Abstract
Variants in PORCN are a cause of Goltz-Gorlin syndrome or Focal Dermal Hypoplasia, an X-linked dominant disorder affecting heterozygous females and until now considered to be embryonic lethal in males. Exome sequencing was performed in a family in which two male siblings were characterized by microphthalmia and additional congenital anomalies including diaphragmatic hernia, spina bifida and cardiac defects. Surprisingly, we identified a maternally inherited variant in PORCN present in both males as well as in two female siblings. This represents the first finding of a PORCN variant in non-mosaic males affected with Goltz-Gorlin syndrome. The apparently asymptomatic mother showed extreme skewing of X-inactivation (90%), an asymptomatic female sibling showed skewing of 88%, and the second female sibling affected with cutis aplasia of the scalp showed X-inactivation considered within the normal range.
Introduction
Microphthalmia occurs with an incidence of 0.9–2.3 per 10 000,1 and is estimated to occur in association with additional congenital malformations in up to 90% of those patients, with malformations in the musculoskeletal, cardiovascular and central nervous systems being the most common anomalies.2 A large number of genes are associated with both isolated and syndromic forms of microphthalmia, with 10 specific OMIM entries for syndromic microphthalmia. Microphthalmia in combination with congenital diaphragmatic hernia (CDH) is infrequent and a clinical diagnosis of Matthew-Wood syndrome (MWS) may be suspected (MCOPS9; OMIM #601186). MWS is caused by variants in the STRA6 gene (OMIM *610745) and is associated with microphthalmia/anophthalmia, CDH, cardiac anomalies and pulmonary defects.3 The combination of microphthalmia with CDH has also been reported because of deletions or variants in other genes or genomic loci.4, 5, 6 Here, we investigated a familial case of syndromic microphthalmia in association with CDH, spina bifida and cardiac anomalies. To identify the cause, we performed exome sequencing on two affected male siblings and both parents, which identified a variant in the X-linked PORCN gene as the underlying cause.
Materials and Methods
Clinical details
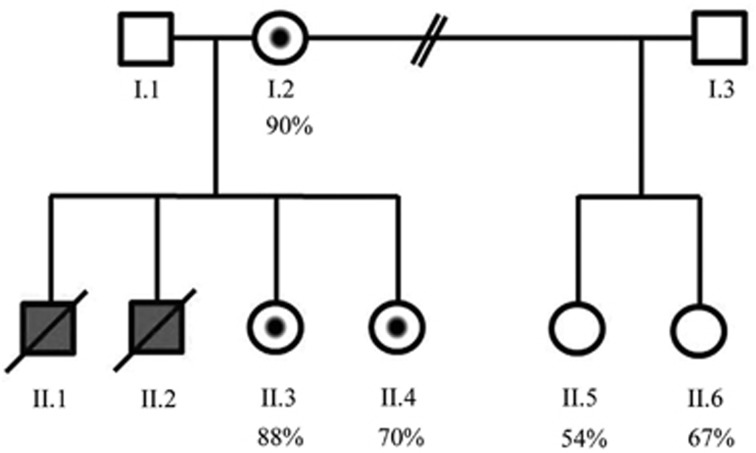
From the first relationship of the mother (I.2), with partner (I.3), two healthy daughters were born (II.5 and II.6). From her current relationship there were two affected male fetuses (II.1 and II.2) and two healthy daughters (II.3 and II.4), see pedigree, Figure 2. Patient II.1 was born at term pregnancy with microphthalmia and coloboma of the retina, a left-sided posterolateral diaphragmatic hernia, and an atrial septal defect. Birth parameters were in the normal range: weight 3010 g, length 49 cm and occipitofrontal circumference 34 cm. Aside from the microphthalmia, no other dysmorphic facial features were observed. Further examination showed bilateral simian creases. The skin was normal and no other digital or skeletal abnormalities were observed. He was deceased at day 0, with confirmation of the clinical findings by pathology examination. Patient II.2 had multiple anomalies that were detected at 28 weeks of gestation: bilateral microphthalmia with dense intra-ocular tissue, a large thoraco-lumbar spina bifida and hydronephrosis of the left kidney. He was born at 38 weeks of gestation, with normal birth parameters: weight 3040 g, length 54 cm, and occipitofrontal circumference 35 cm. In addition to the prenatally detected anomalies, hypospadias and absence of the first radial ray of the right hand were detected postnatally. At day 0, the spina bifida was closed and a ventriculo-peritoneal drain was placed. Cerebral imaging showed enlarged ventricles, partial agenesis of the corpus callosum and several intracerebral haemorrhages. No visible pigmentation anomalies were present upon clinical examination. The boy deceased after 10 days due to respiratory insufficiency. No post-mortem examination was performed. A potential diagnosis of Matthew-Wood syndrome (MWS; MCOPS9; OMIM #601186) was made, but mutation analysis of the STRA6 gene was negative for any causal variants in this patient (performed at the Institute for Human Genetics, Erlangen, Nurnberg, Germany).
