Summary
Each Pseudomonas aeruginosa cell localizes two types of motility structures, a single flagellum and one or two clusters of type IV pili, to the cell poles. Previous studies suggested that these motility structures arrive at the pole through distinct mechanisms. Here we performed a swimming motility screen to identify polar flagellum localization factors and discovered three genes homologous to the TonB/ExbB/ExbD complex that have defects in both flagella-mediated swimming and pilus-mediated twitching motility. We found that deletion of tonB3, PA2983 or PA2982 led to non-polar localization of the flagellum and FlhF, which was thought to sit at the top of the flagellar localization hierarchy. Surprisingly, these mutants also exhibited pronounced changes in pilus formation or localization, indicating that these proteins may co-ordinate both the pilus and flagellum motility systems. Thus, we have renamed PA2983 and PA2982, pocA and pocB, respectively, for polar organelle co-ordinator to reflect this function. Our results suggest that TonB3, PocA and PocB may form a membrane-associated complex, which we term the Poc complex. These proteins do not exhibit polar localization themselves, but are required for increased expression of pilus genes upon surface association, indicating that they regulate motility structures through either localization or transcriptional mechanisms.
Introduction
The list of polarly localized bacterial factors continues to grow, but the mechanisms that regulate their production and localization remain poorly understood. For example, PopZ and HubP spatially co-ordinate specific subsets of polarly localized proteins in Caulobacter crescentus and Vibrio cholerae, but the mechanisms by which the production of polar structures is co-ordinated remains unknown (Bowman et al., 2010; Yamaichi et al., 2012). Furthermore, neither PopZ nor HubP are widely conserved such that the co-ordination of polar structures in most bacteria remains mysterious. To address this topic, we have focused on the motility structures of the Gram-negative bacterium Pseudomonas aeruginosa. P. aeruginosa, a clinically important opportunistic pathogen, produces a single polar flagellum and multiple polar type IV pili which contribute both to motility and to virulence (Amako and Umeda, 1982; Mattick, 2002), presenting excellent targets for understanding the mechanisms of polar protein localization. During the first stages of infection, the flagellum and pili are required for adherence to mammalian host cells (Hahn, 1997; Feldman et al., 1998). P. aeruginosa is predicted to bind to host mucin using surface structures, such as the flagellum, to remain in the lower respiratory tract (Scharfman et al., 2001). Once an infection has become established, chronic P. aeruginosa infection is maintained through strategies that increase resistance to antibiotic therapy, including formation of biofilms (Pier, 2002), a process that requires both flagella and type IV pili (O’Toole and Kolter, 1998).
Assembly of the P. aeruginosa flagellum results from a highly organized series of events, requiring approximately 50 genes. Flagellar gene transcription is divided into a hierarchy of four classes, each expressed in a specific temporal order (Chilcott and Hughes, 2000; Dasgupta et al., 2003). Although transcriptional regulation represents a major control mechanism for organizing flagellum assembly, it does not provide an explanation for the controlled placement of the flagellum at the cell pole. A putative GTP-binding protein FlhF is polarly localized and required for polar localization of the Pseudomonas flagellum (Pandza et al., 2000), such that FlhF has been considered to sit at the top of the flagellar localization genetic hierarchy. Misplacement of the flagellum to the lateral side of the cell, as is seen in a flhF mutant, results in a dramatic decrease in swimming motility despite a fully assembled, rotating flagellum (Pandza et al., 2000; Murray and Kazmierczak, 2006). FlhF has sequence similarity to the receptor protein of the bacterial signal recognition particle (SRP), a ribonucleoprotein complex that contains several proteins and the 4.5S RNA (Fekkes and Driessen, 1999). The factors that regulate FlhF localization remain unknown.
Type IV pili have a multitude of functions including twitching and gliding motility, adhesion, immune escape, DNA uptake, biofilm formation, microcolony formation, secretion, phage transduction and signal transduction. Pilus assembly and disassembly occurs rapidly using molecular motors dependent on ATP, and requires a dozen or more proteins (Mattick, 2002). Previously, we reported that P. aeruginosa type IV pilus formation is dependent on the actin homologue MreB, while flagellum production and FlhF localization are MreB-independent (Cowles and Gitai, 2010). Based on these results, we hypothesized that multiple, potentially independent, pathways are responsible for polar localization of P. aeruginosa pili and flagella: one pathway that requires MreB function and one pathway that is MreB-independent and involves FlhF.
To further elucidate the mechanisms of polar localization and identify additional proteins involved in this process, we identified three mutants with both swimming and twitching defects, suggesting that some proteins may be involved in regulating both flagella and pili. These genes, tonB3, PA2983 and PA2982 are homologous to the TonB–ExbB–ExbD complex and PA2983 and PA2982 associated in co-precipitation experiments, suggesting that, like their Escherichia coli homologues, they form a membrane-associated complex. We refer to this putative complex as the Poc (polar organelle co-ordinator) complex. We found that deletion of any one of these genes leads to random placement of the flagella, as well as defects in pili formation or localization, depending on the mutation. Furthermore, upregulation of pilus gene transcription upon surface association is absent in tonB3 and PA2982 deletion mutants, indicating involvement in pilus gene transcriptional control. These factors thus provide evidence that flagella and pili can be co-ordinated by a common set of proteins.
Results
Polar localization of flagella and type IV pili occurs independently
Typical P. aeruginosa cells contain a single flagellum and multiple type IV pili, both located at the pole. Our previous work suggests that each structure arrives at the pole through separate mechanisms. Pili can be located at the same pole as the flagellum, the opposite pole, or both poles. Furthermore, our previous discovery that pili production is upregulated upon surface association, while flagella are not, as well as the involvement of the actin homologue MreB specifically in pilus and not flagellum localization, suggests that these two structures are localized by separate mechanisms (Cowles and Gitai, 2010).
To test this hypothesis more directly, we used time-lapse microscopy to investigate whether localization of pilus and flagella structures is co-ordinated throughout the cell cycle using the GTPase FlhF and the chemotaxis histidine kinase CheA as markers for flagella localization and a previously characterized pilus retraction ATPase PilT as a marker for pilus localization (Cowles and Gitai, 2010). Previous work had shown that fluorescently labelled FlhF has a bipolar localization pattern with the focus of brightest intensity located at the flagellated pole, while CheA has a unipolar localization pattern at the flagellated pole (Guvener et al., 2006; Murray and Kazmierczak, 2006). We constructed a strain expressing arabinose-inducible C-terminal fusions of both FlhF (to cerulean, FlhF–CER) and CheA (to EGFP, CheA–EGFP), and compared FlhF–CER and CheA–EGFP localization to that of GFP–PilT. Here, we found that FlhF and CheA colocalized in immobile polar puncta throughout the cell cycle, with new puncta forming only at the new pole during cell division (Movies 1 and 2). In contrast, PilT puncta formation followed no predictable pattern, with puncta forming at either or both poles (Movie 3). Furthermore, we observed motile non-polar GFP–PilT puncta that seemed to move along the cell body until they reached a pole (Movie 3).
To examine the role of cell division and pole formation in pilus and flagella localization, we treated P. aeruginosa with cefsulodin to inhibit cell division and examined the localization patterns of FlhF–CER, CheA–EGFP and GFP–PilT in the resulting filamentous cells over time. FlhF–CER and CheA–EGFP began as bipolar puncta, but as the cell elongated, FlhF–CER and CheA–EGFP also became visible at the midpoint of the filament, and then at the quarters (Movies 4 and 5). In contrast, GFP–PilT formed a single unipolar focus that remained static throughout elongation of the filament. As the filament elongated, weak, seemingly mobile, foci simultaneously appeared at the midpoint and other pole (Movie 6). Furthermore, all three markers formed internal foci in filamented cells, splitting the filaments into equal compartments. This indicates that the curvature of the pole was not crucial for localization, and other mechanisms are therefore responsible for achieving polar localization of both flagella and pili.
