Abstract
In recent years, antisense therapy has emerged as an increasingly important therapeutic approach to tackle several genetic disorders, including inborn errors of metabolism. Intronic mutations activating cryptic splice sites are particularly amenable to antisense therapy, as the canonical splice sites remain intact, thus retaining the potential for restoring constitutive splicing. Mutational analysis of Portuguese galactosemic patients revealed the intronic variation c.820+13A>G as the second most prevalent mutation, strongly suggesting its pathogenicity. The aim of this study was to functionally characterize this intronic variation, to elucidate its pathogenic molecular mechanism(s) and, ultimately, to correct it by antisense therapy. Minigene splicing assays in two distinct cell lines and patients' transcript analyses showed that the mutation activates a cryptic donor splice site, inducing an aberrant splicing of the GALT pre-mRNA, which in turn leads to a frameshift with inclusion of a premature stop codon (p.D274Gfs*17). Functional–structural studies of the recombinant wild-type and truncated GALT showed that the latter is devoid of enzymatic activity and prone to aggregation. Finally, two locked nucleic acid oligonucleotides, designed to specifically recognize the mutation, successfully restored the constitutive splicing, thus establishing a proof of concept for the application of antisense therapy as an alternative strategy for the clearly insufficient dietary treatment in classic galactosemia.
Introduction
Over the last years, splicing mutations emerged as an important pathogenic mechanism, underlying 10–30% of genetic diseases (HGMD Professional 2013.1).1 Splicing accuracy depends not only on the recognition of exon–intron junctions, defined by intronic cis-elements: 5′ splice site, 3′ splice site, branch site and poly-pyrimidine tract,2, 3, 4 but also on more discrete elements, entitled splicing regulatory elements, which direct the splicing machinery to use the correct splice sites. Exonic and intronic splicing enhancers stimulate splicing and serve as binding sites mainly for serine/arginine-rich proteins. Exonic and intronic splicing silencers repress splicing, and often function by binding proteins from the heterogenous nuclear ribonucleoprotein family.2, 4, 5, 6 Although most reported splicing mutations directly abolish an authentic splice site or create a novel one, an increasing number of disease-associated variations that alter splicing enhancers or silencers have been reported.2, 7, 8, 9 Each nucleotide modification should be considered a potential candidate for splicing alterations, as not only intronic but also nonsense, missense and silent modifications may impact splicing.7 Accordingly, constitutive and regulated splicing reactions are considered potential therapeutic targets and novel strategies for their correction are evolving. Among these, antisense oligonucleotides display an exquisite specificity, being capable of distinguishing a single nucleotide-mismatch.8 Effectively, spinal muscular atrophy is a candidate for this therapy, with promising results on patient-derived cell lines that have been already described.9, 10, 11 Moreover, many antisense oligonucleotides are already in phase II/III clinical trials, namely AVI4658 (Eteplirsen), a morpholino oligonucleotide for duchenne muscular dystrophy.12
Classic galactosemia (MIM #230400), which affects about 1/47 000 live-births, results from deficient activity of galactose-1-phosphate uridylyltransferase (GALT, EC 2.7.7.12), an ubiquitous key enzyme in galactose metabolism, essential for nursing infants, as lactose represents their primary carbohydrate source.13, 14 The present gold standard of care is a lifelong dietary galactose restriction, which has, however, proven to be insufficient to prevent the long-term sequelae.13, 14, 15, 16, 17 The disorder is caused by mutations in the GALT gene, which profoundly impair GALT enzymatic activity.15, 18, 19 GALT is located in chromosome 9p13, arranged into 11 exons spanning about 4.0 kb of genomic sequence, and encodes a 379 amino-acid polypeptide, which is assembled as a ∼87 kDa homodimer.19, 20, 21, 22
As many other autosomal recessive metabolic disorders, classic galactosemia displays great allelic heterogeneity with >260 variations described (http://arup.utah.edu/database/GALT/GALT_display.php; http://databases.lovd.nl/shared/genes/GALT).23 Although most are missense mutations, other variations have been reported, namely silent, nonsense and noncoding changes.18, 24, 25 Moreover, several intronic variations have been identified in the GALT gene, approximately half of which are known to affect splicing.23 Mutational analysis of 42 Portuguese patients confirmed c.563A>G (p.Q188R) as the prevalent molecular defect (67.1%), and surprisingly revealed an intronic variation, c.820+13A>G (IVS8+13A>G), as the second most frequent mutation, accounting for 8.0% of the mutant alleles.26 Although this transition is currently classified as benign, complete sequence of a galactosemic patient homozygous for c.820+13A>G revealed no other alteration in his genomic DNA.23, 26 Furthermore, this variation has never been identified in controls and was found in Portuguese patients either in homozygous or compound heterozygous state.26
The aim of this study is to confirm that the c.820+13A>G transition is indeed a disease-causing mutation, to elucidate its pathogenic mechanism, by in vivo, ex vivo and in vitro approaches, to functionally characterize the resulting protein and, finally, to use antisense oligonucleotides to modulate the deleterious effect of the mutation.