Figure 2.
Family pedigree. Individuals I.1, II.2, II.1 and II.2 underwent exome sequencing, which revealed the p.(Gly157Asp) variant in the PORCN gene in both affected individuals (II.1 and II.2) as well the asymptomatic mother (I.2) who was shown to have 90% skewing of X-inactivation. Further analysis of additional family members revealed that the two female siblings II.3 and II.4 also carry the variant, with 88% and 70% skewing of X-inactivation, respectively. The two females from the previous relationship (II.5 and II.6) have not inherited the variant and demonstrate 54% and 67% skewing of X-inactivation, respectively. Individual I.3 was not tested.
Chromosomal microarray analysis
Chromosomal microarray analysis was performed using the CytoSure Syndrome Plus 180k array (Oxford Gene Technology,Oxford, UK) for both affected individuals and both parents to exclude any pathogenic CNV(s) as the cause, as previously described.7
Exome sequencing
Written, informed consent was provided by the parents before exome sequencing.
Genomic DNA was sheared by sonication, and whole-genome sequencing libraries were prepared using the TruSeq DNA Library Preparation Kit (Illumina, San Diego, CA, USA) in which platform-specific adaptors and unique DNA indexes were ligated. The gel-free method was performed with no size selection of fragments. DNA sequencing libraries were subsequently enriched with the SeqCap EZ Human Exome Library v2.0 (Roche, NimbleGen, Madison, WI, USA), and 2 × 100-bp paired-end reads were generated on a HiSeq2000 (Illumina) with four exome-seq samples pooled in a single lane of a sequencing flow-cell. Sheared DNA, whole-genome libraries and enriched exome-seq libraries were validated using DNA-1000 chips on the BioAnalyser (Agilent, Santa Clara, CA, USA), and library concentrations were determined using the dsDNA Broad Range Assay on the Qubit (Invitrogen, Life Technologies Europe B.V., Gent, Belgium).
The paired-end sequence reads were aligned to the human genome (hg19) with the Burrows-Wheeler Aligner (version 0.5.9)8 using default settings, and the read trimming parameter was set to 15. SAMtools (version 0.1.12a)9was used for converting (SAM/BAM), sorting and indexing alignments. The quality metrics for mapping were calculated with Picard tools (version 1.38). Duplicate reads were marked with Picard tools and excluded from downstream analysis. The GATK framework (version 1.0.4974)10 was used for performing the local realignment, base call recalibration and SNP calling. Indels were called with Dindel (version 1.01)11 using default parameters. Variants were annotated with ANNOVAR (version 2011),12 including for dbSNP (dbSNP132), and 1000 Genomes data (release May 2011). Functional predictions for the amino-acid changes according to different models (SIFT, Polyphen2, LRT and MutationTaster) were retrieved from dbNSFP (database of human nonsynonymous SNPs and their functional predictions).13
Variant filtering
Variant files annotated by the GATK analysis pipeline were filtered against provided annotations in Excel, and using the web application ‘Annotate-it'14 (http://www.annotate-it.org/). Details of filters applied by each method and the number of variants remaining after filtering are provided as online Supplementary Information Supplementary Tables 1 and 2). Variants of interest were also checked against the Exome Variant Server (NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, USA; http://evs.gs.washington.edu/EVS/).
Variant confirmation by classical Sanger sequencing.
Primers were designed using Primer3 software (http://frodo.wi.mit.edu/), PCR products were purified with ExoSAP-IT (GE Healthcare, Little Chalfont, UK) and sequenced using BigDye Terminator v3.1 chemistry (Life Technologies Europe B.V.) on a 3730 DNA Analyzer (Life Technologies Europe B.V.). Sequence traces were aligned to the reference sequence using Sequence Scanner v1.0 software (Applied Biosystems, Life Technologies Europe B.V.) and CLC Main Workbench v6 (CLC Bio, Aarhus, Denmark). Primer sequences used were forward primer 5′-CATGCTGATCTGCTCTCTGC-3′ and reverse primer 5′-CCACTCAGGACCTCACCAGT-3′.