Three hypothetical proteins are required for swimming motility and polar flagellum localization
Since flagella and pili localization occurred independently, we were interested in finding additional factors that contribute to localization of flagella or pili. To this end, we performed screens to find factors involved in their localization, beginning with flagella. flhF mutants that mislocalize flagella impair swimming such that we reasoned that additional regulators of flagella localization would also exhibit reduced swimming. We thus conducted a motility screen using the University of Washington P. aeruginosa PAO1 transposon mutant library (Jacobs et al., 2003). We screened 9440 mutants for swimming motility defects and found 241 mutants with reduced swimming compared with the wild-type parent. To differentiate mutants that had mislocalized flagella from those where no flagellum was produced, we used transmission electron microscopy (TEM) to determine the cellular location of the flagellum. The majority of mutants had insertions in known structural elements and did not produce a flagellum (data not shown). However, we also isolated mutants in two known regulators of flagellum number and localization, the anti-activator fleN and the GTPase flhF, respectively, confirming the validity of the screen.
Using this approach, we identified three new factors involved in polar flagellum localization, PA0406, PA2983 and PA2982. PA0406 had previously been renamed tonB3 (Huang et al., 2004) due to its similarity to E. coli tonB, a gene involved in energy transduction. PA2983 and PA2982 are predicted to encode hypothetical proteins, are co-transcribed (Fig. S1), and have sequence similarity to ExbB/MotA/TolQ and ExbD/MotB/TolR respectively. The homologues of the three proteins identified in our screen, namely TonB (PA0406 homologue), ExbB (PA2983 homologue) and ExbD (PA2982 homologue), represent a well-characterized complex in E. coli (Postle and Kadner, 2003). Available transposon insertions in the other TonB/ExbB/ExbD orthologues (tonB2, exbB1, exbB2, exbD1 and exbD2) showed no differences in swimming or twitching motility from wild-type (data not shown). Interestingly, a survey of several bacterial genomes suggested that the presence of multiple TonB homologues generally correlates with the production of a single, polar flagellum while peritrichous bacteria tend to have only one TonB (Supplemental Table S1). Thus, it is possible that TonB systems control polar flagellum localization in other bacteria as well.
To confirm the causality of the transposon insertion swimming phenotypes, we constructed in-frame, non-polar deletions of each gene individually and demonstrated that the deletion mutants had swimming motility defects comparable to a flhF mutant in a standard plate assay (Fig. 1). We were unable to construct a PA2983–PA2982 double mutant, suggesting that deletion of both genes together was lethal. To visualize swimming motility on a single cell basis, we used tracking software to follow cells under the microscope. While wild-type cells swam in straight lines and quickly passed through the field of view, the majority of tonB3, PA2983 and PA2982 cells moved in corkscrew-like patterns with little progress made in any one direction (Movies 7–10). As a control, we observed a non-flagellated strain, fliF, and found that these cells did not move at all (Movie 11). To complement the swimming defects, we provided each gene to the respective deletion strain in trans and showed that swimming motility was fully restored (Fig. S2). Additionally, we monitored bacterial growth in liquid media and found that all of the mutants grew like wild-type PAO1, indicating that swimming defects were not due to differences in growth (data not shown).
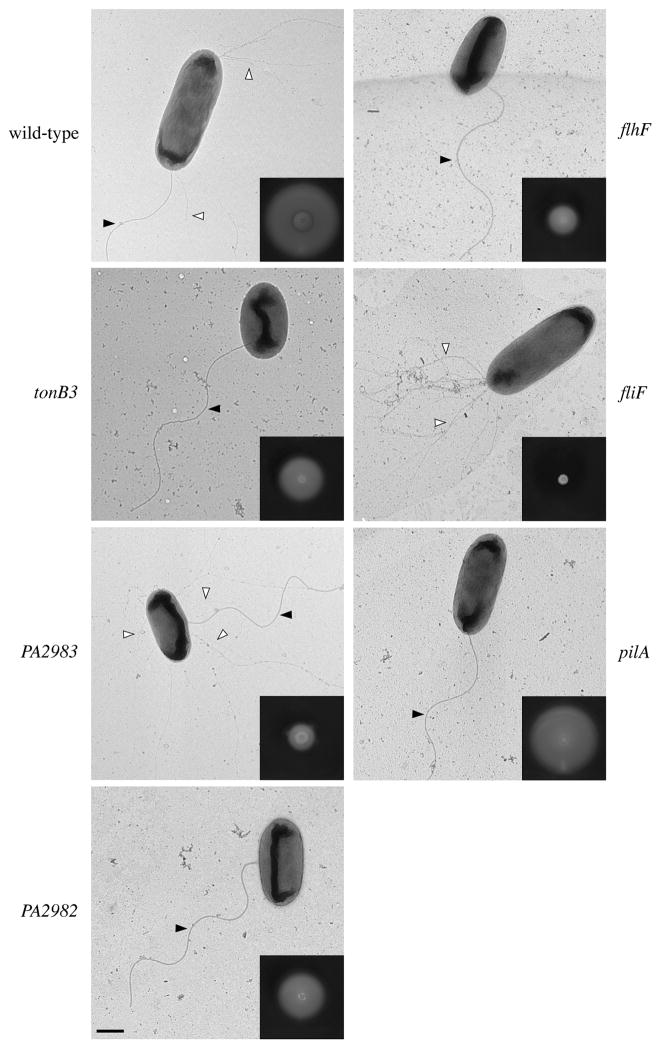
Fig. 1.
Swimming motility and flagella and type IV pili localization are disrupted in tonB3, PA2983 (pocA), PA2982 (pocB) and flhF mutants. Transmission electron microscopy (TEM) images of wild-type and mutant P. aeruginosa strains. fliF and pilA strains were used as controls that lacked flagella and pili respectively. Closed arrowheads denote flagella and open arrowheads indicate representative pili. Scale bar, 500 nm. (Inset) Representative images of swimming motility on 0.3% agar plates.
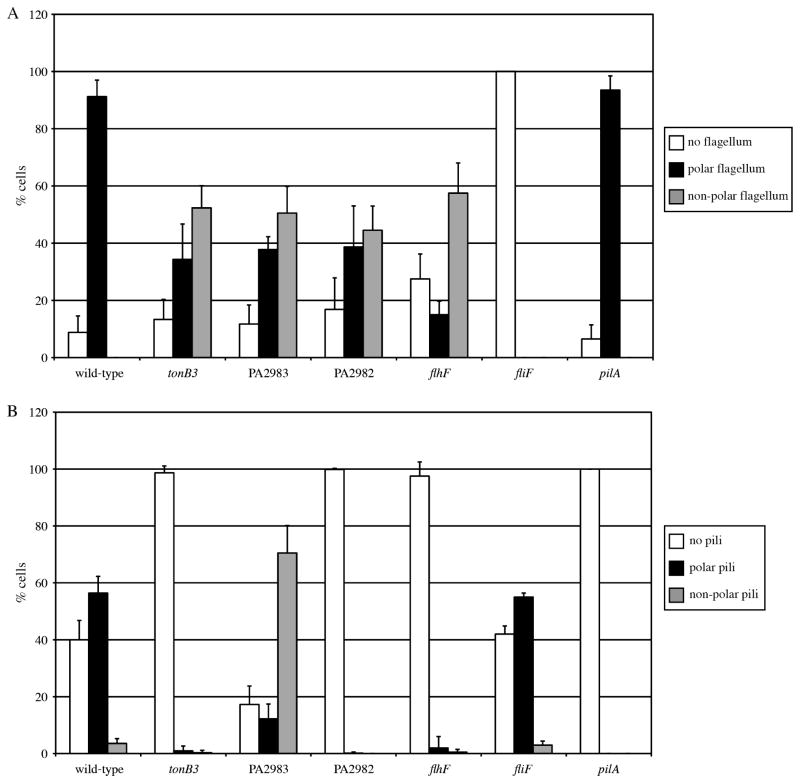
To determine if the swimming motility defects in the deletion mutants were also due to incorrect placement of the flagellum, we examined flagellum localization using TEM (Fig. 1). For these experiments, flagella were designated as non-polar if the base of the flagellum originated from the lateral side of the cell and were considered polar if the flagellum was located anywhere on the curved end of the cell. Using this definition, ~90% of wild-type cells had polar flagella, the remaining cells were non-flagellated, and no cells had non-polar flagella. In contrast, the deletion strains produced flagella at non-polar sites in approximately 50% of cells (Fig. 2A), which is the same proportion of cells with mislocalized flagella as seen in a flhF deletion (Fig. 2A). Under the growth conditions used in these experiments, an average P. aeruginosa cell measured 1.50 ± 0.18 μm long by 0.74 ± 0.07 μm wide (n = 30). We calculated surface areas by considering a cell to be a capsule-shaped object with a cylindrical body capped by two half spheres and determined that random placement of the flagellum would result in 50.6% of cells having the flagellum originate from the lateral sides of the cell and 49.4% of cells with a polar flagellum. Thus, deletion of tonB3, PA2983 or PA2982 results in an apparent random localization of the flagellum. Finally, we observed that tonB3, PA2983 and PA2982 mutant cells were qualitatively shorter than wild-type cells (Fig. 1). Quantification of cell size demonstrated that the mutant cells were ~15% shorter than wild-type (Supplemental Table S2).