Materials and methods
Patients
This study was approved by the local Ethics Committee and written informed consents were obtained from patients and family members. The c.820+13A>G variation was identified in six galactosemic patients: one homozygous (http://arup.utah.edu/database/GALT/GALT_display.php) and five compound heterozygotes, four bearing c.563A>G (p.Q188R) and the other c.883C>A (p.P295T).26
In vivo splicing analysis
Total RNA was isolated from patients and control individuals lymphocytes with TRIZOL (Invitrogen Corporation, Carlsbad, CA, USA). RNA was reverse transcribed using the NZY First-Strand cDNA Synthesis Kit (NZYTech, Lisbon, Portugal) and PCR analysis was performed using the CF and CR primers (Table 1). After separation by 2% NuSieve GTG agarose gel electrophoresis, PCR products were purified (Isolate PCR and Gel Kit, Bioline, London, UK) and analyzed by direct sequencing.
Table 1. Sequence of DNA oligonucleotides used in this study.
| Oligonucleotides | Sequence (5′→3′) |
|---|---|
| CF | GTGAGGAGCGATCTCAGCA |
| CR | GGAGCGGAGGGTAGTAATGA |
| BF | CCCTCTCAACCCTCTGTGTC |
| DR | TTCAAGGCCCTTTCTGCTTA |
| 7F | CACCTTGATGACTTCCTATCC |
| 9R | GAAATGGTGTTGGGGCTAAA |
| SD6 | TCTGAGTCACCTGGACAACC |
| SA2 | GCTCACAAATACCACTGAGAT |
| 6His-Fa | CCAGCGGATCCCCCTCAAAAATGCATCATCACCATCACCACATGTCGCGCAGTGGAACCGATC |
| 6His-Ra | GATCGGTTCCACTGCGCGACATGTGGTGATGGTGATGATGCATTTTTGAGGGGGATCCGCTGG |
| IVS8-Mut-Fb | CCCCTGCTGAGCGTGATGGTCAGTCTCCCAGATCTAGCCTCCATCATG |
| IVS8-Mut-Rb | CATGATGGAGGCTAGATCTGGGAGACTGACCATCACGCTCAGCAGGGG |
Underlined residues encode the hexa-histidyl tag.
Underlined residues represent the first 13 nucleotides of intron 8.
Minigenes construction
Genomic fragments encompassing the GALT region (ID 2592, GenBank accession no. NG_009029.1 / NM_000155.3) containing the c.820+13A>G variation were PCR-amplified from a heterozygous patient carrying this mutation, using the primers 7F and 9R (Table 1). The resulting 399 bp fragment includes the end of intron 7 until the initial portion of intron 9 (exons are numbered as in NG_009029.1). Wild-type and mutant fragments were cloned into pCR2.1TOPO (Invitrogen Corporation), and positively selected by blue/white screening. Fragments were subcloned into the exon-trapping vector pSPL3 (kindly provided by Prof Belén Pérez, Universidad Autónoma de Madrid, Spain). Clones bearing the wild-type and mutant inserts were digested with EcoRI and sequenced for confirmation of their correct orientation.
Ex vivo splicing analysis
Empty pSPL3 vector or minigene constructs containing wild-type (pSPL3.wt) and mutant sequences (pSPL3.mut) were transfected into eukaryotic cell lines (HeLa and COS-7) using Lipofectamine 2000 (Invitrogen Corporation). Twenty-four hours after transfection, total RNA was extracted using TRIZOL, reverse transcribed using the NZY First-Strand cDNA Synthesis Kit and a PCR reaction was performed using the pSPL3-specific primers SD6 and SA2 (Table 1). PCR products were separated by 2% Nusieve GTG agarose gel electrophoresis, purified and sequenced.
Correction of alternative splicing with LNA oligonucleotides
Two antisense locked nucleic acid (LNA) oligonucleotides were designed, synthesized and purified at Exiqon (Vedbaek, Denmark). LNA1 is a 15-mer (5′-CCAGGATCCTACCTG-3′), whereas LNA2 is a 16-mer (5′-GATCCTACCTGGGAGA/3Phos/-3′); mutant nucleotide is underlined.
Cell cultures and minigene transfection followed the protocol as previously mentioned. Five hours after minigene transfection, LNA1, LNA2 or saline solution (control) were also transfected. Three concentration ranges were used, nominally low (0.005–0.1 μM), medium (0.15–0.25 μM) and high (0.5–1.0 μM). Conventional RT-PCR for splicing analysis was performed after 24 h, using the pSPL3-specific primers SD6 and SA2 (Table 1). PCR products were analyzed by 2% Nusieve GTG agarose gel electrophoresis and sequenced.
Production of recombinant wild-type and variant GALT
GALT cDNA, a gift of Prof Judith Fridovich-Keil (Emory University, Atlanta, USA) was cloned into pET24b(+) into the HindIII and SalI restriction sites. The T7 tag was deleted using the NdeI and EcoRI enzymes. By site-directed mutagenesis (QuikChange II XL Mutagenesis kit, Stratagene, La Jolla, CA, USA) using the primers 6His-F and 6His-R (Table 1), six CAT codons were inserted to introduce an N-terminal hexa-histidyl tag. All changes were confirmed by sequencing in both orientations.
The c.820+13A>G mutation was generated by site-directed mutagenesis (NZY mutagenesis kit, NZYTech) using the IVS8-Mut-F and IVS8-Mut-R primers (Table 1), and confirmed by direct sequencing in both forward and reverse orientations. Vectors bearing the wild-type or mutant GALT were transformed into Escherichia coli BL21 (DE3) Rosetta, grown at 37 °C in M9 minimal medium containing iron sulfate (100 μM), zinc sulfate (100 μM), kanamycin (25 μg/ml) and chloramphenicol (34 μg/ml). Protein expression was induced with 400 μM isopropyl β-D-1-thiogalactopyranoside once the Abs600 nm reached 0.3; the cultures were moved to 21 °C and the cells were harvested after 4 h.
