Abstract
Using MIRA-seq, we have characterized the DNA methylome of metastatic melanoma and normal melanocytes. Individual tumors contained several thousand hypermethylated regions. We discovered 179 tumor-specific methylation peaks present in all (27/27) melanomas that may be effective disease biomarkers, and 3,113 methylation peaks were seen in >40% of the tumors. We found that 150 of the approximately 1,200 tumor-associated methylation peaks near transcription start sites (TSS) were marked by H3K27me3 in melanocytes. DNA methylation in melanoma was specific for distinct H3K27me3 peaks rather than for broadly covered regions. However, numerous H3K27me3 peak-associated TSS regions remained devoid of DNA methylation in tumors. There was no relationship between BRAF mutations and the number of methylation peaks. Gene expression analysis showed upregulated immune response genes in melanomas presumably as a result of lymphocyte infiltration. Down-regulated genes were enriched for melanocyte differentiation factors; e.g., KIT, PAX3 and SOX10 became methylated and downregulated in melanoma.
Keywords: DNA methylation, melanoma, melanocytes, Polycomb, H3K27me3, BRAF
1. Introduction
Incidence rates of melanoma have been rising for the past 30 years [1]. Melanoma is the most aggressive form of skin cancer and according to statistics of the American Cancer Society, there are about 10,000 deaths from melanoma each year in the United States. Metastasis is most commonly associated with the demise of the patient. Despite advances in targeted therapy of melanoma, using for example inhibitors of mutant (V600E) BRAF kinase, resistance to therapy develops almost invariably. Melanoma genomes are generally characterized by high frequencies of mutations that carry a signature of ultraviolet B radiation from the sun [2–4]. Among the thousands of mutational events occurring in a melanoma genome, a limited number of specific driver mutation events have been identified [5] suggesting that most sequence alterations found in tumors are inconsequential passenger mutations.
In addition to genetic changes, tumors also carry large numbers of epigenetic alterations. Most notably, DNA cytosine methylation patterns are altered extensively resulting in genome-scale DNA hypomethylation and more localized hypermethylation at CpG-rich genomic loci referred to as CpG islands [6]. Although numerous studies have analyzed gene-specific methylation changes in melanoma, broader investigations that interrogate methylation differences genome-wide have been more limited [7–13].
With regards to DNA hypermethylation in cancer, there are at least two major unresolved general questions: (i) what are the mechanisms leading to DNA methylation changes in tumors, and (ii) which ones of the numerous methylation changes observed are potentially tumor-driving events. Regarding (i), earlier observations have revealed correlations between the presence of the Polycomb mark, histone H3 lysine 27 trimethylation (H3K27me3), and DNA hypermethylation of CpG islands, an observation that has been made for hundreds of developmentally regulated genes including the homeobox genes as the most prevalently affected gene family [14–18]. However, the mechanisms of Polycomb-linked DNA methylation have remained obscure. The identification of driver methylation versus passenger methylation events [19] is similarly unresolved and is reminiscent of the situation with genetic mutations, where only a handful of driver mutations are seen in each individual tumor genome against a background of thousands of passenger mutations [20]. Genes targeted by the Polycomb complex at their promoters are commonly not expressed, or are expressed at very low levels in normal somatic tissues, making it difficult to rationalize why methylation of these particular sequences would provide a strong tumor-driving, selective force.
In this study, we have applied the technology of MIRA-seq [21] to comprehensively identify hypermethylated gene targets in malignant melanoma. We used normal skin melanocytes as the control in these experiments. Melanocytes or their stem cell-like precursors are considered the cells of origin for malignant melanoma, and we are able to use melanocytes as a homogenous cell population. In contrast, most studies of other types of tumors use a normal tissue counterpart that represents a mixed population of cells, and often the cell of origin is undefined. We focused our data analysis on the role of the Polycomb complex in targeting methylation events. Our study provides a large database for potential tumor-specific DNA methylation markers for melanoma, analyzes the relationship between BRAF mutations and methylation changes and also incorporates data on gene expression differences between melanocytes and melanoma tumors.
2. Results
2.1. MIRA-seq identifies numerous tumor-specific DNA methylation peaks and potential DNA methylation markers for melanoma
To characterize the DNA methylation landscape of melanoma, we analyzed 27 metastatic melanoma DNA samples (Table S1) and compared these to DNA from normal melanocytes from three different donors. We used the methylated-CpG island recovery assay (MIRA) [14] as an effective tool to pull down methylated DNA fragments and then analyzed them by high-throughput sequencing (MIRA-seq) [21]. Differentially methylated regions were identified by peak calling and by establishing a threshold of a 3-fold difference between a tumor-specific and background normal-specific peak signal. We identified only few substantially hypomethylated regions in tumors but found 3,113 hypermethylated regions, defined as being hypermethylated in at least 40% of the melanoma cases (Table S2). Individual tumors contained between approximately 1,000 and 3,000 hypermethylation peaks (Table S1). Figures 1 and 2 show snapshots of melanoma-specific peaks. The gene TP73, coding for a TP53 family member, shows methylation of the upstream P1 promoter in melanoma, but constitutive methylation in melanocytes and melanomas of the P2 promoter where the delta-N TP73 transcript is initiated (Fig. 1). This situation is different from that in glioblastoma where the internal TP73 promoter becomes hypomethylated and the oncogenic truncated TP73 transcript gets activated [22]. For SOX10, a gene coding for a master regulator of the melanocyte lineage [23], several tumor-specific methylation peaks surrounding the first two exons and the upstream region of the gene were observed (Fig. 2). We also identified DNA hypermethylation at several gene loci previously analyzed in melanoma by single gene studies, including the genes RASSF1A and RASSF10, for example [24, 25] (data not shown). Several of the tumor-specific methylation peaks were confirmed by standard sodium bisulfite sequencing as exemplified by the 3′ exon of the MYC gene, which becomes hypermethylated in melanomas (Fig. S1). From the hypermethylation peak list shown in Table S2, when sorted by the number of tumors carrying the peak, we identified 179 DNA methylation peaks that were present in all (27 of 27) melanomas. Furthermore, there were 237 methylation peaks present in 26 of 27 melanomas. This list of tumor-specific peaks provides a large data resource for the development of potential DNA methylation biomarkers for malignant melanoma. We next conducted gene ontology analysis of genes with tumor-specific methylation peaks at the promoter and transcription start site (TSS). This analysis showed enrichment for developmental processes and transcription factors involved in cell fate commitment and differentiation (Fig. 3A). Prominent on the identified gene lists were homeobox genes (Fig. 3A), similar as reported for other types of cancer [14]. In fact, when analyzing all 236 homeobox genes present in the human genome [26], we found that 58% of them became methylated in melanoma (Table 1; Table S3). Most of the methylated homeobox genes have multiple CpG islands or large (>2kb) CpG islands. Interestingly, certain homeobox gene families became methylated more frequently than others (Table 1). The NKL subclass (85% of the genes methylated), the LIM class (83%) and the SINE class of homeobox genes (100%) were the subclasses most susceptible to DNA methylation in melanoma (Table 1). The CERS class, and HNF and ZF classes of genes were the classes least prone to methylation.