Fig. 2.
Quantification of flagella and type IV pili localization by TEM. The distribution of flagella (A) and type IV pili (B) collected from populations at the leading edge of motility on the surface of agar plates. fliF and pilA strains were used as controls that lacked flagella and pili respectively. Error bars represent the standard deviation (n = 3 replicates of 100 cells each).
tonB3, PA2983 and PA2982 affect the subcellular localization of known flagellar factors
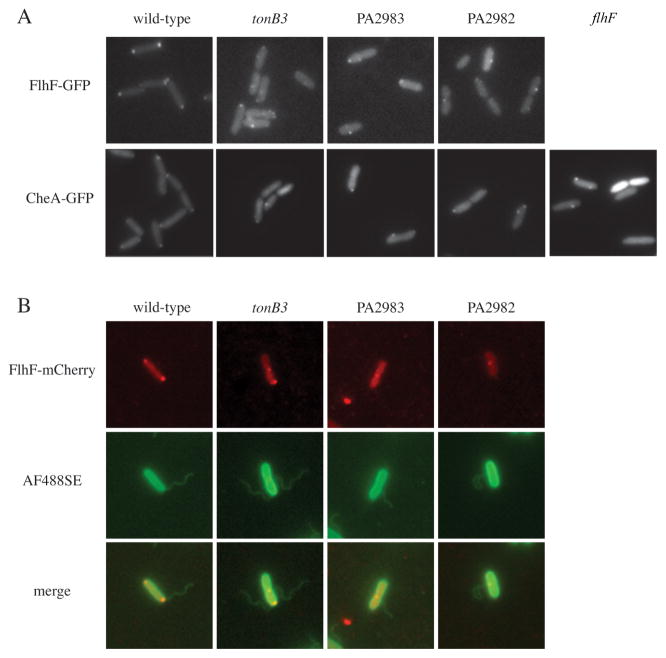
Prior to this work, FlhF was the most upstream factor known to regulate flagellar localization. We therefore examined whether these proteins function upstream or downstream of FlhF. To assess the role of tonB3, PA2983 and PA2982 in FlhF localization, we constructed an arabinose-inducible C-terminal GFP fusion to FlhF. FlhF–GFP was bipolar in wild-type (Fig. 3A) and flhF–gfp complemented the swimming motility defect of a flhF deletion mutant (data not shown), indicating that the fusion was functional. When expressed in tonB3, PA2983 or PA2982 mutant backgrounds, the FlhF–GFP focus was located at non-polar sites (Fig. 3A and Fig. S3). Using an Alexa Fluor 488 carboxylic acid, succinimidyl ester to label the bacterial cell and flagellum, we found that the non-polar FlhF focus (FlhF–mCherry in this experiment) colocalized with the flagellum (Fig. 3B). Together these results demonstrate that tonB3, PA2983 and PA2982 are necessary for the proper polar localization of FlhF.
Fig. 3. tonB3, pocA and pocB mutations disrupt localization of FlhF–GFP and CheA–GFP fluorescent fusions.
A. FlhF–GFP (top) and CheA–GFP (bottom) localization in wild-type, tonB3, PA2983 (pocA), PA2982 (pocB) and flhF strains.
B. Colocalization (bottom) of FlhF–mCherry (top) and Alexa Fluor 488-labelled flagella (middle) in wild-type and mutant strains.
As an additional marker for flagellum protein localization, we created an arabinose-inducible C-terminal GFP fusion to CheA. CheA–GFP was unipolar with one to two foci present at a given pole (Fig. 3A) and cheA–gfp complemented the swimming defect of a cheA mutant (data not shown), showing that the fusion was functional. In tonB3, PA2983 or PA2982 mutants, CheA–GFP foci were non-polar (Fig. 3A and Fig. S3), similar to the pattern seen with FlhF–GFP. In the flhF mutant, CheA–GFP foci were likewise non-polar (Fig. 3A), suggesting that TonB3, PA2983 and PA2982 regulate localization of FlhF, which in turn regulates the localization of CheA. These results implicate TonB3, PA2983 and PA2982 as sitting at the top of the known flagellar localization hierarchy.
tonB3, PA2983 and PA2982 also regulate type IV pilus production
Given the expectation that flagella and pili would be regulated independently, the identification of tonB3 as a flagellar regulator was surprising in light of a previous report that tonB3 is required for pilus-driven P. aeruginosa twitching motility (Huang et al., 2004). We therefore investigated whether our newly discovered flagellar localization factors might also influence type IV pili. We directly imaged pili by TEM and found that deletion of tonB3, PA2982, PA2983 or flhF affects pilus production (Fig. 1). Nearly 100% of tonB3, PA2982 and flhF cells were non-piliated while the PA2983 mutant had a dramatic increase in the number of cells with non-polar pili (70% of cells compared with 3.5% in wild-type) (Fig. 2B). As type IV pili are required for twitching motility, we tested our mutants using a standard plate assay and found that the tonB3, PA2983 and PA2983 mutants were defective for twitching motility, similar to pilA mutants which lack the major pilin protein (Fig. 4).
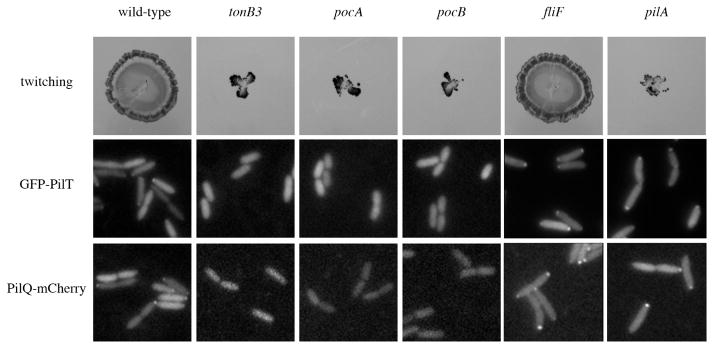
Fig. 4.
Twitching motility and GFP–PilT and PilQ–mCherry localization are impaired in tonB3, PA2983 (pocA) and PA2982 (pocB) mutants. (Top) Twitching motility of wild-type PAO1 and mutant strains. fliF and pilA strains were used as controls that lacked flagella and pili respectively. Localization of GFP–PilT (middle) and PilQ–mCherry (bottom) in cells taken from the leading edge of motility on the surface of agar plates.
The complete lack of population-level twitching motility was particularly surprising for the PA2983 mutant because unlike tonB3 and PA2982 mutants, which exhibited almost no pili, most of the PA2983 mutant cells retained pili. We hypothesized that the lack of twitching motility in the PA2983 mutant could result from these remaining pili being non-functional (not pulling) or from these pili, which were largely non-polar, leading to opposing pulling forces on opposing sides of the cell, thereby resulting in a lack of productive movement in any one direction. To distinguish these two possibilities, we imaged the leading edge of twitching motility in a population of cells. In this assay, wild-type P. aeruginosa moved progressively across the field of view with individual cells moving in a jerking motion that indicated pilus activity (Movie 12). In contrast, non-piliated strains such as pilA, tonB3 and PA2982 mutants did not move, either as a group or when examining the motion of single cells (Movies 13–15). PA2983 cells behaved like the non-piliated cells and did not move (Movie 16), indicating that the non-polar pili produced in this strain were not functional.
