Abstract
The ability of cells to migrate through tissues and interstitial space is an essential factor during development and tissue homeostasis, immune cell mobility, and in various human diseases. Deformation of the nucleus and its associated lamina during 3-D migration is gathering increasing interest in the context of cancer metastasis, with the underlying hypothesis that a softer nucleus, resulting from reduced levels of lamin A/C, may aid tumour spreading. However, current methods to study the migration of cells in confining three dimensional (3-D) environments are limited by their imprecise control over the confinement, physiological relevance, and/or compatibility with high resolution imaging techniques. We describe the design of a polydimethylsiloxane (PDMS) microfluidic device composed of channels with precisely-defined constrictions mimicking physiological environments that enable high resolution imaging of live and fixed cells. The device promotes easy cell loading and rapid, yet long-lasting (>24 hours) chemotactic gradient formation without the need for continuous perfusion. Using this device, we obtained detailed, quantitative measurements of dynamic nuclear deformation as cells migrate through tight spaces, revealing distinct phases of nuclear translocation through the constriction, buckling of the nuclear lamina, and severe intranuclear strain. Furthermore, we found that lamin A/C-deficient cells exhibited increased and more plastic nuclear deformations compared to wild-type cells but only minimal changes in nuclear volume, implying that low lamin A/C levels facilitate migration through constrictions by increasing nuclear deformability rather than compressibility. The integration of our migration devices with high resolution time-lapse imaging provides a powerful new approach to study intracellular mechanics and dynamics in a variety of physiologically-relevant applications, ranging from cancer cell invasion to immune cell recruitment.
Introduction
Cell migration and motility play a critical role in numerous physiological and pathological processes, ranging from development and wound healing to the invasion and metastasis of cancer cells. It is now becoming increasingly apparent that cell migration in 3-D environments imposes additional challenges and constraints on cells compared to migration on 2-D substrates, which can have significant impact on cell motility.1–4 For example, cells migrating through 3-D environments are confined by the extracellular matrix and interstitial space;3 the physical confinement and 3-D environment not only alter the morphology of cells but also their migration mode.1, 2, 5, 6 Furthermore, the deformability of the cell nucleus, the largest and stiffest cell organelle, can become a rate-limiting factor when cells attempt to traverse dense extracellular matrix environments or pores smaller than the nuclear diameter.7–9 Consequently, the composition of the nuclear envelope, particularly the expression levels of lamins A and C, which largely determine nuclear stiffness,10, 11 can strongly modulate the ability of cells to pass through small constrictions.7–9, 12 Collectively, these findings and their implications in various biomedical applications have stimulated an increased interest in 3-D cell migration.
To date, the most common systems to study cell migration in confining 3-D environments fall into two categories, engineered systems and extracellular matrix scaffolds, each with their own limitations. Boyden chambers and transwell migration systems consist of membranes with defined pore sizes, typically 3 to 8 μm in diameter, through which cells migrate along a chemotactic gradient. While these systems can provide precisely-defined and highly uniform pore sizes, imaging the cells during their passage through the constrictions can be challenging, as the cells typically migrate perpendicular to the imaging plane and the membranes are often thick and non-transparent. Furthermore, the chemotactic gradient across the thin membrane may be difficult to control precisely. The second approach, imaging cells embedded in collagen or other extracellular matrix scaffolds, offers a more physiological environment, but the self-assembly of the matrix fibers allows only limited control over the final pore size (e.g., via adjusting the concentration or temperature), and the pore sizes vary widely even within a single matrix.2, 8 Recently, improvements in microfluidic systems have combined well-controlled chemotactic gradients and 3-D structures to study confined migration along a gradient.13 Nonetheless, many of these systems still have inherent limitations, such as the requirement of continuous perfusion to maintain a stable chemotactic gradient. While such a perfusion approach is well-suited for short-term experiments with fast moving cells such as neutrophils or dendritic cells, it proves more challenging for the study of slower cells (e.g., fibroblasts, cancer cells), which often require observation times of many hours to several days.8 Furthermore, current microfluidic devices often face a dichotomy between the low channel heights (3–5 μm), required to fully confine cells in 3-D, and larger feature heights (>10 μm) that facilitate cell loading and nutrient supply but are too tall to confine cells in the vertical direction as they migrate through the constrictions.
To overcome the limitations of current approaches, we identified the following requirements for an improved system to study cell migration in 3-D environments: (1) easy sample preparation and loading of cells, supporting diverse cell lines; (2) precisely-defined channel geometries, relevant to physiological 3-D conditions; (3) rapid and persistent (hours to days) formation of a stable chemotactic gradient without the need for continuous perfusion; and (4) high spatial and temporal resolution for real-time imaging of cell migration through confined spaces. Here we present a novel microfluidic migration device containing several innovative features to fulfil these requirements. We have successfully tested this device with a broad range of cell lines under different chemotactic gradients, demonstrating that we can detect detailed intracellular dynamics during migration through narrow constrictions. Furthermore, we have used the devices to observe dynamic changes in these cells during migration through narrow constrictions, including nuclear lamina buckling, nuclear volume changes, and chromatin strain during nucleus deformation.
Results and Discussion
Overview of microfluidic migration device design
The devices consist of parallel migration channels formed by a series of PDMS pillars, through which cells migrate along a chemotactic gradient (Fig. 1). The channels can be functionalized with various extracellular matrix proteins such as collagen or fibronectin to promote cell adhesion. The narrow constrictions formed by the pillars mimic the 2 to 30 μm wide pores found in extracellular matrices and interstitial spaces.14, 15 The chemotactic gradient is established via diffusion from a large source reservoir across the constriction channels to a sink reservoir. The two reservoirs are also connected by a 250 μm tall and 1000 μm wide bypass channel to provide a low resistance path for fluid flow, allowing for the rapid equilibration of fluid height between the reservoirs in case of uneven filling or mechanical perturbation during handling of the device. Cells are directly seeded into a 250 μm tall area at the mouth of the constriction channels through two 250 μm tall by 300 μm wide ports near the entrance of the constriction channels. The increased height of these areas before and after the constriction channels ensures sufficient space for cells and ample supply of nutrients while facilitating easy loading; if desired, cells can also be collected after having passed through the constrictions, for example, to analyze differences in gene expression between cells that have successfully passed the constrictions and those that remained on the other side.
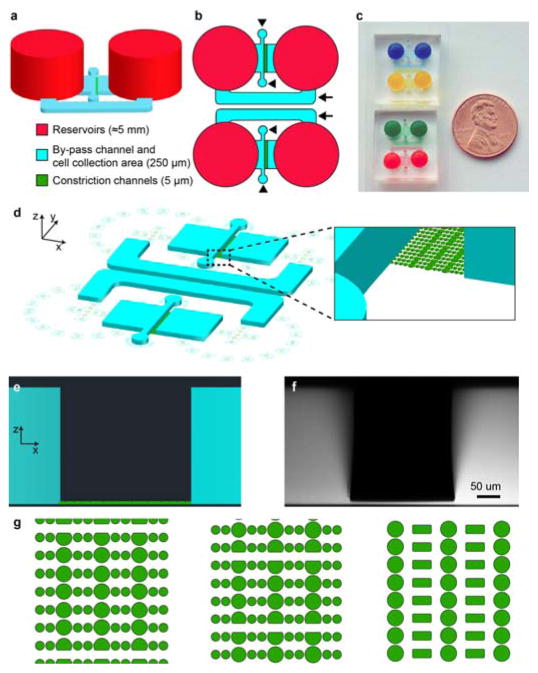
Figure 1. Overview of microfluidic migration device design.
