Abstract
We report progress toward a general strategy for mimicking the recognition properties of specific α-helices within natural proteins through the use of oligomers that are less susceptible than conventional peptides to proteolysis. The oligomers contain both α- and β-amino acid residues, with the density of the β subunits low enough that an α-helix-like conformation can be formed but high enough to interfere with protease activity. Previous studies with a different protein-recognition system suggested ring-constrained β residues can be superior to flexible β residues in terms of maximizing α/β-peptide affinity for a targeted protein surface. Here, we use mimicry of the 18-residue Bim BH3 domain to expand the scope of this strategy. Two significant advances have been achieved. First, we have developed and validated a new ring-constrained β residue that bears an acidic side chain, which complements previously known analogues that are either hydrophobic or basic. Second, we have discovered that placing cyclic β residues at sites that make direct contact with partner proteins can lead to substantial discrimination between structurally homologous binding partners, the proteins Bcl-xL and Mcl-1. Overall, this study helps to establish that α/β-peptides containing ring-preorganized β residues can reliably provide proteolytically resistant ligands for proteins that naturally evolved to recognize α-helical partners.
Introduction
α-Helices play prominent roles in protein associations. In some cases, one partner's contribution to the binding interface is comprised entirely of an α-helical segment, while in other cases the α-helix is part of a more complex recognition surface, as documented in comprehensive structural surveys by Arora et al.1-3 The inherent regularity of helical secondary structure has inspired many efforts to mimic the information content encoded on α-helical surfaces with unnatural oligomers,4 including oligo-aryl compounds,5-8 peptoids,9 peptides comprised of D-α-amino acid residues,10 spiroligomers,11 and amide-sulfonamide oligomers.12 Efforts in a number of groups have focused on peptidic oligomers composed entirely of β-amino acid residues13,14 or containing mixtures of α- and β-amino acid residues.15 Collectively, these β-peptides and α/β-peptides can access diverse helical conformations that offer a variety of side chain display geometries;16,17 the specific conformation adopted can be controlled by modulating the β-amino acid substitution pattern, the arrangement of α and β residues along the backbone, and other molecular parameters.
We have used BH3 domain recognition by anti-apoptotic proteins in the Bcl-2 family, such as Bcl-xL and Mcl-1, as a testbed to compare the α-helix-mimetic competencies of alternative β- and α/β-peptide helices.15 The bioactive BH3 domain conformation is an α-helix with a minimum of four or five turns.18 A set of four hydrophobic side chains is displayed along one side of this helix, and these side chains are accommodated by pockets at the bottom of the BH3-recognition cleft on Bcl-2-family binding partners (Figure 1A). An Asp side chain projects from the opposite side of the BH3 domain helix, relative to the ‘stripe’ of hydrophobic residues; this carboxylate forms a key intermolecular salt bridge with an Arg side chain located on the rim of the BH3-recognition cleft. Our data revealed that neither β-peptide helices nor α/β-peptide helices resulting from a 1:1 α:β pattern are sufficiently faithful mimics of an α-helix to generate high-affinity ligands for Bcl-xL.19,20 α/β-Peptides with smaller β residue proportions, however, proved to be very effective.21-23 For example, homologues of an 18-residue Bim BH3 α-peptide containing α→β3 substitutions in three regular patterns, ααβ αααβ or ααβαααβ, which lead to α/β-peptides containing 25% to 33% β residues, displayed significant affinity for Bcl-xL, Mcl-1 or both (the Bim BH3 domain itself binds to both Bcl-xL and Mcl-1).23 This type of α/β-peptide retains the full complement of side chains relative to the prototype α-peptide, but the backbone contains an extra CH2 unit at the site of each α→β3 replacement (Figure 2). The regular occurrence of β residues along the peptidic backbone usually renders these α/β-peptides much less susceptible to proteolytic cleavage than are homologous α-peptides.15
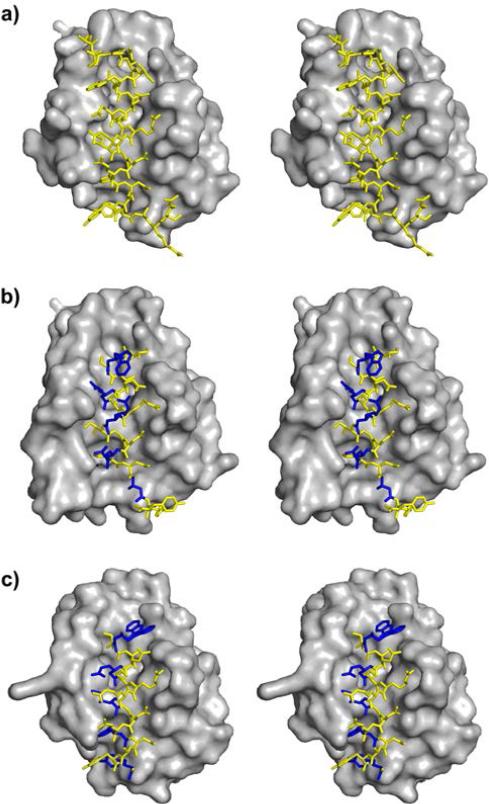
Figure 1.
Comparison of previously reported crystal structures of Bcl-xL bound to each of three BH3-derived peptides (stereo views): (A) 26-residue α-peptide derived from the Bim BH3 domain (PDB 3FDL); (B) 18-residue α/β-peptide B (PDB 4A1U); (C) 18-residue α/β-peptide C (PDB 4A1W).
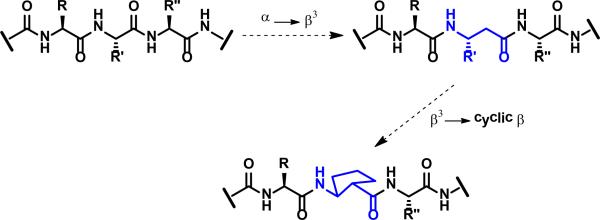
Figure 2.
Illustration of partial α→β3 substitution (step 1), and β3→cyclic β substitution (step 2) starting from an ααα segment and generating an αβα segment.
Crystallographic data demonstrate that α/β-peptides generated via periodic α→β3 substitution, in the ααβ, αααβ or ααβαααβ pattern, can adopt helical conformations that are very similar to an authentic α-helix, despite the presence of at least one additional CH2 unit per helical turn relative to a pure α-peptide backbone (Figure 1B,C).24,25 However, each α→β3 substitution introduces an additional flexible backbone bond relative to the prototype α-peptide; therefore, the energetic cost of helix formation by α/β-peptides generated in this way should be larger than for α-helix formation by homologous α-peptides.26-28 This anticipated difference in helix stability may explain why the affinities for Bcl-xL or Mcl-1 of α/β3 18-mer homologues are uniformly lower than the affinity of the Bim BH3 18-mer α-peptide itself.23
β-Amino acid residues offer opportunities for conformational preorganization that have no parallel among α-amino acid residues, because a ring can be used to constrain the β residue without eliminating a backbone H-bonding site.16 In contrast, ring-based preorganization of an α residue comes at the expense of the H-bond donor site, as illustrated by proline. We have previously shown that β3→cyclic β residue replacements can enhance the affinity of an α-helix-mimetic α/β-peptide for a complementary protein surface when the β-amino acid residues bear a five-membered ring constraint and the amino and carboxyl groups are trans (Figure 3); this earlier work involved 38-residue α/β-peptides that mimic the CHR domain of HIV protein gp41.29,30 Complementary work from Reinert and Horne has demonstrated comparable stabilization effects from β3→cyclic β replacements in an α-helix within a defined tertiary structure.27,28
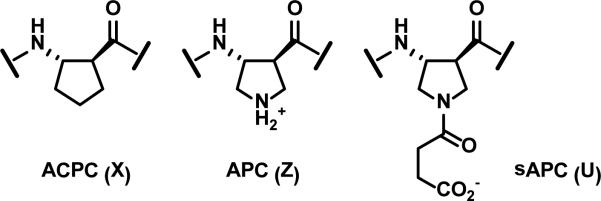
Figure 3.