Bacterial pellets were resuspended in buffer A (50 mM Tris-HCl, pH 7.5, 300 mM KCl, and 10% glycerol) containing 1 mM phenylmethanesulfonyl fluoride and 1 mg/ml lysozyme. After 30-min incubation at 4 °C, cells were disrupted by sonication, centrifuged (10 000 g, 5 min, at 4 °C), and imidazole (20 mM) was added to the soluble fraction. Proteins were purified by immobilized metal affinity chromatography using a HisTrap FF column (GE Healthcare, Uppsala, Sweden), pre-equilibrated with buffer A with 20 mM imidazole. After loading the cleared supernatants, the column was washed with 10 column volumes (Vc) of buffer A containing 20 mM imidazole, and 5 Vc of buffer A with 50 mM imidazole. Pure proteins were obtained by elution with 2.5 ml of buffer A containing 500 mM imidazole, subsequently eliminated with a PD-10 desalting column (GE Healthcare). Protein solutions were concentrated by ultrafiltration using Vivaspin 15R (Sartorius, Goettingen, Germany) 30 kDa-cutoff filters, at 3000 g and 4 °C in a swinging-bucket centrifuge. Immunoblotting and GALT activity determination were performed on the same day as purification; the remaining samples were aliquoted, flash-frozen in liquid nitrogen and stored at −80 °C.
Immunoblotting
Purified proteins were applied to a 12.5% SDS-PAGE and transferred to nitrocellulose membranes (Amersham Hybond ECL, GE Healthcare). After 2-h blocking with 5% milk in PBS-T, the membrane was incubated for 1 h at room temperature with anti-GALT antibody (sc-365577, Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1:1000 dilution). After washes with PBS-T, the membrane was incubated for 1 h at room temperature with peroxidase-conjugated Affinipure goat anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA; 1:1000 dilution). The Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) was used for protein detection.
GALT enzymatic activity
GALT enzymatic activity was measured as previously described.27 All assays were carried out at 37 °C, in a reaction mixture containing 40 mM Tris-HCl, pH 7.5, 2 mM galactose-1-phosphate, 0.5 mM uridine diphosphate (UDP)-glucose, 40 μM dithiothreitol and 125 mM glycine. UDP-glucose and UDP-galactose were separated by HPLC and analyzed by UV detection at 262 nm.26, 27
Far-UV circular dichroism
Far-UV circular dichroism (far-UV CD) spectra and thermal denaturation profiles were recorded in a Jasco J-710 spectropolarimeter, coupled to a Jasco PTC-348WI Peltier temperature controller and a Haake G/D8 water bath. Each protein spectrum (samples at 0.15 mg/ml, wild-type; or 0.25 mg/ml, variant) resulted from six accumulations at 50 nm/min. Thermal denaturation profiles were monitored at 222 nm between 20 and 90 °C (1 °C/min slope; data pitch: 1 °C; delay time: 0 s). Temperature scan curves were fitted according to a two-state model.
Differential scanning fluorimetry
Differential scanning fluorimetry (DSF) was performed in a C1000 Touch thermal cycler equipped with a CFX96 optical reaction module (Bio-Rad, Hercules, CA, USA). All fluorescence measurements were performed at a final protein concentration of 0.1 mg/ml in buffer A, and SYPRO orange (Invitrogen Corporation) at a 5 × working concentration, in a 50 μl total volume. The PCR plate was sealed with Optical-Quality Sealing Tape (Bio-Rad) and centrifuged at 400 g for 1 min. DSF assays were carried out with a 10-min incubation step at 20 °C followed by ramping the temperature from 20 °C to 90 °C at 1 °C/min, with a 1 s hold time every 0.2 °C and fluorescence acquisition using the HEX channel. Data were processed using CFX Manager software V3.0 (Bio-Rad). Temperature scan curves were fit to a biphasic sigmoidal function, Tm values being obtained from the midpoint of the first and second transitions.
Dynamic light scattering
Dynamic light scattering (DLS) analysis was performed on a ZetaSizer Nano-S (Malvern Instrument, Worcestershire, UK) particle size analyzer, coupled to a Peltier temperature control unit; a He-Ne laser was used as the light source (633 nm). Samples were centrifuged at 15 000 g for 30 min at 4 °C, diluted in buffer A to a final concentration of 0.15 mg/ml, and filtered with a 0.22 μm membrane to remove aggregates. Temperature was ramped from 20–70 °C at 0.5 °C/min, with the particle size average, distribution and total-scattering intensity being collected. Data were processed using Zetasizer Nano DTS software v7.01 (Malvern Instrument). The melting temperature (Tagg), at which both size and intensity start to increase significantly, was determined by fitting the obtained data to an equation defined by a plateau followed by one-phase association. The kinetics of thermal aggregation was monitored at 37 and 42 °C for 60 min.