To better understand the mechanism by which tonB3, PA2983 and PA2982 affect pilus production, we examined the localization patterns of two polar pilus proteins, the pilus retraction ATPase PilT and the outer membrane secretin PilQ. Previously, we demonstrated that arabinose-inducible GFP–PilT and PilQ–mCherry fusions were functional and localized to the cell pole in the majority of wild-type PAO1 cells (Cowles and Gitai, 2010). When expressed in tonB3, PA2983 or PA2982 mutants, both GFP–PilT and PilQ–mCherry had a diffuse localization pattern in 100% of cells (Fig. 4). Because these genes are necessary for both flagellum and pilus regulation, we propose to rename PA2983 and PA2982 as pocA and pocB, respectively, for polar organelle co-ordinator.
PocA and PocB form a membrane-associated complex
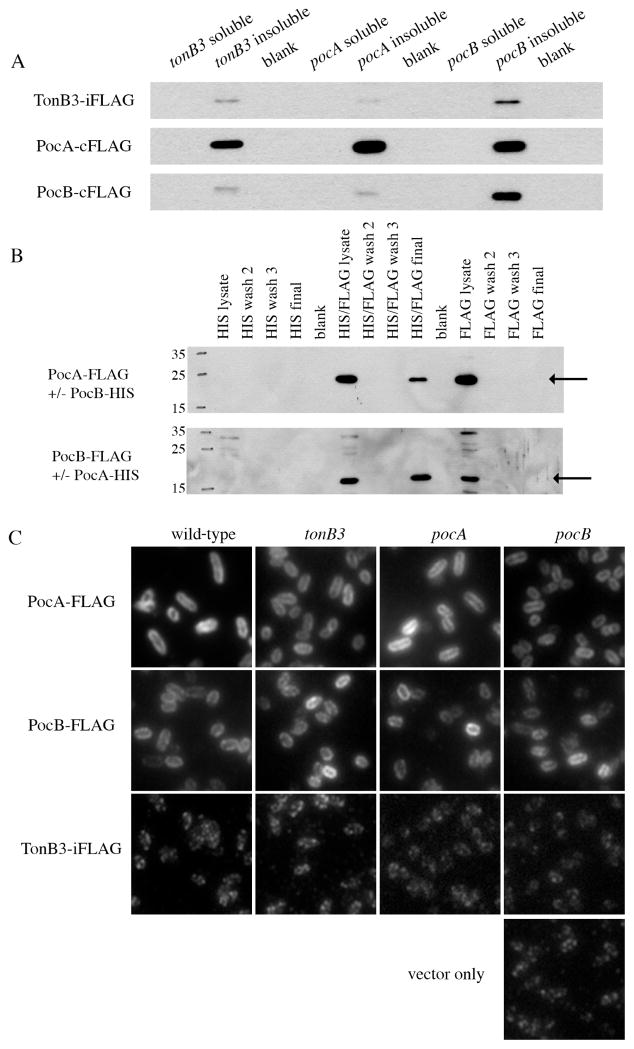
The homology of PocA and PocB to ExbB and ExbD suggested that like their E. coli homologues, PocA and PocB may associate with one another to form a membrane-associated complex. ExbB and ExbD are transmembrane inner membrane proteins with the bulk of ExbB exposed to the cytoplasm and the bulk of ExbD exposed to the periplasm (Postle and Kadner, 2003). Together with TonB, the TonB–ExbB–ExbD complex utilizes the proton motive force across the inner membrane to provide energy for nutrient transport across the outer membrane (Postle and Larsen, 2007). To determine if TonB3 and the Poc proteins are membrane associated, we created arabinose-inducible FLAG fusions to each protein. The arabinose-inducible constructs produced functional fusion proteins that complemented the swimming motility defects of the respective deletion mutants (Fig. S2). By separating soluble and insoluble fractions from clarified cell lysates, we determined that TonB3–iFLAG, PocA–FLAG and PocB–FLAG were located in the insoluble fractions (Fig. 5A), suggesting that, like their E. coli homologues, these proteins are membrane-associated. In addition, we examined the localization of each FLAG fusion in each of the deletion mutants and found that membrane association of each fusion protein is not dependent on the presence of the other proteins (Fig. 5A).
Fig. 5. PocA and PocB form a membrane-associated protein complex.
A. Western blot examining the localization of TonB3–iFLAG, PocA–cFLAG and PocB–cFLAG fusion proteins to soluble or insoluble fractions from clarified cell lysates in tonB3, pocA or pocB mutant strains.
B. Co-precipitation of PocA (22.9 kDa) and PocB (15.7 kDa) using HIS and FLAG tagged proteins. HIS or FLAG labels indicate negative control strains carrying only one fusion protein. HIS/FLAG denotes the strain carrying both PocA–FLAG with PocB–HIS (top) or PocB–FLAG with PocA–HIS (bottom).
C. Immunofluorescence investigating subcellular localization of TonB3–iFLAG, PocA–cFLAG and PocB–cFLAG in wild-type and mutant strains grown to stationary phase in liquid medium. The ‘vector only’ panel is a representative image of background localization patterns in all strains.
To examine the potential protein–protein interactions between TonB3, PocA and PocB, we constructed C-terminal HIS-tagged versions of PocA and PocB for co-precipitation experiments. As with the FLAG fusions, PocA–HIS and PocB–HIS were able to complement the swimming motility defects of their respective deletion strains (Fig. S2), indicating that the fusions were functional. PocA–HIS pulled down PocB–FLAG in a co-precipitation assay using nickel affinity resin (Fig. 5B). We repeated the experiment using PocB–HIS and found that PocB–HIS precipitated PocA–FLAG, as well (Fig. 5B). Negative control strains containing PocA–HIS or PocB–HIS alone had no protein in the α-FLAG Western after precipitation (Fig. 5B). To determine if TonB3 is required for the PocA–PocB interaction, we performed the above co-precipitation experiment in the tonB3 mutant background and found that PocA and PocB still interact in the absence of tonB3 (Fig. S4). Co-precipitation experiments between PocA–HIS or PocB–HIS and TonB3–iFLAG were unable to pull down TonB3 (Fig. S5). Together, these results suggest that PocA and PocB interact while either TonB3 is not a part of the complex or its interaction with PocA and PocB is not detectable by this approach.
PocA and PocB are not polarly localized
We next sought to determine the mechanism by which TonB3 and the Poc proteins impact flagellar and pilus localization and production. These proteins are required for polar FlhF localization and the localization of polar proteins is often mediated by factors that are themselves found at the cell pole. We therefore investigated the localization of TonB3, PocA and PocB using immunofluorescence (IF) with α-FLAG antibodies. Consistent with their predicted membrane association, PocA–FLAG and PocB–FLAG localized to the cell periphery in liquid-grown cells (Fig. 5C) and surface-grown cells (data not shown). Additionally, we determined that the peripheral localization of PocA–FLAG and PocB–FLAG was unaffected by deletion of tonB3, pocA or pocB (Fig. 5C). Attempts to visualize TonB3 localization using IF for TonB3–iFLAG (Fig. 5C) or FlAsH reagent for a tetracysteine-tagged TonB3 (TonB3–iFlAsH) (data not shown) showed no fluorescence over background levels, suggesting that, despite arabinose induction, TonB3 is maintained at levels that remain below the limits of our detection.