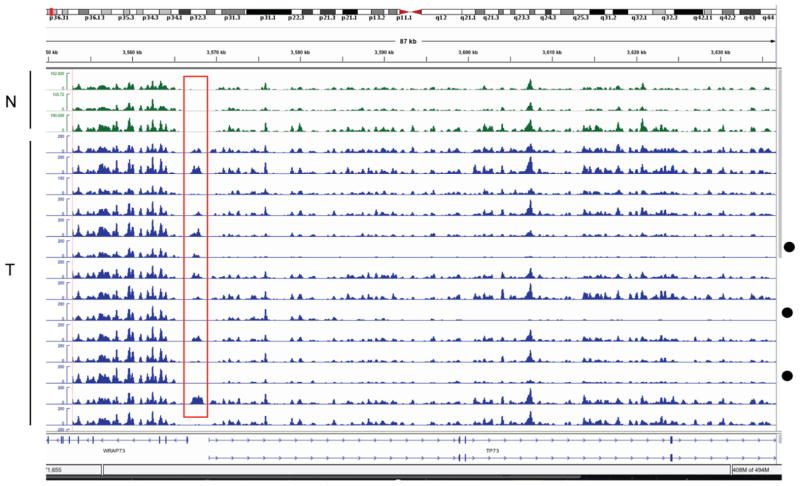
Figure 1. Example of MIRA-seq data showing the TP73 gene at chromosome 1p36.
The top three profiles are DNA from normal melanocytes (green) and the bottom 14 profiles are melanoma samples (blue). The red box indicates tumor-specific methylation just upstream of the TSS. Reduced signal in samples indicated by a black dot indicates potential deletion of TP73 gene sequences in a few samples but retention of signal at upstream noncoding sequences.
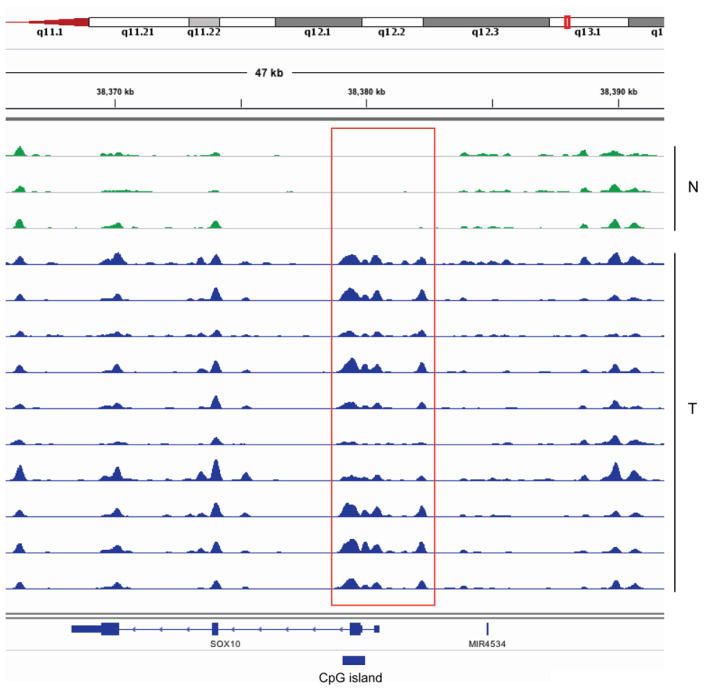
Figure 2. Example of MIRA-seq data showing the SOX10 gene at chromosome 22q13.1.
The top 3 profiles are DNA from normal melanocytes (green) and the bottom 10 profiles are melanoma samples (blue). The red rectangle indicates a tumor-specifically methylated region surrounding the TSS. The blue box shows a CpG island.
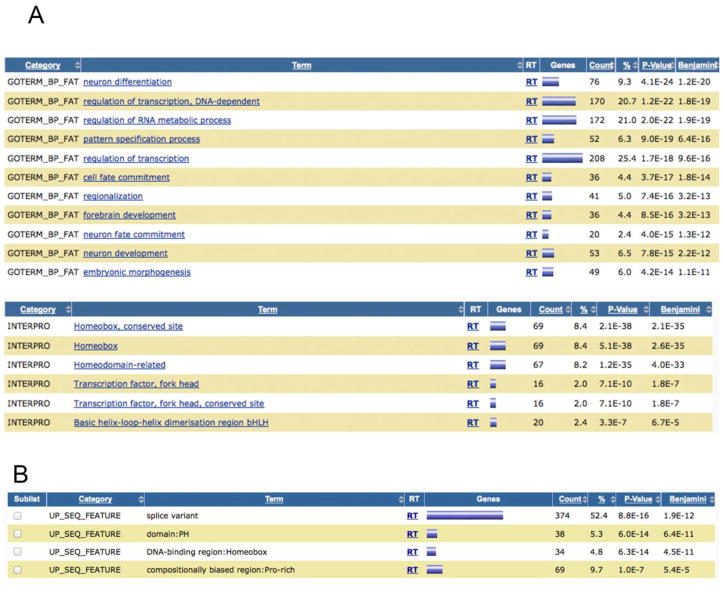
Figure 3. Gene ontology analysis of methylated genes.
A. Enriched gene ontology categories for genes with DNA methylation peaks near the TSS and promoters. Gene ontology analyzing using the DAVID tool [32] was conducted on the list of genes containing a DNA methylation peak near the TSS or promoter (region covered −1.5 to +1.5 kb relative to the start sites) in at least 40% of the melanoma samples.
B. Enriched gene categories for genes with DNA methylation peaks within the gene body in at least 40% of the melanoma samples.
Table 1.