Results
In vivo analysis confirmed that c.820+13A>G is a disease-causing mutation
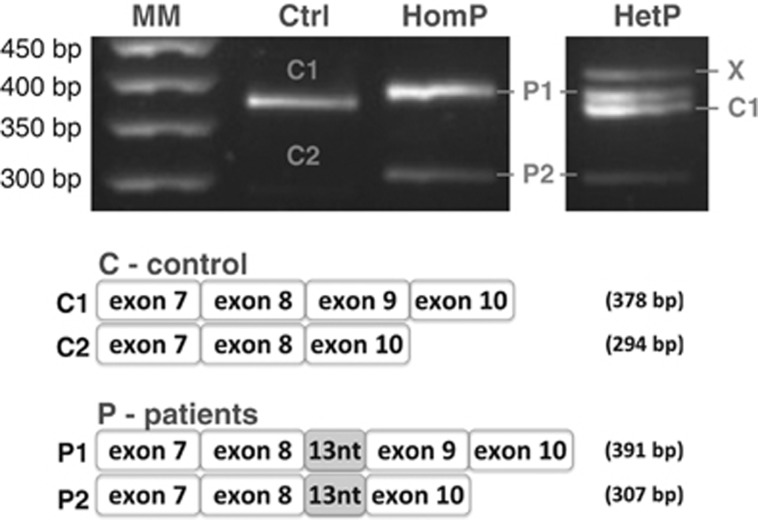
Patients carrying the c.820+13A>G mutation were selected for transcript analysis. RT-PCR analysis of the GALT mRNA revealed the presence of two fragments when analyzing either control individuals or the homozygous patient, and four fragments when analyzing a heterozygous patient.
Control individuals presented the expected fragment (378 bp), resulting from the usage of the canonical splice sites and thus including the end of exon 7, complete exons 8 and 9 and the beginning of exon 10, and a very faint fragment (294 bp), resulting from an alternative splicing event with skipping of exon 9 (Figure 1). This mRNA does not induce a frameshift, rather causing the internal deletion of 28 amino acids, between residues 273 and 302.
Figure 1.
Transcript analysis of control and patients' lymphocytes confirm different splicing patterns. Total RNA was isolated from lymphocytes of control and homozygous and heterozygous patients with the c.820+13A>G mutation. RT-PCR analysis of GALT mRNA revealed the presence of two fragments: control showed the expected fragment (378 bp) and a smaller and very faint fragment (294 bp) that resulted from an alternative splicing event with skipping of exon 9; homozygous patient also showed the two fragments, although both presented the first 13 nucleotides of intron 8 (391 and 307 bp); heterozygous patient showed four fragments corresponding to alleles carrying and not carrying the splicing mutation.
The homozygous patient profile also presented two fragments (391 and 307 bp), which differed from those of control individuals by the additional 13 bp corresponding to the first nucleotides of intron 8, and resulting from transcription of the allele carrying the c.820+13A>G transition. Also, the fragment corresponding to exon 9 skipping is somewhat more intense than that of the control (Figure 1).
The heterozygous patient transcript analysis revealed the presence of the four previously mentioned fragments. Whereas the 391 and 307 bp fragments result from transcription of the allele carrying the c.820+13A>G transition, the 378 bp fragment and a very faint band corresponding to the 294 bp fragment result from transcription of the missense allele yielding c.883C>A (p.P295T). A longer band is also visible, resulting from heteroduplex formation, a common PCR-derived phenomenon (Figure 1).28, 29
In vivo, the c.820+13A>G mutation might induce a frameshift in the open reading frame (ORF) and introduce a PTC 17 amino acids downstream (p.D274Gfs*17) in the exon 9 including transcript, or 55 amino acids downstream (p.D274Gfs*55) in the exon 9 skipping transcript.
Furthermore, and to rule out any PCR-related problem, we performed transcript analyses using additional primer pairs, namely BF+CR and CF+DR (Table 1), obtaining products covering regions upstream and downstream the fragment initially amplified, but always encompassing the variation site. The results always revealed the presence of two transcripts corresponding to the presence and absence of exon 9 (data not shown).
Ex vivo analysis revealed that the intronic mutation c.820+13A>G causes aberrant splicing of the GALT transcript
Our previous in silico analysis26 predicted the activation of a cryptic donor splice site located immediately downstream the variation in intron 8 (c.820+14_820+15). Effectively, in the NetGene2 server, the authentic GT site scores 0.68, whereas the cryptic GT site scores 0.99. Moreover, the authentic acceptor splice site in intron 8 scores very low (0.4) in the same server, revealing its intrinsic weakness.26
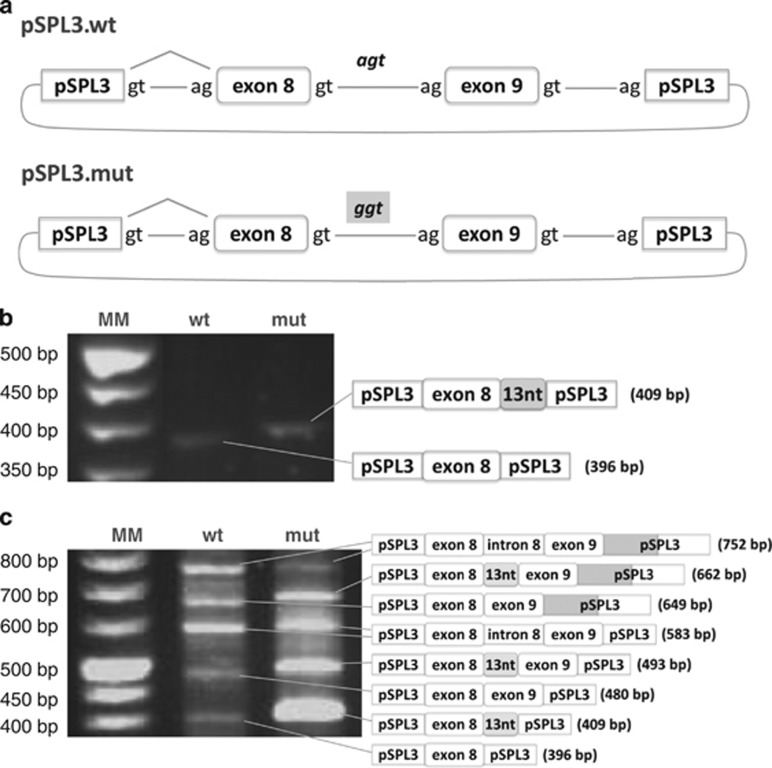
To investigate if this intronic variation was sufficient to cause aberrant splicing, the 399 bp fragment containing the wild-type or the mutant regions of the GALT gene was cloned into the pSPL3 exon-trapping vector. Minigene constructs pSPL3.wt and pSPL3.mut differed exclusively in the 13th nucleotide of intron 8 (Figure 2a). HeLa and COS-7 cells were transiently transfected with each construct or empty vector, and the resulting splicing products were sequenced using the pSPL3-specific primers, thus avoiding the amplification of endogenous GALT transcripts. Transfection with the empty vector showed a single 263 bp fragment according to the pSPL3 canonical splicing sites (Supplementary Figure S1).