X-inactivation analysis
The methylation status of the X-linked androgen receptor gene was assessed by gene methylation assay, as previously described.15, 16 The resulting products were separated on an ABI 3730 (Applied Biosystems) and peak positions and peak intensity areas were further processed using Excel to calculate the percentage inactivation of both alleles.
Submission of variant details to public database
The PORCN variant details and clinical information for individuals I.2, II.1, II.2, II.3, and II.4 have been submitted to the Leiden Open Variation Database v3.0, PORCN gene page; http://databases.lovd.nl/shared/genes/PORCN.
Results
Chromosomal microarray analysis of material from both affected fetuses (II.1 and II.2) revealed no pathogenic CNVs. A possible clinical diagnosis of MWS was excluded in the second affected child (II.2) by mutation analysis for the STRA6 gene by conventional Sanger sequencing, which revealed no pathogenic variants. Given the high likelihood of an autosomal recessive or X-linked mode of inheritance as the cause of the severe phenotypic features, we undertook exome sequencing in the two affected fetuses (II.1 and II.2) and both parents (I.1 and I.2).
Between 52 and 65 million reads were obtained for each of the four samples, with 94.6–96.7% of reads aligned to the reference human genome (hg19). Mean target region coverage of between 43x and 51x was obtained, with 91–92% of target bases having at least 10x coverage.
Variants were filtered on quality parameters, variant type, against the annotated population databases for variants with a MAF <1%, and for functionally damaging predictions. Identification of variants shared by the two affected individuals was aided by the use of ‘annotate-it'. Numbers of variants remaining after filtering is provided in Table 1, and the parameters used are provided in Supplementary Tables 1 and 2.
Table 1. Numbers of shared rare variants remaining after filtering.
| II.1 | II.2 | |
|---|---|---|
| % Captured regions, coverage >10x; Q>30 | 92 | 91 |
| Average coverage of captured region (x) | 51 | 43 |
| Total number of SNPs | 91 412 | 82 221 |
| Total number of indels | 6851 | 5937 |
| Rare homozygous variants | 0 | |
| Rare compound heterozygous variants | 11 (0) | |
| Rare X-linked variants | 4 (2) | |
| Rare de novo variants | 0 | |
| Rare inherited variants (maternal) | 56 (5) | |
| Rare inherited variants (paternal) | 49 (5) | |
Abbreviation: SNP, single-nucleotide polymorphism.
Provides the numbers of rare variants (MAF <1%) remaining after filtering against population databases (1000 genomes and the Exome Variant Server), with the number predicted functionally damaging by all four prediction algorithms within brackets, and listed by potential mechanisms of inheritance.
Of 11 compound heterozygote variants shared by both affected individuals, none were predicted to be functionally damaging, and no shared de novo variants were identified excluding germline mosaicism as a possible cause. Further examination of rare variants shared by both affected individuals identified a likely pathogenic variant; a hemizygous X-linked non-synonymous variant in the PORCN gene (NM_203475.1: ENST00000326194: c.470G>A:p.(Gly157Asp)). Annotate-it provides text-based genotype–phenotype associations (according to HPO and LDDB terms) from ‘A Gene Apart' and respective P-values for this association. Both anophthalmia (P-value=1.348 E-05) and congenital hernia of diaphragm (P-value=7.655 E-03) are listed in association with the candidate variant in PORCN, thus aiding causal variant identification. This novel variant has thus far not been reported in association with Focal Dermal Hypoplasia (FDH)/Goltz-Gorlin syndrome, and has also not been observed in population databases of normal individuals (dbSNP, 1000 genomes or the Exome Variant Server). The PORCN p.(Gly157Asp) variant is predicted to be functionally damaging by four algorithms (SIFT, PolyPhen2, LRT and MutationTaster).