(a) Schematic 3-D model of the device features, showing the large (≈5 mm tall) reservoirs (red), the 250 μm tall chambers and bypass channel (blue), and the constriction channels (green). (b) Top view of two devices side-by-side, demonstrating the small footprint of the device. Arrows indicate the bypass channels; arrowheads the cell loading ports. (c) Photo of two pairs of assembled migration devices. Reservoirs and migration channels have been filled with food coloring to promote visibility. (d) Schematic bird’s eye view of the CAD design, showing the 250 μm tall chambers and bypass channel (blue), the constriction pillars (green), and the alignment markers for hole-punching the PDMS. (e) Side-view of the device design, demonstrating the height difference between the constriction channels (green) and the 250 μm tall chambers (blue). (f) Corresponding 3-D reconstruction of a confocal stack of images obtained using a device filled with 70 kDa dextran conjugated to fluorescent Texas Red. The height measured in the confocal stack is very close to the desired height of 250 μm. (g) Three examples of designs for the constriction channels: (left) channels with three identical constrictions in a row, measuring either 3 μm or 5 μm in width; (center) channels with three sequentially narrower constrictions, decreasing from 5 μm to 3 μm and then 2 μm in width; (right) channels with three identical constrictions in a row and wider spaces between rows of constrictions to avoid deformation of cells in between constrictions. Each of these layouts was designed so that consecutive constrictions were spaced sufficiently far apart to focus on effects during passage through individual constrictions and to minimize confounding effects from residual nuclear deformations. Channels are connected via spaces between the pillars dividing the constriction channels. Each design also includes 15 μm wide through-channels, which serve as a control for migration of cells unimpeded by constrictions.
Design of constriction channels
Current technologies for the observation of 3-D cell migration typically use long, narrow rectangular channels through which cells must travel.6, 16–20 While experiments with such channels have revealed distinct differences in the migration of cells in confining 3-D environments compared to 2-D substrates, their geometries are very different from the constrictions of the physiological micro-environment.14, 15 In our designs we sought to mimic the non-continuous spatial constraints found in the natural microenvironment formed by extracellular matrix fibers and other cells. We devised a design with constrictions formed by adjacent circular PDMS posts that allow high resolution imaging of cell deformation during 3-D migration.
The constriction channels in our devices are collectively composed of dozens of individual channels in parallel, spanning a width of approximately 225 μm. We fabricated several different channel designs for the migration device to suit specific experimental purposes and mimic different physiological environments. The walls of the channels are made up of a series of 15 to 30 μm wide PDMS pillars (Fig. 1g, Supplemental Video 1). In one design, the first constriction in each channel is 5 μm wide, followed by 3 μm, and finally 2 μm wide constrictions (Fig. 1g, right). These constrictions require cells to undergo successively greater deformation during passage through the confined spaces. Such a design can be useful in examining the speed at which cells travel through different sized constrictions, or in determining the smallest pore size a particular cell type can pass through. Wide channels included in the design serve as benchmarks to measure cell migration speed unimpeded by constrictions. Openings across constriction channels allow fluid flow across the constrictions even when some of the channels are blocked by cells, promoting nutrient distribution and uniform gradient formation (see below).
Design of the system to create a stable reproducible gradient
Gradients in microfluidic devices are typically established via the laminar flow of streams of different concentrations of solutes.16 However, fluid flow may cause considerable fluid shear stress to the cells and requires additional external equipment such as pumps and connectors to provide continuous perfusion. Alternative approaches developed to avoid shear stress on the cells have their own limitations: diffusion of chemoattractants across an agarose gel is slow,21 and manual pipetting at the entrance of microchannels results in only very transient gradients.18 A more promising approach is to use diffusion across closely spaced sink and source reservoirs to obtain rapidly formed, stable gradients. This has been previously used in chemotaxis devices to study cell migration in 2-D.22–24
In our system, the gradient is established across two reservoirs separated by constriction channels 5 μm tall and 280 μm wide. The reservoirs are created by punching holes in the PDMS, which are connected to chambers 250 μm tall and end at the constrictions. Thus the shape and the distance between the reservoirs is always the same, regardless of the exact position the holes were punched, resulting in highly reproducible gradients.
Using a bypass channel to prevent convective mass transfer
A major challenge with many diffusion-based microfluidic devices is prevention of fluid flow through the constriction channels caused by uneven fluid levels in the reservoirs. The resulting convective transport prevents the establishment of a linear diffusion-based gradient and can even disrupt an established gradient. Given the high fluidic resistance of micrometer-scale constrictions (Supp. Table 1), equilibration of fluid levels between the reservoirs is very slow, resulting in long delays before stable gradients can form. To address this challenge, we incorporated a larger bypass channel into the device design (Figs. 1b and 1d) capable of rapidly equilibrating differences in fluid height while limiting fluid flow through the constriction channels. To demonstrate the efficacy of the bypass channel design, we modeled the fluid flow between the sink and source reservoirs resulting from uneven filling in the absence or presence of the bypass channel. Without the bypass channel, the characteristic equilibration time constant is on the order of several hours, but is drastically reduced to a few seconds when the bypass channel is included (Supp. Table 1). Accordingly, the time required to equilibrate the fluid levels between the reservoirs given an initial difference of 1 mm is reduced from days to less than a minute when the bypass channel is added (Table 1, Supp. Table 2 and Supp. Fig. 1). This also ensures that the majority of the fluid flow passes through the bypass channel during medium changes or mechanical perturbation of the devices and does not affect the cells in the constriction channels. As a result, our devices are ideally suited for long-term imaging of cell migration.
Table 1.
Results from modeling the fluid exchange between reservoirs in the devices, given an initial fluid height difference of 1 mm between the reservoirs and defining equilibrium as reservoir levels within 1 μm of each other.
| Time to equilibrate without bypass channel | 87 hours |
| Time to equilibrate with bypass channel | 40.3 seconds |
| Total volume exchanged between reservoirs | 9.81 μL |
| Volume exchanged through constriction channels, in presence of bypass channel | 0.0013 μL |
To assess whether diffusion through the large bypass channel could result in equilibration of the concentrations between the sink and source reservoirs, we calculated the diffusion rate of a typical chemoattractant (i.e., platelet derived growth factor, PDGF) across the length of the bypass channel. Over the course of 24 hours, the typical time between medium exchanges, the chemoattractant mass transfer due to diffusion across the bypass channel and the constriction channels was negligible (0.3 % and 0.5% of the initial PDGF concentration, respectively; see Supplementary Information for details on the calculations). These results were confirmed by experiments with fluorescently labeled dextran of comparable molecular weight, which showed that the concentrations of the sink and source reservoirs, and hence the chemotactic gradient, remained stable for at least 24 hours (Supp. Fig. 2).
Validation of rapid and stable gradient formation in device
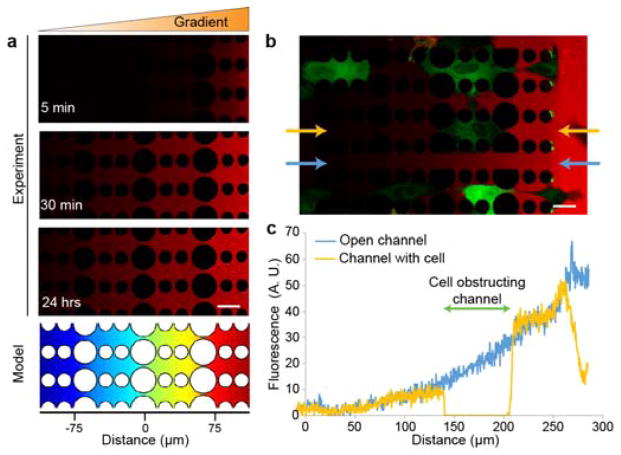
To confirm the rapid formation of a diffusive gradient across the migration device, we conducted experiments with fluorescently-labeled 70 kD dextran placed in the source reservoir. Time-lapse microscopy revealed that a linear gradient formed within 30 minutes and remained stable for at least 24 hours (Fig. 2a). Experiments in which cells were included confirmed that the small openings between the pillars enabled diffusion between individual constriction channels, resulting in a uniform gradient across the entire device (Fig. 2b). For a more detailed analysis of gradient formation, we performed computational modeling (COMSOL) of the device (Fig. 2a and Supp. Fig. 3), showing that stable linear diffusion gradients across the constriction channels were formed within 30 minutes, in strong agreement with imaging results, and indicating that the bypass channel worked as predicted. As fluid flow through the constrictions attenuates within a few minutes, and it takes tens of minutes to form the gradient, we deduce that the gradient forms across the constriction through diffusion without fluid flow.