Figure 2.
Minigene constructs and transcript analysis of transfected cells. (a) schematic representation depicting the minigene constructs. The PCR fragment comprising the last portion of intron 7, exon 8, intron 8, exon 9 and the first portion of intron 9 was cloned in the pSPL3 vector: pSPL3.wt minigene presents the genomic wild-type fragment; pSPL3.mut minigene presents the genomic mutant fragment (c.820+13A>G mutation); (b) transfection of HeLa cells with the wild-type and mutant constructs; RT-PCR pattern and sequence analysis of the corresponding fragments are depicted in the schemes; (c) transfection of COS-7 cells with the wild-type and mutant constructs; RT-PCR pattern and sequence analysis of the corresponding fragments are depicted in the schemes.
Transfection of HeLa cells with pSPL3.wt showed a single 396 bp fragment, resulting from the inclusion of exon 8 and exclusion of exon 9, whereas the pSPL3.mut originated a single 409 bp fragment, also resulting from exon 8 inclusion and exon 9 exclusion, including however the first 13 nucleotides of intron 8 (Figure 2b).
Minigenes transfection in COS-7 yielded several fragments, owing to usage of all splicing sites, including a cryptic donor site in the vector sequence (Figure 2c). Nevertheless, the transcript profile of the mutant minigene was clearly different from that of the wild-type, as all transcripts, except the one including the entire intron 8, presented the additional first 13 nucleotides of intron 8 and the canonical donor site was never used.
Minigene analysis in two different cell lines showed that the c.820+13A>G mutation per se activates a cryptic donor splice site (c.820+14_820+15), leading to inclusion of the first 13 nucleotides of intron 8 in the mature mRNA, thus inducing an aberrant splicing of the GALT transcript.
Antisense therapy allowed the ex vivo reversion of the alternative splicing caused by the c.820+13A>G mutation
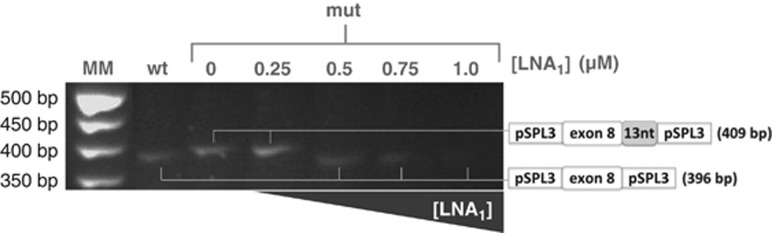
To investigate the possibility of restoring correct splicing by antisense therapy, two LNA oligonucleotides were designed. Both oligonucleotides, with slight differences in sequence coverage (Supplementary Figure S2), were targeted to the cryptic splice site, sterically blocking the access of the spliceosome machinery to this site, thus driving the usage of the canonical donor splice site. HeLa and COS-7 cells were transfected with each minigene constructs and with either LNA at three different concentration ranges. Results revealed that the wild-type minigene was not affected by any of the antisense oligonucleotides, at any concentration (Supplementary Figure S3). Transcripts derived from the mutant minigene revealed that neither LNA at low doses was able to prevent usage of the cryptic splice site. When the concentration of each LNA was raised to the higher range, the aberrant splicing was completely abolished and the natural splicing mechanism was recovered, with no apparent differences between the highest LNA concentrations (Figure 3).
Figure 3.
Antisense nucleotides correct the splicing pattern of mutant minigene. Transcript analysis of HeLa cells expressing the wild-type and mutant minigene constructs and further treated with antisense oligonucleotides. RT-PCR pattern and sequence analysis of the corresponding fragment in untreated (0) and treated (0.25, 0.5, 0.75 and 1 μM) HeLa cells confirmed that both LNA oligonucleotides (herein depicted as LNA1) fully corrected the aberrant splicing. LNA2 showed identical transcript profile, also subsequently confirmed by sequencing.
The truncated variant GALT is stable but inactive and prone to aggregation
Despite the c.820+13A>G mutation inducing a frameshift in the ORF and introducing a PTC (p.D274Gfs*17), reverse transcription analysis detected the aberrant mature transcript in all patients, indicating that the variant protein may be translated in vivo.
