Abstract
The formation of peptide bonds is of great importance from both a biological standpoint and in routine organic synthesis. Recent work from our group demonstrated the synthesis of peptides in the gas-phase via ion/ion reactions with sulfo-NHS reagents, which resulted in conjugation of individual amino acids or small peptides to the N-terminus of an existing ‘anchor’ peptide. Here, we demonstrate a complementary approach resulting in the C-terminal extension of peptides. Individual amino acids or short peptides can be prepared as reagents by incorporating gas phase-labile protecting groups to the reactive C-terminus and then converting the N-terminal amino groups to the active ketenimine reagent. Gas-phase ion/ion reactions between the anionic reagents and doubly protonated “anchor” peptide cations results in extension of the “anchor” peptide with new amide bond formation at the C-terminus. We have demonstrated that ion/ion reactions can be used as a fast, controlled, and efficient means for C-terminal peptide extension in the gas phase.
Keywords: Woodward’s reagent K, ion/ion reaction, peptide bond formation
INTRODUCTION
Peptide bonds play a central role in protein chemistry by providing the link between amino acids in peptides and proteins. As synthetic peptides today are widely used in biological research as well as product and drug development, the generation of peptide bonds is of great interest for synthesis as well. Solid phase peptide synthesis (SPPS), first described in 1963,1 is the widely used peptide synthesis strategy in most labs now. Although SPPS has extraordinary advantages like suitability for combinatorial approaches2 and the ability to synthesize a range of peptide sizes and sequences,3,4 issues related to the time required to carry out reactions, solubility of reagents, and use of cumbersome amount of reagents and solvents remain unsolved. 5, 6
The ability to generate covalent bonds within a mass spectrometer opens up many new possibilities for targeted gas-phase synthesis. Amide bond formation has been reported using various strategies including photoexcitation reactions of proton-bond dimers,7, 8 ion/molecule reactions, 9, 10, 11 and controlled ion/ion reactions using N-hydroxysulfosuccinimide (sulfo-NHS) esters,12, 13, 14 carbodiimide reagents15 or Woodward’s reagent K (wrk).16 Gas-phase peptide extension at the N-terminus of “anchor” peptides has been reported17 using sulfo-NHS esters with reaction efficiencies greater than 30%. McGee and McLuckey generated reagents by activating carboxyl groups in amino acids or peptides with sulfo-NHS esters and protecting reactive amine functionalities. These sulfo-NHS reagents were then covalently attached to the N-terminus of anchor peptides via gas-phase ion/ion reactions.
While most chemical peptide synthesis procedures start at the C-terminus and proceed to the N-terminus, in-cell protein biosynthesis starts at the N-terminus. Here, a reagent capable of covalently attaching to the C-terminus of an anchor peptide is generated in the gas-phase. The amino group of an amino ester (i.e., a tert-butyl (tBu) ester) is ‘activated’ by converting it to a ketenimine (ki, –C=C=N–) in the mass spectrometer. Ketenimines are reactive intermediates18 with an electron-deficient central carbon that can react with various nucleophiles including carboxylic acids, 19 amines,20,21 hydroxyls,22 and thiols.23 Woodward’s reagent K (N-ethyl-3-phenylisoxazolium-3′-sulfonate, wrk)24,25, a ketenimine-based reagent, has been used for the activation of carboxylic acids of peptides and proteins in solution, 19 although reactions with cysteine, histidine, and lysine side-chains have also been reported.26,27 The reaction mechanism involves the irreversible conversion of the isoxazolium group of wrk to a highly reactive N-ethyl ketenimine (ki-Et), followed by nucleophilic attack by a nucleophile.19 Here we use an exchange reaction to switch the N-ethyl groups on wrk to amino acid residues. The reagent synthesis is demonstrated using a variety of amino acids and is extended to small peptides like dialanine, which are then covalently added to the C-terminus of an anchor peptide.
The synthesis and characterization steps employed here are nearly instantaneous and this method does not require the use of deprotecting agents, cleaving agents, or solvents, rendering it a rapid and efficient method compared to SPPS. This strategy leads to small amounts of peptides in the gas phase, which can potentially be collected by soft-landing techniques or further interrogated within the mass spectrometer.28,29 This approach expands gas-phase extension of polypeptide ions to include amino acid or polypeptide addition to the C-terminus of an anchor peptide, which complements the previously described approach for addition of amino acids to the N-terminus of an anchor peptide via sulfo-NHS chemistry.
