signature with better HR and improved RFS in ERα (+) tumors.
In summary, we show that the high expression of estrogen-responsive three-gene signature GATA3/NTN4/MLPH is associated with good prognosis in ERα (+) patients and GATA3/SLC7A8/MLPH is associated with good prognosis in ERα (+) and node (+) patients and are promising novel prognostic biomarkers in breast cancer. Our gene sets may have potential clinical utility since they demonstrated predictive ability in three independent public data sets. Although our results are promising, it needs to be validated in large cohort breast cancer patients. Such signatures will also be the starting point for functional profiling of genes and proteins to understand the biological processes associated with disease formation and progression.
Supplementary Materials
Supplementary Table 1. 76 breast cancer patients collected from Apollo Hospital, Chennai, India.
Supplementary Table 2. Clinicopathological information for three public datasets.
Supplementary Table 3. RT-qPCR data of 195 ER (+) patient samples from public datasets.
Supplementary Table 4. RT-qPCR data of 109 ER (+) and node (+) patient samples from public datasets.
Supplementary Table 5. Three-gene RT-qPCR data of 67 ER (+) patient samples from public datasets.
Supplementary Table 6. Three-gene RT-qPCR data of 43 ER (+) and node (+) patient samples from public datasets.
Acknowledgments
Debarshi Chakrabarti provided scientific input, discussions and deliberations during the time of the study.
Footnotes
ACADEMIC EDITOR: Karen Pulford, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,337 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: AT, MP, AB, HR. Analyzed the data: AT. Wrote the first draft of the manuscript: AT. Contributed to the revising manuscript critically for important intellectual content: AT, AB, BM, MP. All authors made an intellectual input and contributed to writing the paper. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Russo J, Hu YF, Yang X, Russo IH. Developmental, cellular, and molecular basis of human breast cancer. J Natl Cancer Inst Monogr. 2000;(27):17–37. doi: 10.1093/oxfordjournals.jncimonographs.a024241. [DOI] [PubMed] [Google Scholar]
- 2.Lonning PE, Sorlie T, Perou CM, Brown PO, Botstein D, Borresen-Dale AL. Microarrays in primary breast cancer – lessons from chemotherapy studies. Endocr Relat Cancer. 2001;8(3):259–63. doi: 10.1677/erc.0.0080259. [DOI] [PubMed] [Google Scholar]
- 3.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100(18):10393–8. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 5.Huang E, Cheng SH, Dressman H, et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361(9369):1590–6. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 6.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis C, Shah SP, Chin SF, et al. METABRIC Group The genomic and tran-scriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11(16):5678–85. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 12.Rody A, Karn T, Solbach C, et al. The erbB2+ cluster of the intrinsic gene set predicts tumor response of breast cancer patients receiving neoadjuvant chemotherapy with docetaxel, doxorubicin and cyclophosphamide within the GEPAR-TRIO trial. Breast. 2007;16(3):235–40. doi: 10.1016/j.breast.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Ring BZ, Seitz RS, Beck R, et al. Novel prognostic immunohistochemical bio-marker panel for estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(19):3039–47. doi: 10.1200/JCO.2006.05.6564. [DOI] [PubMed] [Google Scholar]
- 14.Bonneterre J, Thürlimann B, Robertson JF, et al. Anastrozole versus tamoxifen as first-line therapy for advanced breast cancer in 668 postmenopausal women: results of the Tamoxifen or Arimidex Randomized Group Efficacy and Toler-ability Study. J Clin Oncol. 2000;18(22):3748–57. doi: 10.1200/JCO.2000.18.22.3748. [DOI] [PubMed] [Google Scholar]
- 15.Colozza M, Azambuja E, Cardoso F, Sotiriou C, Larsimont D, Piccart MJ. Pro-liferative markers as prognostic and predictive tools in early breast cancer: where are we now? Ann Oncol. 2005;16(11):1723–39. doi: 10.1093/annonc/mdi352. [DOI] [PubMed] [Google Scholar]
- 16.Hayes DF. Prognostic and predictive factors revisited. Breast. 2005;14(6):493–9. doi: 10.1016/j.breast.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Mouridsen H, Gershanovich M, Sun Y, et al. Superior efficacy of letrozole versus tamoxifen as first-line therapy for postmenopausal women with advanced breast cancer: results of a phase III study of the International Letrozole Breast Cancer Group. J Clin Oncol. 2001;19(10):2596–606. doi: 10.1200/JCO.2001.19.10.2596. [DOI] [PubMed] [Google Scholar]
- 18.D’Eredita G, Giardina C, Martellotta M, Natale T, Ferrarese F. Prognostic factors in breast cancer: the predictive value of the Nottingham Prognostic Index in patients with a long-term follow-up that were treated in a single institution. Eur J Cancer. 2001;37(5):591–6. doi: 10.1016/s0959-8049(00)00435-4. [DOI] [PubMed] [Google Scholar]
- 19.Olivotto IA, Bajdik CD, Ravdin PM, et al. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol. 2005;23(12):2716–25. doi: 10.1200/JCO.2005.06.178. [DOI] [PubMed] [Google Scholar]
- 20.Thakkar AD, Raj H, Chakrabarti D, et al. Identification of gene expression signature in estrogen receptor positive breast carcinoma. Biomark Cancer. 2010;2:1–15. doi: 10.4137/BIC.S3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh DS, Troester MA, Usary J, et al. Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol. 2006;24(11):1656–64. doi: 10.1200/JCO.2005.03.2755. [DOI] [PubMed] [Google Scholar]