Extended familial PORCN mutation analysis and X-inactivation studies
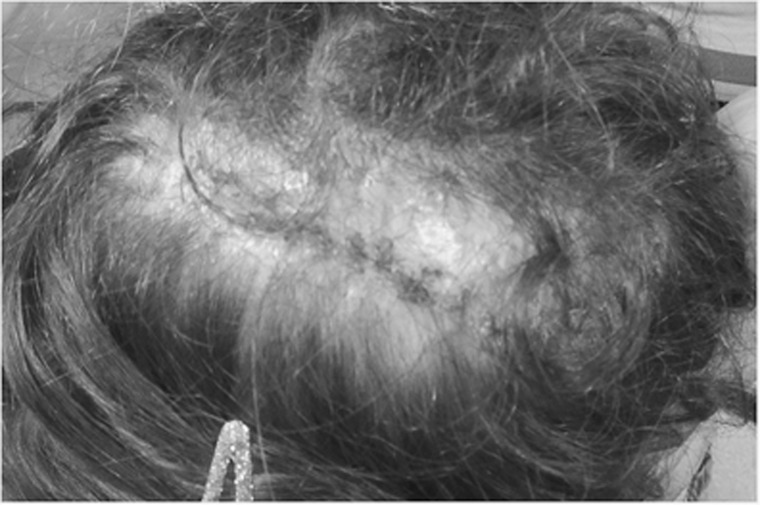
Variants in PORCN are associated with Goltz-Gorlin syndrome or FDH (OMIM #305600), an X-linked dominant disorder in which heterozygous females are affected. Conventional Sanger sequencing of the PORCN gene confirmed the presence of the PORCN variant in the mother (I.2) and both affected males (II.1 and II.2). Analysis of the four daughters revealed the same variant in II.3 and II.4, but not in II.5 and II.6. We therefore investigated the X-inactivation status in genomic DNA from the mother (I.2) and the four daughters from both relationships (II.3 and II.4; and II.5 and II.6). This revealed extreme skewing of X-inactivation (90%/10%) in the heterozygous mother; and levels of 88%/12% and 70%/30% in the two heterozygous daughters (II.3 and II.4, respectively) with the same maternally inherited X chromosome. All daughters were examined by a clinical geneticist upon these results. One of the heterozygous female carriers, initially considered unaffected (II.4), displayed aplasia cutis on the scalp, shown in Figure 1. Interestingly, the skewing of X-inactivation of 70%/30% observed in this girl is considered within the normal range. No additional skin, ocular or skeletal abnormalities were seen in either of the heterozygous female carriers. The two daughters who do not carry the variant (II.5 and II.6) showed random X-inactivation with levels of 54%/46% and 67%/33%, respectively, and inheritance of the alternative maternal X chromosome. The pedigree is shown in Figure 2, including the results of the X-inactivation analysis.
Figure 1.
Cutis aplasia of the scalp in female II.4.
PORCN sequencing in additional unrelated individuals
All coding exons of the PORCN gene were investigated by conventional Sanger sequencing in seven additional unrelated patients with overlapping phenotypic features of eye anomalies in combination with diaphragm or lung defects, or neural tube defects (online Supplementary Information, Supplementary Table 3). However, this did not reveal any pathogenic variants.
Discussion
We describe the first case of non-mosaic males affected with syndromic microphthalmia because of a non-synonymous variant in the PORCN gene. The two fetuses were affected with multiple congenital anomalies including microphthalmia, CDH and an atrial septal defect (II.1); and bilateral microphthalmia and spina bifida (II.2). To our knowledge, there are four previous cases reported of PORCN variants in association with syndromic CDH: a female fetus with multiple congenital anomalies including CDH, limb anomalies, microphthalmia and lung anomalies; a female with phenotypic features consistent with FDH and Pentalogy of Cantrell, including an anterior diaphragmatic hernia; and two unrelated female fetuses born to affected mothers who displayed ectopia cordis, diaphragmatic hernia and abdominal wall defects.5, 17, 18 Our finding thus adds further support for PORCN variants as a cause of syndromic CDH and not a coincidental association. The novel variant that we report affects the fourth transmembrane domain of the PORCN (porcupine) protein and the putative amino-acid change is predicted to be functionally damaging by four algorithms (SIFT, PolyPhen2, LRT and MutationTaster). Pathogenic PORCN variants have previously been reported that affect the same transmembrane domain, which further supports the pathogenicity of the p.(Gly157Asp) variant.5, 19, 20, 21
Variants in PORCN were first reported as a cause of FDH in 2007.19, 22 Froyen et al later identified PORCN variants and gene deletions in a cohort of patients with a clinical diagnosis of FDH.23 FDH is characterized by phenotypic features, including longitudinal striation of the long bones, the combination of split hand with syndactyly and absence of rays (also termed ‘lobster-claw hand'), as well as atrophy and linear pigmentation of the skin, herniation of fat through dermal defects and multiple papillomas of the mucous membranes or skin. Oral anomalies, in addition to lip papillomas, include hypoplastic teeth. Ocular anomalies are another characteristic feature of FDH, including microphthalmia, coloboma of the iris and choroid, and strabismus. Intellectual disability has also been reported, although in a minority of patients. Recently, a ‘mutation update' for PORCN reported a number of new cases and reviewed those reported to date.24
