Abstract
Although 24 Alzheimer’s disease (AD) risk loci have been reliably identified, a large portion of the predicted heritability for AD remains unexplained. It is expected that additional loci of small effect will be identified with an increased sample size. However, the cost of a significant increase in Case-Control sample size is prohibitive. The current study tests whether exploring the genetic basis of endophenotypes, in this case based on putative blood biomarkers for AD, can accelerate the identification of susceptibility loci using modest sample sizes. Each endophenotype was used as the outcome variable in an independent GWAS. Endophenotypes were based on circulating concentrations of proteins that contributed significantly to a published blood-based predictive algorithm for AD. Endophenotypes included Monocyte Chemoattractant Protein 1 (MCP1), Vascular Cell Adhesion Molecule 1 (VCAM1), Pancreatic Polypeptide (PP), Beta2 Microglobulin (B2M), Factor VII (F7), Adiponectin (ADN) and Tenascin C (TN-C). Across the seven endophenotypes, 47 SNPs were associated with outcome with a p-value ≤1x10-7. Each signal was further characterized with respect to known genetic loci associated with AD. Signals for several endophenotypes were observed in the vicinity of CR1, MS4A6A/MS4A4E, PICALM, CLU, and PTK2B. The strongest signal was observed in association with Factor VII levels and was located within the F7 gene. Additional signals were observed in MAP3K13, ZNF320, ATP9B and TREM1. Conditional regression analyses suggested that the SNPs contributed to variation in protein concentration independent of AD status. The identification of two putatively novel AD loci (in the Factor VII and ATP9B genes), which have not been located in previous studies despite massive sample sizes, highlights the benefits of an endophenotypic approach for resolving the genetic basis for complex diseases. The coincidence of several of the endophenotypic signals with known AD loci may point to novel genetic interactions and should be further investigated.
Introduction
All of the common loci that have been linked to late onset Alzheimer’s disease (AD) other than APOE have small effect sizes and a large portion of the predicted heritability for AD remains unidentified[1]. A number of explanations and potential sources have been postulated for this missing heritability, which is observed for many complex human diseases. Examples include rare variants with large effect sizes, epistatic interactions between multiple common alleles, inflated heritability statistics and genetic heterogeneity, among others[2].
Another approach to the identification of genes involved in Alzheimer’s disease pathogenesis is to ascertain quantitative endophenotypes that are associated with AD risk and then look for genetic variants that are associated with those endophenotypes. Endophenotypes are intermediate traits that are closer to the underlying molecular mechanism than the complex phenotype, and are in principle more likely to be affected by the genetic variation. Discovering genetic and environmental factors contributing to complex human diseases, as well as the development of effective therapies often requires understanding endophenotypes of the disease. For example, discovery of genetic factors contributing to coronary artery disease and the eventual development of effective therapies based on HMG-CoA reductase inhibition was made possible by understanding the endophenotype of hypercholesterolemia[3]. Potential endophenotypes of Alzheimer’s disease include quantitative neuroimaging, such as measures of hippocampal atrophy[4–6], or levels of amyloid or tau proteins in the brain or cerebrospinal fluid (CSF)[7–10]. An additional and still evolving source of AD biomarkers is the pool of circulating proteins in the blood[11–14].
Our objective in this project was to identify the genetic variants that impact concentrations of proteins associated with diagnostic status for Alzheimer’s disease. It was expected that genotypes of some variants would be correlated with protein levels and AD status while others would be correlated with protein levels alone. We used conditional regression analysis to assess the relationship between AD risk, biomarkers, SNPs and non-genetic risk factors.
Materials and Methods
Study Cohorts—TARCC and ADNI
TARCC methodologies have been described in detail elsewhere[15]. Criteria for categorizing subjects as probable AD, mild cognitive impairment (MCI) or normal control (NC) are based on neurocognitive evaluations, family and/or caregiver interviews and medical history. NC must have normal psychometric test scores and a clinical dementia rating (CDR) score of 0. MCI subjects are classified based on the Mayo Clinic Alzheimer's Disease Research Criteria[16]. Patients are characterized as probable AD according to the National Institute of Neurological and Communicative Disorders and Stroke (NINCDS) and the Alzheimer’s Disease and Related Disorders Association (ADRDA) criteria[17]. Each participating site that enrolled participants operates with Institutional Review Board (IRB) approval and each of the following IRBs approved this study (University of North Texas Health Science Center IRB, University of Texas Southwestern Medical Center IRB, Baylor College of Medicine IRB, University of Texas Health Science Center at San Antonio IRB, Texas Tech University Health Sciences Center IRB). Written informed consent was obtained for every participant at the site of enrollment. Data that were used in this study as a validation set were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (adni.loni.usc.edu). Details of ADNI clinical evaluation and sample characterization are described elsewhere[18, 19]. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California–San Francisco. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgment_List.pdf. To date these three protocols have recruited over 1500 adults, ages 55 to 90. The follow up duration of each group is specified in the protocols for ADNI-1, ADNI-2 and ADNI-GO. For up-to-date information, see www.adni-info.org. Demographic data for the TARCC and ADNI cohorts are provided in Table 1.
Table 1. Demographic characteristics of the Texas Alzheimer’s Research and Care Consortium (TARCC) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohorts.
| NC | MCI | AD | ||||
|---|---|---|---|---|---|---|
| Variable | TARCC N = 134 | ADNI N = 41 | TARCC N = 0 | ADNI N = 298 | TARCC N = 166 | ADNI N = 84 |
| Age, Mean (SD) | 70 (8.9) | 76 (5.8) | - | 75 (7.6) | 76 (8.5) | 76 (8.0) |
| Years of Education, Mean (SD) | 15 (2.6) | 16 (2.7) | - | 16 (2.9) | 14 (3.3) | 15 (3.1) |
| Race, N (%) | - | |||||
| White | 131 (97.8) | 41 (100) | - | 297 (99.7) | 165 (99.4) | 84 (100) |
| Other* | 3 (2.2) | 0 (0.0) | - | 1 (0.3) | 1 (0.6) | 0 (0.0) |
| Hispanic Ethnicity, N (%) | - | |||||
| Hispanic | 4 (3.0) | 0 (0.0) | - | 10 (3.4) | 4 (2.4) | 0 (0) |
| Non-Hispanic | 130 (97.0) | 41 (100) | - | 284 (95.3) | 162 (97.6) | 83 (98.8) |
| Unknown | 0 (0.0) | 0 (0.0) | - | 4 (1.3) | 0 (0.0) | 1 (1.2) |
| Gender, N (%) | - | |||||
| Female | 99 (73.9) | 21 (51.2) | - | 114 (38.3) | 107 (64.5) | 31 (36.9) |
| Male | 35 (26.1) | 20 (48.8) | - | 184 (61.7 | 59 (35.5) | 53 (63.1) |
| APOE4 Status, N (%) | - | |||||
| εX/ εX | 100 (74.6) | 39 (95.1) | - | 146 (49.0) | 66 (39.8) | 30 (35.7) |
| εX/ε4 and ε4/ε4 | 34 (25.4) | 2 (4.9) | - | 152 (51.0) | 100 (60.2) | 54 (64.3) |
* Other: ADNI = Asian; TARCC = Mixed race
Measurement of Serum/Plasma Proteins
During clinical visits, a blood draw was collected from each subject; both plasma and serum were collected from TARCC subjects, who were non-fasting, whereas only plasma was collected from ADNI subjects, who were fasting. Plasma and serum were isolated from whole blood samples as described previously for each cohort[15], [18, 19]. Frozen specimens (serum for TARCC subjects and plasma for ADNI subjects), either from baseline or from the year-one follow-up exam, were shipped on dry ice to Myriad-Rules Based Medicine (www.rulesbasedmedicine.com, Austin, TX) where protein concentrations were assessed using a multiplex immunoassay panel for human analytes (human Multi-Analyte Profile, humanMAP). Samples were maintained in the frozen state until the time of the assay. Specifics regarding the sensitivity, specificity, range, inter-run variation coefficient, and spike recovery of the assays are available from Myriad-Rules Based Medicine.
