Abstract
Xinjiang is geographically located in central Asia, and it has played an important historical role in connecting eastern Eurasian (EEA) and western Eurasian (WEA) people. However, human population genomic studies in this region have been largely underrepresented, especially with respect to studies of copy number variations (CNVs). Here we constructed the first CNV map of the three major ethnic minority groups, the Uyghur, Kazakh and Kirgiz, using Affymetrix Genome-Wide Human SNP Array 6.0. We systematically compared the properties of CNVs we identified in the three groups with the data from representatives of EEA and WEA. The analyses indicated a typical genetic admixture pattern in all three groups with ancestries from both EEA and WEA. We also identified several CNV regions showing significant deviation of allele frequency from the expected genome-wide distribution, which might be associated with population-specific phenotypes. Our study provides the first genome-wide perspective on the CNVs of three major Xinjiang ethnic minority groups and has implications for both evolutionary and medical studies.
Introduction
Copy number variations (CNVs) are usually defined as DNA segments larger than 1 kb that present different numbers of copies among two or more genomes. By changing gene dosage, CNVs can affect gene expression and phenotype.1 Associations have also been found between CNVs and several neurological diseases and complex traits, including autism, schizophrenia, HIV susceptibility, Crohn's disease, and psoriasis.2 Although it is still unknown how much of the human genome is covered with CNVs, it is evident that genomic regions with CNVs harbor a considerable proportion of genetic diversity both within and among populations.
Xinjiang has played an important role in connecting eastern Eurasia (EEA) and western Eurasia (WEA). It was crossed by the famous Silk Road, which linked the trade among East Asia, Central Asia, and Europe. Many ethnic groups, including Uyghur (UIG), Kazakh (KZK), and Kirgiz (KGZ), have lived there for hundreds of years. Based on the geographic vital role in history and certain phenotypic features of the people living there, it is reasonable to hypothesize that the ethnic groups in Xinjiang share genetic components with EEA and WEA. Previous studies using data from Y-chromosomes have indicated an admixture pattern among Eurasian populations in Central Asia, which was shaped by migration.3 Recently, studies using genomic SNP data have supported the conclusion that UIG is an admixed population incorporating both EEA and WEA ancestries.4, 5 Genetic analysis of Xinjiang ethnic groups may reveal more about human migration history and the mechanisms underlying admixture, which could be useful for gene mapping of disease-gene discovery. As such, admixture mapping has been shown to be of great value in the studies of African Americans.4, 6
Although CNV distribution and properties in several populations have been widely studied,7 those in Xinjiang admixed populations has been largely underrepresented. Here we constructed the first CNV map of the three major Xinjiang populations (KZK, KGZ, and UIG) using Affymetrix Genome-Wide Human SNP Array 6.0. Based on this map, we examined genomic admixture patterns of Xinjiang populations and identified several regions that showed deviation from genome-wide admixture patterns, and regions showing significant frequency differences from those of ancestral source populations. We performed further functional annotation analysis to explore the evolutionary and medical implications of these ‘outlier' CNV regions (CNVRs).
Materials and methods
Samples and CNV detection
We collected 48 samples for each of the three Xinjiang populations. Each individual was the offspring after three generations by non-consanguineous marriage of members in the same ethnicity. Informed consent was obtained from all participants. The procedures followed were in accordance with the ethical standards of the Responsible Committee on Human Experimentation (approved by Ethical Committee of Fudan University) and the Helsinki Declaration of 1975, as revised in 2000. All the samples were assayed using Affymetrix Genome-Wide Human SNP Array 6.0. We used Birdsuite8 to genotype CNVs. The quality control was the same as our previous study.9 Samples which did not pass quality control were removed from subsequent analysis. We also removed the loci that showed difference (FST>0) between combined calling results (all samples) and separate calling results (three ethnic groups). In addition, we included the Affymetrix SNP 6.0 data of 89 Han Chinese from Beijing (CHB) and 110 samples from unrelated Utah residents with northern and western European ancestry (CEU) from HapMap project into analysis and used them as representative ancestral source populations of EEA and WEA, respectively.
The original results of Birdsuite were based on human genome assembly build 36 (hg18), here we converted all the coordinates from hg18 to hg19. The variants those could not be successfully mapped to hg19 were removed. As it is difficult for microarray to determine the exact breakpoints of the variants, the coordinates of variants in this study are the approximate boundaries: the first probe position in CNV and the last probe position in CNV. The individual variants' data are available at dbVar with accession number nstd97.