Differentially methylated homeobox genes in melanoma
| Gene number | Unmethylated genes | Methylated genes* | |
|---|---|---|---|
| Homeobox Genes | 236 | 99 (42%) | 137 (58%) |
| Gene Family | |||
| ANTP class homeoboxes | |||
| HOXL subclass | 52 | 22 (42.3%) | 30 (57.7%) |
| NKL subclass | 47 | 7 (14.9%) | 40 (85.1%) |
| CERS class homeoboxes | 6 | 6 (100%) | 0 (0.0%) |
| CUT class homeoboxes | 7 | 3 (42.9%) | 4 (57.1%) |
| HNF class homeoboxes | 3 | 3 (100%) | 0 (0.0%) |
| LIM class homeoboxes | 12 | 2 (16.7%) | 10 (83.3%) |
| POU class homeoboxes | 17 | 9 (52.9%) | 8 (47.1%) |
| PDR class homeoboxes | 50 | 21 (42.0%) | 29 (58.0%) |
| PROS class homeoboxes | 2 | 1 (50.0%) | 1 (50.0%) |
| SINE class homeoboxes | 6 | 0 (0.0%) | 6 (100%) |
| TALE class homeoboxes | 20 | 13 (65.0%) | 7 (35.0%) |
| ZF class homeoboxes | 14 | 12 (85.7%) | 2 (14.3%) |
A gene was counted as methylated when methylation was found in at least 5 of 27 tumors.
Gene ontology analysis of tumor-associated hypermethylation peaks in gene bodies also revealed homeobox genes (Fig. 3B). Interestingly, the most highly enriched functional category was ‘splice variant.’ This category contains genes that encode at least two different protein isoforms due to alternative splicing. Intragenic DNA methylation may restrict the expression of alternative promoters [27].
2.2. Relationship between DNA methylation and BRAF mutations
In colorectal cancer, there is a strong, direct relationship between the presence of BRAF (V600E) mutations and the CpG island methylator phenotype (CIMP) [28], a process in which a large number of CpG islands become methylated in cancer [29]. We sequenced the BRAF gene in melanomas around codon 600 and found BRAF V600 mutations in 10 of the 24 tumors analyzed (Table S1). Wildtype BRAF sequences were scored in 14 tumors and in all normal melanocyte samples (3 tumors failed PCR). The mean number of methylation peaks was 2,263 in BRAF wildtype melanomas and 2,551 in BRAF V600 mutated tumors. The difference was not statistically significant (P=0.21, Wilcoxon test).
2.3. Mapping of H3K27me3 in melanocytes and its relationship to DNA methylation: three gene categories
Since the Polycomb machinery and its genomic mark, H3K27me3 has been linked to DNA methylation in several tumor types [15, 17, 18], we used ChIP sequencing to map H3K27me3 in normal melanocytes. This analysis revealed two types of distribution of H3K27me3: (i) a broad distribution over large multi-kilobase-size regions, often covering entire genes, and (ii) a more peak-like distribution with an enhanced signal localized over much smaller regions, often less than 1 kb in length. The latter, peak-like pattern was found commonly at the 5′ end of genes encompassing a CpG island. Figure 4A shows a typical H3K27me3 gene pattern with the two modes of H3K27me3 distribution.
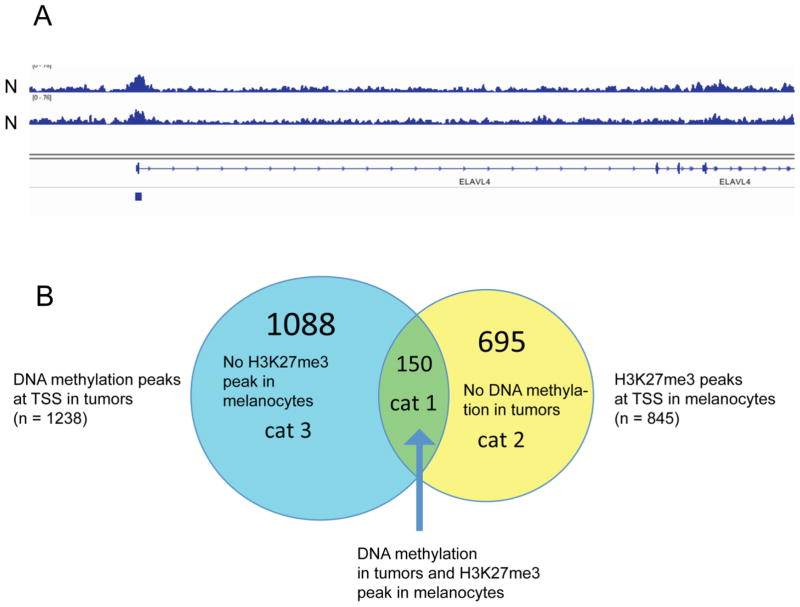
Figure 4. Patterns of H3K27me3 in normal melanocytes and DNA methylation in melanomas.
A. Peak-like and broad distribution patterns of H3K27me3. The H3K27me3 mark is broadly distributed over the gene ELAVL4 but there is a distinct peak at the CpG island (indicated by a blue box) near the TSS.
B. Designation of genes in categories 1, 2 and 3. Green, category 1: genes with H3K27me3 peak in melanocytes and methylated in tumors; yellow, category 2: genes with H3K27me3 peak in melanocytes but not methylated in tumors; blue, category 3: genes without H3K27me3 peak in melanocytes but methylated in tumors.
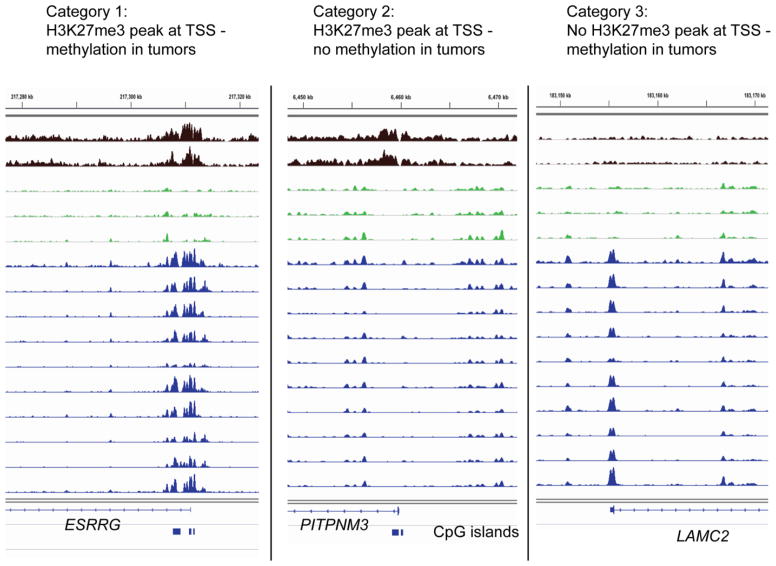
We then specifically examined the relationship between presence of the Polycomb mark in melanocytes and DNA methylation in melanoma. We focused our analysis on the TSS (−500 to +500 base pairs relative to the transcription start sites of Refseq genes) and identified 845 H3K27me3 peaks at the TSS in melanocytes (Table S4). We found that 150 of ~1240 genes with tumor-associated DNA methylation near the TSS showed H3K27me3 peaks at the TSS in melanocytes (Fig. 4B; Table S5). We refer to these genes as category 1 genes. This targeting of DNA methylation towards H3K27me3 peaks at the TSS was statistically highly significant (p = 2.84e–35; hypergeometric probability test). DNA hypermethylation almost exclusively occurred at the peak-like H3K27me3 sites and basically never was found at sites of broader H3K27me3 distribution (e.g., Fig. 5, left panel). We are not aware that this phenomenon has been specifically reported before. However, there were numerous H3K27me3 TSS peak-associated genes (n = 695) that were completely devoid of methylation in tumors (Table S5). We refer to this group of genes as category 2 genes. Figure 5 (middle panel) shows an example of such a gene. Furthermore, there was a rather large third category of genes (category 3; n = 1088) that became methylated in at least 40% of melanomas but completely lacked an H3K27me3 peak in melanocytes at the TSS (Fig. 5, see right panel; Fig. 4B; Table S5). This subdivision of genes into three classes was also observed when looking specifically at homeobox genes (Fig. S2). Of note, mapping of H3K4me3 in melanocytes indicated that the targeting of DNA methylation to H3K27me3 peaks rarely affects bivalent genes carrying overlapping H3K27me3 and H3K4me3 peaks (data not shown). The major reason for that seems to be the fact that bivalent genes were rarely detected in melanocytes.