tonB3 and pocB, but not pocA, are required for surface-regulated transcriptional upregulation of pilus genes
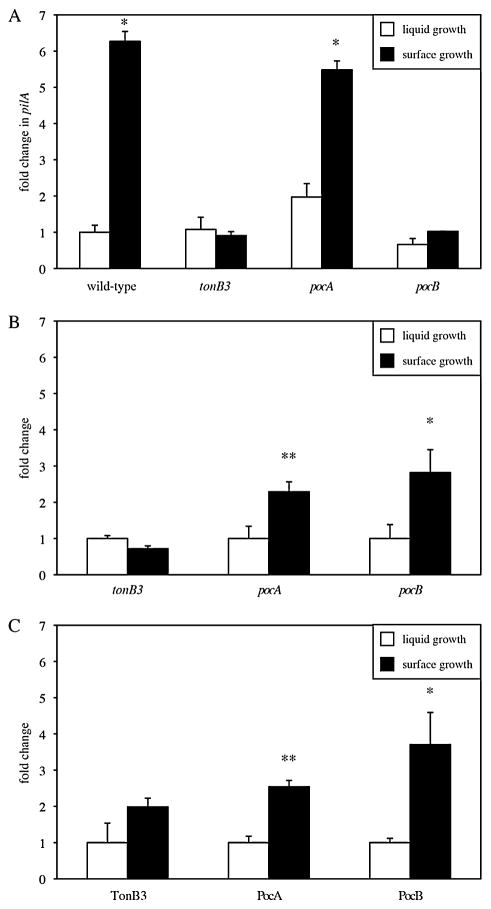
Since their diffuse localization suggests that PocA and PocB do not function as polar anchors, we tested the possibility that they function in regulating gene expression. Previously, we demonstrated that the association of P. aeruginosa cells with solid surfaces promotes pilus production (Cowles and Gitai, 2010). For example, pilus gene expression, as monitored by pilA levels, is markedly increased during growth on surfaces with increasing stiffness. To determine if the Poc proteins are involved in this response to surfaces, we used quantitative PCR (qPCR) to examine pilA expression in the deletion mutants. For tonB3 and pocB, we found that pilA levels were at wild-type levels during growth in liquid media but failed to increase in response to surface association (Fig. 6A), indicating that tonB3 and pocB are important for pilus expression when cells are grown on a solid surface. In contrast, pilA expression was at wild-type levels in the pocA mutant under both conditions (Fig. 6A), suggesting that pocA is not necessary for pilA regulation. These results were consistent with our TEM data where we saw no pili in tonB3 and pocB cells while pocA made non-polar pili (Fig. 2).
Fig. 6. Comparing expression of pilA and localization factors in liquid-grown and surface-grown cells.
A. Relative levels of pilA in wild-type and mutants cells grown to stationary phase in liquid medium (white bars) and in cells taken from the leading edge of motility on agar plates (black bars).
B and C. Relative levels of tonB3 and poc gene (B) and protein (C) expression in wild-type PAO1 grown in liquid (white bars) and surface (black) conditions.
Error bars represent the standard deviation (n = 3) and asterisks denote significant differences from liquid-grown cells (one asterisk indicates P < 0.05 and two asterisks indicate P < 0.01).
One possible mechanism for the environment-specific effects of tonB3 and pocB deletion is that these factors are themselves regulated by surface association. We monitored tonB3, pocA and pocB transcription and found that pocA and pocB increased approximately two- to threefold during growth on a solid surface (Fig. 6B). Unlike pocAB, tonB3 levels were the same in either growth condition (Fig. 6B). To determine if TonB3, PocA or PocB were altered in liquid versus surface growth conditions, we generated natively expressed FLAG-tagged versions of each protein and used Western blots to measure protein levels. C-terminal fusions to PocA (PocA–FLAG) or PocB (PocB–FLAG) complemented the swimming defects of pocA or pocB mutants, respectively, while TonB3 required an internal FLAG fusion (TonB3–iFLAG; between aa 160 and 161) in order to retain functionality (Fig. S2). As was seen with the transcriptional studies, PocA and PocB increased approximately two- to threefold upon surface association while TonB3 levels were statistically unaffected (Fig. 6C).
Because tonB3 and pocB deletion disrupted regulation of pilA expression, we considered the possibility that these factors could be a sensor required for the response to surface association. To test this hypothesis, we performed microarrays comparing gene expression in cells grown in liquid medium to expression from cells at the leading edge of motility on the surface of agar plates. If deletion of tonB3, pocA or pocB prevented P. aeruginosa from differentiating liquid from surface growth, we predicted that the mutant expression profiles would be vastly different from wild-type. Wild-type P. aeruginosa PAO1 had 3247 genes that had more than a twofold change in response to surface association (Fig. 7A). By comparing the gene expression profile of wild-type PAO1 to the profiles of each mutant grown under the same growth conditions, we determined that, although there were some differences from wild-type, general transcription patterns were unaffected by the loss of tonB3, pocA or pocB (Fig. 7A), indicating that these factors do not seem to impact overall P. aeruginosa surface sensing.
Fig. 7. tonB3 and pocB but not pocA affect pilus gene expression in surface-grown cells.
A. Microarray heat map of surface-grown cells in the wild-type and mutant backgrounds (3247 genes with >2-fold change). Yellow indicates increased expression in surface-grown cells compared with liquid-grown cells while blue indicates decreased levels in surface-grown cells.
B and C. Pilus gene expression from the microarray analysis in tonB3 (B, white bars), pocB (B, black bars) and pocA (C) mutants. Fold change denotes expression levels in wild-type PAO1/expression levels in each mutant. Thus, a value of 1 indicates equal expression between strains while values >1 indicate higher expression in wild-type compared with the mutant. Error bars indicate standard error (n = 4).
Despite the relative similarities between the expression profiles of wild-type, tonB3, pocA and pocB strains, we observed that most pilus genes did not respond as strongly to surfaces in tonB3 and pocB. To directly assess these differences, we performed additional microarrays directly comparing wild-type to mutant gene expression after both strains were grown on a solid surface. The data from these arrays indicate that, in general, pilus gene transcripts are lower in the tonB3 and pocB mutants (Fig. 7B), showing that, although dispensable for the overall response to surface association, tonB3 and pocB are involved in the specific upregulation of pilus expression. Notably, known regulators of pilus biogenesis (i.e. PilR/S, FimS/AlgR, Vfr) were not statistically different in the mutant strains. Additionally, although present at lower levels, all known pilus biogenesis genes were transcribed in the mutant strains, indicating that pilus production was not disrupted do to the complete loss of any one transcript. Unlike tonB3 and pocB, pocA had no significant effect on the levels of most pilus genes (Fig. 7C). This result may begin to explain why tonB3 and pocB cells lack pili altogether while pocA cells still produce pili, albeit at non-polar locations.
Because the mutants still produced flagella, we predicted that flagellum gene expression would not be altered in these strains. Indeed, deletion of tonB3, pocA or pocB had no effect on expression of the flagellum subunit gene fliC under liquid or surface growth conditions (Fig. S6). To further investigate a possible transcriptional mechanism for tonB3, pocA and pocB regulation of flagellum localization, we analysed the microarray data from the comparisons of wild-type gene expression to the mutant expression profiles and saw that the vast majority of flagellum genes do not change upon loss of these factors (Fig. S6). Thus, across both pili and flagella the absence of an organelle correlates with reduced transcript levels while the mislocalization of an organelle (namely flagella in all three mutants and pili in pocA) does not.
Discussion
Prior to this work, results from several studies, including our own, suggested that independent processes regulate flagellum and pilus production. The four-tiered hierarchy for flagellum gene transcription (Dasgupta et al., 2003) doesn’t immediately suggest a connection to pilus production and screens for swimming or twitching motility mutants seem to have focused on one form of motility or the other. Our previous results showing that MreB and surface association regulate pilus production but not flagellum formation (Cowles and Gitai, 2010) and that flagella and pili localize at independent subcellular sites and cell cycle time points also led us to believe that these two processes were regulated separately. However, our identification of three proteins that regulate both flagellum and pilus production suggest that these polar structures are actually coupled, with TonB3, PocA and PocB functioning near the top of each structure’s regulatory hierarchy.
Multiple mechanisms contribute to flagella and pili localization
Previously, FlhF was thought to function at the top of the flagellar localization hierarchy. FlhF is mislocalized in the absence of tonB3, pocA or pocB, resulting in non-polar placement of the flagellum at the point where FlhF puncta has formed. This non-polar flagella placement phenocopies the flhF deletion mutant, suggesting that TonB3, PocA and PocB direct polar localization of flagella by directly or indirectly impacting FlhF polar localization. Furthermore, new FlhF puncta form only at the new pole of dividing cells, either following or just prior to division, indicating that regulation of FlhF, and therefore flagella localization, is linked to the cell cycle.