Building the CNV map of Xinjiang population
A CNVR is defined as a union region of overlapping CNVs on the chromosome.10 CNVs are merged from different samples with any amount of overlap by extending the boundaries of the overlapping CNVs. Birdsuite output file (birdseye_canary_calls) was used to generate a CNVR map. Each CNVR was assigned a genotype (ranging from 0-copy to 4-copy state), which was determined using the copy state of the variant with the largest length. Given a set of samples, biallelic CNVs were defined as loci with only two observable alleles (either 0-copy and 1-copy or 1-copy and 2-copy). We compared our Xinjiang CNV map to the Database of Genomic Variants (DGV v10)11 using the criteria of any amount of overlap. Gene functional annotation clustering analysis was performed via DAVID.12
Calculation of allele frequency and FST
At genotype level, there are five copy states generated by Birdsuite: the 0-copy state (homozygous deletion), 1-copy state (heterozygous deletion), 2-copy state (normal state/copy-neutral with LOH), 3-copy state (single copy duplication), and 4-copy state (double copy duplication). At allele level, a three-allele system (0 copy-allele, loss-allele; 1 copy-allele, normal-allele; 2 copy-allele, gain-allele) is sufficient to explain these five genotypic states. We used an Expectation-Maximization algorithm to calculate the allele frequency by assuming that each CNV locus was in Hardy–Weinberg equilibrium, as in our previous study.9 For each deletion or duplication allele in the admixed populations, the expected allele frequency was calculated as the sum of allele frequency of each ancestral source population weighted by its admixture proportion. Population differentiation for each CNV locus between each pair of populations was measured using the widely used pairwise FST13 based on the inferred allele frequency.
Evaluation of the influence of sample size on the estimation of allele frequency
We used HapMap samples to evaluate the influence of sample size on the estimation of allele frequency. A total of 167 CEU, 89 CHB, and 175 Yoruba in Ibadan (Nigeria) from HapMap were included. The allele frequency of the whole data set in each population was used as a standard reference allele frequency. We randomly sampled 10, 20, 30, 40, 50, 60, 70, and 80 individuals from each population. The difference proportions were calculated as the difference between the sampling allele frequency and the standard frequency divided by the standard allele frequency. The sampling process was repeated 100 times, and we took the average difference proportion of these 100 samplings in each frequency bin for common deletions and duplications separately.
Reconstruction of population relationship and population structure analysis
We used the average number of different genotypes between two individuals from different populations to characterize the genetic distance among populations, which was identical to the one described in our previous work.9 Based on the pair-wise distance among populations, we reconstructed the genetic relationship among these populations using Neighbor-joining method. We also performed 100 bootstrap replications and used PHYLIP14 to generate a consensus clustering topology.
Population structure was inferred using two approaches, principal component analysis (PCA, software Eigensoft v2.015) and a model-based clustering method STRUCTURE v2.2.16 Because microarrays only provide information regarding the total number of copies in each CNV, and the allelic copy number states cannot be determined for multiallelic CNVs at individual level, we used biallelic CNVs to analyze the structures of the populations. STRUCTURE was run with 10 000 iterations and 10 000 burn-ins under K=2.
Selection of ancestral informative markers (AIMs) and local ancestry inference
Given that the unknown allelic configuration of the multiallelic CNVs can affect estimates of subsequent ancestral proportions and the relatively less accurate genotyping of duplications using the microarray platform,17 we still focused AIM selection on biallelic CNVs. Biallelic loci with FST values greater than 0.03 (top 9%) between ancestral source populations were selected as AIMs.We tried to identify the ancestral origins of the CNVs in the admixed populations by superimposing the CNVs onto the ancestry map generated by SNPs using software HAPMIX v1.1.18 The ancestral state of the CNV was determined by that of the most adjacent SNP.
Calculation of heterozygosity
We randomly sampled 40 individuals from each population, and calculated the heterozygosity at polymorphic site k as the following:
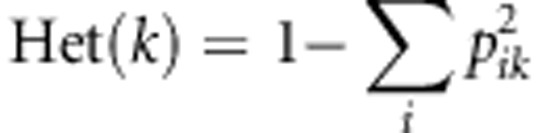 |
pik denotes the ith-allele frequency under a three-allele system. We calculated the average heterozygosity for each population. Statistical test was performed between each admixed population and ancestral source population by Wilcoxon test.
Estimation of linkage disequilibrium (LD) between CNVs and SNPs
We focused on LD between common biallelic CNVs (CNV occurrence more than 10% in each of the five populations: CEU, CHB, KZK, KGZ, and UIG) and their neighboring SNPs extending from 20 kb of CNV boundaries. The CNV and SNP haplotypes were phased using polyHap.19 LD was calculated as r2, and SNPs with maximum r2>0.8 were labeled as CNV tags.