Figure 5. Examples of category 1, 2 and 3 genes.
Category 1: Tumor-specific DNA methylation occurs near the TSS marked by H3K27me3 in melanocytes. Example, ESRRG. Category 2: No tumor-specific DNA methylation occurs near the TSS despite marking with H3K27me3 in melanocytes. Example, PITPNM3. Category 3: Tumor-specific DNA methylation occurs at the TSS in the absence of H3K27me3 marking in melanocytes. Example, LAMC2. The blue boxes below the gene diagrams indicate CpG islands.
In order to understand if specific DNA sequence motifs are present near the TSS of category 1 genes or category 2 genes, we conducted sequence motif analysis. The theory was that specific sequences at category 1 genes would attract DNA methylation and/or that specific sequences at category 2 genes would prevent methylation. We analyzed motifs within 400 bp around the center of H3K27me3 peaks. The most highly enriched (~18-fold) consensus motif in category 1 over category 2 genes was for the HMG-box family member BBX [30]. However, this motif occurred in only 2.7% of the category 1 genes. The most highly enriched (~9-fold) consensus motif enriched in category 2 over category 1 genes was for CCAAT-displacement protein CDP (CUX1) [31]. However, this motif occurred in only 4.5% of the category 2 genes. Therefore, these motifs do not generally explain the differential susceptibility of category 1 and category 2 genes to DNA methylation.
2.4. RNA-seq of melanocytes and melanoma: upregulation of immune genes and downregulation of pigmentation genes in melanoma
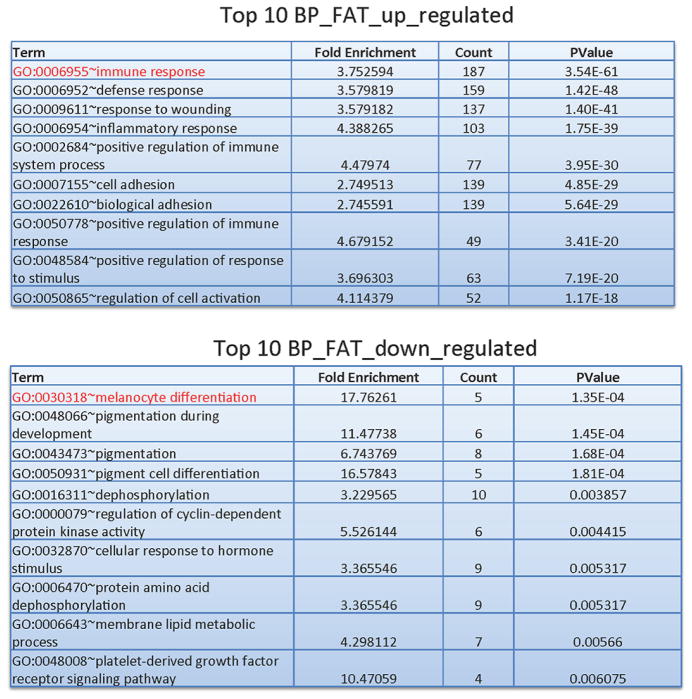
RNA of sufficient quality was available from 17 melanomas and 3 melanocyte samples. These RNA samples were analyzed for gene expression patterns by RNA-seq revealing distinct categories for melanocytes and melanomas as confirmed by hierarchical clustering and principle component analysis (Fig. S3). We identified 1044 upregulated and 702 downregulated genes in melanoma relative to normal melanocytes using a fold-change of >2 and false discovery rate (FDR) of <0.01. Gene ontology analysis using the DAVID tool [32] identified genes of the immune response and inflammatory response as highly significantly upregulated (Fig. 6; Table S6). These immune and inflammatory genes included many cytokines and chemokines (Table S6). The downregulated genes were enriched for genes belonging to the categories of melanocyte differentiation and pigmentation (Table S7). This set of genes representing melanocyte differentiation genes contained the genes MITF, a gene coding for a key melanocyte transcription factor [33, 13], TYRP1 (tyrosinase related protein 1) [34], OCA2 (a tyrosine transporter important for melanin biosynthesis) [35], CITED1 (also known as MSG1, melanocyte-specific gene 1) [36], and RAB33A and RAB27A, two critical regulators of melanosomes [37, 38]. Of particular interest is also the presence of the KIT gene as downregulated in melanoma. KIT mutations are found in a small percentage of melanomas and define a subgroup of melanocytic tumors where this tyrosine kinase appears to play a tumor-driving role [5]. These results suggest that KIT-associated pathways may play opposing roles in melanoma biogenesis.
Figure 6. Gene ontology analysis of differentially expressed genes in melanoma.
A. Gene ontology analysis using the DAVID tool was conducted on the list of upregulated genes in melanoma versus melanocytes.
B. Gene ontology analysis using the DAVID tool was conducted on the list of downregulated genes in melanoma versus melanocytes.