Cyclic β residues that promote an α-helix-like conformation. ACPC and APC were previously described, and sAPC is introduced in the present study.
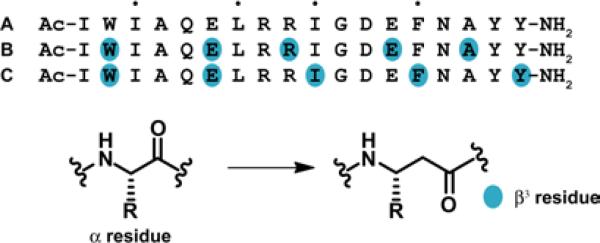
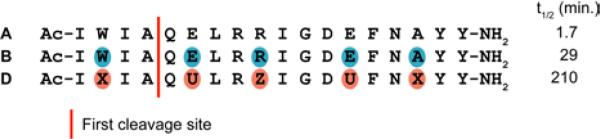
In the present study, we examine the impact of β3→cyclic β residue replacements on the affinities for Bcl-xL and Mcl-1 of α/β-peptides derived from an 18-mer Bim BH3 α-peptide (A; Figure 4).23 This BH3 mimicry testbed is more versatile than the gp41 CHR system for evaluation of alternative α/β-peptide designs because our Bim BH3 sequence is much shorter than the gp41 CHR sequence (18 vs. 38 residues). We have previously conducted a comprehensive survey of the ααβ3, αααβ3 and ααβ3αααβ3 registries (14 α/β variants in total) in terms of binding to both Bcl-xL and Mcl-1;23 in contrast, only a few ααβαααβ registries have been evaluated for mimicry of the much longer gp41 CHR domain.29 The comprehensive survey identified two α/β-peptides that retained the ability of the native Bim BH3 domain to bind to both Bcl-xL and Mcl-1. One of these dual-binding α/β-peptides featured the ααβ3αααβ3 pattern (B), and the other featured the αααβ3 pattern (C). The ααβ3αααβ3 pattern causes the β3 residues to align in a “stripe” upon formation of an α-helix-like conformation, while the αααβ3 pattern causes the β3 residues to spiral around the helix axis. Crystal structures of α/β-peptide+Bcl-xL complexes revealed all of the β3 residues of B to be oriented toward solvent (Figure 1B), while for C, side chains from two β3 residues make critical contacts with the BH3-recognition cleft on the protein (Figure 1C).23 (The small red dots over the sequence shown for A in Figure 4 indicate the positions of the four key hydrophobic side chains that are essential features of BH3 domains.)
Figure 4.
Bim BH3-derived 18-mer α-peptide and selected α/β3 analogues. The dots indicate the four hydrophobic residues that are crucial for binding to antiapoptotic Bcl-2 family proteins.
Replacing an α residue with its β3 homologue is ‘automatic’ because the side chain is defined, but β3→cyclic β replacement requires careful design if the constrained residue is to mimic physicochemical properties of the original β3 residue. Our previous work has been based on just two cyclic β residues, one designated ACPC (Figure 3), which is appropriate for positions that originally had β3 residues with hydrophobic side chains, and another designated APC, which can be used to replace basic residues, β3-hArg or β3-hLys. The experiments described here introduce a new cyclic residue, designated “sAPC” (for “succinyl-APC”), which provides an acidic side chain and can therefore be used to replace β3-hGlu or β3-hAsp.
The BH3 domain mimicry testbed allows us to monitor variations in the responses of different members of the Bcl-2 protein family to specific β3→cyclic β replacement patterns in α/β-peptide binding partners. The data below are interpreted on the assumption that the new α/β-peptides retain the BH3 domain-like helical conformation and binding site established crystallographically for B and C, although altering a ligands structure can lead to changes in binding mode.31 Experiments based on α/β-peptide B involve replacements at sites that are exclusively solvent-exposed upon complex formation. In contrast, experiments based on C explore β3→cyclic β replacements at residues that make direct contact with the partner protein. Since the cyclic β residue cannot perfectly reproduce the steric qualities of the original side chain, replacements at direct contact positions might be highly deleterious to binding. Our data indicate that β3→cyclic β residue replacements at solvent-exposed sites generally improve α/β-peptide affinity for both Bcl-xL and Mcl-1, relative to the analogues that contain exclusively β3 residues. At sites expected to make direct contact, however, β3→cyclic β replacement elicits surprising protein-dependent responses, with high selectivity for Bcl-xL relative to Mcl-1 or vice versa.
Results
Development of a constrained β-amino acid with an acidic side chain
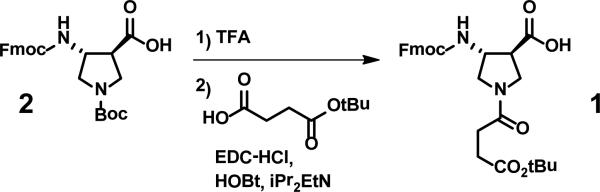
Figure 5 summarizes the preparation of building block 1, which allows incorporation of sAPC residues into α/β-peptides via Fmoc-based solid-phase synthesis. Starting material 2 is used for incorporation of APC residues via solid-phase synthesis; the preparation of this compound in stereoisomerically pure form has been previously described.33 The subunit incorporated during solid-phase peptide synthesis via use of 1 bears a t-butyl ester in the side chain, which is stable during subsequent cycles of Fmoc deprotection and amide bond formation. The t-butyl protecting group is removed along with other side chain protecting groups under the acidic conditions used to detach the polypeptide from the solid support.
Figure 5.
Syntheis of building block 1 for incorporation of sAPC residues via solid-phase methodology, from known APC derivative 2.
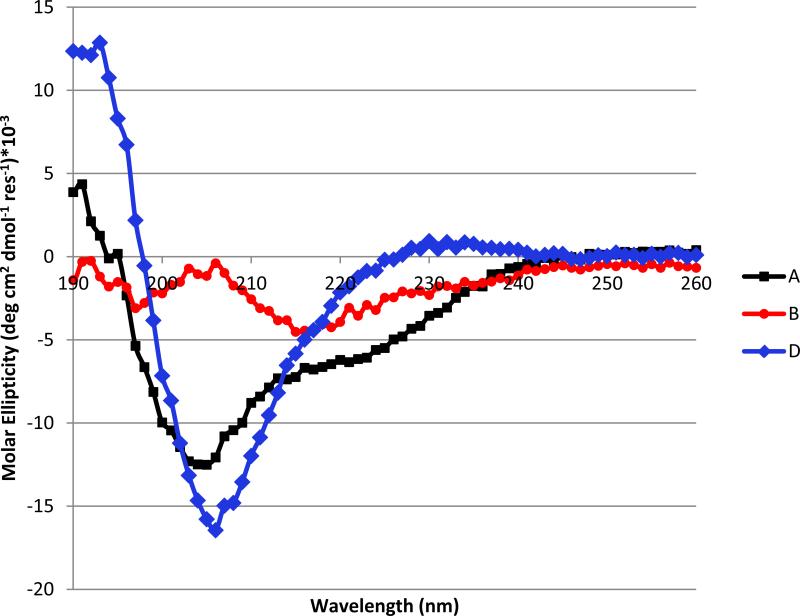
α/β-Peptide D (Figure 6) is the analogue of B in which each β3 residue has been replaced with an appropriate cyclic β residue, including two β3-hGlu→sAPC replacements. Preparation of D proceeded smoothly, which indicates that building block 1 is well-suited for solid-phase synthesis. Figure 7 compares far-UV circular dichroism (CD) data for α-peptide A with CD data for α/β-peptides B and D in aqueous buffer. The data for A, with a minimum at ~208 nm and a shoulder near 220 nm, are consistent with partial α-helix formation, which is common for α-peptides in this length range. Previous work has shown that formation of an α-helix-like conformation by α/β-peptides leads to a single CD minimum at ~208 nm.24 α/β-Peptide D manifests a strong minimum at this characteristic position, suggesting significant population of the helical state. In contrast, α/β-peptide B shows no evidence of helicity. The conformational difference between these two α/β-peptides presumably arises from the stronger local helical propensity of the ring-constrained β residues in D relative to the flexible β3 residues in B.26-28
Figure 6.