The recombinant wild-type and truncated p.D274Gfs*17 GALT were produced in a prokaryotic expression system, purified and analyzed from a functional–structural viewpoint. Immunoblotting analysis of wild-type and variant proteins showed two main bands with molecular masses of ∼45 kDa for the wild-type and ∼34 kDa for the truncated GALT (Supplementary Figure S4A). Besides the main band, both proteins display bands with lower molecular mass and much lower intensity. Recombinant wild-type GALT displayed a specific enzymatic activity of 10.2 μmol.mg−1.h−1, whereas p.D274Gfs*17 had undetectable activity.
The structural impact of the variation was analyzed by different biophysical techniques. By far-UV CD spectroscopy, we observed nearly identical spectra for the wild-type and truncated GALT (Supplementary Figure S5A). Monitoring CD ellipticity at 222 nm as a function of increasing temperature, secondary structure thermal denaturation profiles yielded Tm values of 53.0±1.3 °C and 54.6±2.1 °C, respectively for wild-type and p.D274Gfs*17 GALT. DSF assays were used to obtain the proteins' tertiary structure thermal denaturation profiles. Two melting transitions were observed (Supplementary Figure S5B), with similar Tm values for both proteins: Tm1=43.7±0.6 °C and Tm2=52.5±1.1 °C for wild-type, and Tm1=43.4±0.1 °C and Tm2=51.8±0.1 °C for p.D274Gfs*17 GALT. The propensity to aggregate in solution was analyzed by DLS. Both proteins displayed quite similar Tagg (Supplementary Figure S5C; 41.3±0.1 °C for wild-type and 41.1±0.1 °C for p.D274Gfs*17) and overlapping aggregation kinetics profiles at 37 °C (t1/2=26.6±0.1 min for wild-type and 25.6±2.7 min for p.D274Gfs*17; data not shown), contrasting with a faster aggregation of p.D274Gfs*17 GALT relatively to wild-type at 42 °C (Supplementary Figure S5D; t1/2=7.0±0.2 min for wild-type and 5.0±0.1 min for p.D274Gfs*17).
Discussion
We report the functional characterization of an intronic variation in the GALT gene, and the first description of antisense oligonucleotides as therapeutic agents in classic galactosemia. In silico analysis of the target sequence revealed the presence of a cryptic donor site (c.820+14_820+15) that is activated in the presence of the c.820+13A>G, leading to the exonization of the first 13 nucleotides of intron 8 (r.(936_937ins936+1_936+13; 936+13G>A)).
Patients' transcript analysis confirmed the presence of a fragment containing the first 13 nucleotides of intron 8, in contrast to controls, who presented a fragment resulting from constitutive splicing. Surprisingly, a second fragment was identified, in patients and controls, corresponding to exon 9 skipping, an unprecedented observation that is much more prominent in patients. The occurrence of exon 9 skipping in homozygous wild-type individuals rules out the possibility that this alternative splicing event could result from the c.820+13A>G mutation, but the results strongly suggest that this transition favors it. Previous studies have described transcripts resulting from alternative splicing as a ‘background' noise of the splicing process30 and, accordingly, this GALT alternative splicing should correspond to a physiological event, at least in lymphocytes, that we herein report for the first time.
Minigene analyses in two cell lines, HeLa and COS-7, fully confirmed the patients' genetic analysis. HeLa cells displayed a very simple profile with one single fragment, corresponding to the inclusion of exon 8 and skipping of exon 9, whereas COS-7 cells displayed several fragments corresponding to the usage of all available splicing sites. Nevertheless, a key observation common to both cell lines is that the presence of the c.820+13A>G mutation always induced the usage of the cryptic donor site instead of the canonical one.
The in silico predicted weakness of the acceptor splice site in intron 826 was fully confirmed by the minigene assay. Wild-type minigene analysis in both cell lines also displayed a fragment corresponding to exon 9 skipping, confirming that this splicing event is not related with the variation herein studied.
The splicing process, particularly the recognition of splicing sites, is highly dependent on the availability of tissue-specific factors, which explains the different transcript profiles of HeLa and COS-7 cells.31, 32
Altogether, in vivo and ex vivo analyses demonstrated that the c.820+13A>G variation is actually a disease-causing mutation. Our results, neatly complemented by the in vivo analysis of patients' cells, reiterate the reliability of the ex vivo minigene functional assay to unravel the molecular mechanism(s) whereby a variation affects splicing.33
Once proven the pathogenic effect of the c.820+13A>G mutation, we investigated the possibility of restoring correct splicing by antisense therapy. LNA oligonucleotides were chosen for their high-mismatch discrimination ability34 and their high stability and low toxicity in biologic systems.35 This variation activates a cryptic donor splice site extremely close to the natural one, thus its steric blocking by antisense oligonucleotides could affect the correct splicing. For that reason, two oligonucleotides were designed, LNA1 and LNA2, respectively 10 and 5 nucleotides distant from the canonical GT, and were assayed separately. LNA concentrations in the reported ranges (<0.25 μM) failed to inhibit the alternative splicing process. However, at higher concentrations (0.5–1 μM), each LNA successfully restored the normal splicing profile. Above the lowest LNA concentration at which the normal splicing profile was obtained (0.5 μM), no improvement in efficiency was observed, which is in accordance with previous pharmacological studies showing ‘threshold affinity' for LNA oligonucleotides.36
In patients, the transcript displaying the additional 13 nucleotides of intron 8 induces a frameshift in the ORF, introducing a PTC 17 residues downstream (p.D274Gfs*17). PTC-containing transcripts usually trigger the nonsense-mediated decay (NMD) system, thereby preventing the production of aberrant proteins.2, 37, 38 This system is elicited upon recognition of a PTC located >50–55 nucleotides upstream the next exon–exon junction.38 The PTC induced by the c.820+13A>G mutation is only 44 nucleotides upstream of the exon 9–exon 10 junction, thus preventing the NMD from degrading this transcript which putatively encodes a 290 amino-acids polypeptide, (Supplementary Figure S4B). Moreover, the C-terminal 17 residues of p.D274Gfs*17 differ from the wild-type sequence. In line with the literature, western blot detection of GALT from human samples, even from control individuals, was unsuccessful, which precluded us from demonstrating whether the truncated enzyme is produced in vivo.