Our next goal was to compare the DNA methylation data with gene expression data obtained by RNA-seq. For this purpose, we analyzed genes where the methylation peaks occurred near the TSS. Based on the knowledge that genes marked by the Polycomb complex (H3K27me3) at promoters are generally expressed at very low levels even in the absence of DNA methylation, we primarily focused then on the genes in category 3, those that became methylated in tumors but were free of H3K27me3 in melanocytes. Somewhat unexpectedly, we still found a rather poor correlation between presence of a methylation peak at the TSS and gene downregulation in melanoma even in this category (Table S8). Such data suggest that presumably other, DNA methylation-independent mechanisms play a more critical role in gene control, or alternatively that lymphocyte infiltration compromises these comparisons. However, there were many examples in which downregulation of a gene in melanomas was accompanied by emergence of a methylation peak near the TSS (Table S8). Among this group of genes were KIT, which was the third most significantly downregulated category 3 gene and became methylated in 20 of 27 melanomas. Also SOX10, a critical component of melanocyte lineage determination was moderately downregulated and became methylated at the TSS in 27 of 27 melanomas (Fig. 2; Table S8). The same correlation holds true for PAX3, another transcription factor critical for the proper development of melanocytes [39]. Gene sequences surrounding PAX3 became methylated in 20 of 27 melanomas (Fig. S2) and the gene was downregulated in tumors (Table S8).
3. Discussion
3.1. DNA methylation markers for melanoma
Using MIRA-seq, we established comprehensive methylation profiles of 27 malignant melanomas and identified a large number of tumor-specific peaks. Several large-scale DNA methylation studies in melanoma have been performed previously [7–13, 5]. Most of these reports have used the Illumina bead array platform making it difficult to directly compare these published data with our sequencing-based approach. We catalogued over 400 methylation peaks that occurred tumor-specifically in at least 26 of the 27 melanomas. These gene sequences provide a large resource for developing DNA methylation biomarkers of melanoma. One important future step in this direction will be to determine which ones of the approximately 400 identified hypermethylation peaks occur in benign melanocytic lesions (benign nevi), dysplastic nevi, and primary skin melanomas, in other words during what stage of tumor progression do these changes occur? A methylation biomarker would be particularly useful, if it could distinguish, for example, a dysplastic nevus from a primary superficial spreading melanoma. These types of markers may provide independent verification for histopathological findings.
3.2. DNA methylation and BRAF mutations in melanoma
In colorectal cancers, there is a strong link between BRAF mutations and large numbers of DNA hypermethylation events, the so-called CpG island methylator phenotype (CIMP) [28]. Since about half of human melanomas carry BRAF mutations, it was of interest to know if a similar association exists in this malignancy. However, we have not found such a correlation in our data sets. Our finding of lack of association of BRAF mutations and CIMP is in agreement with recent data from The Cancer Genome Atlas (TCGA), which actually identified a negative correlation between BRAF mutations and methylation changes in melanoma [5]. Similarly, in papillary thyroid cancers, BRAF mutated tumors have fewer DNA methylation changes than BRAF wildtype tumors [40].
Since the underlying mechanisms of CIMP are still unclear, the different findings in different tumors are only adding to the puzzle. Such data suggest a tissue-specific rather than a universal biological mechanism. For example, a specific pathway may operate in colon tumors, which requires BRAF-mutated tumors to engage a mechanism to counteract BRAF oncogene-induced senescence [41], such as activation of the WNT pathway through widespread methylation of WNT inhibitor genes [42]. The same mechanism may be inoperative in thyroid cancer and melanoma.
3.3. Role of H3K27me3 in targeting of DNA methylation in melanoma
About 12% of the promoter and TSS-associated DNA hypermethylation peaks in melanoma were associated with presence of H3K27me3 in melanocytes. This fraction of Polycomb-targeted events seems substantially smaller than those found in colorectal cancers or lung cancers [17, 43]. The reason for these differences is unclear. Formally, we cannot exclude the possibility that H3K27me3 occupies additional genes at other stages of tumor progression, e.g. in dysplastic nevi, and these genes later on become CpG-methylated in melanomas. Interestingly, we observed that melanoma-specific methylation occurred only at peak-like H3K27me3 regions encompassing CpG islands in melanocytes but did not occur at regions of broader but lower levels H3K27me3 coverage. Since these H3K27me3 peaks predominantly reside at CpG islands, this observation points to a potential involvement of CpG island targeting mechanisms for the Polycomb complex which may break down in cancer cells making the exposed DNA susceptible to de novo methylation.
It is curious that a large fraction of H3K27me3-occupied CpG islands do not undergo hypermethylation in tumors. One possibility is that these sequences are shielded from DNA methylation by a specific protective mechanism that is not present at those CpG islands that do undergo methylation. An alternative mechanism may be that the Polycomb complex is more tightly bound at CpG islands that do not undergo DNA methylation thus precluding Polycomb to DNA methylation switching [44]. The three categories of genes that compare Polycomb marking and susceptibility to DNA methylation in tumors (Fig. 4B) were also observed for homeobox genes (Fig. S2) suggesting that this gene family is not noticeably distinct from other genes in the genome, i.e. these genes are not always associated with H3K27me3 and do not always undergo methylation when they are H3K27me3-associated. One hypothesis has been that DNA methylation is linked to presence of a bivalent, H3K4me3 and H3K27me3 containing chromatin configuration. However, we found that bivalent genes were rare in melanocytes. Furthermore, it is unclear why specific families of homeobox genes are more susceptible to methylation than others (Table 1). The grouping of these families is based on protein structure [26] but specific regulatory DNA sequence features that would distinguish these families are not yet identified. Future work will need to focus on specific sequences, transcription factors or chromatin modulators responsible for exclusion or attraction of the DNA methylation machinery to H3K27me3-occupied CpG-rich regions.
3.4. Gene expression changes and DNA methylation
RNA-seq-based gene expression analysis revealed a prominent involvement of immune response genes that are upregulated in melanomas relative to melanocytes. This observation is not unexpected since these metastatic tumors are likely infiltrated by lymphocytes. A similar finding of upregulated immune genes has recently been reported for melanomas by TCGA [5].
Overall, we found a rather limited correlation between tumor-associated DNA methylation at the TSS and suppression of gene expression. This result was somewhat unexpected but similar findings have been made for other types of cancer [45] and may be related to low expression levels of the genes that undergo methylation or are due to other mechanisms. Notably, however, we found significant downregulation of key melanocyte lineage differentiation genes [23, 46], including MITF, PAX3, SOX10, KIT and OCA2. In several cases (SOX10, PAX3, KIT), this downregulation was accompanied by DNA methylation at the TSS. These findings suggest that at least one functional aspect of DNA hypermethylation in melanoma is related to a differentiation defect emerging during malignant transformation of human melanocytes.