Figure 2. Rapid and uniform gradient formation across the constriction channels.
(a) Time-lapse series of 70 kDa dextran conjugated to fluorescent Texas Red showing formation of the dextran-Texas Red gradient. The heat map (bottom) depicts the results of a corresponding COMSOL model of the gradient formation after one hour. Scale bar, 25 μm. (b) Dextran-Texas Red gradient (red) formation after 10 minutes in a device containing fluorescently labeled cells (green). Even when some cells block the channels, the 2 μm side openings between channels allow formation of a uniform gradient. Scale bar, 25 μm. (c) Fluorescence intensity profile of the 70 kDa dextran-Texas Red reveals uniform gradient formation despite partial channel occlusion: a channel that is locally obstructed with a cell (yellow plot) shows a very similar gradient as an adjacent, unobstructed channel (blue plot). The two sets of data were taken from the experiment depicted in panel b, with the analyzed channels indicated by the blue and yellow arrows. The area where the cell is obstructing the channel is marked with a green bar in panel c.
To assess the stability of the chemotactic gradient in response to mechanical perturbation of the device, we imaged a device in which a stable gradient of 70 kDa dextran had been established. The device was then turned upside down several times before being placed back on a confocal microscope and imaged for an additional hour (Supp. Fig. 3). Consistent with the mathematical modeling, we found that the gradient remained unchanged after mechanical perturbation of the device and was stable over the observation period, demonstrating the robustness of the device to normal handling.
Cells migrate through constriction channels along a chemotactic gradient
We validated the effectiveness of the gradient on cell migration by varying the concentrations of the chemoattractant and observing the number of cells that migrated through the constrictions. Mouse embryo fibroblasts displayed strongly concentration-dependent migration along a PDGF gradient, with a more than 10-fold increase in cells that passed through the migration channels in a 24 hour period at the highest PDGF concentration compared to cells crossing in the absence of PDGF (Supp. Fig. 2a and b).
One major motivation for the design of the migration device was to image cellular movement and deformation during migration through narrow constrictions at high temporal and spatial resolution. We performed live-cell imaging of a variety of cell types migrating through the device, including cancerous cells such as breast cancer, fibrosarcoma, osteosarcoma and glioblastoma cell lines, human and mouse fibroblasts (Fig. 3, Supp. Fig. 4c and Supp. Video 1). Cells maintained a healthy phenotype throughout the duration of the experiments (ranging from 3 to 14 hours), as evidenced by their morphology, motility, and undisturbed cell division. Additional experiments revealed that cells remained viable in the channels for more than 5 days, making these devices suitable for long-term endpoint incubator studies. In the time-lapse video microscopy studies, cell migration along the chemotactic gradient through the pillars could be followed over several hours by transmitted light (Supp. Fig. 4c, Supp. Video 2) or fluorescence microscopy (Fig. 3, Supp. Video 3). The devices are also suitable for imaging fixed and stained cells: confocal microscopy showed distinct actin fibers above and below the nucleus of fibroblasts, reaching across the constriction (Supp. Fig. 4d), suggesting a potential mechanism by which the cells might pull the nucleus through the narrow opening.25 In addition, the confocal image stacks confirmed that cells fill the entire height of the channel and take on the typically more rounded morphology of cells in 3-D environments, rather than the flat and well-spread morphology seen on rigid 2-D substrates. The high resolution imaging capabilities of fixed and live (Fig. 3) cells are extremely valuable for studies aimed at comparing cellular structure and organization between 2-D and 3-D migration or to investigate how cells can squeeze through micrometer-sized pores.
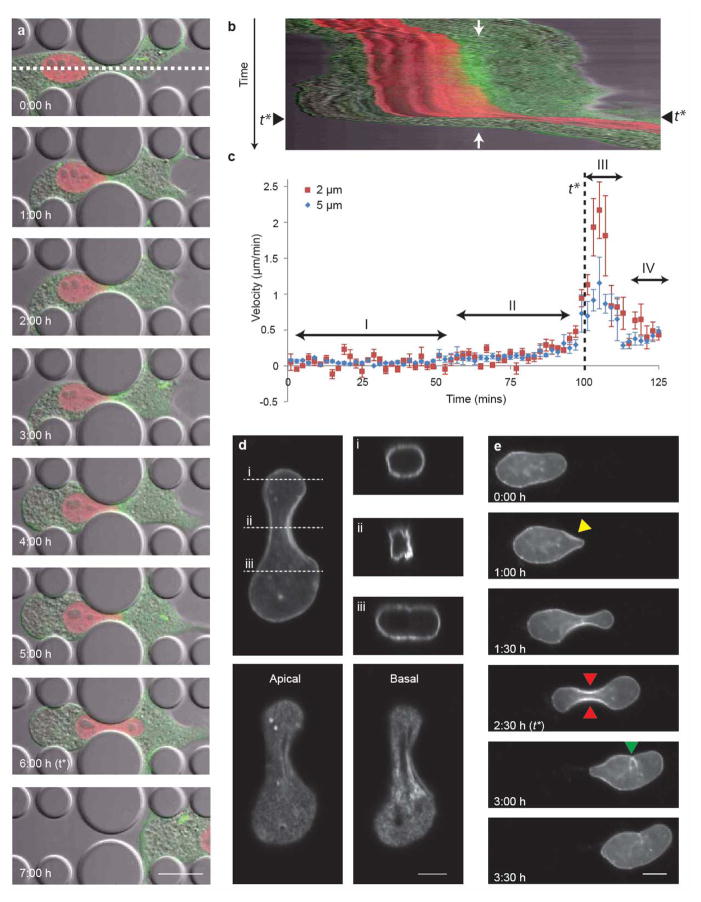
Figure 3. Nuclear deformation during cell migration through narrow constrictions.
(a) Representative time-lapse sequence of a fibroblast expressing mCherry-Histone4 (red) and GFP-actin (green) migrating through a 2 μm-wide constriction. The sequence shows the cell approaching the constriction (0:00 h), stalling as the nucleus encounters the constriction (1:00 h), slowly protruding into the constriction (2:00–5:00 h), and finally succeeding in travelling through the constriction (6:00–7:00 h). The white dashed line indicates the area at the center of the constriction used to generate the kymograph shown in panel b. Scale bar 20 μm. See Supplemental Video 4 for corresponding movie. (b) Kymograph of the cell from panel a moving through the constriction. Each horizontal line in the kymograph corresponds to the image intensity along the white dashed line in panel (a) over time (vertical direction). The position of the center of the constriction is indicated by two white arrows. The time-point at which the center of the nucleus passes the center of the constriction (t*) is marked with black arrowheads. The ‘widening’ of the red signal indicates the elongation of the nucleus into the constriction. (c) Graphical representation of the nuclear velocity for cells migration through 2 μm and 5 μm wide constrictions. The time-point at which the center of the nucleus passes the center of the constriction (t*) is indicated by the black dashed line. The velocity of the nucleus as the cell passes through the constriction can be broken into four phases:3 the nucleus stalls at the entrance of the constriction (I), which is followed by a period of slow increase in speed as the nucleus begins to deform into the constriction (II); once the center of the nucleus passes the obstacle, its speed increases and the nucleus rapidly slips through the constriction (III); subsequently, the cell and nucleus resume their “normal” migration speed (IV). Error bars here and in the subsequent figures represent the standard error of the mean. n = 5 cells for 2 μm constrictions; n = 8 cells for 5 μm constrictions. (d) Confocal 3-D image reconstruction of the nuclear lamina of a human fibroblast expressing GFP-prelamin A migrating through a 3 μm-wide constriction. The side projections, apical and basal images show evidence of folding and buckling of the nuclear lamina in the smallest part of the constriction. Scale bar 5 μm. (e) Time-lapse image series revealing the substantial deformation of the nuclear lamina during migration. Folds and buckling of the nuclear lamina are visible as the nucleus approaches the constriction (yellow arrowhead), as well as within the constriction (red arrowheads) and after the nucleus has exited the constriction (green arrowhead). Scale bar 10 μm. See Supplemental Video 5 for corresponding movie.