EXPERIMENTAL
Materials
Methanol and glacial acetic acid were purchased from Mallinckrodt (Phillipsburg, NJ). The peptides ARAAARA and AKAAAKA were custom synthesized by NeoBioLab (Cambridge, MA). The peptide KGAILDGAILR was custom synthesized by Synpep (Dublin, CA). Woodward’s reagent K, trimethylamine, acetonitrile, Angiotensin II (sequence: DRVYIHPF), L-Valine methyl ester hydrochloride (V-OMe), and L-Asparagine tert-butyl ester (N-OtBu) were obtained from Sigma-Aldrich (St. Louis, MO). The reagent 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC), glycine tert-butyl ester hydrochloride (G-OtBu), L-alanine tert-butyl ester hydrochloride (A-OtBu), L-valine tert-butyl ester hydrochloride (V-OtBu), L-proline tert-butyl ester (P-OtBu), L-tyrosine tert butyl ester (Y-OtBu), O-tert-butyl L-glutamic acid tert-butyl ester hydrochloride (E(OtBu)-OtBu), and L-lysine(boc) tert-butyl ester hydrochloride (K(boc)-OtBu) were purchased from Thermo Fisher Scientific (Rockford, IL). Dialanine tert-butyl ester hydrochloride (AA-OtBu) was obtained from Bachem Americas (Torrance, CA). All materials were used without further purification. All peptide solutions for positive electrospray were prepared in 49.5/49.5/1 (v/v/v) methanol/water/acetic acid (~10 μM).
Ketenimine (ki) reagent preparation
Wrk, tBu protected amino acids (X-OtBu) and small peptides (XX-OtBu) were dissolved in acetonitrile with a final concentration of ~2 mM. 20 μL of the wrk solution and 40 μL of amino acids or small peptides solution were combined. To this, ~2 μL of trimethylamine was added to adjust the pH value to ~7-8. This first allows for the transfer of wrk to the reactive ketenimine, and later enables the reaction between the ketenimine and the amino groups to form the amidine [(ki-Et)+(X-OtBu)].
The ki reagents were generated via a solution-phase nucleophilic addition reaction followed by a gas-phase elimination reaction. In the absence of carboxylic acids, N-alkyl ki reacts with primary amines in solution to form an amidine, 20,21 which tautomerizes and fragments to a new ketenimine reagent when subjected to collisional activation in the gas-phase. The net result is an amine exchange, therefore a series of amidation reagents can be generated via the modification of wrk. Amino acids with protected carboxylic acids can be coupled to a ki reagent via this method. In this work, the tert-butyl (tBu) group is used to protect the acidic end of amino acids.17
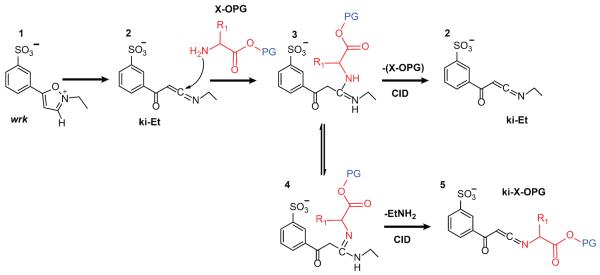
The reaction mechanism for the overall process for reagent ion generation is shown in Scheme 1, which includes the irreversible conversion of the wrk 1 to a reactive N-ethyl keto-ketenimine (ki-Et) 2 by proton abstraction from the 3 position of the isoxazole.19 This intermediate then reacts with a primary amine of an amino acid to create an amidine, 3, 20,21 which can tautomerize to amidine 4. The C-N bonds of the two amidines 3 and 4 are easily cleavable upon collisional activation in gas phase, which either gives rises to the N-amino acid keto-ketenimine (ki-X-OPG) 5 with the loss of an ethylamine, or regenerates the ki-Et 2 by loss of the NH2-X-OPG.
Scheme 1.
Formation of a new ki-based reagent via reaction between wrk and amino acids followed by gas-phase activation. PG is the protecting group (blue) while X represents an amino acid.
Product ion spectra of the [(Et-ki)+(X-OtBu)-H]− complexes for X=leucine (Figure S1) and tyrosine (Figure S2) are provided as Supplementary Information. In each case, a peak corresponding to the mass-to-charge ratio of 3 (or 4), (i.e., the isomeric forms of [(Et-ki)+(X-OtBu)-H]− shown in Scheme 1) can be seen in the mass spectrum when a solution of wrk and X-OtBu is sprayed via nESI. Upon activation, a loss of ethylamine is produced, resulting in the formation of 5. The new reagents ki-X-OtBu were generated with different efficiencies when varying the amino acid X. The loss of the tBu protecting group as neutral 2-methylpropene (56 Da) and the following CO2 loss17 from the deprotected carboxyl end are also observed. For all the amino acids examined here, the yields of the desired ki-X-OtBu were relatively low due to the presence of multiple dissociation pathways of [(Et-ki)+(X-OtBu)-H]−. The secondary amine of proline tert-butyl ester can also react with ki-Et to form 3, however a secondary amine is not capable of forming a C=N double bond, thus the tautomeric form 4 does not exist for [(ki-Et)+(P-OtBu)-H]− and an ethylamine loss is not observed (Figure S2). Hence, a reagent for the addition of proline to the C-terminus cannot be generated via the approach described here.
Solution-phase peptide C-terminal extension
The peptide ARAAARA was dissolved in H2O (1 mg/mL). The C-terminal carboxylic acid group was activated using an equimolar amount of EDC in a mixture of DMF and H2O. To this, an equal volume of X-OtBu in ACN solution (~5 mM) was added, which resulted in the formation ARAAARAX-OtBu. The reaction mixture was then lyophilized and reconstituted in water. The protonated species, [ARAAARAX-OtBu+H]+, was isolated and subjected to collisional induced dissociation (CID) in a mass spectrometer to generate [ARAAARAX+H]+.