4. Materials and Methods
4.1. Samples
Genomic DNAs and total RNAs (5 μg each) from human metastatic melanomas were obtained from OriGene (OriGene, Rockville, MD) (Table S1). Genomic DNAs from normal human melanocytes were obtained from ScienCell (Carlsbad, CA) or were isolated from primary cultured skin melanocytes obtained from Lonza (Allendale, NJ).
4.2. MIRA-seq
Two micrograms of genomic DNA was sonicated to produce DNA fragments with a length distribution of 100 to 300 bp using a Covaris S220 sonicator (Covaris; Woburn, MA) with the following settings: 3 min, 10% duty cycle, 175 Watts peak incident power, and 200 cycles per burst. The sheared DNA was purified using MinElute PCR purification kits (Qiagen; Valencia, CA) following the manufacturer’s instructions and eluted twice with 10 μl Qiagen elution buffer, resulting in a total volume of 20 μl. To prepare Illumina linker-ligated DNA fragments, the sheared DNA was subjected to end repair using the NEBNext® End Repair Module from New England Biolabs (NEB; Ipswich, MA). The reaction mix was incubated at 20°C for 30 min, and the DNA was purified using MinElute PCR purification kit (Qiagen) and eluted twice with 10 μl elution buffer, resulting in a volume of 20 μl. To add 3′-A overhangs to DNA, the end-repaired DNA was incubated in 50 μl of 1x NEBNext® dA-tailing reaction mixture with 3 μl of Klenow (exo−) fragment (NEB) for 30 min at 37°C and purified by using MinElute PCR purification kit (Qiagen). Next, the dA-tailed DNA was ligated with 3 μl of a 10 μM solution of Illumina Tru-seq linkers (Illumina; San Diego, CA) in 50 μl of 1x T4 DNA ligase reaction mix (NEB) containing 1 μl of NEB T4 DNA ligase (400 U/μl) by incubation for 18 hours at 16°C followed by purification using QIAquick PCR purification kit (Qiagen). DNA was eluted with 50 μl elution buffer. Five μl of linker-ligated DNA was saved as a control (input).
Next, methylated DNA fragments were enriched using the methylated-CpG island recovery assay (MIRA) technique as described previously [21]. In brief, 1.5 μg of GST-tagged MBD2b protein and 1 μg of His-tagged MBD3L1 protein were pre-incubated in 355 μl of blocking buffer (40 μl of 10x NEBuffer 2, 1 μl of 10% Triton X-100, and 0.5 μg of sonicated E. coli JM110 genomic DNA) for 20 min at 4°C on a rotating platform and added to each ligated DNA sample, and then the 400 μl of the MIRA reaction mixture was incubated overnight at 4°C on a rotating platform. To allow selective collection of MIRA-captured methyl-CpG-rich DNA fragments, 5 μl of MagneGST magnetic beads (Promega, Madison, WI), pre-blocked with blocking buffer, were added to the MIRA reaction mixture and further incubated for 2 hours at 4°C on a rotating platform. The beads were captured on a magnetic stand and washed three times with 900 μl of cold washing buffer (10 mM Tris-HCl, pH 7.5, 600 mM NaCl, and 0.025% Triton X-100). Next, to purify DNA fragments, the beads were resuspended in 500 μl of Qiagen PB buffer (Qiagen) and incubated for 5 min at room temperature, then purified using MinElute PCR purification kit (Qiagen) according to the manufacturer’s instructions and eluted twice with 10 μl elution buffer, resulting in a volume of 20 μl.
To prepare a sequencing library, the MIRA-enriched DNA fragments and input DNA were PCR-enriched in 100 μl of reaction volume including 50 μl of Phusion High-Fidelity PCR Master Mix (NEB) with 2 μl of 25 μM PE 1.0 primer and 2 μl of 25 μM index primer (Illumina), and 1 M betaine, using the following conditions in a thermal cycler; 98°C for 30 sec and 12 cycles at 98°C for 10 sec, 65°C for 30 sec and 72°C for 30 sec followed by 72°C for 5 min. Next, 100 μl of Ampure XP beads (Beckman Coulter; Brea, CA) was added to the PCR solution to remove DNA fragments smaller than 200 bp, and double bead selection (0.65x) was used to remove fragments larger than 500 bp according to the manufacturer’s instructions. The size-selected sequencing library was sequenced on an Illumina HiSeq instrument.
4.3. ChIP-seq
Five million human melanocytes per ChIP reaction were cross-linked with cross-linking solution (50 mM Hepes pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, and 1% formaldehyde) for 8 min at room temperature with gentle shaking, followed by an additional incubation for 5 min with adding glycine to a final concentration of 125 mM. After washing twice with PBS, the cells were collected and pelleted by centrifugation at 3,500xg for 5 min. The cells were lysed by incubation in lysis buffer containing 50 mM Hepes pH 7.9, 140 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.25 % Triton X-100, 10% glycerol, and 1x complete protease inhibitor cocktail (Roche) for 20 min on ice. After washing twice with cold washing buffer (10 mM Tris-HCl pH 8.1, 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA), the nuclear pellet was resuspended in 1 ml of cold shearing buffer (10 mM Tris-HCl pH 8.1, 1 mM EDTA, and 0.1% SDS) and then sheared to produce chromatin fragments with a length distribution of 100 to 300 bp using a Covaris S220 sonicator (Covaris) with the following settings: 12.5 min, 5% duty cycle, 140 Watts peak incident power, and 200 cycles per burst. After adding 1% Triton X-100 and 150 mM NaCl (final concentrations) to the sheared chromatin and mixing, the solution was cleared by centrifugation at 14,000xg for 10 min at 4°C. Chromatin fragments were incubated with 8 μg of polyclonal anti-H3K27me3 antibody (07-449; Millipore; Billerica, MA) overnight at 4°C with rotation, and then immunoprecipitated using Dynabeads Protein G (Invitrogen; Carlsbad, CA). The libraries for Illumina sequencing of ChIP DNA were prepared and sequenced as described above.
4.4. RNA-seq
RNA samples matching genomic DNA were obtained from Origene (Table S1). RNA was isolated from melanocyte cell cultures as controls. RNA quality was confirmed using Bioanalyzer (Agilent Technologies; Santa Clara, CA). Transcriptome libraries were constructed using TruSeq RNA Sample Preparation Kit V2 (Illumina). Briefly, poly(A)RNA was isolated (using oligo-dT) and reverse-transcribed. After fragmentation, cDNA fragments with sizes between 130–280 bp were isolated from polyacrylamide-urea gels. A 3′ linker with a barcode and a 5′ linker were added to the isolated cDNA fragments. The ligated material was amplified by PCR and then run on an Illumina HiSeq 2500 instrument for parallel sequencing.