The initial list of 11 proteins from the screening algorithm included: NRP, Beta 2 Microglobulin, C-Reactive Protein, Factor VII, Fatty Acid Binding Protein, I.309, Interleukin-18, Monocyte Chemotactic Protein 1, Pancreatic Polypeptide, Tenascin C, and Vascular Cell Adhesion Molecule 1. Proteins were removed from consideration if they failed to contribute to the O’Bryant et al. screening algorithm[12] in the same direction in both the TARCC vs. ADNI cohorts. Based on these criteria, C-Reactive Protein, Fatty Acid Binding Protein, I.309 and Interleukin-18 were excluded. The final list of seven proteins included Adiponectin, Beta 2 Microglobulin, Factor VII, Monocyte Chemotactic Protein 1, Pancreatic Polypeptide, Tenascin C, and Vascular Cell Adhesion Molecule 1.
Genotyping
The TARCC cohort was genotyped using the Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA), which includes 906,600 SNP markers. The ADNI cohort was genotyped using the Illumina 610-Quad BeadChip (Illumina, San Diego, CA), which includes 550,000 SNP markers. Both panels obtain genome-wide coverage. The BirdSeed v2 algorithm[20] was manually optimized and used for genotype calling.
Quality Control Measures
Locally developed Java programs (collectively termed MACHTools) were used to perform critical data quality checking/filtering, imputation analysis, and data restructuring to affect overall computational performance. Participants were excluded from analysis if blood protein concentration data were not available, if the recorded sex did not agree with chromosome markers or if >5% of the markers did not successfully run. Markers were excluded from analysis if 5% of samples were missing, if they were monomorphic (threshold set at 0.01) or if they were out of Hardy-Weinberg Equilibrium (threshold set at 0.000001). In addition, genotype calls for important markers were manually checked independent of phenotype and recalled as necessary in order to account for obvious atypical hybridization intensities (such as discussed in (Didion et al.)[21][21] (21).This checking was conducted on the entire sample, without knowledge of diagnostic status or phenotype. Results were limited to loci with a minor allele frequency greater than 5%.
Data Analysis
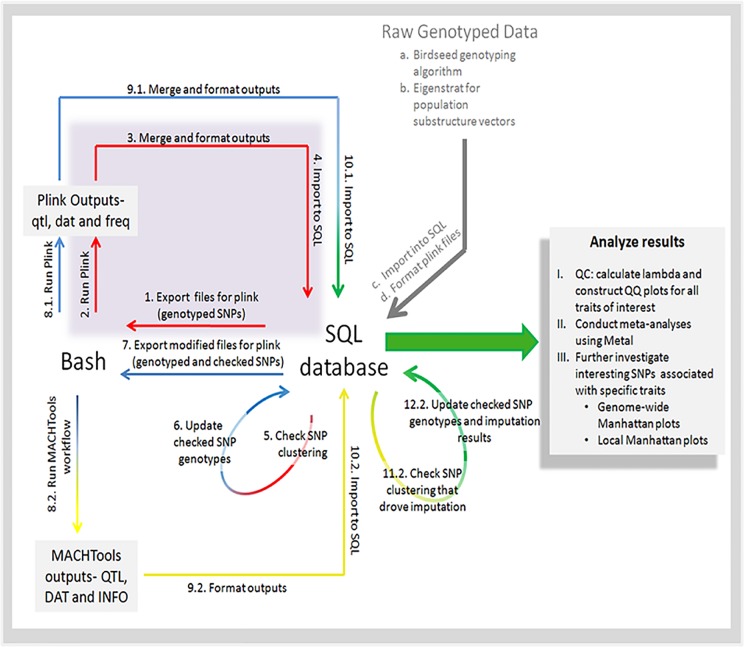
An analysis pipeline was developed to fully analyze these GWA data in association with the quantitative RBM traits (Fig 1). Principle component analysis was performed using the Eigenstrat tool[22] for population substructure covariate determination. Relevant eigenvectors were used as covariates in the analyses, along with sex and education. The following plasma/serum protein concentrations were used as quantitative phenotypes: Adiponectin, Beta 2 Microglobulin, Factor VII, Monocyte Chemotactic Protein 1, Pancreatic Polypeptide, Tenascin C, and Vascular Cell Adhesion Molecule 1 (Table 2). Additionally, age-of-onset and case/control status were analyzed. Preliminary linear mixed model regressions were generated for all quantitative phenotypes using PLINK[23]. Phasing, imputation (using data from the HapMap II, HapMap III and 1000 Genomes databases), and subsequent regressions with the newly imputed GTs were performed using custom applications of the program MaCH[24]. Genotype calls at all significant SNPs were manually checked for proper clustering as described above. After re-clustering, the association regressions and imputation analyses were repeated. This iterative loop was executed three times. Lambda calculations and QQ plots were used to confirm the absence of underlying biases and/or confounders. Manhattan plots were generated for each GWA study in both TARCC and ADNI. Both Manhattan and QQ plots were generated using ggplot2 in R[25]. Final association results for typed and imputed SNPs in both TARCC and ADNI data sets were analyzed in conjunction using Metal[26]. For each quantitative trait, signals with p-values ≤1x10-7 in the meta-analysis were further investigated by plotting the local 1Mbp window (+/-500Kbp) for all three association studies (TARCC, ADNI, and Metal) using LocusZoom[27]. Only signals that were significant in the meta analysis at p ≤1x10-7 and showed evidence of a significant peak in both the TARCC and ADNI cohorts were reported.
Fig 1. Data analysis workflow schematic.
Table 2. Serum protein measures.
| NC | MCI | AD | ||||
|---|---|---|---|---|---|---|
| Protein (Normalized Median (IQR)) | TARCC N = 134 | ADNI N = 41 | TARCC N = 0 | ADNI N = 298 | TARCC N = 166 | ADNI N = 84 |
| Adiponectin | -0.2258 (-0.7516–0.5680) | -0.1664 (-1.3855–0.8740) | - | 0.0759 (-0.6439–0.6987) | 0.0851 (-0.5362–0.8213) | 0.1991 (-0.5519–0.7738) |
| Beta 2 Microglobulin | -0.2030 (-0.7440–0.4490) | 0.0130 (-0.6281–0.4855) | - | 0.0130 (-0.8976–0.4855) | 0.2170 (-0.5470–0.9100) | 0.3422 (-0.3900–1.1211) |
| C-Reactive Protein | 0.2795 (-0.5280–1.0170) | 0.4301 (-0.2202–0.9687) | - | -0.0902 (-0.6694–0.5331) | -0.1870 (-0.8847–0.5227) | -0.0902 (-0.9172–0.8112) |
| Factor VII | 0.0714 (-0.3993–0.6708) | 0.3321 (-0.4641–1.0889) | - | 0.0087 (-0.6516–0.4396) | -0.1235 (-0.6847–0.4562) | 0.0914 (-0.6131–0.8496) |
| Fatty Acid Binding Protein | -0.1886 (-0.8014–0.4985) | 0.1871 (-0.4197–0.9001) | - | -0.1360 (-0.6678–0.5627) | 0.1847 (-0.6659–0.7723) | 0.1581 (-0.4542–0.7569) |
| I.309 | -0.0222 (-0.4888–0.5877) | 0.0819 (-0.5711–0.9290) | - | -0.0442 (-0.6595–0.7106) | 0.0470 (-0.5232–0.5967) | 0.0002 (-0.7757–0.4838) |
| Interleukin-18 | 0.0303 (-0.5095–0.6723) | -0.2648 (-0.9204–0.8005) | - | -0.0881 (-0.6291–0.6027) | -0.2408 (-0.7006–0.4241) | 0.0337 (-0.4914–0.5668) |
| Monocyte Chemotactic Protein 1 | 0.1898 (-0.3829–0.7083) | 0.1413 (-0.3640–0.5187) | - | -0.0488 (-0.5593–0.4246) | -0.0610 (-0.6482–0.4144) | 0.0730 (-0.3076–0.5375) |
| Pancreatic Polypeptide | -0.3350 (-0.9495–0.3015) | -0.3479 (-0.8845–0.2195) | - | -0.0968 (-0.6765–0.6208) | 0.1050 (-0.4080–1.1152) | 0.1147 (-0.5974–0.8456) |
| Tenascin C | -0.3015 (-0.8557–0.2412) | -0.5660 (-1.1650–0.2955) | - | -0.0150 (-0.7108–0.6640) | 0.1450 (-0.4580–0.8990) | 0.3060 (-0.5620–0.7600) |
| Vascular Cell Adhesion Molecule 1 | -0.2350 (-0.9107–0.3712) | -0.0670 (-0.7370–0.5775) | - | -0.0670 (-0.6770–0.6565) | 0.1420 (-0.4917–0.7665) | 0.2470 (-0.4373–1.0150) |
Conditional Regression Methods
Relationships between diagnostic status, each protein biomarker and its associated genotypes were assessed in a series of conditional regressions. In this set of experiments, we included only combinations of proteins and genotypes that were identified as significant in the meta-analyses of both TARCC and ADNI cohorts (Table 3). The conditional regression analyses were conducted using a pair of analytical design models. The first analytical design was to use AD status (AD or NC) as the dependent variable, with either protein concentration or genotype as the independent variable. The resulting residuals from this regression were then used as the dependent variable in a second regression with either genotype or protein concentration as the independent variable. The second design was to use each protein concentration as the dependent variable with either AD status or genotype as the independent variable. The resulting residuals from this regression were then used as the dependent variable in a second regression with either genotype or AD status as the independent variable.