Analogues of α/β3-peptide B that contain exclusively cyclic β residues. The arrows indicate two site at which α residues differ between D and D*. Cyclic β residues U, X and Z are defined in Figure 3.
Figure 7.
Circular dichroism data for α/β-peptides A, B and D, 50 μM each in 10 mM phosphate pH 7.
Binding of BH3-mimetic α/β-peptides to Bcl-xL and Mcl-1
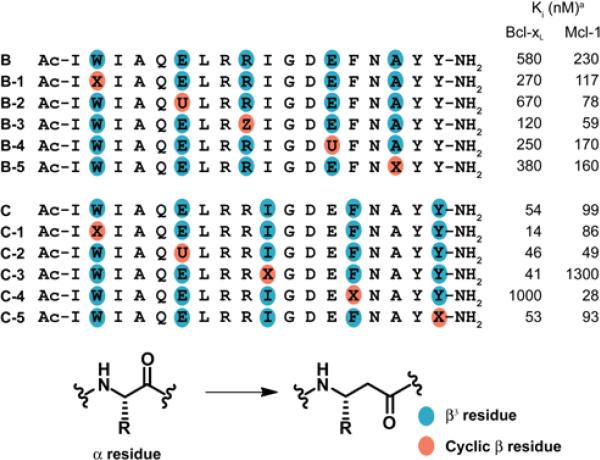
For initial assessment of the impact of replacing β3 residues with cyclic analogues in BH3-mimetic α/β-peptides, we prepared the five derivatives of B in which a single β3 residue was replaced and the five derivatives of C in which a single β3 residue was replaced (Table 1). In each case, the cyclic residue was selected to mimic the properties of the side chain of the original β3 residue, i.e., ACPC (X) was used for hydrophobic side chains, APC (Z) for basic side chains and sAPC (U) for acidic side chains. Binding of these new α/β-peptides to Bcl-xL or Mcl-1 was evaluated with previously described competition fluorescence-polarization (FP) assays.33 Among the derivatives of B (ααβαααβ pattern), each single β3→cyclic β replacement has only a modest effect on affinity for either Bcl-xL or Mcl-1. Individual cyclic replacements are uniformly favorable in terms of binding to Mcl-1. Most replacements are moderately favorable in terms of binding to Bcl-xL, but β3-hGlu-6→sAPC (B-2) is slightly unfavorable.
Table 1.
Effects of Single β3 to cyclic β residue modification of Bim-derived α/β-peptides on binding to anti-apoptotic proteins.
Ki values obtained from competition fluorescence polarization measurements (experimental uncertainty ~2-fold). Previous studies, involving the same FP assays, determined that for α-peptide A, Ki for Bcl-xL = 23 nM, and Ki for Mcl-1 ≤ 2.6 nM (see ref. 23, Table S1). Cyclic β residues U, X and Z are defined in Figure 3.
For single β3→cyclic β replacement derivatives of C (αααβ pattern) involving sites that do not make intimate contacts with the partner protein, small and favorable effects on binding to Bcl-xL and Mcl-1 are observed, as for most single replacements in the context of B. However, β3→cyclic β replacements at the two sites in C that make direct contacts with partner proteins lead to larger and more selective effects. β3-hIle-10→ACPC (C-3) causes a substantial decline in affinity Mcl-1 but has little impact on affinity for Bcl-xL. In contrast, β3-hPhe-14→ACPC (C-4) causes a substantial decline in affinity for Bcl-xL but modestly improves affinity for Mcl-1. In the native Bim BH3 domain, the residues corresponding to β3-hIle-10 and β3-hPhe-14 contribute two of the four crucial hydrophobic side chains to the interface formed with a Bcl-2 family partner. The previously reported crystal structure of α/β-peptide C bound to Bcl-xL shows that the side chains of both β3-hIle-10 and β3-hPhe-14 are buried within the protein's BH3-recognition cleft, as expected (Figure 1C).23 The divergent responses of Bcl-xL and Mcl-1 to β3→cyclic β replacements at these sites suggest that the recognition pockets of these structurally and functionally related proteins differ in their capacity to accommodate local changes in side chain geometry within the α/β-peptide ligands. The pocket that accepts the side chain of β3-hIle-10 seems to be very discriminating in Bcl-xL but less so in Mcl-1, and the situation is reversed for the pocket that accepts the side chain of β3-hPhe-14. The intriguing results of β3→cyclic β replacements at contact residues 10 and 14 could not have been predicted.
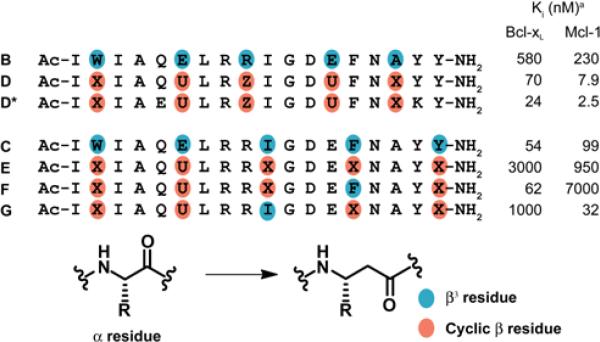
Table 2 provides competition FP assay results for several α/β-peptides that contain multiple β3→cyclic β replacements. Two derivatives were examined for the ααβαααβ pattern of B. α/β-Peptide D, introduced above, contains β3→cyclic β replacements at all five sites. Analogue D* contains the same set of cyclic β residues and two variations among the α residues, Gln-5→Glu and Tyr-17→Lys. The α residue modifications in D* relative to D are based on a previous “hydrophile scan” analysis of the Bim BH3 18-mer sequence,33 which revealed that Gln-5→Glu and Tyr-17→Lys modestly increase α-peptide affinity for both Bcl-xL and Mcl-1. Analogous increases were observed when these two changes were made in α/β3-peptide B.34
Table 2.
Effects of multiple β3 to cyclic β residue modification of Bim-derived α/β-peptides on binding to anti-apoptotic proteins.
Ki values obtained from competition fluorescence polarization measurements (experimental uncertainty ~2-fold). Cyclic β residues U, X and Z are defined in Figure 3.
The FP data for D (Table 2) indicate that replacing all β3 residues with cyclic analogues leads to higher affinity for Bcl-xL and for Mcl-1 than was observed for any of the α/β-peptides containing just a single cyclic residue (B-1 to B-5, Table 1). Small additional improvements are seen for α/β-peptide D* relative to D. α/β-Peptide D* is comparable to the Bim BH3 α-peptide 18-mer (A) in affinity for Bcl-xL and Mcl-1; D* binds moderately more tightly to Bcl-xL and slightly more weakly to Mcl-1 relative to A.