Identification of ancestry-biased CNVs and population-specific CNVs
Ancestry-biased FST was calculated between the observed and expected allele frequencies in admixed populations. Ancestry-biased FST was ranked according to its value and ancestry-biased CNVs were defined as CNVRs in the top 0.5 percentile. We defined population-specific CNVs as those loci showing significant differences (P<0.01) in allele frequency in admixed populations compared with the reference populations at different levels. The sample size were controlled at 40 individuals for each populations and the differences in allele frequency were compared using the χ2-test.
Results
Profiles of CNVs in the three Xinjiang populations
We performed analyses in three Xinjiang ethnic groups (KGZ, KZK, and UIG) with two reference populations (CHB and CEU, as representative populations of EEA and WEA, respectively) from HapMap. Altogether, 46 KGZ, 44 KZK, 41 UIG, 89 CHB, and 110 CEU were included in the analysis and a total of 19 842 CNV events were detected in the three Xinjiang groups. KGZ and UIG carried significantly more CNVs per individual than CHB and CEU (P<0.0002, t-test), whereas KZK showed slightly fewer CNVs (P<0.05, t-test) than the two reference populations (Table 1).
Table 1. CNV detection in Xinjiang populations.
| KGZ | KZK | UIG | CHB | CEU | |
|---|---|---|---|---|---|
| Sample size | 46 | 44 | 41 | 89 | 110 |
| Deletions per individual | |||||
| Avg. | 130.8 | 121.4 | 130.7 | 124.6 | 123.3 |
| Std. | 8.4 | 7.4 | 8.9 | 8.2 | 8.3 |
| Duplications per individual | |||||
| Avg. | 25.2 | 21.7 | 24.6 | 23.2 | 22.6 |
| Std. | 4.2 | 6.0 | 5.5 | 6.3 | 6.0 |
| Total CNV count per individual | |||||
| Avg. | 156.1 | 143.1 | 155.3 | 147.9 | 146.0 |
| Std. | 8.6 | 8.9 | 10.0 | 10.8 | 9.9 |
| Single CNV event length (median bp) | |||||
| Del. | 8969 | 9185 | 8924 | 8924 | 9185 |
| Dup. | 46 869 | 42 154 | 46 796 | 52 119 | 67 092 |
| Total | 10 760 | 10 576 | 10 330 | 10 369 | 11 476 |
| Total CNV length per individual (median bp) | |||||
| Del. | 3 745 555 | 3 770 614 | 3 961 645 | 3 848 095 | 3 922 157 |
| Dup. | 2 925 185 | 1 930 418 | 2 405 381 | 2 281 622 | 2 220 386 |
| Total | 6 656 690 | 5 851 730 | 6 624 935 | 6 288 520 | 6 224 692 |
Abbreviations: Avg, average; CNV, copy number variation; Del., deletion; Dup., duplication; KGZ, Kirgiz; KZK, Kazakh; Std, standard; UIG, Uyghur.
By merging overlapping CNVs into CNVRs (see Methods), we built a CNV map, which consisted of 1436 CNVRs from 131 Xinjiang samples (Supplementary Table 1) with the size ranging from 1.017 to 1906 kb covering 2.5% of the human genome. We determined the genotype and calculated the deletion and duplication allele frequency for each CNVR (Supplementary Table 1, Supplementary Figure 1, see Methods).
We further evaluated the influence of different sample sizes on the estimation of allele frequency. Using random sampling of HapMap individuals as references, which yielded about 90 samples for each of the sampled populations (Methods), we found the difference of allele frequency between 40 and 80 samples to be within the range of approximately 10%, 5%, and 3% of the whole data set sample frequency at ranges of 0.1–0.3, 03–0.5, and 0.5–1, respectively (Supplementary Figure 2). The allele frequency difference between 40 and 80 samples was never observed to be more than 0.03 on average for common CNV loci, which is acceptable because accuracy would not be substantially improved even with a larger sample size.
In addition, we compared the data in this Xinjiang CNV map with those from DGV v10. There were 25 non-singleton regions undiscovered in DGV and the maximum frequency of these regions was 3.1%. These numbers might be underestimated because of many previous studies using lower resolution platforms. There were 581 CNVRs overlapping 986 genes in the Xinjiang CNV map, in which nearly 40% of the genes were duplications. Genes in the CNVRs were enriched in the cluster of peptidase activity, cell adhesion, membrane component, immunoglobulin, olfactory receptor, and defense response.