Table 3. Genome-wide significant signals for each endophenotype.
P-values, chromosomal and gene location are presented for each signal from the meta-analyses (Meta) and from the individual (ADNI) and (TARCC) cohorts. P-values are also shown for the association between each endophenotypic signal and age of onset (AOO) and case-control status (CC).
| Trait | Trait Chromosome | SNP(s) | P-value (TARCC) | P-value (ADNI) | P-value (Meta) | P-value (AOO) | P-value (CC) | SNP Chromosome | SNP Genes |
|---|---|---|---|---|---|---|---|---|---|
| ADN | 3 | chr3:186558403 | 9.56E-04 | 8.99E-06 | 3.16E-08 | 0.4903 | 0.4992 | 3 | MAP3K13 |
| chr3:186562865 | 2.93E-04 | 2.94E-05 | 3.75E-08 | 0.4983 | 0.5105 | ||||
| rs57056768 | 7.40E-04 | 1.97E-05 | 5.53E-08 | 0.5121 | 0.5265 | ||||
| chr3:186552158 | 1.02E-02 | 2.85E-05 | 9.52E-07 | 0.5367 | 0.4756 | ||||
| rs8111139 | 1.79E-05 | 9.47E-04 | 2.21E-07 | 0.0026 | 0.9600 | 19 | ZNF320 | ||
| F7 | 13 | rs561241 | 1.64E-02 | 7.96E-06 | 1.09E-08 | 0.4981 | 0.3392 | 13 | F7 |
| rs3093233 | 1.60E-02 | 2.39E-04 | 1.78E-08 | 0.7954 | 0.2682 | ||||
| rs6039 | 1.63E-02 | 2.24E-04 | 1.87E-08 | 0.7902 | 0.2739 | ||||
| rs2480953 | 1.55E-02 | 1.97E-04 | 2.38E-08 | 0.8831 | 0.2627 | ||||
| rs9670535 | 1.57E-02 | 2.55E-04 | 2.67E-08 | 0.8743 | 0.2589 | ||||
| rs9669828 | 1.57E-02 | 2.49E-04 | 2.71E-08 | 0.8786 | 0.2652 | ||||
| rs9670502 | 1.57E-02 | 2.60E-04 | 3.03E-08 | 0.9689 | 0.9067 | ||||
| rs1046205 | - | 4.99E-08 | 4.99E-08 | 0.0141 | 0.5717 | ||||
| rs3093253 | 5.94E-05 | 6.67E-05 | 1.65E-07 | 0.5414 | 0.4311 | ||||
| rs569557 | 6.36E-06 | 1.46E-05 | 2.67E-07 | 0.5899 | 0.4263 | ||||
| rs2774033 | 3.71E-04 | 1.41E-05 | 2.75E-07 | 0.5621 | 0.4167 | ||||
| rs493833 | 3.96E-04 | 2.12E-05 | 4.13E-07 | 0.1443 | 0.6015 | ||||
| rs7327099 | 5.63E-04 | 1.64E-06 | 4.77E-07 | 0.5073 | 0.5070 | ||||
| rs6042 | 4.42E-04 | 1.76E-05 | 5.89E-07 | 0.9207 | 0.6075 | ||||
| rs11839532 | - | 7.86E-07 | 7.86E-07 | 0.4561 | 0.5065 | ||||
| rs6041 | 1.90E-02 | 9.03E-06 | 9.66E-07 | 0.0146 | 0.5644 | ||||
| MCP-1 | 17 | rs11663180 | 3.37E-05 | 1.03E-03 | 3.88E-07 | 0.5061 | 0.3007 | 18 | ATP9B |
| chr18:76959824 | 3.49E-05 | 1.02E-03 | 3.95E-07 | 0.5075 | 0.2990 | ||||
| rs10468812 | 1.21E-03 | 1.33E-04 | 4.87E-07 | 0.3812 | 0.4848 | ||||
| chr18:76954975 | 2.92E-05 | 1.51E-03 | 5.62E-07 | 0.4298 | 0.2323 | ||||
| rs60585035 | 2.74E-05 | 1.76E-03 | 6.49E-07 | 0.4572 | 0.2203 | ||||
| rs8085999 | 2.70E-04 | 4.51E-04 | 6.79E-07 | 0.3195 | 0.5519 | ||||
| chr18:76891698 | 4.05E-05 | 1.50E-03 | 6.99E-07 | 0.2722 | 0.1835 | ||||
| rs4324200 | 1.45E-04 | 7.18E-04 | 7.23E-07 | 0.1524 | 0.6783 | ||||
| rs59890467 | 3.54E-05 | 1.69E-03 | 7.37E-07 | 0.2713 | 0.1475 | ||||
| rs4471755 | 1.01E-03 | 1.58E-04 | 7.43E-07 | 0.4213 | 0.4191 | ||||
| rs57582689 | 1.49E-04 | 7.35E-04 | 7.54E-07 | 0.1474 | 0.6805 | ||||
| rs4799019 | 2.96E-05 | 1.92E-03 | 7.64E-07 | 0.4017 | 0.2107 | ||||
| rs1942306 | 1.88E-04 | 7.33E-04 | 7.79E-07 | 0.1474 | 0.6806 | ||||
| rs12607019 | 1.46E-04 | 7.33E-04 | 7.79E-07 | 0.1474 | 0.6806 | ||||
| rs1942308 | 1.56E-04 | 7.35E-04 | 7.80E-07 | 0.1475 | 0.6806 | ||||
| rs56894683 | 2.96E-05 | 1.96E-03 | 7.84E-07 | 0.3987 | 0.2088 | ||||
| rs57977665 | 2.97E-05 | 1.96E-03 | 7.87E-07 | 0.3989 | 0.2093 | ||||
| chr18:76891893 | 1.60E-04 | 7.33E-04 | 7.94E-07 | 0.1475 | 0.6806 | ||||
| rs9967354 | 1.61E-04 | 7.33E-04 | 8.00E-07 | 0.1475 | 0.6806 | ||||
| rs9966492 | 2.89E-05 | 2.02E-03 | 8.02E-07 | 0.3736 | 0.1910 | ||||
| chr18:76916979 | 5.83E-05 | 1.39E-03 | 8.21E-07 | 0.3959 | 0.1730 | ||||
| chr18:76895796 | 1.96E-04 | 7.27E-04 | 9.13E-07 | 0.1760 | 0.6762 | ||||
| rs9965326 | 6.55E-05 | 1.50E-03 | 9.70E-07 | 0.3597 | 0.1767 | ||||
| rs35548358 | 7.33E-02 | 1.05E-06 | 5.15E-07 | 0.1802 | 0.4824 | 6 | TREM1 | ||
| rs34689624 | 7.35E-02 | 1.05E-06 | 5.17E-07 | 0.1799 | 0.4830 | ||||
| rs7761652 | 1.58E-01 | 6.05E-07 | 7.92E-07 | 0.1499 | 0.6995 |
All conditional regressions were performed in R. The glm package was used for regressions when the dependent variable was continuous (protein concentration) and the lm package was used when the dependent variables were non-parametric (AD status or residuals). All initial regression equations were adjusted for sex, years of education and population substructure (10 most relevant Eigenvectors from the principal components analysis). For these analyses, an adjusted p-value of ≤0.01 in either cohort, or ≤0.05 in both cohorts was considered significant.