Mass Spectrometry
All experiments were performed using a triple quadrupole/linear ion trap (LIT) mass spectrometer30 (QTRAP 4000, AB Sciex, Concord, ON, Canada), previously modified for ion/ion reactions31 with an alternately pulsed nano-electrospray (nESI) ionization source32. The deprotonated (ki-Et)+(X-OtBu) anions were first isolated in the Q1-mass filter and then subjected to fragmentation via beam-type CID from Q1 to the q2 cell. The fragments were then transferred back to Q1 where the ki-X-OtBu reagent anions were isolated prior to their second injection into the q2 reaction cell. The doubly protonated peptide cations were subsequently isolated in the Q1-mass filter and then injected into q2 that has been modified for mutual storage of ions of opposite polarity.31 After a defined mutual storage reaction time of 100-1000 milliseconds, the product ions were then transferred to Q3 where the electrostatic complexes were mass-selected. A low amplitude ion-trap CID was used to collisionally activate the complex products for 50-2000 ms, which first cleaves the labile protecting group(s) and later induces peptide bond formation. Additional isolations followed by ion-trap CID were performed for all subsequent fragmentation steps. The product ions were then mass analyzed by mass-selective axial ejection.33
RESULTS AND DISCUSSION
Peptide extension using ki reagents
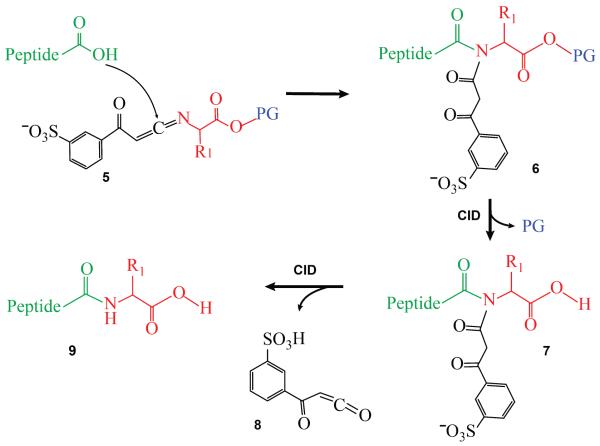
These new ki reagents enable the formation of amide bonds between an amino group of C-terminally protected amino acids and the C-terminus of an anchor peptide via gas-phase ion/ion reactions in a mass spectrometer. The mechanism for peptide bond formation is shown in Scheme 2. The reagent anion [ki-(X-OPG)-H]−, 5, forms an electrostatic complex with a doubly protonated anchor peptide, in which the C-terminal carboxylic acid group adds to the C=N bond of the ki reagent, creating an enol ester 6 which rearranges to an imide 7. An activation step results in the loss of the protecting group, regenerating the initial functional group(s). Upon a second step of CID, one of the amide bonds of the imide 7 can be cleaved. Cleavage at the reagent side gives rise to the signature loss of a ketene 8 (kt, 226 Da) and the extended product 9. Cleavage at the peptide side results in a loss of a diketo derivative and the formation of [peptide-H2O+H]+. 16
Scheme 2.
Formation of the extended peptide via ion/ion reaction between an anchor peptide and ketenimine reagent [(ki-X-OPG)-H]−, where PG is the protecting group (shown in blue). The anchor peptide is shown in green while the amino acid is shown in red.
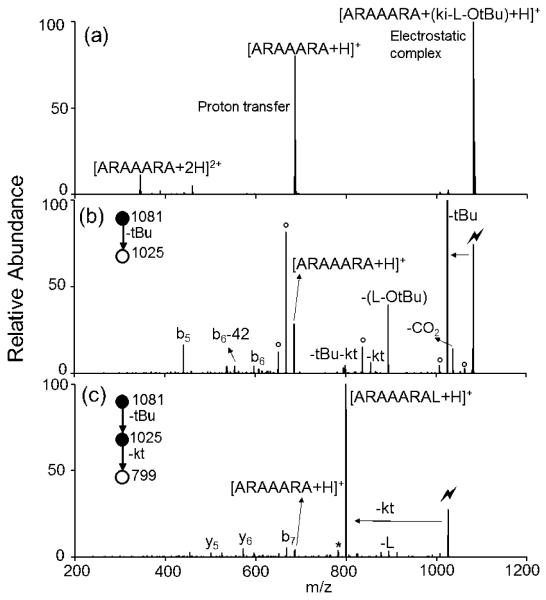
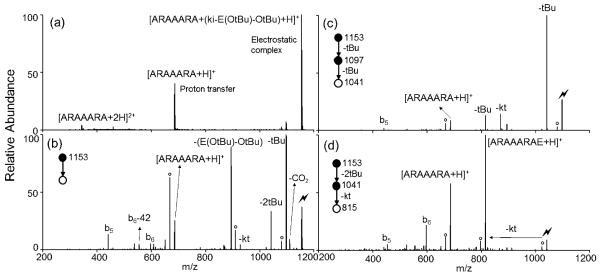
Peptide extension is demonstrated here by adding leucine, L, to the anchor peptide ARAAARA. The spectrum produced via the ion/ion reaction between [ARAAARA+2H]2+ and N-leucine ketenimine anion [(ki-L-OtBu) −H]− is shown in Figure 1a, in which the long-lived electrostatic complex 5, [ARAAARA+ (ki-L-OtBu) +H]+, is observed. Here the sulfonate group on the ki reagent is essential for the electrostatic complex formation between the protonated arginine residue of the anchor peptide and the negatively-charged sulfonate group. Subsequent CID of the complex (Figure 1b) gives rise to multiple dissociation pathways. The first pathway is proton transfer from the peptide to the reagent, resulting in loss of the neutral ki-L-OtBu reagent and generating the [ARAAARA+H]+ species. The second pathway is the loss of L-OtBu from the reagent and the formation of the [ARAAARA+ki+H]+ species, which is thought to result from a side reaction with the N-terminus (Scheme S1). The N-terminal amino group is able to initiate a nucleophilic attack to the ki, resulting in a ring structure formation where the ki is linking the C-terminus and the N-terminus of ARAAARA. The third pathway, which is the main process, proceeds through loss of the protecting tBu group as a neutral 2-methylpropene from the complex, regarded as a deprotection process. There is a sequential ketene (kt) loss from the tBu loss (denoted as –tBu–kt in the Figure 1b), indicating the formation of an amide bond. A peak corresponding to a signature loss of ketene (amide bond formation) without the loss of the protecting group, resulting in the formation of [ARAAARAL-OtBu+H]+ (denoted as –kt in Figure 1b), is also observed. The order in which deprotection and covalent reaction occur is unclear, both processes could also happen simultaneously. Amide bond formation involves multiple rearrangement and tautormerization steps, such that the deprotection can possibly happen at any point. Additional fragmentation of the anchor peptide is also observed as indicated by the presence of the b5 and b6 ions of ARAAARA. Further CID on the tBu loss (Figure 1c) almost exclusively produces a signature loss of ketene, resulting in the extended product [ARAAARAL+H]+. A more detailed mechanism of reaction between ki reagents and carboxylic acids can be found in our previous work using wrk. 16
Figure 1.
Product ion spectra derived from (a) the ion/ion reaction between [ARAAARA+2H]2+ and [(ki-L-OtBu)-H]−, (b) CID of [ARAAARA+ (ki-L-OtBu) +H]+ complex and (c) CID of the tert-butyl loss from [ARAAARA+ (ki-L-OtBu) +H]+ complex. (Lightning bolt denotes ions subjected to collisional activation. Asterisks (*) denote ammonia loss whereas circles (°) denote water losses.)
The overall efficiency, defined as the percentage of initial ARAAARA reactant ions that are converted to product ARAAARAL is determined through the combined efficiencies of three steps, (1) complex formation upon ion/ion reaction, (2) deprotection from activation of the electrostatic complex (MS2) and (3) amide bond formation from activation of the deprotected species (MS3) as shown in Figure 1 a, b and c, respectively. The reaction time in step (1), the amplitude and time of CID in step (2) and (3) were optimized to obtain highest yields of the desired products. (In any case, the efficiencies were not particularly sensitive to CID amplitude or time). The efficiencies (calculated as the peak area of the desired species over the sum of all peak areas) are 56%, 32% and 72%, respectively, giving an overall efficiency of 13%.
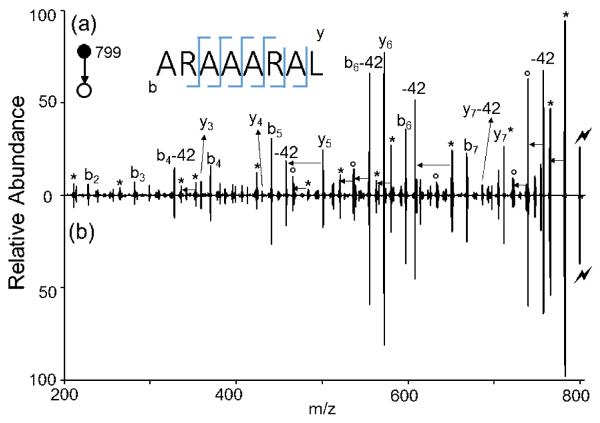
Further isolation and CID was performed to confirm the sequence of the extended peptide noted in Figure 1c. To validate the identity of the extended peptide, its fragmentation spectrum was compared with that of a peptide of the same sequence prepared by solution-phase peptide synthesis using ARAAARA and L-OtBu. Figure 2 compares the dissociation behavior of [ARAAARAL+H]+ generated in the gas phase (Figure 2a) to the fragmentation of the solution-phase analog (Figure 2b). The two spectra are essentially identical. The 42 Da losses are NH=C=NH losses originating from the deguanidination of the arginine side chain.34
Figure 2.