Nuclear deformability as a limiting factor during cell migration through narrow constrictions
It has recently been hypothesized that the nucleus constitutes a limiting factor in cell migration through narrow constrictions3, 8 given its increased stiffness relative to the rest of the cell.26 Supporting this hypothesis, we and others have previously shown that migration efficiency through narrow pores (2–5 μm diameter) is dependent on the levels of the nuclear envelope proteins lamin A/C,7, 9, 12 which largely determine nuclear stiffness.11 In each of the cell lines studied in this manuscript, cells were able to migrate through all of the constrictions, including the smallest (2 × 5 μm2). Previous measurements in cells migrating through collagen pores found the physical limit of cell migration to be around 7 μm2 for tumor cells, corresponding to about 10% of the undeformed nuclear cross-section.8 Our migration devices approach this limit, and wild-type mouse embryo fibroblasts required increasingly longer times to migrate through the constrictions when the size was step-wise reduced from 15 × 5 μm2 to 2 × 5 μm2.12
To investigate the role of the nucleus during cell migration through narrow constrictions in more detail, we imaged nuclear deformation in fibroblasts expressing fluorescently-tagged histone H4 (Fig. 3a–c, Supp. Video 4). As expected, migration temporarily stalled when the nucleus encountered the constriction (Fig. 3a, t = 1:00 h); the nucleus then adopted an hourglass shape as it moved through the constriction (t = 6:00 h); and the cell eventually regained its initial velocity after the nucleus passed the obstacle (t = 7:00 h). The progression of the cell and nuclear deformation can be further visualized by plotting a kymograph (Fig. 3b), which reveals more information on the cell and nuclear velocity at given times and positions (Fig. 3c). Consistent with a model previously postulated by Friedl and colleagues,3 fibroblasts migrating through constrictions smaller than the size of the nucleus display a characteristic velocity profile with distinct phases: first, the nuclear velocity stalls as the constriction impedes the forward movement of the nucleus (Fig. 3c, phase I; Fig. 3a, 1:00 h); the nucleus then slowly elongates and protrudes through the constriction as the cell applies increasing forces to the nucleus (Fig. 3c, phase II; Fig. 3a, 1:00 h to 6:00 h); once the center of the nucleus has passed the mid-point of the constriction, the nucleus rapidly slips through the pore, reaching its peak velocity (Fig. 3c, phase III). Finally, the cell and nuclear movement resumes with its initial velocity (Fig. 3c, phase IV).
Interestingly, when comparing the nuclear transit through different sized constrictions, we found that the peak nuclear velocity was higher for the 2 μm wide constrictions than for the 5 μm wide constrictions. The increased nuclear peak velocity may be due to increased (and prolonged) force/elastic energy built-up by the cells and the subsequent rapid release of elastic energy stored in the interaction between the compressed nucleus, the confining PDMS pillars and cytoskeleton.
Nuclear deformation dynamics
Taking advantage of the high-resolution capabilities of our devices, we observed the deformation of different components of the nucleus during migration through narrow constrictions to understand how differences in nuclear structure and composition can modulate the ability of cells to migrate through tight spaces. Of particular interest is the nuclear lamina, a dense protein meshwork underlying the nuclear membrane composed primarily of lamins that recently emerged as an important modulator of 3-D cell migration.7, 9, 27 Mature neutrophils almost completely lack lamins A and C, which results in more deformable nuclei and enables the cells to more efficiently migrate and perfuse through tight spaces.9 Similar adaptations in cancer cells, which often have altered expression of lamins,28 may facilitate invasion and metastatic.7 We have thus decided to further investigate the role of lamins during migration through narrow constrictions.
We first observed the deformation of the nuclear envelope by imaging cells expressing GFP-prelamin A. These experiments revealed large scale nuclear deformations and evidence of nuclear lamina buckling and folding as the nucleus passed through the constriction (Fig. 3d, see also Supp. Video 5). Such buckling behavior is an indication that, although the lamina displays elastic behavior, it is incompressible below a certain threshold, as shown previously by Dahl et al.10 This effect might have been further amplified by the overexpression of GFP-prelamin A in our cells, which could result in more rigid nuclei.9
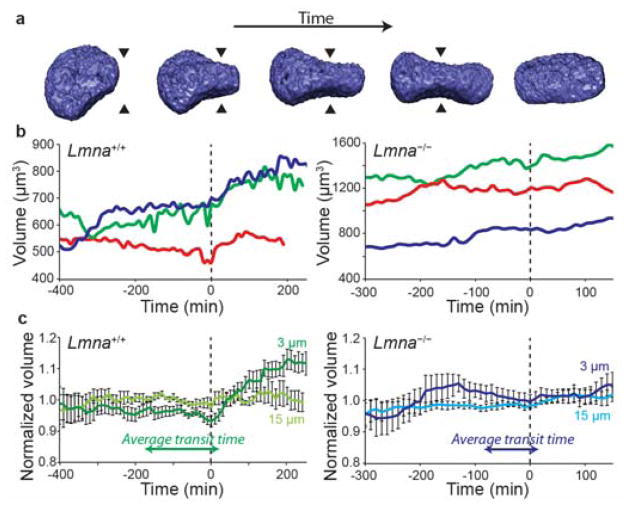
The buckling of the lamina at the constriction could be indicative of a loss of nuclear volume during deformation. In micropipette aspiration experiments, isolated nuclei reduce their volume by as much as 70% by reducing their aqueous fraction and compacting chromatin,29 In fact, it had previously been hypothesized that lamin A/C-deficient cells are able to migrate through narrow constrictions more efficiently than wild-type cells, in spite of their larger nuclei size, because they are able to further compress and reduce their nuclear volume. To address this question, we performed high resolution live confocal microscopy of lamin A/C-deficient and wild-type cells expressing fluorescent histone H2B (Fig. 4a, b). We did not detect any significant reduction in nuclear volume in either cell lines, indicating that nuclear compaction is not a necessary mechanism for passage through narrow constrictions. Interestingly, in wild-type cells we measured a modest (≈15%) increase in nuclear volume immediately after exiting the 3 × 5 μm2 constriction (Fig. 4c), which was not seen in lamin A/C-deficient cells. Our results indicate that nuclear deformability, rather than nuclear compressibility (i.e., reduction in nuclear volume), determines the ability of cells to pass through constrictions that are smaller than the nuclear cross-section. In particular, we found no indication that lamin A/C-deficient cells, which transit through small constrictions significantly faster than wild-type cells,7, 9, 27 show greater changes in nuclear volume than wild-type cells. Instead, the lamin A/C-deficient nuclei exhibit larger and more plastic deformations, enabling them to adjust their shape to the available space and to squeeze through small constrictions faster, despite their larger size. These observations are consistent with previous reports that found that lamin A/C-depleted cells have more deformable nuclei than controls30–32 and undergo plastic deformation, even during non-confined migration.33 Such changes in the mechanical behavior of the nucleus and its interior may be attributed to the role of lamins at the nuclear,10, 34, 35 the nuclear interior,36, 37 or their effect on chromatin organization.32, 38
Figure 4. Volume changes during migration through narrow constrictions.
(a) Representative examples of 3-D reconstructions of the nucleus obtained from confocal image stacks for the volume analysis. The arrowheads indicate the center of the constriction. (b) Representative example traces of the volume of the nucleus of three wild-type (Lmna+/+, left) and three lamin A/C-deficient (Lmna−/−, right) cells during migration through the microfluidic devices. The data was normalized so that time t = 0 min corresponds to the time at which the center of the nucleus passes through the center of the constriction (also indicated with a dashed line). (c) Average volumes of wild-type (left) and lamin A/C-deficient (right) cell nuclei travelling through 3 μm constrictions or 15 μm wide control channels. Values were normalized to the average volume for a given cell during the entire measurement duration. The average transit times for each given cell type through a 3 μm channel are shown to demonstrate that there is no significant loss of volume before passage through a narrow constriction. n = 12 for the wild type cells and n = 13 for the lamin-deficient cells.