Results
Quantile-quantile (QQ) plots showed no evidence of population substructure or inflation due to mistyped SNPs for any trait in either cohort (Figs 2–5). Meta-analyses showed many interesting signals (supplemental data), including a strong replication of the association between AD status and variants in the APOE/TOMM40 region. Conversely, no genome-wide significant (GWS; p<1x10-7) associations were observed for age of onset of disease symptoms. In this paper, we focus on four associations between genetic loci and three of the seven endophenotypes analyzed. In these four instances, the association reached GWS in the meta-analyses and evidence for each of the signals was observed in both the TARCC and ADNI cohorts (Table 3).
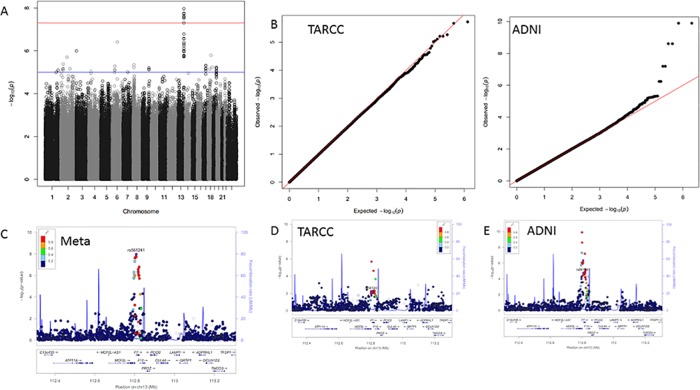
Fig 2. Factor VII results.
Panel A; GWAS Manhattan plot for the meta-analysis of Factor VII. Panel B; QQ plots for the association results for TARCC (left) and ADNI (right). Panel C; LocusZoom plot for the chromosome 13 signal observed in the meta-analysis. Panels D and E; LocusZoom plots for the chromosome 13 signal in TARCC and ADNI, respectively.
Fig 5. Case Control results.
Panel A; GWAS Manhattan plot for Case Control status. Panel B; QQ plots for the association results for TARCC (left) and ADNI (right). Panel C; LocusZoom plot for the chromosome 19 signal from the meta-analysis. Panels D and E; LocusZoom plots for the chromosome 19 signal in TARCC and ADNI, respectively.
The strongest associations were found for blood concentrations of Factor VII (F7). This signal contained 12 SNPs on chromosome 13 within the F7 gene that were associated with serum/plasma concentrations of Factor VII at genome wide significance (Fig 2). Significance within this region ranged from p = 9.66x10-7 to p = 2.67x10-8. The signal within F7 is a sharp peak that roughly corresponds to the width of the F7 gene.
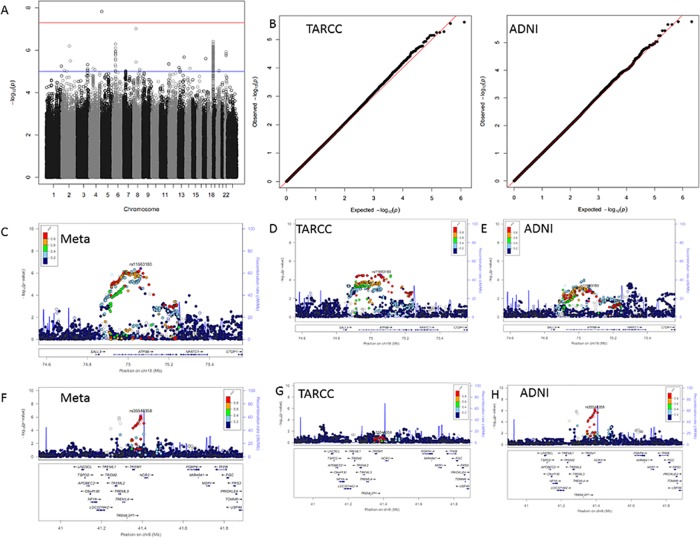
Seventeen SNPs on chromosome 18 were significantly associated with serum/plasma levels of monocyte chemoattractant protein -1 (MCP-1) (Fig 3). These polymorphisms were concentrated within the ATP9B gene. Significance within this signal ranged from p = 9.70x10-7 to p = 3.88x10-7. As with F7, the width of the ATP9B signal corresponds to the length and location of the ATP9B gene. Associations for many SNPs throughout the entire ATP9B gene region are elevated, forming a plateaued signal.
Fig 3. MCP1 results.
Panel A; GWAS Manhattan plot for meta-analysis of MCP1. Panel B; QQ plots for the association results for TARCC (left) and ADNI (right). Panel C; LocusZoom plot for the chromosome 18 signal from the meta-analysis. Panels D and E; LocusZoom plots for the chromosome 18 signal in TARCC and ADNI, respectively. Panel F; LocusZoom plot for the chromosome 6 signal from the meta-analysis. Panels G and H; LocusZoom plots for the chromosome 6 signal in TARCC and ADNI, respectively.
One of the many interesting associations that did not have support in both cohorts was between blood concentrations of MCP-1 and polymorphisms on chromosome 6 within the triggering receptor expressed on monocytes (TREM1) gene (Fig 3). In this case, the meta-analysis showed an association that reached genome wide significance (Table 3), but the signal was only apparent within the ADNI cohort (Fig 3).
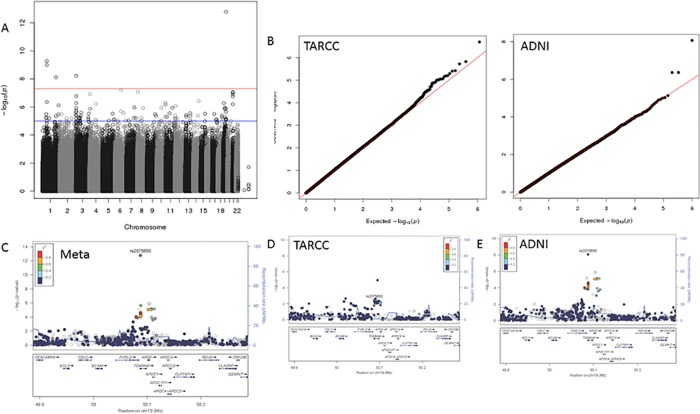
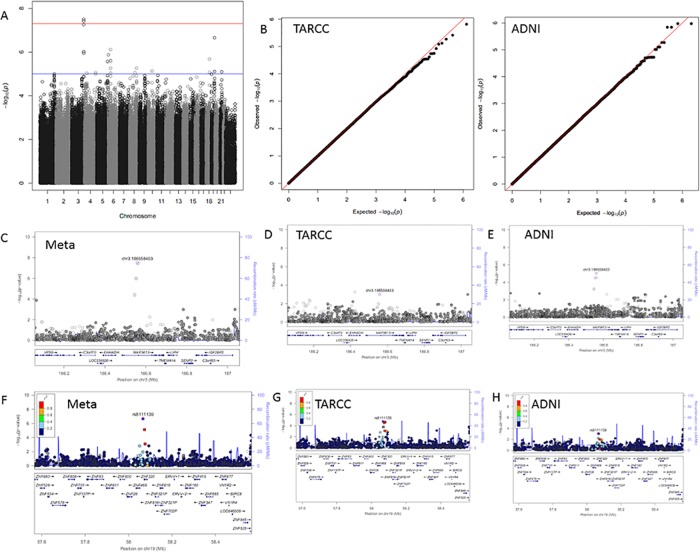
Four SNPs on chromosome 3 were significantly associated with serum/plasma levels of adiponectin (Table 3). These polymorphisms were concentrated within the MAP3K13 gene. Significance within this signal ranged from p = 9.52x10-7 to p = 3.16x10-8. Unlike the signals within F7 and ATP9B, the width of the MAP3K13 signal for adiponectin is much narrower than the MAPK13 gene (Fig 4). In addition, although the signal reaches 10−8 for a pair of SNPs, there are far fewer SNPs in the MAP3K13 signal compared to the signals in F7 and ATP9B.