Product ion spectra derived from CID of (a) [ARAAARAL+H]+ generated via gas phase ion/ion reaction and (b) [ARAAARAL + H]+ from solution phase synthesis. (Lightning bolt denotes ions subjected to collisional activation. Asterisks (*) denote ammonia losses whereas circles (°) denote water losses)
The universality of this peptide extension method is investigated by varying ki-amino acid reagents, protecting groups35 and the sequence of the anchor peptide. Another example using ki-G-OtBu is shown in Figures S3 and S4. To prepare the ki reagents, side-chain reactive functionalities of amino acids, like amino, carboxyl groups, needed to be protected. Carboxylic acid side-chains can be protected with the tBu group, allowing acidic residues to be conjugated to the anchor peptide after the loss of two tBu protecting groups (Figure 3). A partially deprotected (loss of a tBu group) and a fully deprotected (loss of two tBu groups) peak can be seen in the CID spectrum of the complex (Figure 3b). Multiple CID steps can be used to produce the second tBu loss from the partially deprotected species (Figure 3c). Combining the two fully deprotected peaks in Figure 3b and 3c by activating the partially deprotected peak without prior isolation the produces a deprotection step with an efficiency of 27%. This roughly matches the efficiency in the single deprotection scenario (Figure 1).
Figure 3.
Product ion spectra derived from (a) ion/ion reaction between [ARAAARA+2H]2+ and [(ki-E(OtBu)-OtBu)-H]−, (b) CID of [ARAAARA+ (ki-E(OtBu)-OtBu) +H]+ complex, (c) CID of the tert-butyl loss from complex, (d) CID of the second tert-butyl loss from complex (Lightning bolt denotes ions subjected to collisional activation. Circles (°) denote water losses.)
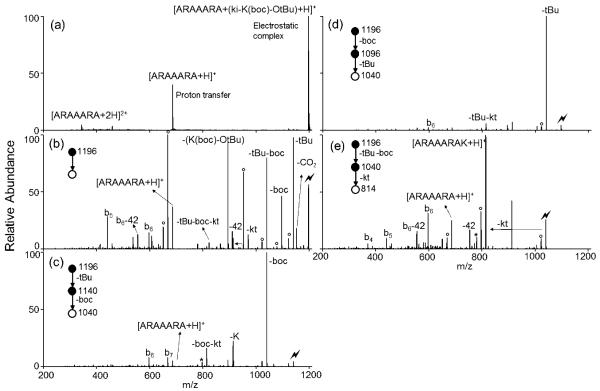
A similar strategy is applied for basic amino acids. The protecting group tertbutyloxycarbonyl (boc) is used to protect the side-chain amine of lysine (Figure 4) and both boc and tBu groups can be cleaved upon MS/MS. 17 Two partially deprotected peaks (loss of a tBu group or loss of a boc group) and a fully deprotected peak (loss of both tBu and boc groups) can be observed in the CID spectrum of the electrostatic complex (Figure 4b). Further activation of the partially deprotected species results in sequential loss of the remaining protecting group (Figure 4c and d). The combined deprotection efficiency is 32%. (We note that peptide methyl esters can also be formed using this strategy when methyl esters are used as protecting groups for carboxylic acids. In this case there is no deprotection step since methyl esters are stable under CID conditions, and the newly formed peptide is a C-terminal methyl ester analogue (Figure S5).) Besides single amino acids, a dipeptide AA can also be conjugated to the C-terminus of ARAAARA via this method (Figure S6).
Figure 4.
Product ion spectra derived from (a) ion/ion reaction between [ARAAARA+2H]2+ and [(ki-K(boc)-OtBu)-H]−, (b) CID of [ARAAARA+ (ki-K(boc)-OtBu) +H]+ complex, (c) CID of the tert-butyl loss from complex, (d) CID of the Boc loss from complex and (e) CID of the Boc+tBu loss from complex and (Lightning bolt denotes ions subjected to collisional activation. Asterisks (*) denote ammonia loss whereas circles (°) denote water losses.)
The anchor peptide is required to contain at least one basic residue (arginine or lysine) in order to be doubly or multiply protonated. An experiment using a lysine-containing anchor peptide, AKAAAKA, is shown in Figure S7 where a lower efficiency is observed. The proton transfer pathway is more competitive in all three steps for this anchor peptide due to the lower proton affinity of lysine residue than arginine residues.
A list of reagents investigated is provided in Table 1. Aliphatic (G, A, V, L), basic (K), acidic (E) amino acids as well as ones with a hydroxyl side-chain (Y) and an amide side-chain (N) are used to represent different amino acid classes. It can be anticipated that all amino acids, with the exception of proline (P), can be added to the C-terminus of an anchor peptide as long as proper protecting group(s) is used. For example, the sulfhydryl group of cysteine (C) can be protected with acetamidomethyl (acm); the imidazole group of histidine (H) can be protected with boc.17, 35 The reagent precursors shown in Table 1 are picked based on their commercial availability. The overall efficiencies were calculated by multiplication of efficiencies of the three steps listed in Table 1 proceeding from the reactants to the final products. The efficiencies of the complex formation varies from 52% percent to 78%, in which the general trend is that more abundant complex can be formed when the reagent ion is larger.36 The yields of the deprotection step are 24~32% for single amino acids and 9% for dialanine. The yields of the amide bond formation are around 60-70% for aliphatic amino acids, and decrease with amino acids containing a side-chain functional group. An ammonia loss from the side-chain of asparagine dominates the CID spectra (Figure S8(c)), which lowers the efficiency greatly for this reagent. With the exception of asparagine, the overall yields of single amino acid are around 8-13%. The increased structural complexity of the dipeptide AA-OtBu over a single amino acid may contribute to the lower efficiency for peptide conjugation.