Comprehensive replacement of the β3 residues in C with cyclic residues, to generate E (Table 2) leads to substantial reduction in affinity for both proteins. This result can be rationalized based on data in Table 1, which show that binding to Bcl-xL is diminished by ACPC replacement for β3-hIle-10 (C-3), and binding to Mcl-1 is diminished by ACPC replacement for β3-hPhe-14 (C-4). We therefore examined α/β-peptides F and G, which each contain only four β3→cyclic β replacements relative to C; F retains β3-hIle-10 and G retains β3-hPhe-14. These α/β-peptides manifest the expected qualitative preferences based on results in Table 1, with F binding preferentially to Bcl-xL and G binding preferentially to Mcl-1. It is noteworthy that F binds to Bcl-xL with >100-fold selectivity relative to Mcl-1, which stands in contrast to the >100-fold selectivity of native Bim BH3 18-mer A for Mcl-1 relative to Bcl-xL.23
We conducted competition surface plasmon resonance (SPR) measurements to evaluate the binding of α/β-peptides B, D and D* to Bcl-xL and Mcl-1, as a complement to FP measurements. Previous SPR studies showed that B binds moderately to Bcl-xL and Mcl-1, which is consistent with FP results; α/β-peptide B was re-analyzed alongside D and D* to allow direct comparison. The results (Table 3) are consistent with the conclusion drawn from FP assays (Table 2) in that the α/β-peptides containing cyclic residues, D and D*, bind to both pro-survival proteins substantially more tightly than does B, the analogue containing β3 residues. In addition, the SPR data suggest a small increase in Mcl-1 affinity for D* relative to D.
Table 4.
Susceptibility of the Bim BH3 domain and selected α/β analogues to degradation by proteinase K.a
Cyclic β residues U, X and Z are defined in Figure 3.
Engagement of an apoptosis signaling network by BH3-mimetic α/β-peptides
Bcl-xL and Mcl-1, along with other members of the pro-survival Bcl-2 protein family, inhibit apoptosis by binding tightly to other family members, such as Bak and Bax, that can permeabilize mitochondrial membranes and thereby initiate the apoptotic signaling cascade. Wild-type mouse embryonic fibroblasts (MEFs) are protected from apoptosis by Bcl-xL and Mcl-1;35 therefore, molecules that bind tightly to both of these pro-survival proteins, such as α/β-peptides D and D*, should induce apoptotic signaling in this cell type by causing release of Bak and Bax. Conventional BH3 domain α-peptides and analogous α/β-peptides do not spontaneously cross cell membranes. However, the ability of such peptides to engage the MEF apoptotic control network can be assessed with well-established assays involving cells pre-treated with digitonin, which permeabilizes the plasma membrane but does not damage the mitochondrial membrane.20,23
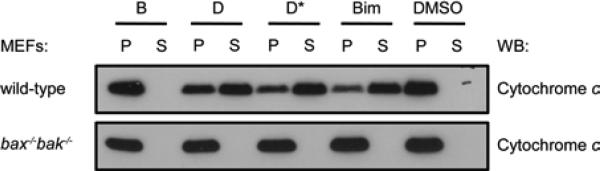
Results obtained with wild-type and Bax/Bak-doubly deficient MEFs, after permeabilization, are summarized in Figure 8. A key step in the early stages of apoptosis signaling is the release of cytochrome c from mitochondria, a process that is mediated by proapoptotic proteins such as Bax and Bak. Thus, cells deficient in Bax and Bak do not possess the machinery to permeabilize the outer mitochondrial membrane, and these cells serve as negative controls that enable us to detect any non-specific effects that the peptides might exert on the mitochondrial membrane. Cytochrome c is normally not found in the cytoplasm, and this protein is therefore undetectable by western blot analysis in the soluble fraction from permeabilized MEFs that have not been treated with any of the peptides (see “DMSO” data at the right side of Figure 8; DMSO was the solvent used to prepare peptide stock solutions). However, when permeabilized MEFs are treated with Bim BH3 domain 18-mer A (10 μM), cytochrome c appears in the cytoplasm. Comparable cytochrome c release is observed upon treatment with α/β-peptide D or D*. In contrast, as previously reported, treatment with B does not lead to cytochrome C release from mitochondria; in this case, all of the cytochrome c remains in the insoluble fraction, which contains the mitochondria. The lack of cytochrome c release for B is presumably explained by inadequate affinity of this α/β-peptide for Bcl-xL and Mcl-1. α/β-Peptides D and D* can induce cytochrome c release because of their tighter binding to both pro-survival proteins, which enables them to displace Bak and Bax. As predicted, neither D nor D* induces cytochrome c release from permeabilized MEFs derived from embryos in which the bak and bax genes have been knocked out (Figure 8). This control study supports our conclusion that α/β-peptides D and D* induce cytochrome c release via a mechanism that requires Bak or Bax, rather than through a non-specific disruption of the mitochondrial membrane.
Figure 8.
Cytochrome c release assay. α/β-peptides D and D*, but not B, elicit cytochrome c release from mitochondria into the cytosol of wild-type but not bax/bak-deficient MEFs. Peptide Bim, which represents the native Bim BH3 domain 26-mer, was included as a positive control. Each peptide was used at 10 μM. P: pellet fraction containing mitochondria, S: soluble fraction containing cytosol. DMSO was used as the solvent for all peptide stock solutions.
Proteolytic susceptibilities
We have previously shown that α/β-peptides containing ≥ 25% β residues uniformly distributed along the sequence tend to be much poorer substrates for proteases than are comparable peptides comprised exclusively of α residues.22,23,29,30 Table 4 compares the effect of proteinase K, an aggressive and relatively non-sequence-selective enzyme, on three molecules: (1) the Bim BH3 18-mer (A); (2) α/β analogue B, which contains exclusively β3 residues; and (3) α/β analogue D, which contains exclusively cyclic β residues. Periodic α→β3 replacement significantly hinders proteinase K activity, as indicated by the 17-fold greater half-life of α/β3-peptide B relative to α-peptide A. However, cyclic β residues provide stronger protection from proteolysis than do β3 residues: the half-life of D is 120-fold greater than that of α-peptide A and 7-fold greater than that of α/β3-peptide B. The enhanced protection conferred by cyclic relative to acyclic β residues is consistent with previous observations in a different sequence context29,30 and presumably arises from differences in conformational propensities. Proteases generally bind substrates in extended conformations, while the cyclic β residues strongly favor helical conformations.
Discussion
We previously explored the effects of β3→cyclic β replacements in the context of α/β-peptide inhibitors of HIV infection.29,30 This activity required mimicry of a long α-helix formed by the CHR domain of viral protein gp41. Fusion of the viral envelope with the target cell membrane, essential for HIV propagation, is mediated by gp41,36,37 and conventional CHR-derived peptides block fusion by interfering with helix-helix interactions within gp41 trimers.38 Efforts to optimize CHR-mimetic α/β-peptides revealed that replacing β3 residues with cyclic analogues enhanced affinity for the targeted protein surface.29,30 The BH3 domain mimicry testbed has now enabled us to test the generality of this beneficial effect and to broaden our evaluation of the impact of β3→cyclic β replacements. The gp41-based efforts were limited in scope because each candidate contained 38 residues, which prevented broad exploration of alternative α/β patterns. Thus, only the ααβαααβ backbone pattern was considered, and only registries that oriented the β residue ‘stripe’ toward the solvent were evaluated. This α/β arrangement is analogous to that in Bim BH3 analogue B. As with the Bim BH3 domain, the gp41 CHR-derived sequence we used contained hydrophobic, basic and acidic side chains at the positions selected for α→β substitution; however, we could evaluate β3→cyclic β replacements at only hydrophobic and basic sites because a cyclic β residue bearing an acidic side chain had not yet been developed.