Fig 4. Adiponectin results.
Panel A; GWAS Manhattan plot for meta-analysis of Adiponectin. Panel B; QQ plots for the association results for TARCC (left) and ADNI (right). Panel C; LocusZoom plot for the chromosome 3 signal from the meta-analysis. Panels D and E; LocusZoom plots for the chromosome 3 signal in TARCC and ADNI, respectively. Panel F; LocusZoom plot for the chromosome 19 signal from the meta-analysis. Panels G and H; LocusZoom plots for the chromosome 19 signal in TARCC and ADNI, respectively.
Despite the limited samples size in the present study, several previously reported associations for case control status were replicated at p≤0.05 and all published SNPs that were in the dataset showed a trend for association (Table 4). The most strongly associated SNP (rs2075650), which reached GWS for association with case control status (Fig 5) is located within the intron of the translocase of outer mitochondrial membrane 40 (TOMM40) gene[28]. TOMM40 is in the same region as the APOE gene, but has been reported to contribute additional genetic risk for AD[28].
Table 4. Associations between published AD SNPs and diagnostic status.
| Gene | Marker | p-value |
|---|---|---|
| NME8 | rs2718058 | 0.02 |
| HKA DRB1-5 | rs9271192 | 0.05 |
| INPP5D | rs35349669 | 0.06 |
| ZCWPW1 | rs1476679 | 0.06 |
| BIN1 | rs6733839 | 0.07 |
| CD2AP | rs10948363 | 0.07 |
| EPHA1 | rs11771145 | 0.08 |
| CASS4 | rs7274581 | 0.08 |
| PTK2B | rs28834970 | 0.09 |
| SORL1 | rs11218343 | 0.09 |
| ABCA7 | rs4147929 | 0.09 |
| CLU | rs9331896 | 0.09 |
| MEF2C | rs190982 | 0.10 |
| CR1 | rs6656401 | 0.12 |
| CLEF1 | rs10838725 | 0.12 |
| FERMT2 | rs17125944 | 0.12 |
| SLC24A4 | rs10498633 | 0.12 |
| PICALM | rs10792832 | 0.13 |
| MS4A6A | rs983392 | 0.13 |
Conditional regression analyses recapitulated the GWAS results for F7, MCP-1 and adiponectin (Table 5). In addition, there were significant associations between AD status and blood concentrations of F7, MCP-1 and adiponectin, which were expected, given the membership of these proteins in the AD biomarker panel[12]. Finally, conditional regression analyses suggested that the SNPs contributed to variation in protein concentration independent of AD status. It was not possible to determine whether genotypes also contributed directly to disease risk.
Table 5. Results of conditional regression analyses.
For each regression equation, dependent variables are listed in the top row, independent variables in the second row. For conditional regressions (two columns to the far right) the dependent variables were the residuals from an initial regression and the independent variables were the genotypes of candidate SNPs. [Protein~Disease status] indicates that protein concentration was the dependent variable and disease status was the independent variable in the initial regression. Similarly, [Disease status~ Protein] indicates that disease status was the dependent variable and protein concentration was the independent variable in the initial regression. Correlation statistics indicating the amount of variance explained by the independent variable is presented where appropriate for each regression. Results of regression analyses recapitulated the GWAS results for F7, MCP-1 and adiponectin (column one). In addition, there were significant associations between AD status and blood concentrations of F7, MCP-1 and adiponectin (column two). Conditional regression analyses suggested that candidate SNPs contributed to variation in protein concentration independent of AD status (column three). None of the results of regressions interrogating whether genotypes contributed directly to disease risk independent of protein endophenotype were significant (column four). However, due to insufficient statistical power, it was not possible to determine the true relationship between these factors.
| Dependent Variable | Protein concentration | Disease status | Residuals of Protein Concentration Regressed on Disease Status | Residuals of Disease Status Regressed on Protein Concentration | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Independent Variable | Genotype | Protein concentration | Genotype | Genotype | |||||||
| Cohort | Protein | Marker | Gene | p | r2 | p | r2 | p | r2 | p | r2 |
| TARCC | Adipo | chr3.186558403 | MAP3K13 | 6.31E-10 | 24% | 0.016 | NA | 0.001 | 4% | 0.574 | 0% |
| ADNI | Adipo | chr3.186558403 | MAP3K13 | 5.09E-05 | 31% | 0.023 | NA | 0.017 | 5% | 0.244 | 1% |
| TARCC | Adipo | rs8111139 | ZNF320 | 3.91E-10 | 22% | 0.026 | NA | 3.24E-04 | 4% | 0.208 | 1% |
| ADNI | Adipo | rs8111139 | ZNF320 | 3.58E-04 | 28% | 0.023 | NA | 0.982 | 0% | 0.653 | 0% |
| TARCC | F7 | rs561241 | F7 | 0.001 | 13% | 0.001 | NA | 0.011 | 2% | 0.386 | 0% |
| ADNI | F7 | rs561241 | F7 | 0.002 | 24% | 0.069 | NA | 0.006 | 6% | 0.998 | 0% |
| TARCC | MCP-1 | rs11663180 | ATP9B | 0.022 | 9% | 0.014 | NA | 3.03E-05 | 6% | 0.959 | 0% |
| ADNI | MCP-1 | rs11663180 | ATP9B | 0.010 | 21% | 0.092 | NA | 0.003 | 7% | 0.390 | 1% |
| TARCC | MCP-1 | rs35548358 | TREM1 | 0.759 | 3% | 0.009 | NA | 0.054 | 1% | 0.350 | 0% |
| ADNI | MCP-1 | rs35548358 | TREM1 | 0.031 | 19% | 0.092 | NA | 0.012 | 5% | 0.161 | 2% |
None of the exact SNPs previously reported to be associated with Alzheimer’s disease were associated with blood concentrations of the proteins investigated in these experiments. However, signals were observed in the vicinity of CR1 (endoPT: Factor VII), MS4A6A/MS4A4E (endoPT: B2M), PICALM (endoPT: B2M and VCAM1), CLU (endoPT: Tenascin C), and PTK2B (endoPT: B2M).
Discussion
The use of quantitative endophenotypes as outcome variables in genome-wide association studies has proven to be useful for identifying the genetic basis of complex disease[29–34]. This method is likely to be maximally effective for diseases that exhibit significant phenotypic heterogeneity, such as Alzheimer’s. The use of endophenotypes presumably provides increased statistical power due to greater proximity of the outcome variable to functional genetic variants, which reduces the impact of confounding non-genetic factors.
The strongest overall signal in the meta-analysis was between diagnostic status for Alzheimer’s disease and a group of SNPs in the region of the APOE gene. Given the well-replicated strength of the APOE signal, this result was not surprising even with the small sample size that was employed in the present study.
The strongest signal observed in the meta-analysis was between the serum/plasma concentration of F7 and a group of SNPs within the F7 gene on chromosome 13. Factor VII is a serine protease that is a key member of the coagulation cascade[35]. Along with tissue factor, F7 is responsible for initiating the coagulation cascade. The process begins with release of tissue factor from the external wall of blood vessels following vascular injury. Once inside the circulation, tissue factor binds to F7, which is converted to F7a, leading to conversion of factors IX and X into active proteases; factors IXa and Xa[35]. Factor VII is a vitamin K dependent enzyme and the target of warfarin and other anticoagulants that are used to prevent thrombosis and thromboembolism[36]. Serum concentrations of F7 were negatively associated with AD status in prior work[12].
