Abstract
The function of human regulatory regions depends exquisitely on their local genomic environment and cellular context, complicating experimental analysis of the expanding pool of common disease- and trait-associated variants that localize within regulatory DNA. We leverage allelically resolved genomic DNaseI footprinting data encompassing 166 individuals and 114 cell types to identify >60,000 common variants that directly impact transcription factor occupancy and regulatory DNA accessibility in vivo. The unprecedented scale of these data enable systematic analysis of the impact of sequence variation on transcription factor occupancy in vivo. We leverage this analysis to develop accurate models of variation affecting the recognition sites for diverse transcription factors, and apply these models to discriminate nearly 500,000 common regulatory variants likely to affect transcription factor occupancy across the human genome. The approach and results provide a novel foundation for analysis and interpretation of noncoding variation in complete human genomes, and for systems-level investigation of disease-associated variants.
INTRODUCTION
The regulatory DNA compartment of complex metazoan genomes collectively instructs the gene expression programs underlying development, differentiation, and environmental responses. The information encoded in regulatory DNA is actuated through the cooperative binding of sequence-specific transcription factors (TFs) in place of a canonical nucleosome, resulting in focal alteration of chromatin structure that is detectable through markedly increased nuclease sensitivity1. Comprehensive detection of DNaseI hypersensitive sites (DHSs) enables delineation of all recognized functional classes of regulatory elements, and, applied systematically across hundreds of cell and tissue types and states2, has yielded deep catalogues of human regulatory DNA. Common variants associated with diverse human diseases and phenotypic traits are concentrated in regulatory DNA marked by DHSs3, as are expression quantitative trait loci (eQTLs)4, implicating regulatory variation as an important mediator of quantitative human phenotypes.
Assessment of the functional consequences of regulatory variation is complicated by several factors. It has long been recognized that regulatory elements are finely tuned for their native chromatin and chromosomal environments within specific cell types5. Regulatory elements interact with cognate target gene(s) typically located at some distance (tens to hundreds of kilobases)2,6; these interactions may in turn be influenced by interposing genes7. Regulatory DNA function also critically depends on the identity and precise configuration of TF recognition sites8, together with the modification state of immediately flanking chromatin9,10. As such, accurate assessment of the potential impact of genetic variation on a given regulatory region should be made within its native context in vivo, in a cognate cell type.
The state of chromatin remodeling (i.e., nuclease sensitivity11) of regulatory DNA is exquisitely sensitive to the occupancy of individual TFs. High sequencing depth at DHSs results in an effective resequencing of regulatory regions, in turn enabling de novo identification of genotypes directly from the DNase I reads3,12,13. Thus perturbation of TF occupancy by genetic variants that impact the DNA recognition interface within their endogenous site in vivo can be accurately detected by allele-specific DNase-seq, with the sensitivity dependent on the number of DNaseI cleavages (i.e., sequencing depth). To date, however, this approach has only been applied in a limited fashion, with delineation of a relatively small number of regulatory variants4,14–20.
Here we systematically combine regulatory DNA genotyping with allelically resolved DNase-seq to over 114 cell and tissue types and states sampled from 166 individuals. We uncover an expansive trove of regulatory DNA variants that directly impact the chromatin architecture of individual regulatory regions in an allele-specific fashion. While imbalanced variants are concentrated at sites of TF-DNA recognition, a substantial fraction of variation within regulatory DNA regions is buffered in a context-dependent manner. By creating dense in vivo profiles of variation affecting diverse TF families, we further identify nearly 500,000 common variants strongly predicted to affect TF activity. Collectively, our results reveal genetic effects on TF activity at unprecedented scale.