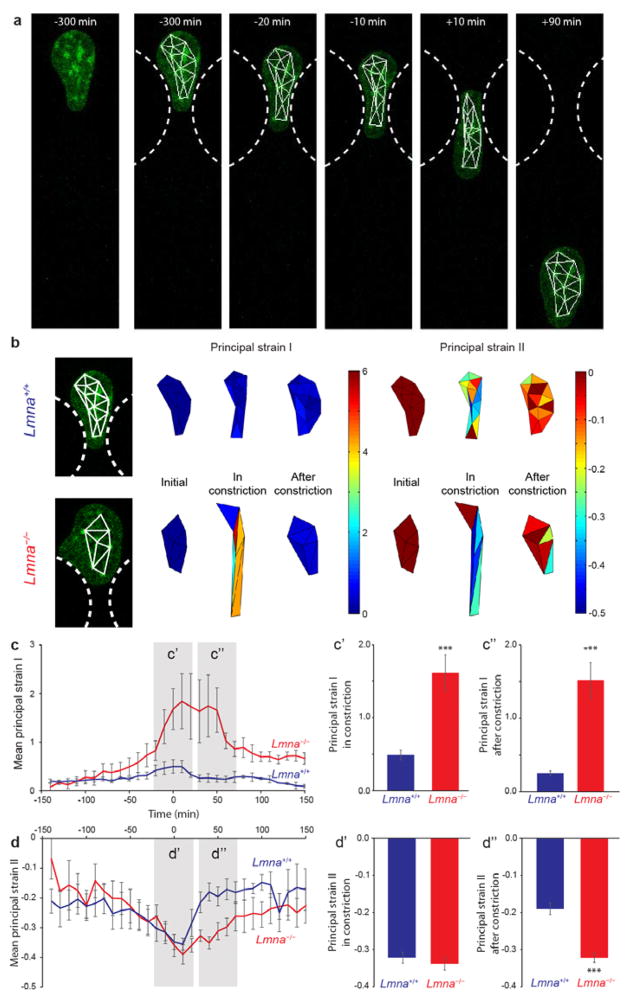
For a more detailed analysis of nuclear deformation, we tracked individual naturally-present dense chromatin foci (i.e., chromocenters) during passage through the constriction (Fig. 5a) and then computed nuclear strain maps by triangulation between these foci, as well as the direction and magnitude of the principal strains for each of these triangles (Fig. 5b–d and Supp. Fig. 5). Lamin A/C-deficient cells exhibited significantly larger nuclear strains as they passed through the constrictions than wild-type cells. In particular, lamin A/C-deficient cells showed larger extension in the migration direction (principal strain EI) than wild-type cells (Fig. 5c), while lateral compression (principal strain EII) inside the constriction was comparable for both cell types (Fig. 5d). Furthermore, the nucleus of the lamin A/C-deficient cells behaved more like a viscoelastic material compared to the mostly elastic nuclei of wild-type cells. This behavior was clearly visible in the time-course of both principal strains: in the wild-type cells, the principal strains return back to pre-deformation values within 30 minutes after transit through the constriction (Fig. 5c, d); conversely, the principal strain values of lamin A/C-deficient cells did not recover within the measurement time-frame, consistent with earlier reports of increased and more plastic nuclear deformability in lamin A/C-deficient cells during migration on 2-D surfaces.11
Figure 5. Strain maps and quantitative strain analysis of nuclear deformation during cell migration through narrow constrictions.
(a) Representative example of a cell expressing fluorescently labeled histones (green) with vertices used for strain calculation superimposed (white). Constrictions are indicated by white dashed lines. Left, initial image, Right, tracked images and triangulation of tracked points. (b) Representative examples of strain maps derived from the deformation of the triangles in a wild-type (top) and lamin A/C-deficient (bottom) cell. Shown are the results for the principal strains EI and EII that represent the maximal and minimal strains in the orthogonal directions at which the shear strain is zero. Principal strain EI typically represented positive strain (extension) and aligned approximately with the cell migration direction; Principal strain EII was negative (compression) and typically perpendicular to the migration direction. For a more detailed explanation of principal strains, see Supp. Fig. 5 (c) Mean area-weighted average principal strain values of wild-type and lamin-deficient nuclei migrating through 3 μm-wide constrictions. Average principal strain EI during the deformation (t = −20 min to +20 min) and after the deformation (t = 30 min to 70 min) was compared in c′ and c″. The lamin A/C-deficient cells had significantly larger principal strains both during and after nucleus passage through the constriction. (d) Average orthogonal principal strain EII of wild-type and lamin-deficient nuclei migrating through 3 μm-wide constrictions. The average principal strain EII during the deformation (t = −20 min to +20 min) and after the deformation (t = 30 min to 70 min) was compared in d′ and d″. The orthogonal principal strain EII (i.e., compression) during migration through the constriction (d′) was not significantly different between the two cell types, indicating that the two cell types underwent similar compressive strain during nuclear deformation. The nuclear strain in the wild-type cells immediately after deformation decreased quickly (d″), indicating that wild-type nuclei behave as elastic bodies, whereas the lamin A/C-deficient nuclei exhibit more viscoelastic behavior (slower recovery) with plastic deformations. (***, P < 0.001, compared to wild-type. n = 4 for wild type cells and n = 5 for lamin-deficient cells.)
To assess whether successful passage through narrow constrictions could condition the cells to migrate through subsequent constrictions more easily, we compared the times it takes for human skin fibroblasts to migrate through a device with multiple constrictions of the same size (Fig. 1g, right). Cells that passed through two consecutive constrictions showed a trend towards faster transit times for the second constriction (Supp. Fig. 6a). However, this effect was not statistically significant (P = 0.29, n = 12), possibly due to the fact that only few cells moved through multiple constrictions within the 24 hour observation period. Therefore, we also analyzed the average transit times of all cells migrating through the first or second row of constrictions, without requiring individual cells to pass through consecutive constrictions during the observation window. In this case, there was a significant decrease in the transit time for the second row of constrictions compared to the first row of constriction (Supp. Fig. 6a). However, these results may be biased towards faster migrating cells, which would be more likely to have reached the second constriction by the time we performed the experiments (i.e., 24 hours after seeding). The existence of such a potential bias is supported by the overall faster transit times of the cells that migrated through multiple constrictions compared to cells that only cross a single constriction during the experimental observation period (Supp. Fig. 6a). Nonetheless, both analysis approaches showed similar trends of a 25–50% reduction in transit time for the second constriction relative to the transit time for the first constriction (Supp. Fig. 6b). Furthermore, we observed similar results when studying invasive breast cancer cells (Supp. Fig. 6c), suggesting that cells do become better at passing through subsequent constrictions.
The above studies were carried out with migration devices in which consecutive constrictions were separated by ≈100 μm, so that the cells required several hours to cover the distance between the constrictions. Human and mouse fibroblasts typically took 400 – 500 minutes to pass from one constriction to the next, which is several times longer than the recovery time of wild-type nuclei, i.e. ≈30 min (Fig. 5c). As such, we conclude that the observed increase in migration efficiency through consecutive constrictions is not due to residual changes in nuclear shape, but likely arises from cellular adaptations that occurred during or after the passage through the first constriction. Such adaptations could include changes in the structure and composition of the nuclear envelope, for example by phosphorylation or degradation of lamins A/C,39 which would render the nucleus more deformable, or by re-organization of cytoskeletal elements to facilitate squeezing the nucleus through narrow constrictions, such as recruitment of non-muscle myosin IIB to the back of the nucleus.40
Taken together, these results suggest that increased deformability, rather than increased compressibility, is responsible for the ability of lamin A/C-deficient cells to move through narrow constrictions more easily than wild-type cells,12 despite their larger nuclear size. This is particularly relevant in cancer research, where cancer cell invasion requires passage through dense extracellular matrices and tight interstitial spaces, which may be aided by reduced expression of lamin A/C as reported in breast cancer and other tumors.28 Identifying the molecular mechanisms that facilitate cancer cell migration through narrow constrictions may offer new avenues for the development of therapeutic approaches to combat cancer metastasis.