Genetic relationships between Xinjiang populations and reference populations
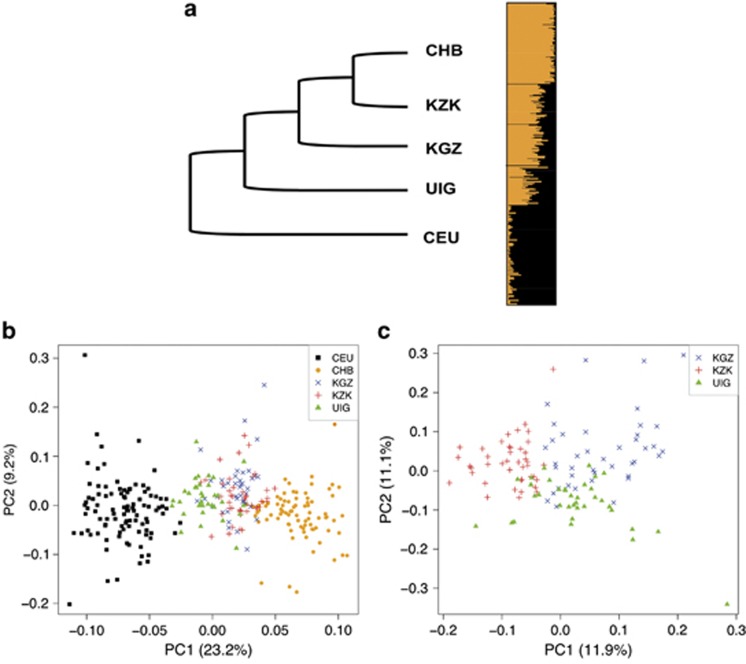
Population relationships can be characterized and measured using genetic distance (Methods). KZK showed the smallest average intra-population distance and UIG showed the largest. KZK and CHB had the smallest inter-population distance, whereas CHB and CEU showed the largest. Based on this distance, we did clustering analysis to study the relationship among these populations (Methods).The topology of the clustering graph, which was supported by 100 bootstrapping replications, showed that all the three Xinjiang populations were in the middle of the two reference populations. Of these, KZK was closer to CHB and UIG was closer to CEU (Figure 1a). This pattern was confirmed by PCA based on biallelic CNVs (Figure 1b). The three Xinjiang groups could also be distinguished without the two reference populations, where the first PC separated KZK from KGZ and UIG, and the second PC separated the latter two groups (Figure 1c). Furthermore, we used a model-based clustering algorithm STRUCTURE to infer population structure, the consistent result (Figure 1a) supported that Xinjiang ethnic groups are admixed populations with genetic components source from both EEA and WEA.
Figure 1.
Population relationship and structure of Xinjiang populations with ancestral source populations inferred from bilallelic CNVs. (a) The left panel shows the topology of the clustering graph of KZK, KGZ, UIG, CHB, and CEU based on average pairwise genetic population distance; the right panel shows the population structure of admixed and ancestral source populations inferred by STRUCTURE with K=2. (b) Population structure of admixed populations and ancestral source populations inferred by PCA. (c) Population structure of admixed populations inferred by PCA.
Population differentiation among populations can be characterized by FST. Although the majority of FST values of CNVRs were very small (<0.1), for each admixed population, we observed that the FST values between the admixed population and one ancestral populations were different from FST values between the admixed population and the other ancestral population (Supplementary Figure 3). For comparisons among Xinjiang populations, pairwise FST was much smaller than that between admixed and ancestral source populations (P<10−15, Wilcoxon rank sum test, Supplementary Figure 3).
Genetic architecture and admixture properties of Xinjiang populations
The average population admixture proportion inferred from biallelic CNVs by STRUCTURE was 65:35, 63:37, 45:55 (EEA/WEA) for KZK, KGZ, and UIG, respectively. The estimated WEA ancestry of UIG was very similar to that in previous study using random SNPs on chromosome 21 (56.2%).20 The admixture proportion between any two individuals from the same population also varied to some extent—8.2%, 9.3%, and 13.7% for KZK, KGZ, and UIG, respectively.
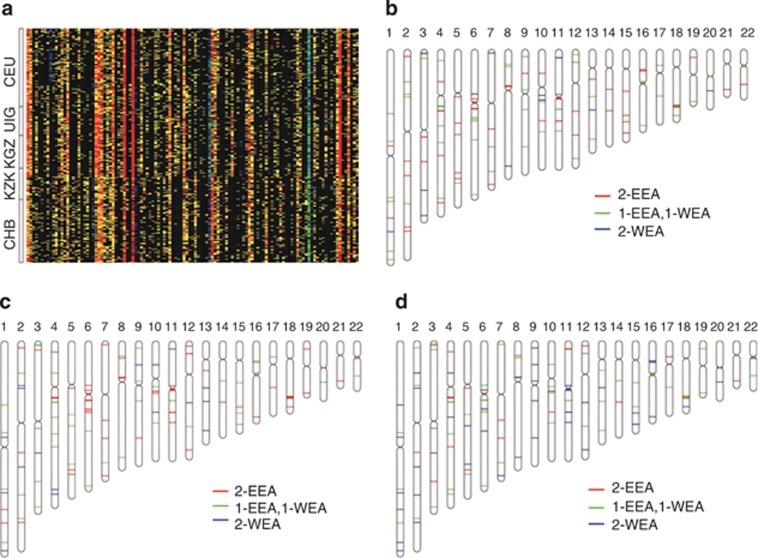
Previous studies have identified that SNPs showing pronounced differences in allele frequency between CHB and CEU could serve as AIMs for UIG.20 Here we selected 115 biallelic CNVs as AIMs for Xinjiang admixed populations (Figure 2a, Supplementary Table 2, Methods). These selected AIMs were able to retrieve the average admixture proportion of EEA versus WEA as 67:33, 66:34, and 46:54 for KGZ, KZK, and UIG, respectively, which showed no significant deviation from the results based on the whole data set (P>0.26, t-test), and would also facilitate the estimation of the CNV admixture proportion of samples from the three populations as well as from other admixed populations with genetic origins from EEA and WEA.