RESULTS
Profiling of variation impacting chromatin accessibility
We collected 493 high-resolution DNase-seq profiles of genome-wide regulatory activity including both previously published and novel data, all generated through a uniform pipeline (Fig. 1a and Supplementary Tables 1–4). Each profile was sequenced to a median depth of 75 * 106 nonredundant autosomal reads and total sequencing comprised 26.2 * 109 reads. These samples comprise diverse cultured primary cells, cultured multipotent and pluripotent progenitor cells, and fetal tissues. We specifically excluded low-quality and potentially aneuploid samples to avoid artificial bias (Online Methods). We developed a pipeline using SAMtools21 to identify single nucleotide polymorphisms (SNPs) directly from the DNase I sequencing reads for each individual represented. We found an average of 26,176 heterozygous sites per individual, depending largely on total sequencing depth (Supplementary Table 3). We validated our genotypes against Illumina 1M Duo array data available from the ENCODE project for 23 individuals in common22. At SNPs represented in both data sets, we measured an average specificity of 99.7% and sensitivity of 99.4% at genotypes passing our filters (Supplementary Table 5), and a raw sensitivity of up to 73% at sites of high (>32×) sequencing (Supplementary Fig. 1 and Supplementary Table 5).
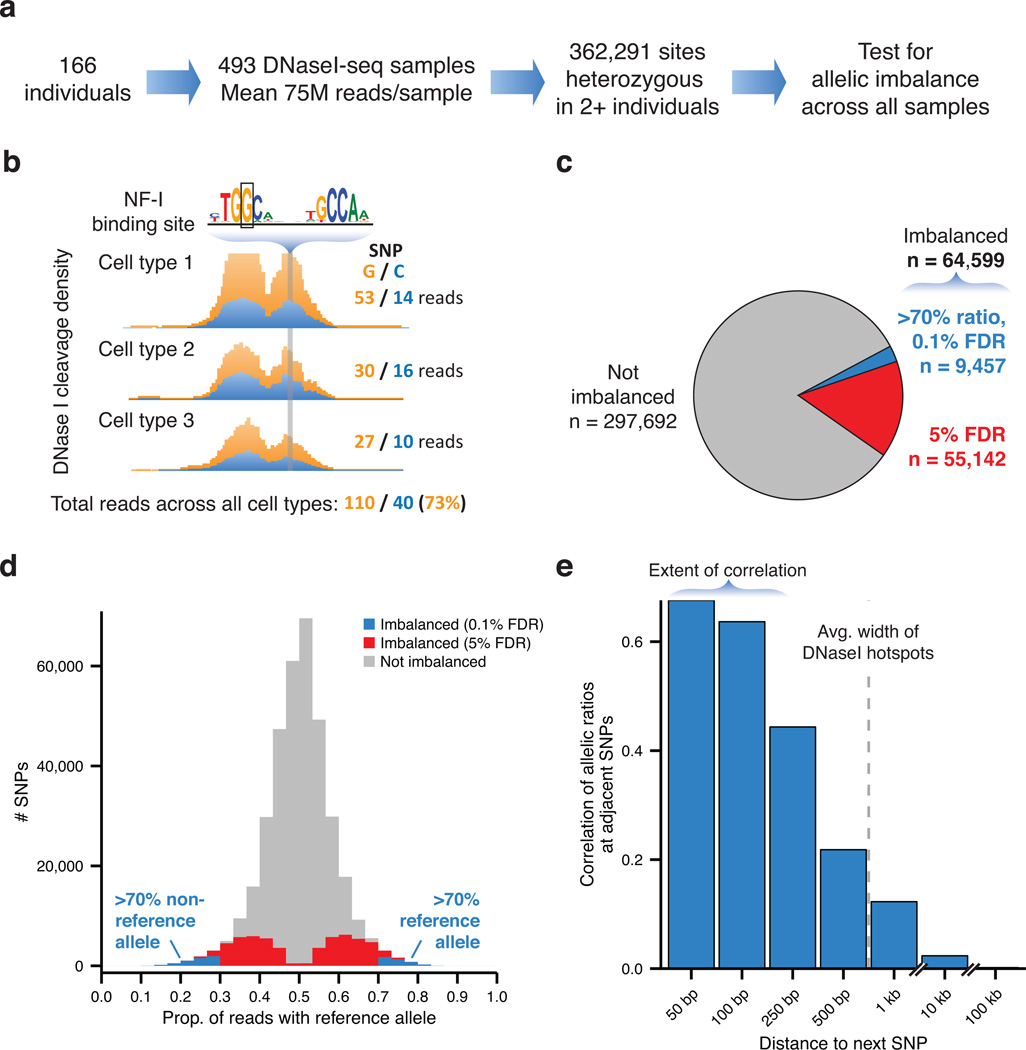
Figure 1. Identification of regulatory variants impacting DNA accessibility.