Structural models of wild-type and p.D274Gfs*17 GALT (Supplementary Figure S4C) were obtained based on the structure of bacterial GALT (PDB code: 1GUP).39 The missing residues in the truncated protein include (i) a helix that protrudes into the other monomer to assemble the functional homodimer, and (ii) three of the four non-heme mononuclear iron ligands in bacterial GALT.21, 40, 41 These observations led us to anticipate considerable structural–functional perturbations in p.D274Gfs*17 and prompted us to characterize recombinant wild-type and variant GALT by different biophysical methods, which showed that p.D274Gfs*17 GALT does not display significant alterations at the secondary and tertiary structure levels, although it has a higher propensity than wild-type GALT for aggregation upon thermal insult (Supplementary Figure S5D). This observation, together with the null activity exhibited by p.D274Gfs*17, suggests that, in a cellular context, the truncated GALT will most likely aggregate and will be prematurely marked for degradation. Therefore, despite the possibility that the stable mutant mRNA is translated into p.D274Gfs*17 GALT in the patients' cells, it should be completely devoid of any functional ability. These observations reinforce the importance of antisense therapy in correcting splicing mutations and, particularly, in restoring GALT function.
In conclusion, this study constitutes the first functional characterization of an intronic variation in classic galactosemia by two distinct yet complementary approaches: patients' genetic analysis and minigene assays. Furthermore, two LNA oligonucleotides, specifically designed to recognize the c.820+13A>G, have successfully restored the splicing profile, thus establishing a proof of concept for the application of antisense therapy for mis-splicing mutations in classic galactosemia.
Acknowledgments
We wish to acknowledge the patients enrolled in this study, and Prof Bélen Pérez and Dr Sandra Brasil (Centro de Biologia Molecular Severo Ochoa, Universidad Autónoma de Madrid, Spain) for their assistance with the minigene assays. This work was supported by Fundação para a Ciência e Tecnologia SFRH/BD/48259/2008 PhD Grant to AIC, FEBS Short-term Fellowship to JBV, SPDM Grant to IR, and PEst-OE/SAU/UI4013/2011.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- 1Krawczak M, Ball EV, Fenton I et al: Human gene mutation database—a biomedical information and research resource. Hum Mutat 2000; 15: 45–51. [DOI] [PubMed] [Google Scholar]
- 2Cartegni L, Chew SL, Krainer AR: Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 2002; 3: 285–298. [DOI] [PubMed] [Google Scholar]
- 3Sironi M, Menozzi G, Riva L et al: Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res 2004; 32: 1783–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Black DL: Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 2003; 72: 291–336. [DOI] [PubMed] [Google Scholar]
- 5Buratti E, Baralle FE: Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 2004; 24: 10505–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Roscigno RF, Weiner M, Garcia-Blanco MA: A mutational analysis of the polypyrimidine tract of introns. J Biol Chem 1993; 268: 11222–11229. [PubMed] [Google Scholar]
- 7Houdayer C, Caux-Moncoutier V, Krieger S et al: Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat 2012; 33: 1228–1238. [DOI] [PubMed] [Google Scholar]
- 8Bennett CF, Swayze EE: RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol 2010; 50: 259–293. [DOI] [PubMed] [Google Scholar]
- 9Marquis J, Meyer K, Angehrn L, Kampfer SS, Rothen-Rutishauser B, Schumperli D: Spinal muscular atrophy: SMN2 pre-mRNA splicing corrected by a U7 snRNA derivative carrying a splicing enhancer sequence. Mol Ther 2007; 15: 1479–1486. [DOI] [PubMed] [Google Scholar]
- 10Baughan TD, Dickson A, Osman EY, Lorson L: Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum Mol Genet 2009; 18: 1600–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN: A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol 2009; 6: 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Malik R, Roy I: Making sense of therapeutics using antisense technology. Expert Opin Drug Discov 2011; 6: 507–526. [DOI] [PubMed] [Google Scholar]
- 13Leslie ND: Insights into the pathogenesis of galactosemia. Annu Rev Nutr 2003; 23: 59–80. [DOI] [PubMed] [Google Scholar]
- 14Waisbren SE, Potter NL, Gordon CM et al: The adult galactosemic phenotype. J Inherit Metab Dis 2012; 35: 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Fridovich-Keil JL, Walter JH: Galactosemia; in Valle D, Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A (eds): The Online Metabolic and Molecular Bases of Inherited Disease. New York: Mc-Graw Hill, 2008, pp 1–92. [Google Scholar]
- 16Fridovich-Keil JL: Galactosemia: the good, the bad, and the unknown. J Cell Physiol 2006; 209: 701–705. [DOI] [PubMed] [Google Scholar]