Polymorphisms within the F7 gene have not been suggested previously as contributing to AD risk, despite multiple large-scale studies. Nevertheless, a SNP within this region (rs6046) has been associated with variation in risk for cardiovascular disease, venous thrombosis and stroke[37–41]; conditions that are associated with risk for AD and other forms of dementia. The rs6046 polymorphism, which is located in exon 9 and is predicted to cause the substitution of glutamine in place of arginine at amino acid position 353 (R353Q), has been shown to result in reduced levels of F7 activity[39]. The haplotype containing this SNP has been reported as both protective and a risk factor for coagulation related disease phenotypes[39–41]. The rs6046 SNP was associated with a later age of AD onset in our study.
Monocyte Chemoattractant Protein -1 (MCP-1) is one of the key chemokines involved in the regulation of monocyte and macrophage migration during the inflammatory response (see Deshmane et al. 2009 for review[42]). A variety of cells produce MCP-1, including epithelial, smooth muscle, astrocytes and microglial cells[42]. Increased MCP-1 has been shown to contribute to a variety of disease states, including Alzheimer’s disease,[43] atherosclerosis[44, 45], increased risk for AD following traumatic brain injury[46], insulin resistance[47], and neuronal death following ischemia[48]. Serum concentrations of MCP-1 were negatively associated with AD status in prior work[12].
We observed a significant signal in association with MCP-1 levels located on chromosome 18 in the coding region for ATP9B (ATPase, class II, type 9B). ATP9B is a class 2 P4-ATPase. Generally speaking, the P4-ATPases orchestrate phospholipid translocation from the exoplasmic to cytoplasmic leaflet which is critical for the maintenance of biological membrane characteristics and protein trafficking through vesicular transport. Alterations in the functionality of this family of flippases have been associated with multiple diseases and disorders (e.g., variants in ATP8B4 have been associated with Alzheimer’s disease[49, 50]). The ATP9B gene product has recently been shown to function independent of the CDC50 subunit complex, a characteristic unique to the class 2 P4-ATPases, and localize specifically to the trans-Golgi network[51]. The implied relationship between MCP-1 levels and the function of ATP9B gene products is not clear.
The adipocyte-derived hormone adiponectin (also known as 30-kDa adipocyte complement-related protein; Acrp30) has been mapped to a susceptibility locus for type 2 diabetes within the AdipoQ gene[52]. AdipoQ is shown to be dysregulated in obesity, metabolic syndrome, and cardiovascular disease[53–55]. Adipose tissues secrete many factors into the bloodstream, such as leptin, TNF-α, adipsin, and adiponectin. These proteins are referred to as adipocytokines, and are secreted to sensitize the tissues to insulin[52]. Levels of adiponectin in the blood are lower in individuals with diabetes, insulin resistance, and obesity.[56] Serum concentrations of adiponectin were positively associated with AD status in prior work[12].
We observed two significant signals in association with adiponectin levels. The first signal is located on chromosome 3 within the MAP3K13 gene. The MAP3K13 gene encodes a protein kinase that is expressed most strongly in the pancreas, brain, and liver, but not detected in heart, lung, skeletal muscle, or kidney. Protein kinases are involved in a litany of pathways, however MAP3K13 has been shown to be involved in the stress-activated JNK1 pathway[57]. This gene’s specific involvement with adiponectin expression is unclear.
The second signal associated with adiponectin levels was located on chromosome 19 within a zinc finger gene, ZNF320. Zinc fingers are a heterogeneous class of protein structural motifs that are involved in the expression or repression of genes. Specifically, ZNF320 has been shown to be implicated in glioblastoma. The ZNF320 gene’s involvement with adiponectin expression is also unclear.
Results of the current study appear to confirm the utility of an endophenotypic approach to the analysis of GWAS data from complex traits. Four novel associations were observed between genetic variants and endophenotypes for AD. Polymorphisms within the F7, ATP9B, MAP3K13 and ZNF320 genes have not been suggested previously as contributing to AD risk, despite multiple large-scale GWAS studies[1, 58–65]. Interestingly, meta-analysis showed that two SNPs within the F7 gene (including rs6046) showed a trend for association with age at disease onset in our sample (Table 3). Unsurprisingly due to the relatively small sample, none of the SNPs were associated with diagnostic status.
In an attempt to determine the specifics of the most likely biological relationship model for AD, a series of conditional regressions were performed that assessed the relationships between diagnostic status and non-genetic factors as well as specific protein biomarkers and their associated genotypes. The majority of significant associations recapitulated either current GWAS relationships between specific SNPs and blood protein concentrations, or relationships inherent to the AD biomarker panel between blood protein concentrations and disease status. In addition, associated genotypes explained a significant amount of the variation that remained in protein concentration after disease status and covariates (sex, population substructure eigenvectors) were accounted for. It was not possible to determine conclusively whether associated SNPs also contributed directly to AD risk, independent of protein concentration.
It was interesting that several AD loci that have been reliably reported in the literature were associated with our AD endophenotypes. If these associations are confirmed in independent cohorts, they may help to explain the etiological mechanisms and functional variants that are responsible for these previously published associations.
There a number of caveats to these results. First, although the observations reported were derived from a meta-analysis of two entirely independent cohorts of study participants, the sample sizes were small. This is particularly true in light of recent publications by the IGAP group, which were based upon an international sample of nearly 75,000 individuals. However, the analytical approach that we adopted provided much greater statistical power than would have been possible with a traditional GWAS, where diagnostic status is used as the outcome variable.
In summary, the use of endophenotypes for Alzheimer’s disease in the place of diagnostic status as the outcome variable in GWAS analysis overcame sample size constraints and allowed the identification and independent replication of two putative novel genetic loci that appear to impact risk for AD. Polymorphisms in F7 and ATP9B may impact the risk or development of AD and should be studied in a larger, independent cohort.
Acknowledgments
This study was made possible by the Texas Alzheimer’s Research and Care Consortium (TARCC), which is funded by the state of Texas through the Texas Council on Alzheimer’s Disease and Related Disorders.
Investigators from the Texas Alzheimer’s Research and Care Consortium: Baylor College of Medicine: Rachelle Doody MD, PhD, Mimi M. Dang MD, Valory Pavlik PhD, Wen Chan PhD, Paul Massman PhD, Eveleen Darby, Tracy Evans RN, Aisha Khaleeq; Texas Tech University Health Sciences Center: Chuang-Kuo Wu MD, PhD, Matthew Lambert PhD, Victoria Perez, Michelle Hernandez; University of North Texas Health Science Center: Thomas Fairchild PhD, Janice Knebl DO, James R. Hall PhD, Leigh Johnson PhD, Douglas Mains, Lisa Alvarez, Rosemary McCallum; University of Texas Southwestern Medical Center: Perrie Adams PhD, Munro Cullum PhD, Roger Rosenberg MD, Benjamin Williams MD, PhD, Mary Quiceno MD, Joan Reisch PhD, Natalie Martinez, Janet Smith; University of Texas Health Science Center–San Antonio: Donald Royall MD, Raymond Palmer PhD, Marsha Polk; Texas A&M University Health Science Center: Farida Sohrabji PhD, Steve Balsis PhD, Rajesh Miranda, PhD;
The replication data set was funded by the Alzheimer′s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904; RC2 AG036535). ADNI is also funded by Department of Defense (award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare;; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer′s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. Investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. ADNI principal investigators and executive committee include; Michael W. Weiner MD, Paul Aisen MD, Ronald Petersen MD PhD, Clifford R. Jack, Jr. MD, William Jagust MD, John Q. Trojanowski MD PhD, Arthur W. Toga PhD, Laurel Beckett PhD, Robert C. Green MD MPH, Andrew J. Saykin PsyD, John Morris MD, Leslie M. Shaw.
Partial support and computer resources were provided by the Renaissance Computing Institute (RENCI) at the University of North Carolina at Chapel Hill.
SJS, NRP, JSM acknowledge NIA Training grant T32 AG020494. RMH acknowledges a grant from NIGMS P30AG12300-20. KCW acknowledges grants 5R01DA030976, 5U01HG006487, 1U19HD077632, 1U01HG007437, 5P01-DK058335, 5U24 AA020024 and 1UL1TR001111.