Table 1.
Efficiencies of reagent amino acids or peptides added and the reagents for successful coupling (efficiency calculated using anchor peptide ARAAARA)
| Reagent precursor |
Activated precursor |
Stepwise efficiencies/ % |
Overall efficiency/ % |
||
|---|---|---|---|---|---|
| Complex formation |
Deprotection | Amide bond formation |
|||
| Glycine | ki-G-OtBu | 52 | 27 | 63 | 9 |
| Alanine | ki-A-OtBu | 54 | 29 | 62 | 10 |
| Valine | ki-V-OtBu | 54 | 24 | 58 | 8 |
| Leucine | ki-L-OtBu | 56 | 32 | 72 | 13 |
| Tyrosine | ki-Y-OtBu | 76 | 25 | 66 | 13 |
| Asparagine | ki-N-OtBu | 60 | 26 | 24 | 4 |
| Lysine | ki-K(boc)-OtBu | 73 | 32‡ | 42 | 10 |
| Glutamic acid | ki-E(OtBu)-OtBu | 65 | 27‡ | 43 | 8 |
| Dialanine | ki-AA-OtBu | 78 | 9 | 6 | 4 |
Combined efficiency of multiple CID steps for two protecting groups.
Side-chain extension
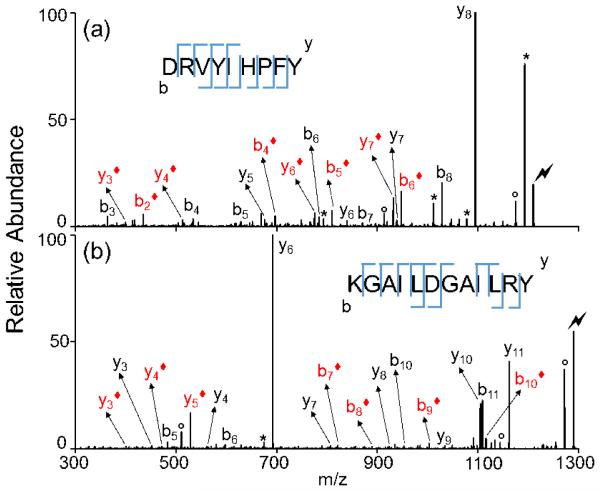
When an aspartic acid or glutamic acid residue is present on the anchor peptide, the extension reaction can happen at the side-chain carboxylic acid as well as the C-terminus, resulting in the formation of a mixture of linear and branched extended products. This is demonstrated in Figure 5. MS/MS was performed on the peptide [DRVYIHPFY+H]+ generated via gas-phase peptide extension on doubly protonated DRVYIHPF (Figure 5a). The main fragment is the y8 ion originating from the aspartic acid effect,37 which is consistent with the C-terminal extended sequence. However, the presence of b- and y- ions labeled with a solid diamond suggests modification at the aspartic acid side chain as a minor reaction channel. It is likely that the mixture of isomers consists of mainly DRVYIHPFY with only a small amount (<10%) of the product with the Y added to the D residue. For the branched peptide, fragments coming from the aspartic acid effect are largely suppressed due to the modification on the acidic side chain. The same phenomenon is also observed on reactant peptide KGAILDGAILR (Figure 5b). A similar side-chain reaction is also observed in the gas-phase N-terminal peptide extension method,17 in which less than 10% of the products were modified at the side-chain amino group of a lysine residue.
Figure 5.
Product ion spectra derived from CID of (a) [DRVYIHPFY+H]+ generated via gas phase ion/ion reaction between [DRVYIHPF+2H]2+ and [(ki-Y-OtBu)-H]−; (b) [KGAILDGAILRY+H]+ generated via gas phase ion/ion reaction between [KGAILDGAILR+2H]2+ and [(ki-Y-OtBu)-H]− (Lightning bolt denotes ions subjected to collisional activation. Asterisks (*) denote ammonia losses whereas circles (°) denote water losses. The red labels with a solid diamond represent fragments corresponding to a modification on an aspartic acid side chain).