- 23.Perreard L, Fan C, Quackenbush JF, et al. Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. Breast Cancer Res. 2006;8(2):R23. doi: 10.1186/bcr1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox C. Multinomial regression models based on continuation ratios. Stat Med. 1988;7(3):435–41. doi: 10.1002/sim.4780070309. [DOI] [PubMed] [Google Scholar]
- 25.Bardou VJ, Arpino G, Elledge RM, Osborne CK, Clark GM. Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol. 2003;21(10):1973–9. doi: 10.1200/JCO.2003.09.099. [DOI] [PubMed] [Google Scholar]
- 26.Althuis MD, Fergenbaum JH, Garcia-Closas M, Brinton LA, Madigan MP, Sherman ME. Etiology of hormone receptor-defined breast cancer: a systematic review of the literature. Cancer Epidemiol Biomarkers Prev. 2004;13(10):1558–68. [PubMed] [Google Scholar]
- 27.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 28.Vielh P, Chevillard S, Mosseri V, Donatini B, Magdelenat H. Ki67 index and S-phase fraction in human breast carcinomas. Comparison and correlations with prognostic factors. Am J Clin Pathol. 1990;94(6):681–6. doi: 10.1093/ajcp/94.6.681. [DOI] [PubMed] [Google Scholar]
- 29.Dumay A, Feugeas JP, Wittmer E, et al. Distinct tumor protein p53 mutants in breast cancer subgroups. Int J Cancer. 2013;132(5):1227–31. doi: 10.1002/ijc.27767. [DOI] [PubMed] [Google Scholar]
- 30.Ebeling FG, Stieber P, Untch M, et al. Serum CEA and CA 15–3 as prognostic factors in primary breast cancer. Br J Cancer. 2002;86(8):1217–22. doi: 10.1038/sj.bjc.6600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vizcarra E, Lluch A, Cibrián R, et al. Value of CA 15.3 in breast cancer and comparison with CEA and TPA: a study of specificity in disease-free followup patients and sensitivity in patients at diagnosis of the first metastasis. Breast Cancer Res Treat. 1996;37(3):209–16. doi: 10.1007/BF01806502. [DOI] [PubMed] [Google Scholar]
- 32.Senst N, Llacuachaqui M, Lubinski J, et al. Hereditary Breast Cancer Study Group Parental origin of mutation and the risk of breast cancer in a prospective study of women with a BRCA1 or BRCA2 mutation. Clin Genet. 2013;84(1):43–6. doi: 10.1111/cge.12037. [DOI] [PubMed] [Google Scholar]
- 33.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 34.Albain KS, Barlow WE, Shak S, et al. Breast Cancer Intergroup of North America. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11(1):55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mook S, Schmidt MK, Viale G, et al. TRANSBIG Consortium The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1–3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat. 2009;116(2):295–302. doi: 10.1007/s10549-008-0130-2. [DOI] [PubMed] [Google Scholar]
- 36.Mittempergher L, de Ronde JJ, Nieuwland M, et al. Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PLoS One. 2011;6(2):e17163. doi: 10.1371/journal.pone.0017163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theriault RL, Carlson RW, Allred C, National Comprehensive Cancer Network Breast cancer, version 3. featured updates to the NCCN guidelines. J Natl Compr Canc Netw. 2013;2013;11(7):753–60. doi: 10.6004/jnccn.2013.0098. quiz 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma XJ, Hilsenbeck SG, Wang W, et al. The HOXB13:IL17BR expression index is a prognostic factor in early-stage breast cancer. J Clin Oncol. 2006;24(28):4611–9. doi: 10.1200/JCO.2006.06.6944. [DOI] [PubMed] [Google Scholar]
- 39.Ma XJ, Salunga R, Dahiya S, et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res. 2008;14(9):2601–8. doi: 10.1158/1078-0432.CCR-07-5026. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Dahiya S, Provencher H, et al. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007;13(21):6327–34. doi: 10.1158/1078-0432.CCR-07-0310. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 42.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239–46. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 43.Fisher B, Bauer M, Wickerham DL, et al. Relation of number of positive axillary nodes to the prognosis of patients with primary breast cancer. An NSABP update. Cancer. 1983;52(9):1551–7. doi: 10.1002/1097-0142(19831101)52:9<1551::aid-cncr2820520902>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 44.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67(13):6477–83. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 45.Mehra R, Varambally S, Ding L, et al. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res. 2005;65(24):11259–64. doi: 10.1158/0008-5472.CAN-05-2495. [DOI] [PubMed] [Google Scholar]
- 46.Hinck L. The versatile roles of “axon guidance” cues in tissue morphogenesis. Dev Cell. 2004;7(6):783–93. doi: 10.1016/j.devcel.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Yamamura J, Miyoshi Y, Tamaki Y, et al. mRNA expression level of estrogen-inducible gene, alpha 1-antichymotrypsin, is a predictor of early tumor recurrence in patients with invasive breast cancers. Cancer Sci. 2004;95(11):887–92. doi: 10.1111/j.1349-7006.2004.tb02198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esseghir S, Kennedy A, Seedhar P, et al. Identification of NTN4, TRA1, and STC2 as prognostic markers in breast cancer in a screen for signal sequence encoding proteins. Clin Cancer Res. 2007;13(11):3164–73. doi: 10.1158/1078-0432.CCR-07-0224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. 76 breast cancer patients collected from Apollo Hospital, Chennai, India.
Supplementary Table 2. Clinicopathological information for three public datasets.
Supplementary Table 3. RT-qPCR data of 195 ER (+) patient samples from public datasets.
Supplementary Table 4. RT-qPCR data of 109 ER (+) and node (+) patient samples from public datasets.
Supplementary Table 5. Three-gene RT-qPCR data of 67 ER (+) patient samples from public datasets.
Supplementary Table 6. Three-gene RT-qPCR data of 43 ER (+) and node (+) patient samples from public datasets.