Experimental Methods
Cell lines
NIH 3T3 cells (a kind gift from Dr. Anthony Bretscher) were cultured in D10 medium, consisting of high glucose DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Fisher Scientific) and penicillin/streptomycin (Lonza). MEFs from wild-type mice were generously provided by Dr. Colin Stewart41. Human SV40 virus-transformed skin fibroblasts (GM00637J), originally obtained from Coriell Cell Repositories, were stably modified with a tetracyclin-regulated (Tet-off) GFP-lamin A construct42 provided by Dr. Tom Glover and maintained as described previously.43 Glioblastoma cells were a kind gift from Dr. Steven S. Rosenfeld (Cleveland Clinic). 293TN cells were cultured in D10 media supplemented with 110 mg/L sodium pyruvate.
Plasmids
mCherry-Histone4 was generated in two steps. First cDNA for mCherry (kind gift from Dr. P. Patwari44 was PCR amplified (Phusion, NEB) introducing the restriction enzyme site for EcoRI and the Kozak sequence on the 5′ site of mCherry and on the 3′ site a linker (5′-GGTGGAGGCGGAGGT-3′) and the restriction sites for AvrII and NotI (Fwd-mCherry 5′-GTAGAATTCTAACCATGGTGAGCAAGGGCG-3′ and Rev-mCherry 5′-CTAGCGGCCGCTACCCTAGGACCTCCGCCTCCACCCTTGTACAGCTCGTCCATGCCG-3′). The PCR product and the vector, pCDH-CMV-MCS-EF1-blasti (a kind gift from Dr. R. Kakkar, a modified version of SBI product, in which the puromycin resistance gene had been replaced with a blasticidin S resistance gene, were digested with EcoRI and NotI (NEB) and then ligated. Histone4 (RefSeq NM_001034077.4) was amplified from human cDNA using the primers Fwd-H4: 5′-CCTAGGTCCGGCAGAGGAAAGGGC-3′ introducing an AvrII restriction site and Rev-H4 5′-GCGGCCGCCTAGCCTCCGAAGCCGTACAG-3′ introducing a NotI restriction site. PCR product and mCherry containing vector were digested with AvrII and NotI and ligated into the final construct: pCDH-CMV-mCherry-Histone4-EF1-blasti. copGFP-beta-actin was generated in a similar procedure. copGFP was PCR amplified with the primers Fwd-copGFP 5′-GAATTCGCCACCATGACCGGTGGCGGAGAGAGCGACGAGAGCGGCC-3′ introducing EcoRI and Rev-copGFP 5′-GCGGCCGCTACCCTAGGTCCGCCGCGAGATCCGGTGGAGCCG-3′ introducing a linker (GGCGGA) and the restrictions sites for AvrII and NotI. The amplicon and vector (pCDH-CMV-MCS-EF1-puro, SBI) were digested with EcoRI and NotI and ligated. Human beta actin (RefSeq NM_001101.3) was PCR amplified using the primers Fwd-ACTB 5′-CCTAGGGATGATGATATCGCCGCGCTCG 3′ introducing AvrII and Rev-ACTB 5′-GCGGCCGCCTAGAAGCATTTGCGGTGGACGATG-3′. Amplicon and copGFP vector were digested with AvrII and NotI and ligated to the final construct pCDH-CMV-copGFP-hBetaActin-EF1-puro.
Lentiviral infection
NIH 3T3 fibroblasts were modified with lentiviral particles to stably express mCherry-Histone4 or copGFP-beta-actin. Pseudoviral particles were generated by transfection of 293TN cells with the cloned pCDH vectors and the second generation lentiviral vectors psPAX2 and pMD2.G (provided by Dr. Didier Trono) using PureFection transfection reagent following the manufacturer’s instruction. The medium was changed the next day and virus-containing supernatant was collected the following two days, centrifuged at 2,000 × g for 15 min at 4 °C and filtered through a 0.45 μm pore syringe filter. The filtrate was supplemented with PEG-it following the manufacturer’s instructions (SBI). Briefly, PEG-it was added to the filtrate, stored at 4 °C for 1–2 days, and centrifuged at 3,000 × g for 30 min at 4 °C. The pellet was suspended in PBS (Invitrogen) as a 100× stock and stored at −80 °C. For transduction of NIH 3T3 cells, D10 was supplemented with concentrated pseudoviral particles and 8 μg/mL Polybrene (Sigma). For selection of virally transduced cell lines, 5 μg/mL puromycin and/or 8 μg/mL blasticidin S (both from Sigma) were added to the culture medium 48 h post transduction.
Device fabrication
Molds of the final device design were fabricated using 2-layer SU-8 photolithography. In this process, a 5 μm thick layer of SU-8 photoresist was first patterned using one mask, and a second layer of SU-8, 250 μm thick, was patterned on top with a different mask as described below, resulting in SU-8 features with two different heights. Device designs were drawn in AutoCAD (Autodesk, San Rafael, CA) and photolithography masks were fabricated using a Heidelberg DWL 2000 Mask Writer.
SU-8 2010 photoresist (MicroChem, Newton, MA) was spun onto a clean, dehydrated (baked at 200 °C for 30 minutes) 4-inch silicon wafer to achieve a height of approximately 5 μm. The photoresist-coated wafer was then baked at 60 °C for 10 minutes and exposed to ultraviolet light on an ABM contact aligner (Scotts Valley, CA). The wafer was baked once again at 95 °C for 3.5 minutes. The wafer was developed in SU-8 developer (Microchem, Newton, MA); tape was placed over the alignment marks and a 250 μm tall layer of SU-8 2075 was spun on top of the first layer. After removing the tape, the wafer was baked at 50 °C for 24 hours and exposed using the mask for the 250 μm features and aligning the mask to the wafer with the optical alignment marks. The wafer was then baked at 95 °C for 30 minutes, cooled down and developed in SU-8 developer. Finally, the wafer was coated with (tridecafluoro-1,1,2,2-tetrahydrooctyl) trichlorosilane, providing an anti-stiction layer to facilitate demolding.
A sample of two-component Sylgard 184 PDMS kit (Dow Corning, Midland, MI), was mixed in a 10:1 ratio, following manufacturer’s recommendations, and then degassed in a desiccator under vacuum to remove air bubbles. The solution was then poured onto the SU-8 master in a petri dish and placed in an oven at 65 °C for 2 hours to cure the PDMS. After curing, the PDMS was peeled from the wafer and cut to size. The reservoirs for each device were cut out using a 6 mm diameter Harris Uni-Core biopsy punch (Harris Uni-Core, Ted Pella, Inc.), while the perfusion channel inlet and outlet for cell seeding were cut out with a 0.75 or 1.2 mm diameter punch. Coverslips were cleaned with 0.2 M HCl, rinsed with water and isopropanol, dried, and placed into a plasma cleaner (Harrick Plasma, Ithaca, NY) along with PDMS devices. The coverslips and PDMS pieces were treated with air plasma for 5 minutes at 200 Watts. The PDMS and glass coverslips were then placed into contact with each other and baked on a hotplate at 95 °C for 5 minutes to improve bonding. Devices were then cooled at room temperature and filled with 70% ethanol inside a tissue-culture hood to sterilize the migration device. The devices were then rinsed with sterile water by aspirating the ethanol solution and filling one well with sterile water to flush out the ethanol, repeating once.
Experimental validation of gradient formation
Devices with a constriction channel height of 5 μm were prepared as described above and further incubated overnight with 3% bovine serum albumin (BSA) to prevent adsorption of dye to the PDMS surfaces. Prior to the experiments, the BSA solution was removed from both reservoirs of each device and replaced with fresh PBS; the PBS in the source reservoir was supplemented with Texas Red-labeled 70 kDa dextran. A glass coverslip was placed on top of the reservoirs to minimize evaporative loss of fluid over the course of the experiment. The devices were imaged on a confocal microscope (Zeiss LSM 700) and images of the constriction channels were taken at regular intervals, starting as soon as the device was filled and placed on the microscope.