While TonB3, PocA and PocB affect flagellar localization, their effects on type IV pili are more complex. Deletion of tonB3 or pocB resulted in the absence of pili formation, while pocA deletion led to the formation of mislocalized, non-functional pili. Thus, even within the hypothesized Poc complex, the individual genes may have distinct roles in regulating pili formation. One such mechanism appears to be transcriptional, since we did not observe the expected wild-type increase in pili gene transcription upon surface association in the absence of tonB3 and pocB. However, the pocA deletion did not have transcriptional defects, but rather defects in pilus localization, suggesting a second mechanism regulating pili localization independent of transcription.
Membrane topology of the putative TonB3–PocA–PocB complex may contribute to function
How a single putative TonB3–PocA–PocB complex can use several different methods (i.e. localization versus transcript levels) to impact both flagella and pili remains to be determined. In fact, we cannot rule out the possibility that these three proteins do not form a single complex or that they may interact with other factors or in different combinations with each other for their distinct effects. We hypothesize that cellular location and membrane topology contribute to the functions of each individual factor. TonB3 and PocB homologues TonB and ExbD have single transmembrane regions with the majority of the protein in the periplasm, whereas the PocA homologue ExbB has three transmembrane regions with the majority forming a large cytoplasmic loop (Postle and Kadner, 2003). Interestingly, this spatial grouping within the putative complex members is consistent with differences in phenotype. While all three proteins regulate flagellum localization, the largely periplasmic TonB3 and PocB regulate pilus production transcriptionally, while the mainly cytoplasmic PocA regulates pili localization independent of transcription. Since the pocA mutant still makes pili, it may have an association with unique factors, potentially located in the cytoplasm, distinct from TonB3 and PocB. Additionally, downstream factors unique to either the flagella or pili may determine the functions of each individual complex. Alternatively, separate complexes may exist that affect the flagella and pili in different ways. It will be important to determine the identity of additional downstream factors, and to investigate whether this regulation occurs as part of a TonB3–PocA–PocB complex or in isolation.
PocAB impact polar localization of flagella without being polarly localized themselves
While the importance of tonB3, pocA and pocB for both P. aeruginosa polar motility structures is clear, the mechanism(s) by which these factors function remains unclear. Homologues of TonB3, PocA and PocB provide the energy for transporting large molecules across the outer membrane using the proton motive force (Postle and Larsen, 2007). It is possible that this energy transduction aspect is important for their function in localization; however, while Pseudomonas TonB1 and TonB2 have been implicated in iron and haem uptake (Zhao and Poole, 2000), a transport role for TonB3 has not been established. Furthermore, it is unclear why energy transduction would be needed for flagellum localization. Reports have indicated that membrane potential is important for cell division in Bacillus subtilis, and perturbations of the proton motive force modulate localization of several cell division proteins including MinD, FtsA and MreB (Strahl and Hamoen, 2010). Since MreB is important for pili formation in P. aeruginosa, membrane potential could provide a mechanism whereby MreB localization affects pili localization in the pocA mutant. Additionally, we found that tonB3, pocA and pocB cells are somewhat shorter than wild-type, suggesting a possible connection to cell size maintenance during division or, at least, more pleotropic effects of these mutations. Furthermore, since flagella localization is tied to the cell cycle, perhaps it is also dependent on membrane potential. Additional studies are needed to determine if other proteins are mislocalized in the absence of tonB3, pocA and pocB.
Alternatively, it is possible that these proteins may have adopted an independent and previously unknown function in localization and formation of flagella and pili. For example, they could serve as anchors to bring additional factors to the poles. We have shown that PocA and PocB are membrane associated, but have no discrete localization within the membrane, so it remains unclear how a complex that is not localized could direct polar localization of additional proteins. We were unable to examine the localization of TonB3 or precipitate it with either PocA or PocB. This is consistent with studies with E. coli homologues, where TonB does not pull down without cross-linking (Ollis et al., 2009). Therefore it remains possible that TonB3 could be localized to the poles and that transient interactions with PocAB could direct polar localization of flagella.
TonB3 and PocB contribute to upregulation of pilus genes in response to surface association
One way in which non-localized proteins can regulate the assembly of a polar motility complex is by regulating motility protein production. Consistent with this model, we found that TonB3 and PocB affected pilus gene expression in an environment-specific manner. In particular, we previously described an increase in pilus gene transcription upon surface association and this surface-dependent pilus upregulation was less robust in tonB3 and pocB mutants. These mutants did not affect the overall expression profile in response to surface growth but rather altered pilus transcript levels. Thus, TonB3 and PocB specifically function in pilus regulation as opposed to surface sensing itself. How surface recognition occurs and leads to TonB3- and PocB-dependent upregulation of pilus genes remains an open question. Multiple sensors may contribute to global expression profile changes in response to surface association, which would lead to little difference detected in single mutants. A screen for sensors using fusions to the pilus gene promoters could prove illuminating.
While transcriptional regulation can partially explain the absence of pili in tonB3 and pocB, these factors also appear to have a transcription-independent role in subcellular localization. Specifically, pocA mutants disrupt pilus localization (but not assembly) without significantly perturbing pilus gene regulation, and pocA, pocB and tonB3 all disrupt flagellar and FlhF localization without disrupting flagellar gene regulation. Future studies of the protein interaction partners of TonB3, PocA and PocB will help dissect if these proteins form one or multiple complexes and will provide insight into the mechanism(s) by which this predicted complex co-ordinates pili and flagella formation and directs proteins such as FlhF to the cell poles.
Experimental procedures
Bacterial strains and growth conditions
Pseudomonas aeruginosa PAO1 (C. Manoil, University of Washington) was used as wild-type for all experiments. All strains are listed in Supplemental Table S3. All bacterial cultures were grown in Luria–Bertani (LB) broth (Miller, 1972) at 37°C. Plasmids were conjugated from E. coli S17-1 (λpir) or electroporated (25 μF; 200 Ω; 2.5 kV) into P. aeruginosa strains and were maintained with the following antibiotic concentrations: tetracycline (Tet), 25 μg ml−1 for E. coli and 100 μg ml−1 for P. aeruginosa; gentamicin (Gent), 15 μg ml−1 for E. coli and 30 μg ml−1 for P. aeruginosa.
Molecular biological methods
Standard molecular biological methods were used for this study (Sambrook et al., 1989). Restriction enzymes (NEB, Ipswich, MA), plasmid preparation, gel extraction and PCR purification kits (Qiagen, Valencia, CA) were all used according to manufacturer’s recommendations. Constructs were confirmed by sequencing (Genewiz, South Plainfield, NJ). PCR amplification of DNA was performed using Platinum Pfx according to manufacturer’s directions (Invitrogen, Carlsbad, CA). Primers (0.2 μM) used in this study (Integrated DNA Technologies, Coralville, IA) are described in Supplemental Table S4.
Motility assays
To assess gross motility, overnight cultures of wild-type P. aeruginosa were spotted onto 0.3% agar LB plates (for swimming motility) and stabbed into 1% agar LB plates (for twitching motility). Swimming motility was measured as the distance moved from the point of inoculation on the surface of the agar plate after 20 h. Twitching motility was observed by removing the agar and staining cells attached to the Petri dish with 1% crystal violet after 48 h.
Deletion construction
Non-polar, in-frame deletions of tonB3, pocA and pocB were made by PCR-amplifying regions upstream and downstream of each gene, performing sewing PCR to fuse those products together, cloning the final deletion fragment into pEX18Tc (Hoang et al., 1998), and using conjugation to transfer the deletion construct into wild-type PAO1. A 924 bp fragment of tonB3 was deleted using 0406UpFOREcoRI and 0406UpREV to amplify the region upstream of tonB3 and 0406DnFOR and 0406DnREVXbaI to amplify the region downstream. A 594 bp fragment of pocA was deleted using 298382UpFOREcoRI and 2983UpREV to amplify the region upstream of pocA and 2983DnFOR and 298382DnREVXbaI to amplify the region downstream. A 414 bp fragment of pocB was deleted using 298382UpFOREcoRI and 2982UpREV to amplify the region upstream of pocB and 2982DnFOR and 298382DnREVXbaI to amplify the region downstream. Selected exconjugants were grown overnight in LB broth, streaked to LB 5% sucrose plates for counterselection, and patched to LB ± Tet plates to confirm Tet sensitivity. PCR was used to identify deletion mutants.