Interestingly, FDH is mainly observed in heterozygous females, whereas the few reports of PORCN variants in affected males are limited to cases of somatic mosaicism. Variants in PORCN thus cause an X-linked dominant condition, which, until now, was considered to be embryonic lethal in males. Our finding thus represents the first report of affected non-mosaic males, adding evidence for PORCN variants in males as a cause of phenotypic features, including eye anomalies, diaphragm defects, spina bifida, cardiac defects, kidney defects and structural brain anomalies. This is in contrast to some of the characteristic phenotypic features more commonly seen in females affected with Goltz-Gorlin syndrome or FDH, especially the skin anomalies. Of particular, interest is the difference in phenotypic severity seen between the male patients we report and the female sibling with cutis aplasia for whom skewing of X-inactivation of 70%/30% would be considered in the normal range. Possible explanations for this disparity are differing degrees of X-inactivation in different tissues in this female, which although not investigated, but have been reported to occur,25 combined with the likelihood that the non-synonymous mutation observed encodes a hypomorphic allele with reduced function only and not complete loss of protein function. A possible clinical diagnosis of MWS/Microphthalmia, Syndromic 9 (MCOPS9; OMIM #601186) in one of our patients because of the phenotypic features of CDH and microphthalmia may suggest that some male fetuses without a genetic diagnosis of MWS owing to variants in STRA6 may actually harbour variants in PORCN. However, we did not identify any damaging PORCN variants in a screen of seven additional unrelated patients with eye anomalies in combination with diaphragm or lung abnormalities, or neural tube defects.
PORCN is involved in the trafficking of Wnt proteins between the endoplasmic reticulum and Golgi (reviewed in Clements26). Defective PORCN impairs transfer of Wnt proteins through the cell leading to reduced Wnt secretion affecting downstream pathways reliant on Wnt signalling. Wnt signalling is essential for many aspects of embryonic development and this explains the constellation of congenital anomalies in multiple organs. A conditional mouse model of PORCN loss of function was used to study its requirement in Wnt signalling and embryonic development.27 Consistent with the female-specific inheritance pattern of FDH, Porcn hemizygous male murine embryos arrest during early embryogenesis and fail to generate mesoderm, a phenotype previously associated with loss of Wnt activity. Heterozygous Porcn mutant murine females exhibit a spectrum of limb, skin and body patterning abnormalities resembling those observed in human patients with FDH. In a conditional mouse model of PORCN, loss of function defects were observed in ectodermal- and mesenchymal-derived structures.28 It has been proposed that CDH originates from a defect in the mesenchymal cells of the developing pleuroperitoneal folds, and aberrant Wnt signalling provides a plausible mechanism for the association with CDH.
Acknowledgments
This work has been made possible by the Agency for Innovation by Science and Technology (IWT; SBO-60848 to JRV); Research Foundation Flanders (FWO; FWO grant G.0320.07. to JRV); University of Leuven (KU Leuven) SymBioSys (PFV/10/016 and GOA/12/015 to JRV and KD). HVE and KD received fund as a ‘Clinical Researcher' of the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO). The Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO, 1.8.012.07.N.02) and the Instituut voor Wetenschap en Technologie (IWT/070715) fund JDP as a ‘Clinical Researcher'. The programme is further supported by the Industria-Academia Partnership Marie Curie Grant of the European Commission (http://www.endovv.com; PIAP-GA-2009-251356). This research was supported by funding from the Belgian Science Policy Office Interuniversity Attraction Poles (BELSPO-IAP) programme through the project IAP P7/43-BeMGI.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- 1Kallen B, Robert E, Harris J: The descriptive epidemiology of anophthalmia and microphthalmia. Int J Epidemiol 1996; 25: 1009–1016. [DOI] [PubMed] [Google Scholar]
- 2Stoll C, Dott B, Alembik Y, Roth MP: Associated malformations among infants with anophthalmia and microphthalmia. Birth Defects Res A Clin Mol Teratol 2012; 94: 147–152. [DOI] [PubMed] [Google Scholar]