Figure 2.
AIMs and ancestry inference. (a) Copy number states of 115 ancestry informative markers in Xinjiang populations and ancestral source populations. Each row represents an individual and each column represents a locus. Red, yellow, black, blue, and green color corresponds to 0-, 1-, 2-, 3- and 4-copy state, respectively. (b–d) Examples of one sample genome with inferred ancestry of AIMs in KGZ (b), KZK (c), and UIG (d). Red, green, and blue color corresponds to both alleles from EEA, one allele from EEA and the other from WEA, and both alleles from WEA, respectively.
At the individual level, we tried to identify the ancestry of the AIMs in admixed populations (see Methods). We showed in Figures 2b–d the examples of each individual from one admixed group. In addition, we calculated the ancestral proportion in each individual based on the superimposed ancestry-assignments of the AIMs. The results had no significant difference from the ancestral proportions inferred directly from biallelic CNVs (P>0.84, paired t-test).
Increased heterozygosity and extended LD have been observed in the admixed populations, such as the UIG population, using SNP data.20 In terms of CNV data, we calculated the expected heterozygosity using allele frequency (see Methods). On average, all the three admixed populations have increased heterozygosity (0.055) than the two ancestral source populations (0.052) at 1872 polymorphic loci (Supplementary Table 3). Although the admixed groups showed slightly higher CNV taggability than two ancestral source populations at 95 biallelic loci (only 26, 27, 30, 32, and 27 CNVRs were well-tagged by SNPs (r2>0.8, see Methods) in CHB, KZK, KGZ, UIG, and CEU, respectively). Furthermore, there were no significant differences in the overall LD pattern between admixed groups and ancestral source populations when comparing all the LDs of 1241 SNPs located adjacent to 20 kb of the 95 CNVRs (P>0.27, paired Wilcoxon test) except for UIG and KGZ, which showed higher LD than CEU (P<0.024, paired Wilcoxon test).
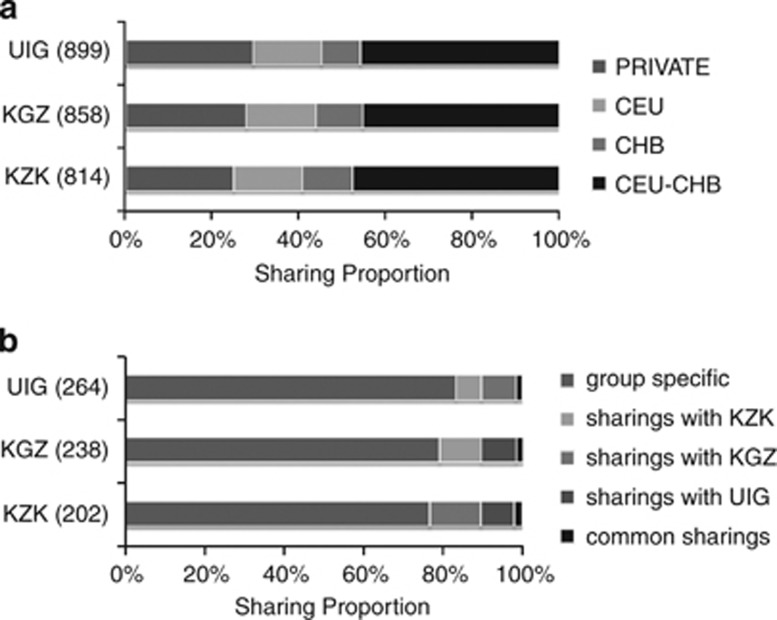
We performed a CNV sharing analysis to evaluate the CNV distribution pattern between Xinjiang ethnic groups and ancestral source populations. More than 40% of the CNVRs in Xinjiang groups were also detected in both CHB and CEU, and about 16% of the CNVRs were observed only in CEU. CNVRs that were detected only in CHB varied from 9.2 to 11.5% (Figure 3a). For the comparisions excluding the CNVs found in either ancestral population, all three Xinjiang groups showed a large proportion of specific CNVRs (Figure 3b).