Transmission electron microscopy
To prepare samples, overnight cultures were inoculated to 1% agar LB plates and incubated at 37°C for 24 h. Cells were collected from the leading edge of surface motility and gently resuspended in PBS. For TEM images and flagellum and pilus counts in a population, 100 cells from three independent replicates were collected and samples were processed as described (Cowles and Gitai, 2010) (Confocal and Electron Microscopy Core Facility Laboratory, Princeton University). Samples were blinded to assure objective scoring.
Construction of fluorescent fusions
The Gateway® Technology (Invitrogen, Carlsbad, CA) was utilized to construct C-terminal fluorescent fusions to flhF and cheA as described (Cowles and Gitai, 2010). Briefly, flhF and cheA genes were PCR-amplified using primers FlhFGWUp, FlhFGWDn, CheAGWUp and CheAGWDn, respectively, and cloned into pDONR223. flhF and cheA were then transferred into the destination vectors pJN(Gateway–GFP) and pJN(Gateway–mCherry) for flhF and pJN(Gateway–GFP) for cheA. pJN(Gateway–GFP) and pJN(Gateway–mCherry) were constructed by PCR-amplifying GFP or mCherry in conjunction with the Gateway® cassette using the primers GWr-fpFORSpe and GWrfpREVSpe and cloning the products into pJN105 (Newman and Fuqua, 1999; Baynham et al., 2006). The expression vectors pJN(flhF–gfp), pJN(flhF–mCherry) and pJN(cheA–gfp) were transformed into wild-type, tonB3, pocA and pocB mutants. Plasmid maintenance was confirmed using primers AraCAmp and GWrfpREVSpe. pSW(gfp–pilT) and pSW(pilQ–mCherry) fusions were created previously (Cowles and Gitai, 2010) and conjugated into the tonB3, pocA, pocB, flhF, fliF and pilA deletion strains.
FlhF-CERC was created by PCR-amplifying flhF from genomic DNA with FlhFfowardSpeI and FlhFreverseMluI primers and digesting the product with SpeI and MluI. CERC was removed from pVCERC-5 (Thanbichler et al., 2007) with MluI and SacI and ligated to flhF. The resulting fusion was used as template for PCR using FlhFforwardSpeI and CER-CreverseEcoRISacI primers. flhF-CERC was then cloned into pSW196 (Baynham et al., 2006) using SpeI and SacI enzymes.
CheA–GFP was created by PCR-amplifying cheA from genomic DNA with CheAfowardEcoRI and CheAreverseMluI primers and digesting the product with EcoRI and MluI. EGFP was removed from pVGFP5 (Thanbichler et al., 2007) with MluI and SacI and ligated to cheA. The resulting fusion was used as template for PCR using CheAforwardEcoRI and EGFPreverseXbaISacI primers. cheA-CERC was then cloned into pSC8(flhF-CERC) using EcoRI and SacI enzymes.
Microscopy of fluorescent fusions
Pseudomonas aeruginosa carrying pJN(flhF–gfp), pJN(flhF–mCherry), pJN(cheA–gfp), pSW(gfp–pilT) and pSW(pilQ–mCherry) were grown overnight in LB broth with the appropriate antibiotic, subcultured 1:100 into fresh LB broth containing 0.02% arabinose, and grown for 3 h at 37°C. To collect images, bacterial cells were placed on pads made from 1% agarose in PBS and visualized using a 100× 1.4NA objective on a Nikon 90i microscope equipped with a Rolera XR camera and NIS Elements software. For studies examining colocalization of FlhF–mCherry and the flagellum, overnight cultures were subcultured with arabinose as described above and grown for 3 h at 37°C. Two hundred and fifty microlitres of cells were pelleted at 2000 g for 5 min, and the pellets were resuspended gently in 250 μl of DB buffer [(5 mM KH2PO4, 5 mM Na2HPO4, adjusted to pH 6.5), 1 mM CaCl2, 2 mM MgCl2]. Cells were washed in DB buffer before being resuspended in 20 μl DB buffer. One microlitre of the Alexa Fluor 488 carboxylic acid, succinimidyl ester dye (10 mg ml−1 in DMSO) (Invitrogen, Carlsbad, CA) was added and samples were incubated with shaking (~ 100 r.p.m.) for 1 h at room temperature in the dark. Cells were pelleted, washed twice with 250 μl of DB buffer, and resuspended in a final volume of 250 μl DB buffer. Images were collected as described above. For cefsulodin experiments, strains were prepared as described above (grown overnight, subcultured 1:100 with 0.02% arabinose and incubated for 3 h to log phase) and spotted to agarose pads containing 30 μg ml−1 cefsulodin. Time-lapse images were collected every 15 min for 8 h.
Real-time PCR
Total RNA from wild-type P. aeruginosa PAO1 was isolated from cells grown in LB broth to an OD600 of 5.0 (liquid-grown cells) and from cells collected at the leading edge of motility from the surface of LB agar plates after 24 h (surface-grown cells) with three replicates per growth condition for each strain examined. RNA was processed as described (Cowles and Gitai, 2010). Reactions for real-time PCR were performed in duplicate in 10 μl reactions with SYBR® Green PCR Master mix (Applied Biosystems, Foster City, CA), cDNA template, and appropriate primers as described (Cowles and Gitai, 2010). Transcript levels of pilA, fliC, tonB3, pocA and pocB were measured with primers listed in Supplemental Table S4. Cycle threshold results for each sample were normalized according to rpoD levels (amplified with RpoDForRT and RpoDRevRT) and then converted to relative values factoring in a twofold change in product amount per cycle.
Western blot analysis
Whole-cell lysates from P. aeruginosa strains were isolated from cells grown in LB broth to an OD600 of 5.0 (liquid-grown cells) and from cells collected at the leading edge of motility from the surface of LB agar plates (surface-grown cells) for 24 h with three replicates per growth condition. Cell samples were processed for immunoblot quantification as described (Cowles and Gitai, 2010). Samples were separated on 15% SDS/PAGE gels and transferred to nitrocellulose membranes. For immunoblot analysis, membranes were blocked with 5% milk (w/v) in TBS-T, then washed and immunodetected in TBS-T. Primary anti-FLAG antiserum (Invitrogen, Carlsbad, CA) was used at 1:2000 and secondary anti-rabbit-HRP IgG conjugate (GE Healthcare, NA934V) was used at 1:8000. ECL Western Blotting Reagent (GE Healthcare, RPN2106) and film were used for signal detection; ImageJ software was used to quantify signal intensity (Abramoff et al., 2004).
Microarrays
Total RNA from wild-type, tonB3, pocA and pocB was isolated from cells grown in LB broth to an OD600 of 5.0 (liquid-grown cells) and from cells collected at the leading edge of motility from the surface of LB agar plates after 24 h (surface-grown cells) with three replicates per growth condition for each strain examined. RNA was processed using the Trizol extraction method, cDNA synthesis and labelling, and microarray hybridization and analysis as described (Rutherford et al., 2011). Microarrays were designed using Agilent software in an 8× 15 grid format. Each array contained two 60-mer probes per open reading frame in the P. aeruginosa PAO1 genome (Agilent custom array 028678). Data were extracted with Agilent Feature Extractor and analysed on the Princeton University Microarray Database (PUMAdb, http://puma.princeton.edu) based on Gollub et al. (2003).