- 17Bosch AM: Classical galactosaemia revisited. J Inherit Metab Dis 2006; 29: 516–525. [DOI] [PubMed] [Google Scholar]
- 18Tyfield L, Reichardt JK, Fridovich-Keil JL et al: Classical galactosemia and mutations at the galactose-1-phosphate uridyl transferase (GALT) gene. Hum Mutat 1999; 13: 417–430. [DOI] [PubMed] [Google Scholar]
- 19Leslie ND, Immerrman EB, Flach JE, Florez M, Fridovich-Keil JL, Elsas LJ II: The human galactose-1-phosphate uridyltransferase gene. Genomics 1992; 14: 474–480. [DOI] [PubMed] [Google Scholar]
- 20Elsas LJ, Lai K, Saunders CJ, Langley SD: Functional analysis of the human galactose-1-phosphate uridyltransferase promoter in Duarte and LA variant galactosemia. Mol Genet Metab 2001; 72: 297–305. [DOI] [PubMed] [Google Scholar]
- 21Williams VP: Purification and some properties of galactose 1-phosphate uridylyltransferase from human red cells. Arch Biochem Biophys 1978; 191: 182–191. [DOI] [PubMed] [Google Scholar]
- 22Shih LY, Suslak L, Rosin I, Searle BM, Desposito F: Gene dosage studies supporting localization of the structural gene for galactose-1-phosphate uridyl transferase (GALT) to band p13 of chromosome 9. Am J Med Genet 1984; 19: 539–543. [DOI] [PubMed] [Google Scholar]
- 23Calderon FR, Phansalkar AR, Crockett DK, Miller M, Mao R: Mutation database for the galactose-1-phosphate uridyltransferase (GALT) gene. Hum Mutat 2007; 28: 939–943. [DOI] [PubMed] [Google Scholar]
- 24Reichardt JK: Genetic basis of galactosemia. Hum Mutat 1992; 1: 190–196. [DOI] [PubMed] [Google Scholar]
- 25Reichardt JK: Molecular basis of galactosemia: mutations and polymorphisms in the gene encoding human galactose-1-phosphate uridylyltransferase. Proc Natl Acad Sci USA 1991; 88: 2633–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Coelho AI, Ramos R, Gaspar A et al: A frequent splicing mutation and novel missense mutations color the updated mutational spectrum of classic galactosemia in Portugal. J Inherit Metab Dis 2013; 37: 43–52. [DOI] [PubMed] [Google Scholar]
- 27Lindhout M, Rubio-Gozalbo ME, Bakker JA, Bierau J: Direct non-radioactive assay of galactose-1-phosphate:uridyltransferase activity using high performance liquid chromatography. Clin Chim Acta 2010; 411: 980–983. [DOI] [PubMed] [Google Scholar]
- 28Freeman WM, Walker SJ, Vrana KE: Quantitative RT-PCR: pitfalls and potential. Biotechniques 1999; 26: 112–125. [DOI] [PubMed] [Google Scholar]
- 29Eckhart L, Ban J, Ballaun C, Weninger W, Tschachler E: Reverse transcription-polymerase chain reaction products of alternatively spliced mRNAs form DNA heteroduplexes and heteroduplex complexes. J Biol Chem 1999; 274: 2613–2615. [DOI] [PubMed] [Google Scholar]
- 30Rincon A, Aguado C, Desviat LR, Sanchez-Alcudia R, Ugarte M, Perez B: Propionic and methylmalonic acidemia: antisense therapeutics for intronic variations causing aberrantly spliced messenger RNA. Am J Hum Genet 2007; 81: 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Grosso AR, Gomes AQ, Barbosa-Morais NL et al: Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res 2008; 36: 4823–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Zhang C, Zhang Z, Castle J et al: Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev 2008; 22: 2550–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Baralle D, Baralle M: Splicing in action: assessing disease causing sequence changes. J Med Genet 2005; 42: 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34You Y, Moreira BG, Behlke MA, Owczarzy R: Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res 2006; 34: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Jepsen JS, Sorensen M, Wengel J: Locked nucleic acid: a potent nucleic acid analog in therapeutics and biotechnology. Oligonucleotides 2004; 14: 130–146. [DOI] [PubMed] [Google Scholar]
- 36Straarup EM, Fisker N, Hedtjarn H et al: Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res 2010; 38: 7100–7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Wilkinson MF: The cycle of nonsense. Mol Cell 2003; 12: 1059–1066. [DOI] [PubMed] [Google Scholar]
- 38Maquat LE: Nonsense-mediated mRNA decay in mammals. J Cell Sci 2005; 118: 1773–1776. [DOI] [PubMed] [Google Scholar]
- 39Thoden JB, Ruzicka FJ, Frey PA, Rayment I, Holden HM: Structural analysis of the H166G site-directed mutant of galactose-1-phosphate uridylyltransferase complexed with either UDP-glucose or UDP-galactose: detailed description of the nucleotide sugar binding site. Biochemistry 1997; 36: 1212–1222. [DOI] [PubMed] [Google Scholar]
- 40Geeganage S, Frey PA: Significance of metal ions in galactose-1-phosphate uridylyltransferase: an essential structural zinc and a nonessential structural iron. Biochemistry 1999; 38: 13398–13406. [DOI] [PubMed] [Google Scholar]
- 41DeLano WL PyMOL: an open-source molecular graphics tool 2002.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.