Data Availability
Due to third party restrictions data are available upon request from the following institutions: Data from TARCC participants are available for researchers who meet eligibility criteria for access to data generated with state of Texas funds. Please contact Dr. Joan Reisch (Joan.Reisch@utsouthwestern.edu) regarding access to TARCC data. Additional information is available on the TARCC website (http://www.txalzresearch.org/). Data from ADNI participants are available to all researchers meeting ADNI eligibility criteria. ADNI data are available from Dr. Michael Weiner (adni@loni.usc.edu). Additional information is available on the ADNI website (http://adni.loni.usc.edu/data-samples/access-data/).
Funding Statement
The Texas Alzheimer’s Research and Care Consortium (TARCC), which is funded by the state of Texas through the Texas Council on Alzheimer’s Disease and Related Disorders funded data generation for the TARCC cohort. The replication data set was funded by the Alzheimer′s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904; RC2 AG036535). ADNI is ALSO funded by the Department of Defense (award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer′s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. Investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. Partial support and computer resources were provided by the Renaissance Computing Institute (RENCI) at the University of North Carolina at Chapel Hill. SJS, NRP, JSM acknowledge a National Institute of Aging Training grant (T32AG020494). RMH acknowledges a grant from the National Institute of General Medical Sciences (P30AG12300-20). KCW acknowledges grants from the National INstitutes of Health (5R01DA030976, 5U01HG006487, 1U19HD077632, 1U01HG007437, 5P01DK058335, 5U24AA020024 and 1UL1TR001111).
References
- 1. Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45(12):1452–8. Epub 2013/10/29. 10.1038/ng.2802 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68(2):270–81. Epub 2010/10/20. 10.1016/j.neuron.2010.10.013 . [DOI] [PubMed] [Google Scholar]
- 3. Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232(4746):34–47. Epub 1986/04/04. . [DOI] [PubMed] [Google Scholar]
- 4. den Heijer T, van der Lijn F, Koudstaal PJ, Hofman A, van der Lugt A, Krestin GP, et al. A 10-year follow-up of hippocampal volume on magnetic resonance imaging in early dementia and cognitive decline. Brain. 133(Pt 4):1163–72. Epub 2010/04/09. doi: awq048 [pii] 10.1093/brain/awq048 . [DOI] [PubMed] [Google Scholar]
- 5. Murray AD, Staff RT, McNeil CJ, Salarirad S, Ahearn TS, Mustafa N, et al. The balance between cognitive reserve and brain imaging biomarkers of cerebrovascular and Alzheimer's diseases. Brain. 2011;134(Pt 12):3687–96. Epub 2011/11/22. 10.1093/brain/awr259 . [DOI] [PubMed] [Google Scholar]
- 6. Melville SA, Buros J, Parrado AR, Vardarajan B, Logue MW, Shen L, et al. Multiple loci influencing hippocampal degeneration identified by genome scan. Ann Neurol. 2012;72(1):65–75. Epub 2012/06/30. 10.1002/ana.23644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagy Z, Esiri MM, Jobst KA, Morris JH, King EM, McDonald B, et al. Relative roles of plaques and tangles in the dementia of Alzheimer's disease: correlations using three sets of neuropathological criteria. Dementia. 1995;6(1):21–31. . [DOI] [PubMed] [Google Scholar]
- 8. Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol. 2009;65(4):403–13. Epub 2009/03/20. 10.1002/ana.21610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blennow K. Cerebrospinal fluid protein biomarkers for Alzheimer's disease. NeuroRx. 2004;1(2):213–25. Epub 2005/02/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Trojanowski JQ, Vandeerstichele H, Korecka M, Clark CM, Aisen PS, Petersen RC, et al. Update on the biomarker core of the Alzheimer's Disease Neuroimaging Initiative subjects. Alzheimers Dement. 2010;6(3):230–8. 10.1016/j.jalz.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S, Swaminathan S, Inlow M, Risacher SL, Nho K, Shen L, et al. Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel. PLoS ONE. 2013;8(7):e70269 Epub 2013/07/31. 10.1371/journal.pone.0070269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Bryant SE, Xiao G, Barber R, Huebinger R, Wilhelmsen K, Edwards M, et al. A blood-based screening tool for Alzheimer's disease that spans serum and plasma: findings from TARC and ADNI. PLoS ONE. 2011;6(12):e28092 Epub 2011/12/14. 10.1371/journal.pone.0028092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Bryant SE, Xiao G, Barber R, Reisch J, Doody R, Fairchild T, et al. A serum protein-based algorithm for the detection of Alzheimer disease. Arch Neurol. 2010;67(9):1077–81. Epub 2010/09/15. 10.1001/archneurol.2010.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Bryant SE, Xiao G, Barber R, Reisch J, Hall J, Cullum CM, et al. A blood-based algorithm for the detection of Alzheimer's disease. Dement Geriatr Cogn Disord. 2011;32(1):55–62. Epub 2011/08/26. 10.1159/000330750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Waring S, O'Bryant SE, Reisch JS, Diaz-Arrastia R, Knebl J, Doody R, et al. The Texas alzheimer's Research Consortium longitudinal research cohort: Study design and baseline characteristics. Texas Public Health Journal. 2008;60(3):9–13. [Google Scholar]
- 16. Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256(3):183–94. Epub 2004/08/25. 10.1111/j.1365-2796.2004.01388.x . [DOI] [PubMed] [Google Scholar]
- 17. McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34(7):939–44. Epub 1984/07/01. . [DOI] [PubMed] [Google Scholar]
- 18. Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG, et al. Clinical Core of the Alzheimer's Disease Neuroimaging Initiative: progress and plans. Alzheimers Dement. 2010;6(3):239–46. Epub 2010/05/11. 10.1016/j.jalz.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, et al. Alzheimer's Disease Neuroimaging Initiative (ADNI) Clinical characterization. Neurology. 2010;74(3):201–9. 10.1212/WNL.0b013e3181cb3e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40(10):1253–60. Epub 2008/09/09. 10.1038/ng.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Didion J, Yang H, Sheppard K, Fu C-P, McMillan L, de Villena F, et al. Discovery of novel variants in genotyping arrays improves genotype retention and reduces ascertainment bias. BMC Genomics. 2012;13(1):34 10.1186/1471-2164-13-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38(8):904–9. [DOI] [PubMed] [Google Scholar]
- 23. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. Epub 2007/08/19. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–34. Epub 2010/11/09. 10.1002/gepi.20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wickham H. ggplot2: elegant graphics for data analysis New York, NY: Springer; 2009. [Google Scholar]
- 26. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. Epub 2010/07/10. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. Epub 2010/07/17. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS ONE. 2009;4(8):e6501 Epub 2009/08/12. 10.1371/journal.pone.0006501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gorman KF, Julien C, Moreau A. The genetic epidemiology of idiopathic scoliosis. European Spine Journal. 2012;21(10):1905–19. 10.1007/s00586-012-2389-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gorwood P, Le Strat Y, Ramoz N, Dubertret C, Moalic JM, Simonneau M. Genetics of dopamine receptors and drug addiction. Human Genetics. 2012;131(6):803–22. 10.1007/s00439-012-1145-7 [DOI] [PubMed] [Google Scholar]
- 31. Ertekin-Taner N. Gene expression endophenotypes: A novel approach for gene discovery in Alzheimer's disease. Molecular Neurodegeneration. 2011;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin PI, Shuldiner AR. Rethinking the genetic basis for comorbidity of schizophrenia and type 2 diabetes. Schizophrenia Research. 2010;123(2–3):234–43. 10.1016/j.schres.2010.08.022 [DOI] [PubMed] [Google Scholar]
- 33. Weidinger S, Baurecht H, Naumann A, Novak N. Genome-wide association studies on IgE regulation: Are genetics of IgE also genetics of atopic disease? Current Opinion in Allergy and Clinical Immunology. 2010;10(5):408–17. 10.1097/ACI.0b013e32833d7d2d [DOI] [PubMed] [Google Scholar]