4.5. Bioinformatics analysis
Raw data files were deposited into the GEO database (accession numbers GSE71938, GSE71747 and GSE71692). For RNA-seq, the 40-bp long, single-ended sequence reads were mapped to the human genome (hg19) using TopHat, and the frequency of Refseq genes detected was counted with customized R scripts. The raw counts were then normalized using the trimmed mean of M values (TMM) method implemented in the Bioconductor package “edgeR”. Heat maps were created by hierarchical clustering using Cluster v3.0. Differential expression analysis was conducted by “edgeR” and gene functional analysis was performed using DAVID [32].
For MIRA-seq, reads were aligned to the hg19 genome using Novoalign (www.novocraft.com). Only uniquely aligned reads were kept for further analysis. Peak calling was performed using MACS 2.0. Methylation peaks were identified using R scripts and bioconductor package “edgeR.” Common peak regions were defined as peaks present in at least 40% of tumor samples. Reads falling into these common peak regions in tumor and normal tissues were counted and compared using “edgeR.” Peak regions with FC >= 3 in tumor compared to control and P value <= 0.05 were selected as hypermethylated peaks. Methylation peaks were annotated using the Refseq annotation database to TSS (transcription start site +/− 500 bp), extended promoter (500 bp upstream or downstream from the TSS to 1500 bp upstream or downstream from the TSS), and gene body (transcription start + 1500 bp to transcription end). Motif analysis was done using Biobase Transfac.
For ChIP-seq of H3K27me3, peak calling at the TSS was done using Bioconductor package “chipseq” and in-house developed R scripts. Annotation to the TSS was done as described in MIRA-seq analysis.
Supplementary Material
Supplementary Figure 1: MIRA-seq and bisulfite sequencing of the MYC gene
A. Genome browser view of a melanocyte sample (N) and two melanomas (T).
B. Bisulfite sequencing. After conversion with sodium bisulfite, the hypermethylated MYC exon region indicated by the arrow was sequenced in the same samples shown in panel A. Open circles represent unmethylated CpG sequences and closed circles show methylated CpG sequences. The fraction of methylated CpG sites is shown.
Supplementary Figure 2: Examples of the three gene categories including homeobox genes.
Category 1: Tumor-specific DNA methylation occurs at sequences marked by H3K27me3 in melanocytes. Example, SHOX2. Category 2: No tumor-specific DNA methylation occurs near the TSS despite marking with H3K27me3 in melanocytes. Example, PAX4. Category 3: Tumor-specific DNA methylation occurs in the absence of H3K27me3 marking in melanocytes. Examples, PAX3 and IRX2.
Supplementary Figure 3: RNA-seq analysis.
A. Heat map of upregulated (red) and downregulated (green) genes in melanomas (T) and melanocytes (N). The three melanocyte samples are on the right.
B. Principal component analysis of melanocytes and melanomas.
Highlights.
We discovered 179 tumor-specific methylation peaks present in all (27/27) melanomas.
We describe 3 gene categories for the interplay between H3K27me3 and DNA methylation.
We found no relationship between BRAF mutations and the number of methylation peaks.
Gene expression analysis showed upregulated immune response genes in melanomas.
Melanocyte differentiation factors KIT, PAX3 and SOX10 were methylated and silenced in tumors.
Acknowledgments
We acknowledge Jinhui Wang and the City of Hope Integrative Genomics Core for conducting Illumina sequencing. This work was supported by NIH grants ES06070 and CA084469 to G.P.P.
Footnotes
Author contributions
GPP designed the study. SGJ and WX conducted the experiments. XW and LY analyzed the data. GPP wrote the manuscript. All authors read and accepted the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leiter U, Eigentler T, Garbe C. Epidemiology of skin cancer. Adv Exp Med Biol. 2014;810:120–140. doi: 10.1007/978-1-4939-0437-2_7. [DOI] [PubMed] [Google Scholar]
- 2.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, Stratton MR. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeifer GP. How the environment shapes cancer genomes. Curr Opin Oncol. 2015;27:71–77. doi: 10.1097/CCO.0000000000000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.TCGA. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuta J, Nobeyama Y, Umebayashi Y, Otsuka F, Kikuchi K, Ushijima T. Silencing of Peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006;66:6080–6086. doi: 10.1158/0008-5472.CAN-06-0157. [DOI] [PubMed] [Google Scholar]
- 8.Koga Y, Pelizzola M, Cheng E, Krauthammer M, Sznol M, Ariyan S, Narayan D, Molinaro AM, Halaban R, Weissman SM. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res. 2009;19:1462–1470. doi: 10.1101/gr.091447.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao L, Smit MA, van den Oord JJ, Goeman JJ, Verdegaal EM, van der Burg SH, Stas M, Beck S, Gruis NA, Tensen CP, Willemze R, Peeper DS, van Doorn R. Genome-wide promoter methylation analysis identifies epigenetic silencing of MAPK13 in primary cutaneous melanoma. Pigment Cell Melanoma Res. 2013;26:542–554. doi: 10.1111/pcmr.12096. [DOI] [PubMed] [Google Scholar]
- 10.Li JL, Mazar J, Zhong C, Faulkner GJ, Govindarajan SS, Zhang Z, Dinger ME, Meredith G, Adams C, Zhang S, Mattick JS, Ray A, Perera RJ. Genome-wide methylated CpG island profiles of melanoma cells reveal a melanoma coregulation network. Sci Rep. 2013;3:2962. doi: 10.1038/srep02962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ecsedi S, Hernandez-Vargas H, Lima SC, Vizkeleti L, Toth R, Lazar V, Koroknai V, Kiss T, Emri G, Herceg Z, Adany R, Balazs M. DNA methylation characteristics of primary melanomas with distinct biological behaviour. PloS one. 2014;9:e96612. doi: 10.1371/journal.pone.0096612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzese DM, Scolyer RA, Huynh JL, Huang SK, Hirose H, Chong KK, Kiyohara E, Wang J, Kawas NP, Donovan NC, Hata K, Wilmott JS, Murali R, Buckland ME, Shivalingam B, Thompson JF, Morton DL, Kelly DF, Hoon DS. Epigenome-wide DNA methylation landscape of melanoma progression to brain metastasis reveals aberrations on homeobox D cluster associated with prognosis. Hum Mol Genet. 2014;23:226–238. doi: 10.1093/hmg/ddt420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauss M, Haq R, Cirenajwis H, Phung B, Harbst K, Staaf J, Rosengren F, Holm K, Aine M, Jirstrom K, Borg A, Busch C, Geisler J, Lonning PE, Ringner M, Howlin J, Fisher DE, Jonsson G. Genome-Wide DNA Methylation Analysis in Melanoma Reveals the Importance of CpG Methylation in MITF Regulation. J Invest Dermatol. 