(a) Outline of experimental procedure and data set. (b) Allelic analysis of DNA accessibility at heterozygous sites. Imbalance manifests as a deviation from 50:50 in the ratio of reads mapping to two homologous chromosomes, potentially due to alteration of TF binding by the sequence variant itself. (c) Extent of imbalanced variants discovered. A strict set of imbalanced variants were identified at 0.1% FDR and >70% imbalance (blue). (d) Allelic ratios of sequencing reads relative to reference allele. A ratio of 70% represents a 2.3-fold difference in accessibility between the two alleles. (e) Pearson correlation of allelic ratios at adjacent SNPs broken down by distance to next SNP. Dashed line represents the median width of DHS hotspots overlapping SNPs in this study. Shown are SNPs in high linkage disequilibrium (r2 > 0.8) in our samples (see Supplementary Fig. 5).
We tested the SNPs we identified for allelic imbalance in chromatin accessibility (Supplementary Fig. 2a). We restricted our analysis to 362,291 SNPs with high power, requiring at least two heterozygous individuals, sufficient total read depth (>50 reads) and good mappability for both alleles (Supplementary Fig. 2b and Online Methods). At each SNP, we quantified the relative proportion of reads mapping to each allele totaled across all heterozygous cell types (Fig. 1b and Online Methods). This revealed 64,599 imbalanced SNPs where the ratio of sequencing reads mapping to the two alleles significantly deviated from 50:50 at a 5% false discovery rate (FDR) (Fig. 1c). These variants exhibited a broad spectrum of effect sizes as measured by the allelic ratio and a subset of 9,457 variants exhibited extremely strong (>70%) imbalance at a strict FDR cutoff of 0.1% (Fig. 1d, Supplementary Fig. 2c, and Supplementary Fig. 3). The proportion of imbalanced sites remained the same when restricting to the ENCODE Illumina genotypes, confirming the accuracy of our genotyping approach (Supplementary Table 6). The majority of variants were located in intronic or intergenic regions outside of the transcription start site (Supplementary Table 7). Fully 19% of DHSs surveyed in 114 cell and tissue types overlapped a SNP tested for imbalance (counting a DHS once per cell type it appears in), and 5.6% of DHSs overlapped imbalanced variants, emphasizing the unprecedented extent of our data set. Fully 47% of dsQTLs4 and 81% of CTCF QTLs17 also examined in the present study were imbalanced, a 2.7-fold and 4.5-fold enrichment, respectively. Furthermore, imbalance was concentrated at sites of TF occupancy marked by DNase I footprints, suggesting a tight relationship between imbalance in chromatin accessibility and TF activity (Supplementary Fig. 4).
We then examined the co-occurrence of imbalance at nearby SNPs in our data. Although nearby SNPs are known to demonstrate correlation in the presence of certain alleles (i.e., linkage disequilibrium, or LD), we reasoned that imbalance in chromatin accessibility will only be correlated at two sites if they additionally occupy a common regulatory domain within the nucleus. We found that allelic ratios at nearby polymorphic sites were strongly correlated at distances less than 100 bp, well below the median width of a DHS hotspot (751 bp) (Fig. 1e). Importantly, there was little correlation found for SNPs unlikely to be found on the same haplotype in our samples (r2<0.20), even at close range. Conversely, SNPs in high LD separated by >250 bp showed no correlation in imbalance (Supplementary Fig. 5). The narrow range of correlation of imbalance for linked SNPs thus likely reflects focal alteration of TF binding within composite binding elements.