The Bim BH3-based studies reported here complement the gp41 CHR-based findings in several important ways. First, we have now generated a β residue with the ring constraint appropriate for α-helix mimicry that contains an acidic side chain (sAPC), and we have shown that this residue can be used to generate α/β-peptides that bind tightly to a protein partner. Second, we have evaluated all possible single-site β3→cyclic β replacements in the context of the ααβαααβ pattern of Bim BH3 analogue B. These individual replacements (B-1 to B-5) almost always improve binding to protein partners Bcl-xL or Mcl-1, but the effects are generally modest. Global β3→cyclic β replacement (D), generates a very effective ligand for both proteins, as demonstrated not only by binding experiments (FP and SPR assays) but also by the ability of D to engage the apoptotic signaling network in permeabilized MEFs. Third, we have shown that replacing β3 residues with cyclic analogues enhances resistance to enzymatic degradation. In concert with previous findings in the gp41 CHR system,29,30 this observation suggests that proteolytic stabilization is a general benefit of β3→cyclic β replacement.
The Bim BH3 domain testbed has enabled the unexpected discovery that β3→cyclic β replacement at sites making direct contact with partner proteins can lead to highly selective ligands. Although α/β3-peptide C binds with comparable affinities to Bcl-xL and Mcl-1, β3→ACPC replacement at either of the positions that makes direct contact with the partner protein, β3-hIle-10 or β3-hPhe-14 (C-3 or C-4), generates a highly selective ligand. The binding preferences of C-3 and C-4 are complementary, with ACPC at position 10 causing >30-fold selectivity for Bcl-xL and ACPC at position 14 causing >30-fold selectivity for Mcl-1. These results suggest that β3→cyclic β replacement at different protein-contact sites has identified a subtle distinction between Bcl-xL and Mcl-1 in terms of local adaptability within these proteins’ BH3-recognition grooves. Such variation is not evident from comparison of conventional structural data (e.g., crystal structures). Molecules with strong binding preferences among related proteins can be very useful from a biomedical perspective,39,40 and observations in the BH3 domain mimicry testbed encourage future studies to determine whether β3→cyclic β replacements at contact sites lead to comparable selectivities within other protein families. Our observations regarding Bcl-xL vs. Mcl-1 selectivity complement other recent reports of comparable selectivity in within different ligand families.41,42
Conclusions
Our results broaden understanding of an emerging methodology for α-helix mimicry and strengthen the prospect that this approach will prove to be of general utility. The strategy is based on peptidic oligomers that have unnatural backbones in which some residues are derived from β-amino acids. These α/β-peptides can be readily prepared via conventional solid-phase synthesis; many of the necessary β-amino acid building blocks are commercially available. Thus, it is straightforward to prepare sets of α/β-peptides based on a prototype α-helix-forming sequence via α→β3 replacement in patterns such as ααβαααβ or αααβ, and to evaluate these α/β-peptides for functional α-helix mimicry. The data provided here indicate that the α-helix-mimetic properties α/β3-peptides can be generally improved via replacement of some or all β3 residues, which are inherently flexible, with analogues containing a five-membered ring constraint and bearing appropriate side chain functionality. In addition to providing high affinity for protein partners, β3→cyclic β replacements can confer binding selectivity among related proteins. α/β-Peptides containing cyclic β residues benefit from improved helical stability relative to analogous α/β3-peptides and from decreased susceptibility to proteolysis relative the prototype α-helix-forming peptide and analogous α/β3-peptides. This combination of properties makes α/β-peptides containing cyclic β residues attractive for biological applications.
Methods
Materials
Fmoc-L-α-amino acids, O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU), and NovaPEG Rink amide resin were purchased from NovaBiochem (San Diego, CA). Fmoc-L-β-amino acids were purchased from Peptech (Burlington, MA). 6-((4,4-Difluoro-1,3-dimethyl-5-(4-methoxyphenyl)-4-bora-3a,4a-diaza-s-indacene-2-propionyl)amino)hexanoic acid, succinimidyl ester (BODIPY-TMR-X-SE) was purchased from Invitrogen. Piperidine, 1-hydroxybenzotriazole hydrate (HOBt), trifluoroacetic acid (TFA), HPLC-grade acetonitrile (MeCN), dimethylformamide (DMF), dichloromethane (DCM), and all other chemical reagents were purchased from Sigma-Aldrich (Milwaukee, WI) or Fisher Scientific (Pittsburgh, PA).
Fmoc-APC(t-butylsuccinyl)-OH (1)
(1S,2S)-Fmoc-APC(Boc)-OH43 (2) (298 mg, 0.66 mmol) was dissolved in 6.6 mL 25 vol %TFA in DCM without a stir bar in the flask. The reaction was allowed to proceed at r.t., with periodic flask agitation, for 4 h. TFA and DCM were removed by evaporation under a stream of N2 in a hood. The oily solid residue was dissolved in 5 mL DCM, and the solvent was evaporated again in a hood under a stream of N2. The product was dissolved in 3 × 5 mL DCM, each time removing solvent under rotary evaporation, until the brownish oil bubbled under vacuum during rotary evaporation (indicating that the TFA had been removed).
While DCM from the previous step was removed, the following materials were combined in 3 mL DCM: mono-tert-butylsuccinate44 (162.2 mg, 0.72 mmol), EDC·HCl (139.1 mg, 0.73 mmol), and HOBt (100 mg, 0.74 mmol). This solution was cooled to 0°C in an ice bath. Boc-deprotected Fmoc-APC (preceding paragraph) was dissolved in 3 mL DCM, cooled to 0°C, and 346 μL diisopropylethlamine (DIEA) was added. The preactivated succinate solution was transferred to the deprotected APC solution via syringe, and the reaction mixture was allowed to sit in the ice bath stirring overnight, during which time the mixture warmed to room temperature. The solvent was removed under rotary evaporation, and the residue was dissolved in 50 mL EtOAc. The organic layer was extracted with 2 × 50 mL 5% NaHSO4 and 1 × 50 mL brine. The organic layer was dried with MgSO4 and filtered, and the filtrate was concentrated. The residue was purified by column chromatography (column loaded in 1% MeOH/DCM + 1% HOAc, eluted with 1-4% MeOH/DCM + 1% HOAc). The fractions containing the product were combined, toluene was added to form an azeotrope with HOAc, and the solvents were removed under rotary evaporation. The oily product was dissolved in a small amount of EtOAc and precipitated with pentane. A white solid (173 mg, 51%) was obtained. Rf = 0.18, 4% MeOH/DCM + 1% HOAc. MS-ESI: m/z = 531.2102 (M+Na)+. mp 166-167°C; 1H-NMR (CD3OD, 300 MHz) δ 7.80 (d, JHH 7.2 Hz, 2H), 7.65 (d, JHH 7.5 Hz, 2H), 7.39 (t, JHH 7.4 Hz, 2H), 7.31 (t, JHH 7.4 Hz, 2H), 4.49-4.33 (m, 3H), 4.22 (t, JHH 6.4, 1H), 3.90-3.63 (m, 3H), 3.43-3.380 (m, 1H), 3.17-3.00 (m, 1H), 2.55 (s, 2H), 2.52 (s, 2H), 1.45 (s, 9H); 13C NMR (CD3OD, 75.4 MHz, 24 °C) δ172.81, 172.75, 171.531, 156.97, 144.10, 144.026, 141.448, 127.61, 126.97, 124.99, 124.92, 119.75, 80.58, 66.50, 53.79, 52.51, 50.87, 49.98, 31.05, 30.03, 29.67, 28.37.