CONCLUSIONS
A novel method for the rapid formation of amide bonds in peptides within the mass spectrometer is described. The present approach complements a previously described approach that formed amide bonds at the N-terminus of an anchor peptide in that it generates amide linkages at the C-terminus of an anchor peptide. Amino acids with a variety of side-chains have been demonstrated to form peptide bonds to the C-terminal carboxyl group of anchor peptides. When the C-terminus and nucleophilic side chains of the reagent ions are protected with gas phase-labile protecting groups (boc, tBu, etc.), all amino acids can, in principle, can be added to the C-terminus of anchor peptides, provided the anchor peptide has more than one basic site to be doubly or multiply protonated. Proline, which is an imino acid, cannot be conjugated to the C-terminus of an anchor peptide with this approach. Wrk provides the functional ketenimine group for activation of the amino group of amino acids, and a charge-bearing sulfonate group for complex formation in ion/ion reactions. This method does not use significant amount of solvents and deprotecting agents that are required in most current peptide synthesis methods. An overall reaction yield of ~10% can be achieved for adding an amino acid (except asparagine) to the anchor peptide ARAAARA. Branched peptides can be formed in this approach when a carboxylic acid group exists in the side chain of the anchor peptides.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health under Grant GM 45372.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Merrifield RB. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 1963;85:2149–2154. [Google Scholar]
- 2.Jayawichreme CK, Graminski GE, Quillan JM, Lerner MR. Creation and function screening of a multiuse peptide library. Proc. Natl. Acad. Sci. USA. 1994;91:1614–1618. doi: 10.1073/pnas.91.5.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott WL, Martynow JG, Huffman JC, O’Donnell MJ. Solid-phase synthesis of multiple classes of peptidomimetics from versatile resin-bound aldehyde intermediates. J. Am. Chem. Soc. 2007;129:7077–7088. doi: 10.1021/ja069188y. [DOI] [PubMed] [Google Scholar]
- 4.Nandy JP, Prakesch M, Khadem S, Reddy PT, Sharma U, Arya P. Advances in solution- and solid-phase synthesis toward the generation of natural product-like libraries. Chem. Rev. 2009;109:1999–2060. doi: 10.1021/cr800188v. [DOI] [PubMed] [Google Scholar]
- 5.Pattabiraman VR, Bode JW. Rethinking amide bond synthesis. Nature. 2011;480:471–479. doi: 10.1038/nature10702. [DOI] [PubMed] [Google Scholar]
- 6.Coin I, Beyermann M, Bienert M. Solid-phase peptide synthesis: From standard procedures to the synthesis of difficult sequences. Nat. Protoc. 2007;2:3247–3256. doi: 10.1038/nprot.2007.454. [DOI] [PubMed] [Google Scholar]
- 7.Lee S, Valentine SJ, Reilly JP, Clemmer DE. Controlled formation of peptide bonds in the gas phase. J. Am. Chem. Soc. 2011;133:15834–15837. doi: 10.1021/ja205471n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Julian RR, Valentine SJ, Reilly JP, Clemmer DE. Biomolecular condensation via ultraviolet excitation in vacuo. Int. J. Mass Spectrom. 2012;316:6–11. [Google Scholar]
- 9.Wincel H, Fokkens RH, Nibbering NMM. Peptide bond formation in gas-phase ion/molecule reactions of amino acids: A novel proposal for the synthesis of prebiotic oligopeptides. Rapid Commun. Mass Spectrom. 2000;14:135–140. doi: 10.1002/(SICI)1097-0231(20000215)14:3<135::AID-RCM851>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 10.O’Hair RAJ, Androutsopoulos NK. Can transacylation reactions occur via S(N)2 pathways in the gas phase? Insights via ion-molecule reactions of N-acylpyridinium ions and ab initio calculations. Org. Lett. 2000;2:2567–2570. doi: 10.1021/ol006060r. [DOI] [PubMed] [Google Scholar]
- 11.O’Hair RAJ, Androutsopoulos NK, Reid GE. Do amines react with protonated peptides in the gas phase via transacylation reactions to induce peptide bond cleavage? Rapid Commun. Mass Spectrom. 2000;14:1707–1716. doi: 10.1002/1097-0231(20000930)14:18<1707::AID-RCM83>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Mentinova M, McLuckey SA. Covalent modification of gaseous peptide ions with N-Hydroxysuccinimide ester reagent ions. J. Am. Chem. Soc. 2010;132:18248–18257. doi: 10.1021/ja107286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mentinova M, McLuckey SA. Intra- and inter-molecular cross-linking of peptide ions in the gas phase: reagents and conditions. J. Am. Soc. Mass Spectrom. 2011;22:912–921. doi: 10.1007/s13361-011-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb IK, Mentinova M, McGee WM, McLuckey SA. Gas-phase intramolecular protein crosslinking via ion/ion reactions: Ubiquitin and a homobifunctionalsulfo-NHS Ester. J. Am. Soc. Mass Spectrom. 