Fluorescence intensities across the constriction channels, the 15 μm wide channels, and in the 250 μm tall sections of the devices were analyzed in ImageJ. Briefly, a “plot profile” was measured along a section spanning the entire length of the channel but excluding the constriction pillars to compute the average fluorescence intensity distribution along the channel length. In addition, we determined the fluorescence intensity values inside the sink and source reservoirs at the same time-points. These data were then binned and expressed as a normalized concentration, where 1 corresponds to the maximum observed concentration and 0 to the minimum observed concentration:
| (16) |
Where Intensity(x) represents the local fluorescence intensity, IntensityMax the maximal fluorescence intensity value in the device, and IntensityMin the minimal fluorescence intensity value measured in the device (after binning).
Device preparation and cell seeding
Following the sterile water rinse, devices that were destined for cell migration studies were filled with 0.2 mg/mL fibronectin (Millipore) in phosphate buffered saline (PBS) and incubated for at least 2 hours at 37 °C. Cells were trypsinized, counted and suspended in complete medium (DMEM containing 10% fetal bovine serum) at a density of 5 × 106 cells/mL. The migration devices were rinsed with complete medium and aliquots of 4 μL were added to the cell inlet ports, corresponding to 20,000 cells. The cells were then allowed to attach overnight before changing the medium.
Migration studies to assess the effect of the chemoattractant were carried out in the incubator. Twenty four hours after seeding NIH 3T3 mouse embryonic fibroblasts (MEF), images were taken of the migration devices. The medium was then changed in both reservoirs simultaneously: the medium on the side of the cells was replaced with complete medium, whereas the reservoir towards which the cells migrated was replaced with complete medium containing concentrations of PDGF varying from 0 to 200 ng/mL. Migration devices destined for live cell imaging were treated similarly: the medium was replaced 24 hours after seeding with phenol-red free medium containing 25 mM HEPES (Gibco). Immediately following medium change, the devices were covered with a coverslip seal the device and limit medium evaporation.
Cell migration speed analysis
To assess the effectiveness of the chemoattractive gradient on migration, pictures of the migration devices were taken 24 and 48 hours after seeding NIH 3T3 fibroblasts and the media were changed every day. Images were acquired with a Zeiss epifluorescence microscope using a 10× objective. Results were reported as the average number of cells that had migrated through the devices, over the course of two experiments.
Devices destined for live cell imaging were placed on the heated stage of a Zeiss LSM700 confocal microscope (AxioObserver) equipped with a CCD camera (CoolSNAP EZ, Photometrics), and a motorized stage (Zeiss). Images were acquired either in confocal or epifluorescence modes, using 20× air or 63× oil immersion objectives. Positions were set and imaged every 2 to 10 minutes. Image stabilization was achieved using a focus stabilizer and x-y-drift was corrected after the experiment using a cross-correlation algorithm in MATLAB.
The position of the front and back of the nucleus were measured in time-lapse experiments in which images were taken every 2 minutes using a 63× objective. The velocity of the nucleus was then determined as the average of the front and back positions over time. Kymographs were generated in MATLAB along the migration direction by averaging a 7 pixel wide line to a single value and plotted over time. The centre of the constriction was recorded at this point. The time values were normalized to the time at which the centre of the nucleus passes the centre of the constriction and data from multiple cells were then averaged.
Immunohistochemistry
Migration devices were prepared as mentioned above for live cell imaging. Cells were fixed in cytoskeletal buffer (10 mM MES pH 6.1, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, and 5 mM MgCl2) containing 0.3% glutaraldehyde and 0.25% Triton X-100 for 30 min at room temperature and incubated overnight at 4 °C in cytoskeletal buffer containing 2% glutaraldehyde. The samples were incubated with 0.1% NaBH4 (freshly-prepared in PBS) for 30 minutes and washed three times with PBS, 8 hours each at 4 °C. (Adapted from a previous protocol.45) The samples were blocked overnight in blocking solution (10% horse serum (vector), 3% BSA (Sigma) in PBS) and stained with primary antibody (rabbit anti-vimentin, a kind gift from Dr. Thomas Magin) for a minimum of 18 hours at 4 °C. Following three washes with PBS, (8 hours each) samples were incubated with secondary antibody in blocking solution (Invitrogen) for 18 h at 4 °C. After two 8 h PBS washes the samples were stained with fluorescent conjugated phalloidin (Invitrogen) overnight followed by a Hoechst 33342 (Invitrogen) stain for 3 h at 4 °C. The samples were stored in 0.05% NaN3 (Sigma) in PBS at 4 °C and imaged the same day.
Volume measurements
Confocal images of cells in migration channels expressing histone H2B mNeonGreen were obtained at planes 0.33 μm apart and every 10 minutes. These images were saved in ZEN software and analyzed using Volocity software. To ensure that the reconstructions reflected the true shape of the nucleus, settings were used in the software such as filling of holes and noise removal. The software calculated volume, surface area and centroid position measurements for each time point. Only cells that had not yet entered the constrictions were picked for these measurements, resulting in 12 cells for the wild-type cells and 13 cells for the lamin-deficient cells. For comparison, the volume values were normalized to the average volume during the entire measurement. These were then averaged and plotted.
Nuclear deformation measurements
Confocal images containing several easily identifiable chromocenters (bright H2B-mNeonGreen foci) were chosen for analysis. The individual chromocenters were tracked using the Manual Tracking ImageJ plugin. As foci showed little displacement in Z-direction, tracking was limited to in-plane displacements. Intranuclear deformation maps were computed using the Green-Lagrange strain definition based on the change in length of the vertices of triangles defined by the chromatin foci, following procedures previous established for cytoskeletal deformation fields.46 In brief, Delaunay triangulation was performed to generate a set of triangles from the tracked points in which the circumcircle associated with each triangle contains no other point in its interior. The Lagrangian strain tensor was then computed for each triangle at each time point based on the following equation, ds2 − dS2 = 2(Exx dax2 + 2 Exy dax day + Eyy day2), where ds is the distance between adjacent vertices of interest in the deformed state, dS the initial distance between the same vertices, dax and day are the projected initial distances on the x- and y-axis, respectively, and Exx, Exy, and Eyy the three components of the symmetric strain tensor, E. The above relationship can be applied to the three sides of each triangle, resulting in a system of three equations that can be solved for Exx, Exy, and Eyy. The resulting strain tensor then describes the average strain inside the triangle formed by three adjacent vertices over time. The magnitude and orientation of the principal strains EI and EII were computed from the eigenvalues an eigenvectors of the strain tensor E. The average magnitude for each principal strain was calculated for each cell at each time point, using the (initial) area of each triangle to weigh the contribution of each triangle to the overall nuclear strain. These were then averaged for each time point and plotted.
Statistical analysis
All cell migration data are based on at least three independent experiments. Statistical analysis was performed with PRISM 3.0 software (GraphPad). Groups were compared by unpaired Student’s t-test with Welch’s correction to allow for different variances. Data are expressed as mean ± S.E.M. For all experiments, a two-tailed P-value of less than 0.05 was considered significant. Error bars represent the standard error of the mean.
Conclusions
A better understanding of how cells overcome the mechanical constraints and confinement of their 3-D surroundings is highly relevant for a variety of physiological phenomena and diseases that require cell migration through dense tissues or interstitial space, including embryonic development, leukocyte migration to sites of inflammation, or cancer metastasis. We have designed a microfluidic device capable of imaging live cell migration at high resolution in physiologically-relevant 3-D environments. Using this device, we have quantified the dynamic deformations as cells squeeze through tight spaces. We identified specific differences in nuclear mechanics in cells lacking lamins A/C, which is poor prognosticator in certain types of cancer.47, 48 Future applications will further investigate the molecular mechanisms by which (cancer) can overcome the resistance of the large nucleus to migrate through narrow constrictions, assess the consequences of the considerable forces exerted on the nucleus on nuclear and genomic integrity, as well as further minimize the size of the constrictions to test the physical limits of 3-D cell migration.