Construction of FLAG and HIS proteins
C-terminal FLAG-tagged PocA and PocB were made by PCR-amplifying pocA using primers 2983cFLAGUpEcoRI with 2983cFLAGDnXbaI (for inducible expression) or 2983cFLAGUpEcoRI2 with 2983cFLAGDnSpeI (for native expression) and pocB using primers 2982cFLAGUpEcoRI with 2982FLAGDnXbaI (for inducible expression) or 2983cFLAGUpEcoRI2 with 2982cFLAGDnSpeI (for native expression). Resulting PCR products were cloned into pJN105 for arabinose-inducible fusions or miniCTX1 (Hoang et al., 2000) for natively expressed fusions. C-terminal, arabinose-inducible HIS-tagged PocA and PocB were similarly constructed by PCR-amplifying pocA (2983cFLAGUpEcoRI with 2983cHISDnSpeI) and pocB (2982cFLAGUpEcoRI with 2982cHISDnSpeI) and cloning the products into pSW196 (Baynham et al., 2006). An arabinose-inducible FLAG tag was inserted into tonB3 by PCR-amplifying upstream and downstream halves of tonB3 using 0406UpFLAGEcoRI2 with 0406UpFLAG (519 bp) and 0406DnFLAG with 0406UpFLAGXbaI2 (504 bp) primers respectively. The two resulting products were sewn together using 0406UpFLAGEcoRI2 and 0406UpFLAGXbaI2 primers and cloned into pJN105. Similarly, a natively expressed internal TonB3–FLAG fusion was constructed using 0406UpFLAGEcoRI2 with 0406UpFLAG and 0406DnFLAG with 0406DnFLAGSpeI and cloning the resulting product into miniCTX1. Sequence-confirmed vectors were transformed into PAO1 strains of interest. Plasmid maintenance or integration at the attP site was confirmed by PCR.
Isolation of soluble and insoluble fractions
Samples were prepared by diluting overnight cultures 1:500 in fresh medium and incubation at 37°C to OD600 = 0.5. Cells were pelleted at 5000 r.p.m. for 10 min at 4°C and resuspended in lysis buffer (50 mM PO4, pH 8.0, 150 mM NaCl, 10 mM MgCl2), normalizing to OD600 = 20. Cells were lysed using a French pressure cell (at 20 000 psi) followed by centrifugation at 2500 r.p.m. to remove unbroken cells. Soluble and insoluble fractions were isolated by ultracentrifugation (100 000 r.p.m.) for 30 min. FLAG-tagged protein levels were measured by Western blot as described above.
Immunofluorescence
Overnight cultures were diluted into fresh medium and grown to OD600 = 0.5. Cells were mixed gently with 5× fix solution [12.5% paraformaldehyde, 150 mM NaPO4 (pH 7.5)] and incubated at room temperature for 15 min. Samples were then incubated on ice for 20 min before being washed three times in PBS and resuspended in 75 μl GTE [50 mM glucose, 10 mM EDTA (pH 8.0), 20 mM Tris-HCl (pH 7.5)] containing lysozyme (10 μg ml−1). Fixed cells were spotted to poly-Lysine-coated slides, dried, and immersed in cold methanol for 5 min and cold acetone for 5 min. Samples were rehydrated and blocked in 2% BSA in PBS and incubated with anti-FLAG antibody (Invitrogen, Carlsbad, CA) diluted 1:250 in PBS with 2% BSA for 1 h. Slides were then washed 10 times with PBS and incubated for 1 h in the dark with goat anti-rabbit Alexa Fluor 488 antibody (Invitrogen) diluted 1:200. Cells were washed 10 times with PBS and visualized by microscopy as described above.
Co-precipitation of PocA and PocB
To investigate protein–protein interactions between TonB3, PocA and PocB, overnight cultures carrying pSW (PocA–HIS) or pSW (PocB–HIS) alone, pJN (TonB3–iFLAG), pJN (PocA–FLAG) or pJN (PocB–FLAG) alone, or a combination of HIS and FLAG constructs were subcultured 1:500 into LB gentamicin and incubated at 37°C to OD600 = 0.5. Cells were pelleted at 5000 r.p.m. for 10 min at 4°C and resuspended in lysis buffer (50 mM PO4, pH 8.0, 150 mM NaCl, 10 mM MgCl2), normalizing to OD600 = 20. Clarified lysates were made using a French pressure cell (at 20 000 psi) followed by centrifugation at 2500 r.p.m. to remove unbroken cells. Lysates were combined with 1% Thesit (Sigma, St. Louis, MO), 10 mM imidazole and 100 μl nickel resin (Qiagen, Valencia, CA) and incubated at 4°C for 90 min with gentle shaking. Resin was pelleted, washed in lysis buffer + Thesit and imidazole, and incubated at room temperature for 5 min with gentle shaking. This wash step was repeated three times with samples collected for immunoblots after each one. The final resin pellet was resuspended in 2× SDS-PAGE loading dye. Western blots to detect FLAG protein were performed as described above.
Statistical analysis
P-values were calculated using a Student’s t-test.
Supplementary Material
Acknowledgments
This work was supported in part by a NIH New Innovator Award (Number 1DP2OD004389-01), a Beckman Young Investigator Award, and a Grand Challenges Seed Program from Princeton University awarded to Z.G., Award Number F32AI074271 from the National Institute of Allergy and Infectious Diseases given to K.N.C., and Award Number F32AI095002 from the National Institute of Allergy and Infectious Diseases given to A.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
We would like to thank M. Bisher for technical assistance with transmission electron microscopy, C. Cowles for technical assistance with co-precipitations and B. Boles for plasmids.
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Amako K, Umeda A. Flagellation of Pseudomonas aeruginosa during the cell division cycle. Microbiol Immunol. 1982;26:113–117. doi: 10.1111/j.1348-0421.1982.tb00160.x. [DOI] [PubMed] [Google Scholar]
- Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J Bacteriol. 2006;188:132–140. doi: 10.1128/JB.188.1.132-140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong SH, Jones Y, et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol. 2010;76:173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles KN, Gitai Z. Surface association and the MreB cytoskeleton regulate pilus production, localization and function in Pseudomonas aeruginosa. Mol Microbiol. 2010;76:1411–1426. doi: 10.1111/j.1365-2958.2010.07132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- Fekkes P, Driessen AJM. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub J, Ball CA, Binkley G, Demeter J, Finkelstein DB, Hebert JM, et al. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 2003;31:94–96. doi: 10.1093/nar/gkg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- Hahn HP. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa – a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer H. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Hoang TT, Kutchma AJ, Becher A, Schweizer H. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- Huang B, Ru K, Yuan Z, Whitchurch CB, Mattick JS. tonB3 is required for normal twitching motility and extracellular assembly of type IV pili. J Bacteriol. 2004;186:4387–4389. doi: 10.1128/JB.186.13.4387-4389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. p. 466. [Google Scholar]
- Murray TS, Kazmierczak BI. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J Bacteriol. 2006;188:6995–7004. doi: 10.1128/JB.00790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Fuqua C. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Ollis AA, Manning M, Held KG, Postle K. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol Microbiol. 2009;73:466–481. doi: 10.1111/j.1365-2958.2009.06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandza S, Baetens M, Park CH, Au T, Keyhan M, Matin A. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol Microbiol. 2000;36:414–423. doi: 10.1046/j.1365-2958.2000.01859.x. [DOI] [PubMed] [Google Scholar]
- Pier GB. CFTR mutations and host susceptibility to Pseudomonas aeruginosa lung infection. Curr Opin Microbiol. 2002;5:81–86. doi: 10.1016/s1369-5274(02)00290-4. [DOI] [PubMed] [Google Scholar]
- Postle K, Kadner RJ. Touch and go: tying TonB to transport. Mol Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- Postle K, Larsen RA. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals. 2007;20:453–465. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- Rutherford ST, van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scharfman A, Arora SK, Delmotte P, Brussel EV, Mazurier J, Ramphal R, Roussel P. Recognition of Lewis x derivatives present on mucins by flagellar components of Pseudomonas aeruginosa. Infect Immun. 2001;69:5243–5248. doi: 10.1128/IAI.69.9.5243-5248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci USA. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Bruckner R, Ringgaard S, Moll A, Cameron DE, Briegel A, et al. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 2012;26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Poole K. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol Lett. 2000;184:127–132. doi: 10.1111/j.1574-6968.2000.tb09002.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.