Peptide Synthesis
Peptides were synthesized using standard Fmoc-solid phase methods in 4.0-mL solid-phase extraction tubes from Alltech (Deerfield, IL) on NovaPEG Rink Amide resin, to afford C-terminal amides upon cleavage from the resin. Microwave irradiation was used as previously described43-45 to synthesize all Bim-derived α/β-peptides. Briefly, Fmoc-amino acids were activated with HBTU and HOBt in the presence of DIEA in NMP for coupling reactions. Fmoc deprotection was accomplished with 20% (v/v) piperidine in DMF. Acetylation of the N-terminus was accomplished after the final Fmoc deprotection with 8:2:1 DMF:DIEA:Ac2O at room temperature. Peptides were cleaved from the resin with 95% TFA, 2.5% H2O, and 2.5% triisopropylsilane (TIS), except peptides containing β3-hTrp, which required 81.5% TFA, 5% thioanisole, 5% phenol, 5% H2O, 2.5% ethanedithiol (EDT), and 1% TIS. After the TFA was evaporated under a stream of nitrogen, the crude α/β-peptide was dissolved/suspended in TFA and precipitated with cold ether. α/β-Peptides were purified using preparative, reverse-phase HPLC performed with a C4 or C18 column (Vydac, Anaheim, CA) and eluting with gradients of MeCN w/0.1% TFA (B solvent) in water w/0.1% TFA (A solvent). MALDI-TOF mass spectrometry was used to establish α/β-peptide identity. The purity of the α/β-peptides was assessed by analytical HPLC; in all cases, purity was ≥ 95%.
All α/β-peptides to be tested for binding to a Bcl-2 family protein were dissolved in DMSO. The concentration of the DMSO stock solution was measured by UV spectroscopy, where the molar extinction coefficient at 280 nm for each α/β-peptide was calculated based on the chromophores present (Trp or Tyr).43 α/β-Peptides with a single tryptophan were predicted to have an extinction coefficient of 5690 M−1cm−1, and α/β-peptides with a single tyrosine were predicted to have an extinction coefficient of 1280 M−1cm−1.48 α/β-Peptides containing more than one chromophore were predicted to have an extinction coefficient corresponding to the sum of the single chromophore extinction coefficients.
Competition fluorescence polarization (FP) assays
Expression and purification of the proteins Bcl-xL and Mcl-1 were performed as previously described.49 FP assays were performed in a 384-well black polystyrene plate. A BODIPYTMR-Bak tracer peptide (Kd = 1.2 nM)49,50 was used for Bcl-xL binding assays, and a fluorescein-Bim tracer for Mcl-1 binding assays (Kd = 1.4 nM).51 Kd was recalculated for each new synthesis of tracers and proteins to account for slight variations between preparations. For Bcl-xL binding assays, wells of a 384-well plate contained 3 nM BODIPYTMR-Bak tracer, 2 nM Bcl-xL protein, and 2 μL DMSO solution of α/β-peptide (final concentration from 4.2 pM to 25 μM) in a final volume of 50 μL in FP Buffer (50 mM NaCl, 16.2 mM Na2HPO4, 3.8 mM KH2PO4, 0.15 mM NaN3, 0.15 mM EDTA, 0.5 mg/mL Pluronic; pH 7.4).52 For Mcl-1 binding assays, wells of a 384 plate contained 10 nM Flu-Bim tracer, 10 nM Mcl-1 protein, and 2 μL DMSO solution of α/β-peptide (final concentration from 4.2 pM to 25 μM) in a final volume of 50 μL in FP Buffer. Plates were read after a 5 h incubation at room temperature, the time necessary for complete equilibration. Experiments were performed in duplicate. The equilibrium dissociation constant (Ki)52,53 or IC50 was calculated using GraphPad Prism.54
Circular dichroism
All CD data were acquired using an Aviv 420 circular dichroism spectrophotometer. Peptide solutions were prepared in 10 mM phosphate buffer, pH 7, and the concentration was determined by UV absorbance. Data were acquired at 20 °C with a step value of 1 nm from 260 to 190 nm and an averaging time of 5.0 s. A 0.1-mm path length cell was used for all spectra. We used data only for wavelengths at which the dynode voltage was < 400 V. Mean residue ellipticity (θ) was calculated using the following equation.
where δS = sample signal, δR = reference signal, n = # amides in the backbone, l = path length (in cm) and c = concentration in dmol*cm−3.
Proteolysis
Stock solutions of each peptide were prepared in a TBS solution, pH 7.5, with 10% DMSO (for solubility) at 100 μM peptide, as determined by UV absorbance. A 25 μg/mL stock solution of proteinase K was prepared in TBS. For each proteolysis reaction, 25 μL of peptide stock solution was mixed with 15 μL TBS. A 10 μL aliquot of proteinase K stock solution was added to the mixture, and the reaction was allowed to proceed at room temperature. A 100 μL aliquot of 1% TFA in 50:50 acetonitrile/H2O was added to quench the reaction at the desired time point. A 125 μL aliquot of the resulting solution was injected onto an analytical reverse-phase HPLC column, and the amount of full-length peptide remaining was quantified using the absorbance at 220 nm of this peptide. Duplicate reactions were run for each time point. Half-life values were determined by plotting the percent remaining peptide versus time and fitting the data to an exponential decay using GraphPad Prism. Amide bond cleavage sites were identified by MALDI-TOF-MS analysis of crude reaction mixtures at various time points.
Surface plasmon resonance
All recombinant pro-survival proteins used for binding studies, which have N- and/or C-terminal truncations (Bcl-2ΔC22, Bcl-xLΔC24, Bcl-w C29S/A128EΔC29, Mcl-1ΔN170ΔC23), were expressed and purified exactly as described previously.55,56 SPR competition assays were performed using a Biacore 3000 instrument exactly as described previously.57 Briefly, pro-survival proteins were incubated with α/β-peptides for 2 hr prior to the solution being passed over a CM5 chip on which was immobilized either a wild-type 26-mer Bim BH3 peptide (DMRPEIWIAQELRRIGDEFNAYYARR) or an inert quadruple-variant peptide (Bim4E: DMRPEIWEAQEERREGDEENAYYARR; note that the four key hydrophobic residues necessary for binding to pro-survival proteins have been changed to glutamic acid residues). The signal from the Bim4E channel was subtracted from the signal from the wild-type channel to provide the binding fraction that arises from specific BH3-mediated binding.
Cytochrome c release assays
The cytochrome c assay was performed as described previously.57 Briefly, wild-type or bax−/−/bak−/− MEFs were permeabilized in digitonin-containing buffer (20 mM HEPES pH 7.2, 100 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1250 mM sucrose and 0.05% (w/v) digitonin) and then incubated with peptides (10 μM, dissolved in DMSO) for 1 hour at 30°C before pelleting via centrifugation. The supernatant was retained (soluble fraction), and the pellet was lysed in Triton-X100-containing buffer (20 mM Tris pH 7.4, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 10% (v/v) glycerol, 1% (v/v) Triton-X100 and protease inhibitors) to generate the pellet fraction. Both soluble and pellet fractions were analyzed for cytochrome c by Western blotting using an anti-cytochrome c antibody (clone 7H8.2C12; BD Biosciences).