Supplementary Material
Acknowledgments
The authors thank Kelly Karman, and Jinwoo Choi for their help with the device design and fabrication, and Chin Yee Ho for movies showing glioblastoma migration. We thank Drs. Anthony Bretscher, Colin Stewart, Tom Glover and Steven S. Rosenfeld for supplying cells, as well as Dr. Thomas Magin for antibodies. We thank Drs. Parth Patwari and Rahul Kakkar for providing plasmids. We acknowledge Ileana D’Aloisio and Kathy Zhang for their help with file formatting and image analysis. This work was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (Grant ECCS-0335765). This work was supported by National Institutes of Health awards [R01 NS059348 and R01 HL082792]; the Department of Defense Breast Cancer Idea Award [BC102152]; a National Science Foundation CAREER award to J Lammerding [CBET-1254846]; and a Pilot Project Award by the Cornell Center on the Microenvironment & Metastasis through Award Number U54CA143876 from the National Cancer Institute. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Notes and References
- 1.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, Martin SS, Konstantopoulos K. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedl P, Sahai E, Weiss S, Yamada KM. Nature reviews Molecular cell biology. 2012;13:743–747. doi: 10.1038/nrm3459. [DOI] [PubMed] [Google Scholar]
- 3.Friedl P, Wolf K, Lammerding J. Current opinion in cell biology. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrie RJ, Yamada KM. Journal of cell science. 2012;125:5917–5926. doi: 10.1242/jcs.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tozluoglu M, Tournier AL, Jenkins RP, Hooper S, Bates PA, Sahai E. Nature cell biology. 2013;15:751–762. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- 6.Stroka KM, Jiang H, Chen SH, Tong Z, Wirtz D, Sun SX, Konstantopoulos K. Cell. 2014;157:611–623. doi: 10.1016/j.cell.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE. The Journal of cell biology. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. The Journal of cell biology. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J. The Journal of biological chemistry. 2013;288:8610–8618. doi: 10.1074/jbc.M112.441535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahl KN, Kahn SM, Wilson KL, Discher DE. Journal of cell science. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 11.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. The Journal of biological chemistry. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 12.Davidson PM, Denais C, Bakshi MC, Lammerding J. Cellular and Molecular Bioengineering. 2014 doi: 10.1007/s12195-014-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim BJ, Wu M. Annals of biomedical engineering. 2012;40:1316–1327. doi: 10.1007/s10439-011-0489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoitzner P, Pfaller K, Stossel H, Romani N. The Journal of investigative dermatology. 2002;118:117–125. doi: 10.1046/j.0022-202x.2001.01631.x. [DOI] [PubMed] [Google Scholar]
- 15.Weigelin B, Bakker GJ, Friedl P. IntraVital. 2012;1:32–43. doi: 10.4161/intv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong Z, Balzer EM, Dallas MR, Hung WC, Stebe KJ, Konstantopoulos K. PloS one. 2012;7:e29211. doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson K, Lewalle A, Fritzsche M, Thorogate R, Duke T, Charras G. Nature communications. 2013;4:2896. doi: 10.1038/ncomms3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jowhar D, Wright G, Samson PC, Wikswo JP, Janetopoulos C. Integrative biology : quantitative biosciences from nano to macro. 2010;2:648–658. doi: 10.1039/c0ib00110d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao VLS, Tata U, Raj G, Hsieh JT, Nguyen K, Chiao JC. J Nanotechnol Eng Med. 2010;1:021003-021003-021006. [Google Scholar]
- 20.Mak M, Reinhart-King CA, Erickson D. Lab on a chip. 2013;13:340–348. doi: 10.1039/c2lc41117b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haessler U, Pisano M, Wu M, Swartz MA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5614–5619. doi: 10.1073/pnas.1014920108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berthier E, Surfus J, Verbsky J, Huttenlocher A, Beebe D. Integrative biology : quantitative biosciences from nano to macro. 2010;2:630–638. doi: 10.1039/c0ib00030b. [DOI] [PubMed] [Google Scholar]
- 23.Muinonen-Martin AJ, Veltman DM, Kalna G, Insall RH. PloS one. 2010;5:e15309. doi: 10.1371/journal.pone.0015309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amadi OC, Steinhauser ML, Nishi Y, Chung S, Kamm RD, McMahon AP, Lee RT. Biomedical microdevices. 2010;12:1027–1041. doi: 10.1007/s10544-010-9457-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isermann P, Lammerding J. Current biology : CB. 2013;23:R1113–1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowat AC, Lammerding J, Herrmann H, Aebi U. BioEssays : news and reviews in molecular cellular and developmental biology. 2008;30:226–236. doi: 10.1002/bies.20720. [DOI] [PubMed] [Google Scholar]
- 27.Davidson PM, Denais C, Bakshi MC, Lammerding J. Cell Mol Bioeng. 2014;7:293–306. doi: 10.1007/s12195-014-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denais C, Lammerding J. Advances in experimental medicine and biology. 2014;773:435–470. doi: 10.1007/978-1-4899-8032-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowat AC, Lammerding J, Ipsen JH. Biophysical journal. 2006;91:4649–4664. doi: 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. The Journal of clinical investigation. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broers JL, Peeters EA, Kuijpers HJ, Endert J, Bouten CV, Oomens CW, Baaijens FP, Ramaekers FC. Human molecular genetics. 2004;13:2567–2580. doi: 10.1093/hmg/ddh295. [DOI] [PubMed] [Google Scholar]
- 32.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. The Journal of cell biology. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwerger M, Roschitzki-Voser H, Zbinden R, Denais C, Herrmann H, Lammerding J, Grutter MG, Medalia O. Journal of cell science. 2015;128:3607–3620. doi: 10.1242/jcs.171843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. Molecular biology of the cell. 2015 doi: 10.1091/mbc.E15-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moir RD, Yoon M, Khuon S, Goldman RD. The Journal of cell biology. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hozak P, Sasseville AM, Raymond Y, Cook PR. Journal of cell science. 1995;108(Pt 2):635–644. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- 38.Nikolova V, Leimena C, McMahon AC, Tan JC, Chandar S, Jogia D, Kesteven SH, Michalicek J, Otway R, Verheyen F, Rainer S, Stewart CL, Martin D, Feneley MP, Fatkin D. The Journal of clinical investigation. 2004;113:357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buxboim A, Swift J, Irianto J, Spinler KR, Dingal PC, Athirasala A, Kao YR, Cho S, Harada T, Shin JW, Discher DE. Current biology : CB. 2014;24:1909–1917. doi: 10.1016/j.cub.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas DG, Yenepalli A, Denais CM, Rape A, Beach JR, Wang YL, Schiemann WP, Baskaran H, Lammerding J, Egelhoff TT. The Journal of cell biology. 2015;210:583–594. doi: 10.1083/jcb.201502039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. The Journal of cell biology. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glynn MW, Glover TW. Human molecular genetics. 2005;14:2959–2969. doi: 10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- 43.Verstraeten VL, Peckham LA, Olive M, Capell BC, Collins FS, Nabel EG, Young SG, Fong LG, Lammerding J. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4997–5002. doi: 10.1073/pnas.1019532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patwari P, Chutkow WA, Cummings K, Verstraeten VL, Lammerding J, Schreiter ER, Lee RT. The Journal of biological chemistry. 2009;284:24996–25003. doi: 10.1074/jbc.M109.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu K, Babcock HP, Zhuang X. Nature methods. 2012;9:185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helmke BP, Rosen AB, Davies PF. Biophysical journal. 2003;84:2691–2699. doi: 10.1016/S0006-3495(03)75074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Z, Wu L, Weng D, Xu D, Geng J, Zhao F. Journal of experimental & clinical cancer research : CR. 2009;28:8. doi: 10.1186/1756-9966-28-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willis ND, Wilson RG, Hutchison CJ. Biochemical Society transactions. 2008;36:1350–1353. doi: 10.1042/BST0361350. [DOI] [PubMed] [Google Scholar]
- 49.McGinty B. Pnncipal Stresses & Strains. [accessed 19 October 2015];Continuum Mechanics. < http://www.continuummechanics.org>.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.