- 3Pasutto F, Sticht H, Hammersen G et al: Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet 2007; 80: 550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Sharma VM, Ruiz de Luzuriaga AM, Waggoner D, Greenwald M, Stein SL: Microphthalmia with linear skin defects: a case report and review. Pediatr Dermatol 2008; 25: 548–552. [DOI] [PubMed] [Google Scholar]
- 5Maas SM, Lombardi MP, van Essen AJ et al: Phenotype and genotype in 17 patients with Goltz-Gorlin syndrome. J Med Genet 2009; 46: 716–720. [DOI] [PubMed] [Google Scholar]
- 6Zayed H, Chao R, Moshrefi A et al: A maternally inherited chromosome 18q22.1 deletion in a male with late-presenting diaphragmatic hernia and microphthalmia-evaluation of DSEL as a candidate gene for the diaphragmatic defect. Am J Med Genet A 2010; 152A: 916–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Brady PD, Dekoninck P, Fryns JP, Devriendt K, Deprest JA, Vermeesch JR: Identification of dosage-sensitive genes in fetuses referred with severe isolated congenital diaphragmatic hernia. Prenat Diagn 2013; 33: 1283–1292. [DOI] [PubMed] [Google Scholar]
- 8Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Li H, Handsaker B, Wysoker A et al: The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Albers CA, Lunter G, Macarthur DG, McVean G, Ouwehand WH, Durbin R: Dindel: accurate indel calls from short-read data. Genome Res 2011; 21: 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Wang K, Li M, Hakonarson H: ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Liu X, Jian X, Boerwinkle E: dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 2011; 32: 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Sifrim A, Van Houdt JK, Tranchevent LC et al: Annotate-it: a Swiss-knife approach to annotation, analysis and interpretation of single nucleotide variation in human disease. Genome Med 2012; 4: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW: Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992; 51: 1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 16Froyen G, Van EH, Bauters M et al: Detection of genomic copy number changes in patients with idiopathic mental retardation by high-resolution X-array-CGH: important role for increased gene dosage of XLMR genes. Hum Mutat 2007; 28: 1034–1042. [DOI] [PubMed] [Google Scholar]
- 17Smigiel R, Jakubiak A, Lombardi MP et al: Co-occurrence of severe Goltz-Gorlin syndrome and pentalogy of Cantrell - Case report and review of the literature. Am J Med Genet A 2011; 155A: 1102–1105. [DOI] [PubMed] [Google Scholar]
- 18Dias C, Basto J, Pinho O et al: A nonsense porcn mutation in severe focal dermal hypoplasia with natal teeth. Fetal Pediatr Pathol 2010; 29: 305–313. [DOI] [PubMed] [Google Scholar]
- 19Wang X, Reid SV, Omar Peraza-Llanes J et al: Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat Genet 2007; 39: 836–838. [DOI] [PubMed] [Google Scholar]
- 20Bornholdt D, Oeffner F, Konig A et al: PORCN mutations in focal dermal hypoplasia: coping with lethality. Hum Mutat 2009; 30: E618–E628. [DOI] [PubMed] [Google Scholar]
- 21Harmsen MB, Azzarello-Burri S, Garcia Gonzalez MM et al: Goltz-Gorlin (focal dermal hypoplasia) and the microphthalmia with linear skin defects (MLS) syndrome: no evidence of genetic overlap. Eur J Hum Genet 2009; 17: 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Grzeschik KH, Bornholdt D, Oeffner F et al: Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nat Genet 2007; 39: 833–835. [DOI] [PubMed] [Google Scholar]
- 23Froyen G, Govaerts K, Van EH et al: Novel PORCN mutations in focal dermal hypoplasia. Clin Genet 2009; 76: 535–543. [DOI] [PubMed] [Google Scholar]
- 24Lombardi MP, Bulk S, Celli J et al: Mutation update for the PORCN gene. Hum Mutat 2011; 32: 723–728. [DOI] [PubMed] [Google Scholar]
- 25Sharp A, Robinson D, Jacobs P: Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet 2000; 107: 343–349. [DOI] [PubMed] [Google Scholar]
- 26Clements SE: Importance of PORCN and Wnt signaling pathways in embryogenesis. Am J Med Genet A 2009; 149A: 2050–2051. [DOI] [PubMed] [Google Scholar]
- 27Barrott JJ, Cash GM, Smith AP, Barrow JR, Murtaugh LC: Deletion of mouse Porcn blocks Wnt ligand secretion and reveals an ectodermal etiology of human focal dermal hypoplasia/Goltz syndrome. Proc Natl Acad Sci USA 2011; 108: 12752–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Liu W, Shaver TM, Balasa A et al: Deletion of Porcn in mice leads to multiple developmental defects and models human focal dermal hypoplasia (Goltz syndrome). PLoS One 2012; 7: e32331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.