2013;24:733–743. doi: 10.1007/s13361-013-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prentice BM, Gilbert JD, Stutzman JR, Forrest WP, McLuckey SA. Gas-phase reactivity of carboxylic acid functional groups with carbodiimides. J. Am. Soc. Mass Spectrom. 2013;24:30–37. doi: 10.1007/s13361-012-0506-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Z, Pilo AL, Luongo CA, McLuckey SA. Gas-phase amidation of carboxylic acids with Woodward’s reagent K ions. J. Am. Soc. Mass Spectrom. doi: 10.1007/s13361-015-1209-8. In press. DOI: 10.1007/s13361-015-1209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGee WM, McLuckey SA. Efficient and directed peptide bond formation in the gas phase via ion/ion reactions. Proc. Natl. Acad. Sci. USA. 2014;111:1288–1292. doi: 10.1073/pnas.1317914111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu P, Wang Y. The thriving chemistry of ketenimines. Chem. Roc. Rev. 2012;41:5687–5705. doi: 10.1039/c2cs35159e. [DOI] [PubMed] [Google Scholar]
- 19.Hermanson GT. Bioconjugation Techniques. 2nd ed Academic Press; Amsterdam: 2008. [Google Scholar]
- 20.DeKorver KA, Johnson WL, Zhang Y, Hsung RP, Dai H, Deng J, Lohse AG, Zhang Y. N-allyl-N-sulfonyl ynamides as synthetic precursors to amidines and vinylogous amidines. An unexpected N-to-C 1,3-sulfonyl shift in nitrile synthesis. J. Org. Chem. 2011;76:5092–5103. doi: 10.1021/jo200780x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yavari I, Ahmadian S, Ghazanfarpur-Darjani M, Solgi Y. Formation of N-sulfonylamidines by copper-catalyzed coupling of sulfonyl azides, terminal alkynes, and trialkylamines. Tetrahedron Lett. 2011;52:668–670. [Google Scholar]
- 22.Yoo EJ, Bae I, Cho SH, Han H, Chang S. A facile access to N-sulfonylimidates and their synthetic utility for the transformation to amidines and amides. Org. Lett. 2006;8:1347. doi: 10.1021/ol060056j. [DOI] [PubMed] [Google Scholar]
- 23.Larksarp C, Sellier O, Alper H. Palladium-catalyzed cyclization reactions of 2 Vinylthiiranes with heterocumulenes. Regioselective and enantioselective formation of thiazolidine, oxathiolane, and dithiolane derivatives. J. Org. Chem. 2001;66:3502–3506. doi: 10.1021/jo010037h. [DOI] [PubMed] [Google Scholar]
- 24.Woodward RB, Olafson RA, Mayer H. A new synthesis of peptide. J. Am. Chem. Soc. 1961;83:1010–1012. [Google Scholar]
- 25.Woodward RB, Olafson RA. A useful synthesis of peptides. Tetrahedron. 1966;(Suppl. 8):321–346. [Google Scholar]
- 26.Bustos P, Gajardo MI, Gómez C, Glodie H, Cardemil E, Jabalquinto AM. Woodward’s reagent K reacts with histidine and cysteine residues in Escherichia coli and Saccharomyces cerevisiae phosphoenolpyruvate carboxykinases. J. Protein Chem. 1996;15:467–472. doi: 10.1007/BF01886854. [DOI] [PubMed] [Google Scholar]
- 27.Llamas K, Owens M, Blakeley RB, Zerner B. N-ehtyl-5-phenylisoxazolium-3′-sulfonate (Woodward’s reagent K) as a reagent for nucleophilic side chains of proteins. J. Am. Chem. Soc. 1986;108:5543–5548. [Google Scholar]
- 28.Ouyang Z, Takáts Z, Blake TA, Gologan B, Guymon AJ, Wiseman JM, Oliver JC, Jo Davisson V, Cooks RG. Preparing protein microarrays by soft-landing of mass-selected ions. Science. 2003;301:1351–1354. doi: 10.1126/science.1088776. [DOI] [PubMed] [Google Scholar]
- 29.Laskin J, Wang P, Hadjar O. Soft-landing of peptide ions onto self-assembled monolayer surfaces: An overview. Phys. Chem. Chem. Phys. 2008;10:1079–1090. doi: 10.1039/b712710c. [DOI] [PubMed] [Google Scholar]
- 30.Hager JW. A new linear ion trap mass spectrometer. Rapid Commun. Mass Spectrom. 2002;16:512–526. doi: 10.1002/rcm.1020. [DOI] [PubMed] [Google Scholar]
- 31.Xia Y, Wu J, Londry FA, Hager JW, McLuckey SA. Mutual storage mode ion/ion reactions in hybrid linear ion trap. J. Am. Soc. Mass Spectrom. 2005;16:71–81. doi: 10.1016/j.jasms.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Liang XR, Xia Y, McLuckey SA. Alternatively pulsed nanoelectrosptray ionization/atmospheric pressure chemical ionization for ion/ion reactions in an electrodynamic ion trap. Anal. Chem. 2006;78:3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Londry FA, Hager JW. Mass selective axial ion ejection from a linear quadrupole ion trap. J. Am. Soc. Mass Spectrom. 2003;14:1130–1147. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- 34.McGee WM, McLuckey SA. The ornithine effect in peptide cation dissociation. J. Mass Spectrom. 2013;48:856–861. doi: 10.1002/jms.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isidro-Liobet A, Álvarez M, Albericio F. Amino acid-protecting groups. Chem. Rev. 2009;109:2455–2504. doi: 10.1021/cr800323s. [DOI] [PubMed] [Google Scholar]
- 36.Prentice BM, McLuckey SA. Gas-phase ion/ion reactions of peptides and proteins: acid/base, redox, and covalent chemistries. Chem. Commun. 2013;49:947–965. doi: 10.1039/c2cc36577d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paizs B, Suhai A. Fragmentation pathways of protonated peptides. Mass Spectrom. Rev. 2005;24:508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.