Figure 3.
CNV sharing analysis of Xinjiang populations and ancestral source populations. (a) CNV sharing between admixed populations and ancestral source populations. (b) CNV sharing among admixed populations excluding regions shared by both sets of ancestral source populations. The numbers in the parentheses represent the total number of the CNV regions.
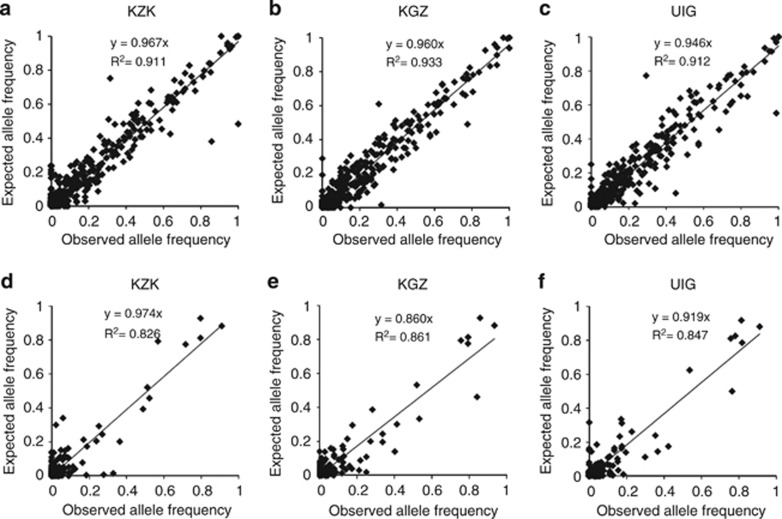
To investigate the relationships between admixed and ancestral source populations with respect to the CNV allele frequency spectrum, we compared the observed deletion and duplication allele frequencies in the admixed populations with their respective expected allele frequencies (see Methods). Excluding loci with rare variants (allele frequency <0.025), the observed allele frequency matched the expected allele frequency on the genome-wide scale (Pearson correlation coefficient r2>0.8; Figure 4). Furthermore, we determined whether the observed and expected data could be explained by regression model y=x. Although the duplication alleles of KGZ deviated from the null hypothesis (H0: β=1; t-test), neither the deletion alleles of any of the three admixed groups nor the duplication alleles of KZK and UGR showed any significant difference from the model y=x (P>0.1), indicating that the overall allele frequency spectrum of the admixed population followed a two-way admixture scenario that could explain the majority of the genetic architectures.
Figure 4.
Observed and expected allele frequency of Xinjiang admixed populations. Observed and expected frequencies of (a–c) deletion alleles and (d–f) duplication alleles in (a, d) KZK, (b, e) KGZ, and (c, f) UIG. Expected allele frequency was calculated as the sum of allele frequency in ancestral source populations weighted by their contributions.
Population-specific CNVs identified in the Xinjiang admixed populations
Although the overall frequency spectra of Xinjiang populations showed a pattern typical of admixture, the allele frequency of some regions under different evolutionary forces might deviate from such expected distribution. To identify these outliers, we applied a strategy analogous to one that was recently used to assess natural selection among African Americans.21 We calculated the ancestry-biased FST between the expected and observed allele frequency for each admixed population (Supplementary Figure 4). CNVRs in the top 0.5 percentile were regarded as ancestry-biased CNVs (Table 2). For example, at chr22: 22315089-23258615, which contains several genes like GGTLC2 (gamma-glutamyltransferase light chain 2) and PRAME (preferentially expressed antigen in melanoma), deletions and duplications could hardly be observed in all three admixed populations, but could be observed with certain frequencies in the ancestral populations.
Table 2. Ancestry-biased CNVs in Xinjiang populations.