Supplementary Material
Table 3.
| B | D | D* | |
|---|---|---|---|
| Bcl-xL | 1470 (370) | 98 (7) | 80 (13) |
| Mcl-1 | 151 (21) | 19 (3) | 11 (2) |
Competition SPR assays for relative binding of α/β-peptides to Bcl-xL or Mcl-1. Standard deviations were derived from two or three measurements.
Acknowledgements
This work was supported by the NIH (GM056414) and grants and fellowships from the NHMRC of Australia (Project Grant 1041936 to W.D.F. and Career Development Fellowship 1024620 to E.F.L). Infrastructure support from NHMRC IRIISS grant #361646 and the Victorian State Government OIS grant is gratefully acknowledged. K. J. P.-K. was supported in part by an Chemistry-Biology Training Grant from NIGMS (T32 GM008505). We thank W. Seth Horne and Lindsay Fay for assistance with Mcl-1 expression, and James Checco and Ross Cheloha for assistance with graphics.
Footnotes
Supporting Information. Supplementary Tables, Supplementary Figures, and further details of peptide characterization are available free of charge at http://pubs.acs.org
References
- 1.Jochim AL, Arora PS. Assessment of helical interfaces in protein-protein interactions. Mol. Biosyst. 2009;5:924. doi: 10.1039/b903202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jochim AL, Arora PS. Systematic analysis of helical protein interfaces reveals targets for synthetic inhibitors. ACS Chem. Biol. 2010;5:919. doi: 10.1021/cb1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullock BN, Jochim AL, Arora PS. Assessing helical protein interfaces for inhibitor design. J. Am. Chem. Soc. 2011;133:14220. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of α-helix-mediated protein-protein interactions using designed molecules. Nat. Chem. 2013;5:161. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- 5.Yin H, Lee GI, Park HS, Payne GA, Rodriguez JM, Sebti SM, Hamilton AD. Terphenyl-based helical mimetics that disrupt the p53/HDM2 interaction. Angew. Chem. Int. Ed. 2005;44:2704–2707. doi: 10.1002/anie.200462316. [DOI] [PubMed] [Google Scholar]
- 6.Plante JP, Burnley T, Malkova B, Webb ME, Warriner SL, Edwards TA, Wilson AJ. Oligobenzamide proteomimetic inhibitors of the p53-hDM2 protein-protein interaction. Chem. Comm. 2009:5091–5093. doi: 10.1039/b908207g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee T-K, Ahn J-M. Solid-phase synthesis of tris-benzamides as α-helix mimetics. ACS Comb. Sci. 2010;13:107–111. doi: 10.1021/co100056c. [DOI] [PubMed] [Google Scholar]
- 8.Whitby LR, Boger DL. Comprehensive peptidomimetic libraries targeting protein-protein interactions. Acc. Chem. Res. 2012;45:1698–1709. doi: 10.1021/ar300025n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara T, Durell SR, Myers MC, Appella DH. Probing the structural requirements of peptoids that inhibit HDM2-p53 interactions. J. Am. Chem. Soc. 2006;128:1995–2004. doi: 10.1021/ja056344c. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Pazgier M, Li J, Li C, Liu M, Zou G, Li Z, Chen J, Tarasov SG, Lu W-Y, Lu W. Limitations of peptide retro-inverso isomerization in molecular mimicry. J. Biol. Chem. 2010;285:19572. doi: 10.1074/jbc.M110.116814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown ZZ, Akula K, Arumanyan A, Alleva J, Jackson M, Bichenkov E, Sheffield JB, Feitelson MA, Schafmeister CE. A spiroligomer α-helix mimic that binds HDM2, penetrates human cells and stabilizes HDM2 in cell culture. PLoS One. 2012;7:e45948. doi: 10.1371/journal.pone.0045948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Qiao Q, Hu Y, Teng P, Gao W, Zuo X, Wojtas L, Larsen RW, Ma S, Cai J. Sulfono-γ-AApeptides as a new class of nonnatural helical foldamer. Chem. Eur. J. 2015 doi: 10.1002/chem.201406112. in press (DOI: 10.1002/chem.201406112) [DOI] [PubMed] [Google Scholar]
- 13.Werder M, Hauser H, Abele S, Seebach D. β-peptides as inhibitors of small-intestinal cholesterol and fat absorption. Helv. Chim. Acta. 1999;82:1774–1783. [Google Scholar]
- 14.Kritzer JA, Lear JD, Hodsdon ME, Schepartz A. Helical β-peptide inhibitors of the p53-hDM2 interaction. J. Am. Chem. Soc. 2004;126:9468–9469. doi: 10.1021/ja031625a. [DOI] [PubMed] [Google Scholar]
- 15.Johnson LM, Gellman SH. α-Helix Mimicry with α/β-Peptides. Meth. Enzymol. 2013;523:407. doi: 10.1016/B978-0-12-394292-0.00019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng RP, Gellman SH, DeGrado WF. β-Peptides: From Structure to Function. Chem. Rev. 2001;101:3219. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 17.Horne WS, Gellman SH. Foldamers with Heterogeneous Backbones. Acc. Chem. Res. 2008;41:1399. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petros AM, Olejniczak ET, Fesik SW. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Sadowsky JD, Schmitt MA, Lee H-S, Umezawa N, Wang S, Tomita Y, Gellman SH. Chimeric (α/β+α)-Peptide Ligands for the BH3-Recognition Cleft of Bcl-xL: Critical Role of the Molecular Scaffold in Protein Surface Recognition. J. Am. Chem. Soc. 2005;127:11966. doi: 10.1021/ja053678t. [DOI] [PubMed] [Google Scholar]
- 20.Sadowsky JD, Fairlie WD, Hadley EB, Lee H-S, Umezawa N, Nikolovska-Coleska Z, Wang S, Huang DCS, Tomita Y, Gellman SH. (α/β+α)-Peptide Antagonists of BH3 Domain/Bcl-xL Recognition: Toward General Strategies for Foldamer-Based Inhibition of Protein-Protein Interactions. J. Am. Chem. Soc. 2007;129:139. doi: 10.1021/ja0662523. [DOI] [PubMed] [Google Scholar]
- 21.Horne WS, Boersma MD, Windsor MA, Gellman SH. Sequence-based design of α/β-peptide foldamers that mimic BH3 domains. Angew. Chem. Int. Ed. 2008;47:2853–2856. doi: 10.1002/anie.200705315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee EF, Smith BJ, Horne WS, Mayer KN, Evangelista M, Colman PM, Gellman SH, Fairlie WD. Structural basis of Bcl-xL recognition by a BH3-mimetic α/β-peptide generated by sequence-based design. ChemBioChem. 2011;12:2025–2032. doi: 10.1002/cbic.201100314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boersma MD, Haase HS, Peterson-Kaufman KJ, Lee EF, Clarke OB, Colman PM, Smith BJ, Horne WS, Fairlie WD, Gellman SH. Evaluation of diverse α/β-backbone patterns for functional α-helix mimicry: analogues of the Bim BH3 domain. J. Am. Chem. Soc. 2012;134:315–323. doi: 10.1021/ja207148m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horne WS, Price JL, Gellman SH. Interplay among side chain sequence, backbone composition, and residue rigidification in polypeptide folding and assembly. Proc. Natl. Acad. Sci. U S A. 2008;105:9151–9156. doi: 10.1073/pnas.0801135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price JL, Horne WS, Gellman SH. Structural consequences of β-amino acid preorganization in a self-assembling α/β-peptide: fundamental studies of foldameric helix bundles. J. Am. Chem. Soc. 2010;132:12378–12387. doi: 10.1021/ja103543s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price JL, Hadley EB, Steinkruger JD, Gellman SH. Detection and Analysis of Chimeric Tertiary Structure via Backbone Thioester Exchange: Packing of an α-Helix against an α/β-Peptide Helix. Angew. Chem. Int. Ed. 2010;49:368. doi: 10.1002/anie.200904714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinert ZE, Horne WS. Folding thermodynamics of protein-like oligomers with heterogeneous backbones. Chem. Sci. 2014;5:3325. doi: 10.1039/C4SC01094A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinert ZE, Horne WS. Protein backbone engineering as a strategy to advance foldamers toward the frontier of protein-like tertiary structure. Org. Biomol. Chem. 2014;12:8796. doi: 10.1039/c4ob01769b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Structural and biological mimicry of protein surface recognition by α/β-peptide foldamers. Proc. Natl. Acad. Sci. U S A. 2009;106:14751–14756. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson LM, Mortenson DE, Yun HG, Horne WS, Ketas TJ, Lu M, Moore JP, Gellman SH. Enhancement of α-helix mimicry by an α/β-peptide foldamer via incorporation of a dense ionic side-chain array. J. Am. Chem. Soc. 2012;134:7317–7320. doi: 10.1021/ja302428d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wysoczanski P, Mart RJ, Loveridge EJ, Williams C, Whittaker SB-M, Crump MP, Allemann RK. NMR solution structure of a photoswitchable apoptosis activation Bak peptide bound to Bcl-xL. J. Am. Chem. Soc. 2012;134:7644–7647. doi: 10.1021/ja302390a. [DOI] [PubMed] [Google Scholar]
- 32.Lee H-S, LePlae PR, Porter EA, Gellman SH. An Efficient Route to Either Enantiomer of Orthogonally Protected Trans-3-Aminopyrrolidine-4-Carboxylic Acid. J. Org. Chem. 2001;66:3597. doi: 10.1021/jo001534l. [DOI] [PubMed] [Google Scholar]
- 33.Boersma MD, Sadowsky JD, Tomita Y, Gellman SH. Hydrophile-Scanning as a Complement to Alanine-Scanning for Exploring and Manipulating Protein-Protein Recognition: Application to the Bim BH3 Domain. Protein Sci. 2008;17:1232. doi: 10.1110/ps.032896.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boersma MD. PhD thesis. University of Wisconsin – Madison; 2008. [Google Scholar]
- 35.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DC. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-x(L), but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 37.Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Adv. Vir. Res. 2005;64:231. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dwyer JJ, Wilson KL, Davison DK, Freel SA, Seedorff JE, Wring SA, Tvermoes NA, Matthews TJ, Greenberg ML, Delmedico MK. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc. Natl. Acad. Sci. U S A. 2007;104:12772. doi: 10.1073/pnas.0701478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DCS, Hymowitz SG, Jin S, Khaw SL, Kovar PJ, Lam LT, Lee J, Maecker HL, Marsh KC, Mason KD, Mitten MJ, Nimmer PM, Oleksijew A, Park CH, Park C-M, Phillips DC, Roberts AW, Sampath D, Seymour JF, Smith ML, Sullivan GM, Tahir SK, Tse C, Wendt MD, Xiao Y, Xue JC, Zhang H, Humerickhouse RA, Rosenberg SH, Elmore SW. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013;19:202. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 40.Foight GW, Ryan JA, Gulla SV, Letai A, Keating AE. Designed BH3 peptide with high affinity and specificity for targeting Mcl-1 in cells. ACS Chem. Biol. 2014;9:1962. doi: 10.1021/cb500340w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnard A, Long K, Martin HL, Miles JA, Edwards TA, Tomlinson DC, Macdonald A, Wilson AJ. Selective and potent proteomimetic inhibitors of intracellular protein-protein interactions. Angew. Chem. Int. Ed. 2015;54:2960–2965. doi: 10.1002/anie.201410810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moon H, Lee WS, Oh M, Lee H, Lee JH, Im W, Lim H-S. Design, solid-phase synthesis, and evaluation of a phenyl-piperazine-triazine scaffold as α-helix mimetics. ACS Comb. Sci. 2014;16:695–701. doi: 10.1021/co500114f. [DOI] [PubMed] [Google Scholar]
- 43.LePlae PR, Umezawa N, Lee H-S, Gellman SH. An efficient route to either enantiomer of trans-2-aminocyclopentanecarboxylic acid. J. Org. Chem. 2001;66:5629. doi: 10.1021/jo010279h. [DOI] [PubMed] [Google Scholar]
- 44.Guzzo PR, Miller MJ. Catalytic, asymmetric synthesis of the carbacephem framework. J. Org. Chem. 1994;59:4862. [Google Scholar]
- 45.Murray JK, Farooqi B, Sadowsky JD, Scalf M, Freund WA, Smith LM, Chen J, Gellman SH. Efficient synthesis of a β-peptide combinatorial library with microwave irradiation. J. Am. Chem. Soc. 2005;127:13271. doi: 10.1021/ja052733v. [DOI] [PubMed] [Google Scholar]
- 46.Murray JK, Gellman SH. Microwave-assisted parallel synthesis of a 14-helical β-peptide library. J. Comb. Chem. 2006;8:58. doi: 10.1021/cc0501099. [DOI] [PubMed] [Google Scholar]
- 47.Murray JK, Gellman SH. Parallel synthesis of peptide libraris using microwave irradiation. Nat. Protoc. 2007;2:624. doi: 10.1038/nprot.2007.23. [DOI] [PubMed] [Google Scholar]
- 48.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acids sequence data. Anal. Biochem. 1989;182:319. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 49.Horne WS, Boersma MD, Windsor MA, Gellman SH. Sequence-based design of á/β-peptide foldamers that mimic BH3 domains. Angew. Chem. Int. Ed. 2008;47:2853. doi: 10.1002/anie.200705315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadowsky JD, Schmitt MA, Lee HS, Umezawa N, Wang S, Tomita Y, Gellman SH. Chimeric (á/β+á)-peptide ligands for the BH3-recognition cleft of Bcl-xL: Critical role of the molecular scaffold in protein surface recognition. J. Am. Chem. Soc. 2005;127:11966. doi: 10.1021/ja053678t. [DOI] [PubMed] [Google Scholar]
- 51.Boersma MD, Sadowsky JD, Tomita YA, Gellman SH. Hydrophile scanning as a complement to alanine scanning for exploring and manipulating protein-protein recognition: Application to the Bim BH3 domain. Protein Sci. 2008;17:1232. doi: 10.1110/ps.032896.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Assay Guidance Manual, version 4.1. Eli Lilly and NIH Chemical Genomics Center; 2005. [PubMed] [Google Scholar]
- 53.Roehrl MHA, Wang JY, Wagner G. A general famework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein-protein interactions by fluorescence polarization. Biochemistry. 2004;43:16056. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- 54.GraphPad, Sigmoidal dose-response curve fitting. http://www.graphpad.com%22.
- 55.Lee EF, Czabotar PE, van Delft MF, Michalak EM, Boyle MJ, Willis SN, Puthalakath H, Bouillet P, Colman PM, Huang DCS, Fairlie WD. A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation. J. Cell. Biol. 2008;180:341–355. doi: 10.1083/jcb.200708096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DCS. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 57.Smith BJ, Lee EF, Checco JW, Evangelista M, Gellman SH, Fairlie WD. Structure-Guided Rational Design of α/β-Peptide Foldamers with High Affinity for Bcl-2 Family Pro-Survival Proteins. ChemBioChem. 2013;14:1564. doi: 10.1002/cbic.201300351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.