- 34. Potkin SG, Turner JA, Guffanti G, Lakatos A, Torri F, Keator DB, et al. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: Methodological considerations. Cognitive Neuropsychiatry. 2009;14(4–5):391–418. 10.1080/13546800903059829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mackman N, Tilley RE, Key NS. Role of the extrinsic pathway of blood coagulation in hemostasis and thrombosis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2007;27(8):1687–93. [DOI] [PubMed] [Google Scholar]
- 36. Broderick J, Connolly S, Feldmann E, Hanley D, Kase C, Krieger D, et al. Guidelines for the management of spontaneous intracerebral hemorrhage in adults: 2007 Update. Guideline from the American Heart Association/American Stroke Association Stroke Council, high blood pressure research council, and the quality of care and outcomes in research interdisciplinary working group. Stroke. 2007;38(6):2001–23. [DOI] [PubMed] [Google Scholar]
- 37. Drenos F, Talmud PJ, Casas JP, Smeeth L, Palmen J, Humphries SE, et al. Integrated associations of genotypes with multiple blood biomarkers linked to coronary heart disease risk. Hum Mol Genet. 2009;18(12):2305–16. Epub 2009/04/02. 10.1093/hmg/ddp159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fujimaki T, Kato K, Yoshida T, Oguri M, Watanabe S, Metoki N, et al. Association of genetic variants with myocardial infarction in Japanese individuals with chronic kidney disease. Thromb Haemost. 2009;101(5):963–8. Epub 2009/05/01. . [DOI] [PubMed] [Google Scholar]
- 39. Ken-Dror G, Drenos F, Humphries SE, Talmud PJ, Hingorani AD, Kivimaki M, et al. Haplotype and genotype effects of the F7 gene on circulating factor VII, coagulation activation markers and incident coronary heart disease in UK men. J Thromb Haemost. 2010;8(11):2394–403. Epub 2010/08/26. 10.1111/j.1538-7836.2010.04035.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zakai NA, Lange L, Longstreth WT Jr., O'Meara ES, Kelley JL, Fornage M, et al. Association of coagulation-related and inflammation-related genes and factor VIIc levels with stroke: the Cardiovascular Health Study. J Thromb Haemost. 2011;9(2):267–74. Epub 2010/12/01. 10.1111/j.1538-7836.2010.04149.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zee RY, Glynn RJ, Cheng S, Steiner L, Rose L, Ridker PM. An evaluation of candidate genes of inflammation and thrombosis in relation to the risk of venous thromboembolism: The Women's Genome Health Study. Circulation Cardiovascular genetics. 2009;2(1):57–62. Epub 2009/12/25. 10.1161/CIRCGENETICS.108.801969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29(6):313–26. Epub 2009/05/16. 10.1089/jir.2008.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galimberti D, Fenoglio C, Lovati C, Venturelli E, Guidi I, Corra B, et al. Serum MCP-1 levels are increased in mild cognitive impairment and mild Alzheimer's disease. Neurobiol Aging. 2006;27(12):1763–8. Epub 2005/11/26. doi: S0197-4580(05)00336-2 [pii] 10.1016/j.neurobiolaging.2005.10.007 . [DOI] [PubMed] [Google Scholar]
- 44. Ylä-Herttuala S, Lipton BA, Rosenfeld ME, Särkioja T, Yoshimura T, Leonard EJ, et al. Expression of monocyte chemoattractant protein 1 in macrophage-rich areas of human and rabbit atherosclerotic lesions. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(12):5252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gosling J, Slaymaker S, Gu L, Tseng S, Zlot CH, Young SG, et al. MCP-1 deficiency reduces susceptibility to atherosclerosis in mice that overexpress human apolipoprotein B. Journal of Clinical Investigation. 1999;103(6):773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ho L, Zhao W, Dams-O'Connor K, Tang CY, Gordon W, Peskind ER, et al. Elevated plasma MCP-1 concentration following traumatic brain injury as a potential "predisposition" factor associated with an increased risk for subsequent development of Alzheimer's disease. J Alzheimers Dis. 2012;31(2):301–13. Epub 2012/05/01. 10.3233/JAD-2012-120598 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kanda H, Tateya S, Tamori Y, Kotani K, Hiasa KI, Kitazawa R, et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. Journal of Clinical Investigation. 2006;116(6):1494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sakurai-Yamashita Y, Shigematsu K, Yamashita K, Niwa M. Expression of MCP-1 in the hippocampus of SHRSP with ischemia-related delayed neuronal death. Cellular and Molecular Neurobiology. 2006;26(4–6):823–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65(1):45–53. Epub 2007/11/14. 10.1001/archneurol.2007.3 . [DOI] [PubMed] [Google Scholar]
- 50. van der Mark VA, Elferink RP, Paulusma CC. P4 ATPases: Flippases in Health and Disease. International journal of molecular sciences. 2013;14(4):7897–922. Epub 2013/04/13. 10.3390/ijms14047897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takatsu H, Baba K, Shima T, Umino H, Kato U, Umeda M, et al. ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J Biol Chem. 2011;286(44):38159–67. Epub 2011/09/15. 10.1074/jbc.M111.281006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Balendran N, Clough RL, Arguello JR, Barber R, Veal C, Jones AB, et al. Characterization of the major susceptibility region for psoriasis at chromosome 6p21.3. J Invest Dermatol. 1999;113(3):322–8. Epub 1999/09/01. 10.1046/j.1523-1747.1999.00710.x . [DOI] [PubMed] [Google Scholar]
- 53. Barber R, Gholkar A, Scheltens P, Ballard C, McKeith IG, Morris CM, et al. Apolipoprotein E epsilon4 allele, temporal lobe atrophy, and white matter lesions in late-life dementias. Arch Neurol. 1999;56(8):961–5. Epub 1999/08/17. . [DOI] [PubMed] [Google Scholar]
- 54. Mansourian M, Javanmard SH. Adiponectin gene polymorphisms and susceptibility to atherosclerosis: A meta-analysis. Journal of research in medical sciences: the official journal of Isfahan University of Medical Sciences. 2013;18(7):611–6. Epub 2014/02/12. [PMC free article] [PubMed] [Google Scholar]
- 55. Wassel CL, Pankow JS, Rasmussen-Torvik LJ, Li N, Taylor KD, Guo X, et al. Associations of SNPs in ADIPOQ and subclinical cardiovascular disease in the multi-ethnic study of atherosclerosis (MESA). Obesity (Silver Spring). 2011;19(4):840–7. Epub 2010/10/12. 10.1038/oby.2010.229 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clark RB, Knoll BJ, Barber R. Partial agonists and G protein-coupled receptor desensitization. Trends Pharmacol Sci. 1999;20(7):279–86. Epub 1999/07/03. . [DOI] [PubMed] [Google Scholar]
- 57. Barber R, Scheltens P, Gholkar A, Ballard C, McKeith I, Ince P, et al. White matter lesions on magnetic resonance imaging in dementia with Lewy bodies, Alzheimer's disease, vascular dementia, and normal aging. J Neurol Neurosurg Psychiatry. 1999;67(1):66–72. Epub 1999/06/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41(10):1094–9. Epub 2009/09/08. 10.1038/ng.439 . [DOI] [PubMed] [Google Scholar]
- 59. Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41(10):1088–93. Epub 2009/09/08. 10.1038/ng.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–40. Epub 2010/05/13. 10.1001/jama.2010.574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43(5):436–41. Epub 2011/04/05. 10.1038/ng.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43(5):429–35. Epub 2011/04/05. 10.1038/ng.803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368(2):107–16. Epub 2012/11/16. 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013;368(2):117–27. Epub 2012/11/16. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, et al. SORL1 is genetically associated with late-onset Alzheimer's disease in Japanese, Koreans and Caucasians. PLoS ONE. 2013;8(4):e58618 Epub 2013/04/09. 10.1371/journal.pone.0058618 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Due to third party restrictions data are available upon request from the following institutions: Data from TARCC participants are available for researchers who meet eligibility criteria for access to data generated with state of Texas funds. Please contact Dr. Joan Reisch (Joan.Reisch@utsouthwestern.edu) regarding access to TARCC data. Additional information is available on the TARCC website (http://www.txalzresearch.org/). Data from ADNI participants are available to all researchers meeting ADNI eligibility criteria. ADNI data are available from Dr. Michael Weiner (adni@loni.usc.edu). Additional information is available on the ADNI website (http://adni.loni.usc.edu/data-samples/access-data/).