| CNVRa | Ancestry-biased FST | Populations | RefSeq genes |
|---|---|---|---|
| chr1:(106002162_106012754)_(106024798_106028674)del | 0.065 | KZK | |
| chr1:(210071062_210081613)_(210083984_210086552)del | 0.07 | KZK | |
| chr2:(180061655_180067192)_(180079144_180083591)del | 0.076 | UIG | SESTD1 |
| chr3:(98937784_98944458)_(98949421_98950964)del | 0.076 | KGZ | |
| chr5:(97930840_97933671)_(97944946_97954791)del | 0.078,0.058,0.053 | KGZ,KZK,UIG | |
| chr5:(151514171_151515386)_(151520497_151521211)del | 0.052 | KGZ | |
| chr6:(32587457_32593021)_(32602107_32602269)del | 0.06 | UIG | |
| chr6:(65710729_65711399)_(65713792_65717384)del/dup | 0.074,0.091 | KZK,UIG | EYS |
| chr6:(165724052_165727817)_(165731928_165734652)del | 0.075 | UIG | |
| chr7:(6838724_6854110)_(6864083_6864247)del/dup | 0.153,0.118 | KZK,UIG | C7orf28B |
| chr7:(64554240_64565585)_(64595698_64637128)del | 0.1 | KGZ | |
| chr7:(143213351_143216729)_(143568059_143572166)dup | 0.061 | KGZ | CTAGE6,FAM115A,FAM115C,LOC154761,LOC441294 |
| chr8:(2133375_2141462)_(2393160_2395110)del/dup | 0.069 | KZK | |
| chr9:(29091097_29094549)_(29097680_29100799)del/dup | 0.065 | KGZ | |
| chr9:(68183981_68186051)_(68188547_68209916)del/dup | 0.069 | UIG | |
| chr9:(135933784_135934431)_(135965662_135967602)del/dup | 0.057 | KZK | CEL,CELP |
| chr10:(107056279_107057038)_(107059170_107062476)del/dup | 0.052 | KZK | |
| chr10:(114111348_114112075)_(114116839_114118417)dup | 0.085 | UIG | |
| chr11:(5519684_5520062)_(5523593_5525152)del | 0.064 | KZK | |
| chr16:(69988386_70024589)_(70272708_70277632)del/dup | 0.152,0.068 | KGZ,UIG | CLEC18C,PDPR,PDXDC2 |
| chr17:(43574921_43585756)_(43650957_43655090)del/dup | 0.055 | KGZ | LRRC37A4 |
| chr21:(23646523_23651134)_(23674393_23674592)del | 0.262,0.093 | KGZ,UIG | |
| chr22:(22307519_22315089)_(23258615_23258909)del/dup | 0.111,0.069,0.130 | KGZ,KZK,UIG | GGTLC2,LOC96610,POM121L1P,PRAME,TOP3B,VPREB1,ZNF280A,ZNF280B |
Abbreviations: CNVR, copy number variation region; KGZ, Kirgiz; KZK, Kazakh; UIG, Uyghur.
The coordinates were mapped to reference genome assembly hg19. The CNV is in the following format: chromosome:(outer-start_inner-start)-(inner-end_outer-end)CNV-type.
On the other hand, admixture event might cause the difference in allele frequency between ancestral source populations and admixed populations as well as among the admixed populations. We defined population-specific CNVs as regions with significant differences in frequency from reference populations. Here we identified the population-specific CNVs for each of the Xinjiang populations at two levels using different reference populations: (1) ancestral source populations and (2) the other two admixed populations. These population-specific CNVs were highlighted in Supplementary Table 4. At the first level with ancestral source populations as reference, the three admixed have three population-specific CNVRs in common. Whereas at the second level with admixed populations as reference, we found that the region on chromosome 1 overlapping with Rh blood system genes (RHCE, RHD, TMEM50A) had a higher deletion frequency in UIG (31.7%) than in KGZ (9.9%), KZK (12.5%), or CHB (7.9%), but a lower frequency than in CEU (43.2%). In addition, we identified the non-singleton CNVs that were observed only in admixed populations but not present in the ancestral source populations. The functional annotation analysis of the genes overlapping with all private CNVs showed enrichment in response to wounding (genes: TPST1, LPA, and NINJ2; P<0.04, Benjamini correction: P<0.1).
Discussion
In this study, we investigated CNVs in three major ethnic minority groups, UIG, KZK, and KGZ, using Affymetrix Genome-Wide Human SNP Array 6.0. On a genome-wide scale, the fitness of the admixture model supported the previous conclusion that the UIG gene pool was probably a recipient of EEA and WEA rather than a donor.5 The admixture proportions inferred here were the most reliable estimations, as indicated by the fact that the correlation coefficients between observed and expected allele frequency of admixed populations were larger at our inferred admixture proportion than at any other values of the proportion (Supplementary Figure 5).