2015;135:1820–1828. doi: 10.1038/jid.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 15.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, Berman DM, Jenuwein T, Pruitt K, Sharkis SJ, Watkins DN, Herman JG, Baylin SB. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, Bergman Y, Simon I, Cedar H. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 18.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 19.Kalari S, Pfeifer GP. Identification of driver and passenger DNA methylation in cancer by epigenomic analysis. Adv Genet. 2010;70:277–308. doi: 10.1016/B978-0-12-380866-0.60010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu ET. Functional genomics of cancer. Curr Opin Genet Dev. 2008;18:251–256. doi: 10.1016/j.gde.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Jung M, Kadam S, Xiong W, Rauch TA, Jin S-G, Pfeifer GP. MIRA-seq for DNA methylation analysis of CpG islands. Epigenomics. 2015 doi: 10.2217/epi.15.33. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagarajan RP, Zhang B, Bell RJ, Johnson BE, Olshen AB, Sundaram V, Li D, Graham AE, Diaz A, Fouse SD, Smirnov I, Song J, Paris PL, Wang T, Costello JF. Recurrent epimutations activate gene body promoters in primary glioblastoma. Genome Res. 2014;24:761–774. doi: 10.1101/gr.164707.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris ML, Baxter LL, Loftus SK, Pavan WJ. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010;23:496–513. doi: 10.1111/j.1755-148X.2010.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003;63:1639–1643. [PubMed] [Google Scholar]
- 25.Helmbold P, Richter AM, Walesch S, Skorokhod A, Marsch W, Enk A, Dammann RH. RASSF10 promoter hypermethylation is frequent in malignant melanoma of the skin but uncommon in nevus cell nevi. J Invest Dermatol. 2012;132:687–694. doi: 10.1038/jid.2011.380. [DOI] [PubMed] [Google Scholar]
- 26.Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 29.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen T, Zhou L, Yuan Y, Fang Y, Guo Y, Huang H, Zhou Q, Lv X. Characterization of Bbx, a member of a novel subfamily of the HMG-box superfamily together with Cic. Dev Genes Evol. 2014;224:261–268. doi: 10.1007/s00427-014-0476-x. [DOI] [PubMed] [Google Scholar]
- 31.Ramdzan ZM, Nepveu A. CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers. Nat Rev Cancer. 2014;14:673–682. doi: 10.1038/nrc3805. [DOI] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Hsiao JJ, Fisher DE. The roles of microphthalmia-associated transcription factor and pigmentation in melanoma. Arch Biochem Biophys. 2014;563:28–34. doi: 10.1016/j.abb.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghanem G, Fabrice J. Tyrosinase related protein 1 (TYRP1/gp75) in human cutaneous melanoma. Mol Oncol. 2011;5:150–155. doi: 10.1016/j.molonc.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellono NW, Escobar IE, Lefkovith AJ, Marks MS, Oancea E. An intracellular anion channel critical for pigmentation. eLife. 2014;3:e04543. doi: 10.7554/eLife.04543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shioda T, Fenner MH, Isselbacher KJ. MSG1, a novel melanocyte-specific gene, encodes a nuclear protein and is associated with pigmentation. Proc Natl Acad Sci USA. 1996;93:12298–12303. doi: 10.1073/pnas.93.22.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng E, Trombetta SE, Kovacs D, Beech RD, Ariyan S, Reyes-Mugica M, McNiff JM, Narayan D, Kluger HM, Picardo M, Halaban R. Rab33A: characterization, expression, and suppression by epigenetic modification. J Invest Dermatol. 2006;126:2257–2271. doi: 10.1038/sj.jid.5700386. [DOI] [PubMed] [Google Scholar]
- 38.Booth AE, Seabra MC, Hume AN. Rab27a and melanosomes: a model to investigate the membrane targeting of Rabs. Biochem Soc Trans. 2012;40:1383–1388. doi: 10.1042/BST20120200. [DOI] [PubMed] [Google Scholar]
- 39.Medic S, Ziman M. PAX3 across the spectrum: from melanoblast to melanoma. Crit Rev Biochem Mol Biol. 2009;44:85–97. doi: 10.1080/10409230902755056. [DOI] [PubMed] [Google Scholar]
- 40.TCGA Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riemer P, Sreekumar A, Reinke S, Rad R, Schafer R, Sers C, Blaker H, Herrmann BG, Morkel M. Transgenic expression of oncogenic BRAF induces loss of stem cells in the mouse intestine, which is antagonized by beta-catenin activity. Oncogene. 2015;34:3164–3175. doi: 10.1038/onc.2014.247. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, Toyota M, Tokino T, Hinoda Y, Imai K, Herman JG, Baylin SB. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 43.Rauch TA, Wang Z, Wu X, Kernstine KH, Riggs AD, Pfeifer GP. DNA methylation biomarkers for lung cancer. Tumour Biol. 2012;33:287–296. doi: 10.1007/s13277-011-0282-2. [DOI] [PubMed] [Google Scholar]
- 44.Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sproul D, Nestor C, Culley J, Dickson JH, Dixon JM, Harrison DJ, Meehan RR, Sims AH, Ramsahoye BH. Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proc Natl Acad Sci USA. 2011;108:4364–4369. doi: 10.1073/pnas.1013224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Fukunaga-Kalabis M, Li L, Herlyn M. Developmental pathways activated in melanocytes and melanoma. Arch Biochem Biophys. 2014;563:13–21. doi: 10.1016/j.abb.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: MIRA-seq and bisulfite sequencing of the MYC gene
A. Genome browser view of a melanocyte sample (N) and two melanomas (T).
B. Bisulfite sequencing. After conversion with sodium bisulfite, the hypermethylated MYC exon region indicated by the arrow was sequenced in the same samples shown in panel A. Open circles represent unmethylated CpG sequences and closed circles show methylated CpG sequences. The fraction of methylated CpG sites is shown.
Supplementary Figure 2: Examples of the three gene categories including homeobox genes.
Category 1: Tumor-specific DNA methylation occurs at sequences marked by H3K27me3 in melanocytes. Example, SHOX2. Category 2: No tumor-specific DNA methylation occurs near the TSS despite marking with H3K27me3 in melanocytes. Example, PAX4. Category 3: Tumor-specific DNA methylation occurs in the absence of H3K27me3 marking in melanocytes. Examples, PAX3 and IRX2.
Supplementary Figure 3: RNA-seq analysis.
A. Heat map of upregulated (red) and downregulated (green) genes in melanomas (T) and melanocytes (N). The three melanocyte samples are on the right.
B. Principal component analysis of melanocytes and melanomas.