One of the practical applications for the admixed populations is admixture mapping. It has been shown the success in other admixed populations like African Americans. Unlike African Americans, the Xinjiang admixed populations have longer admixture time and have ancestral source populations with much shorter divergence time. Admixture mapping still holds in Xinjiang admixed populations as there are a large number of markers (AIMs) available with high-differentiated frequency between WEA and EEA populations, and the previous study has also demonstrated the effectiveness of the admixture mapping for localization of disease genes in UIG populations.4 However, the long admixture time in Xinjiang populations has two sides for admixture mapping: on one hand, it is a challenge as weaker LD between markers and the causal variants as a consequence of more recombination events occurred due to longer admixture history; on the other hand, it could help fine mapping of disease-associated genes, again, because of the shorter LD between AIMs and potential causal variants. CNVs usually affect genome more than hundreds and thousands of base pairs; they are not only the markers but also likely the variants that make the impact directly. The AIMs we identified here are a number of CNVs showing high-differentiated frequency between ancestral source populations, which have potential utility for admixture mapping. For example, the region of chr17: 44161867-44364214 overlapping with gene KANSL1 has no duplication in EEA, but has a duplication frequency of 0.44 in WEA. This duplication copy, which was found to be associated with novo transcript and is likely to be underlying positive selection,22 could also be observed in Xinjiang admixed populations at a frequency of ∼0.13 with a clear background of complete WEA-inheritance. In addition, although the deletion of this gene was absent in the normal samples here, it was found to be associated with the 17q21.31 microdeletion syndrome.23
In summary, we constructed the first CNV map of three admixed populations in Xinjiang and provided a first landscape of population genomics based on CNVs in this region. Our analysis showed that the genomic CNV distribution of the three representative populations, UIG, KZK, and KGZ, each followed an admixture pattern with an ancestral genetic pool from both EEA and WEA. Our study advanced our understanding of CNV distribution and admixture history of populations in Xinjiang and may serve as a useful resource for further medical and evolutionary studies.
Acknowledgments
These studies were supported by the Strategic Priority Research Program the Chinese Academy of Sciences (XDB13040100), by the National Science Foundation of China (NSFC) grants (91331204 and 31171218). SX is Max-Planck Independent Research Group Leader and member of CAS Youth Innovation Promotion Association. SX also gratefully acknowledges the support of the National Program for Top-notch Young Innovative Talents of The ‘Ten-Thousand-Talents' Project and the support of K.C.Wong Education Foundation, Hong Kong. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Author contributions
SX conceived and designed the study. HL, WJ, RF, and DL performed data analysis. XP contributed to sample collection. SL, HZ, and YP contributed to the experimental work. SX and HL interpreted the data and wrote the paper. LJ contributed to reagents and materials. All authors have read and approved the final version of the manuscript.
Supplementary Material
References
- 1Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO: Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res 2011; 21: 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Zhang F, Gu W, Hurles ME, Lupski JR: Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 2009; 10: 451–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Wells RS, Yuldasheva N, Ruzibakiev R et al: The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 2001; 98: 10244–10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Xu S, Jin L: A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am J Hum Genet 2008; 83: 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Xu S, Jin W, Jin L: Haplotype-sharing analysis showing Uyghurs are unlikely genetic donors. Mol Biol Evol 2009; 26: 2197–2206. [DOI] [PubMed] [Google Scholar]
- 6Reich D, Patterson N, De Jager PL et al: A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet 2005; 37: 1113–1118. [DOI] [PubMed] [Google Scholar]
- 7Mills RE, Walter K, Stewart C et al: Mapping copy number variation by population-scale genome sequencing. Nature 2011; 470: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Korn JM, Kuruvilla FG, McCarroll SA et al: Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008; 40: 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Lou H, Li S, Yang Y et al: A map of copy number variations in chinese populations. PLoS One 2011; 6: e27341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Shaikh TH, Gai X, Perin JC et al: High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res 2009; 19: 1682–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951. [DOI] [PubMed] [Google Scholar]
- 12Dennis G 1, Sherman BT, Hosack DA et al: DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003; 4: P3. [PubMed] [Google Scholar]
- 13Weir BS, Hill WG: Estimating F-statistics. Annu Rev Genet 2002; 36: 721–750. [DOI] [PubMed] [Google Scholar]
- 14Felsenstein J: PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 1989; 5: 164–166. [Google Scholar]
- 15Patterson N, Price AL, Reich D: Population structure and eigenanalysis. PLoS Genet 2006; 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Falush D, Stephens M, Pritchard JK: Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 2003; 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA: Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet 2008; 40: 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Price AL, Tandon A, Patterson N et al: Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet 2009; 5: e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Su SY, Asher JE, Jarvelin MR et al: Inferring combined CNV/SNP haplotypes from genotype data. Bioinformatics 2010; 26: 1437–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Xu S, Huang W, Qian J, Jin L: Analysis of genomic admixture in Uyghur and its implication in mapping strategy. Am J Hum Genet 2008; 82: 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Jin W, Xu S, Wang H et al: Genome-wide detection of natural selection in African Americans pre- and post-admixture. Genome Res 2011; 22: 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Boettger LM, Handsaker RE, Zody MC, McCarroll SA: Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet 2012; 44: 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Cooper GM, Coe BP, Girirajan S et al: A copy number variation morbidity map of developmental delay. Nat Genet 2011; 43: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.