Broad cell-type sampling dramatically increases detection power
Power to detect imbalance at individual sites depended strongly on sequencing depth, as expected from the binomial distribution, and power calculations indicated that additional sequencing was likely to uncover novel moderate-effect size variants (Supplementary Fig. 6). We therefore applied a targeted footprinting method17,23–25 to enrich DNase I libraries from abdominal skin (AG10803) and mammary stromal (HMF) fibroblasts (Supplementary Fig. 7a). Sequencing depth in the two targeted cell types was enriched up to 5-fold (Supplementary Fig. 7b), with coverage at targeted sites approaching that of the full genomic data set (Supplementary Fig. 6a). Allelic ratios were highly reproducible between genomic and targeted samples (Supplementary Fig. 7c). We did observe a slight bias for the reference allele at SNPs directly overlapping capture probes (Supplementary Fig. 7d and Supplementary Table 8). We attributed this to decreased hybridization energy for DNA fragments containing a mismatch to the probe sequence and compensated by adjusting the expected allelic ratio in the binomial test accordingly. Enrichment of the targeted sites enabled the discovery of 1,174 novel imbalanced SNPs (Supplementary Fig. 7e–f). We measured a high replication rate of imbalance calls from the full genomic data set against these calls (Supplementary Fig. 7g), suggesting that targeted enrichment of sequencing libraries can efficiently reveal novel alterations in DNA accessibility.
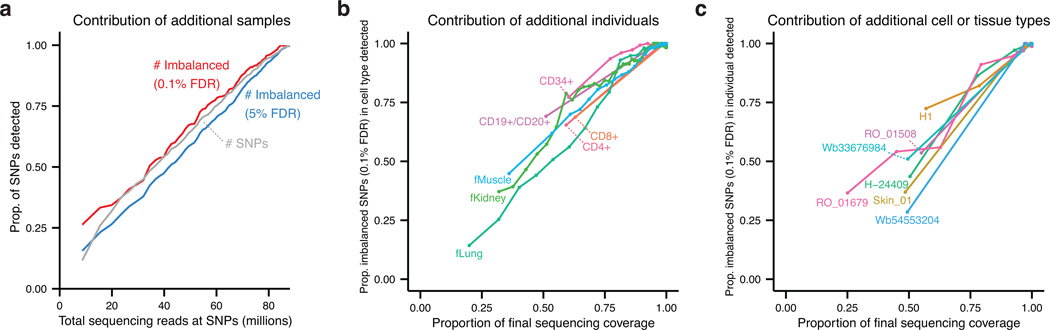
The breadth of cell types surveyed also provides access to both novel regulatory compartments and individual sequence diversity. We computed the cumulative contribution of additional cell types to the discovery of imbalanced variants, and found that iterative incorporation of subsequent samples continued to reveal novel imbalanced SNPs (Fig. 2a). We broke down this increased discovery power in both terms of the contribution of additional individuals for a given cell type and additional cell types (Fig. 2b) for a given individual (Fig. 2c), and found a continued yield of imbalanced SNPs with each additional sample.
Figure 2. Effect of sampling depth on detection of imbalance.
The discovery of imbalanced variants is depicted when considering: (a) additional samples; (b) additional individuals for a given cell type; or (c) additional cell types for a given individual. Imbalanced SNPs were identified in an increasing subset of the data, adding one sample at a time (starting with the most deeply sequenced). Proportions were computed as the number of SNPs identified at intermediate data points divided by the total number of SNPs from the full data set for that series. Imbalance was established using the P-value cutoff corresponding to a 0.1% FDR in the total data set and required at least 70% imbalance. Sequencing coverage was measured as total reads over all SNPs passing filters. Shown in b and c are subsets of highly sampled cell types or individuals.
Cellular context sensitivity of imbalance
We analyzed the consistency of allelic imbalance across different individuals and cellular contexts. To reduce the confounding effect of detection power, we focused on a subset of high-depth samples with multiple samples per cell type and individual (Supplementary Table 9). We limited our analysis to sites with at least 3 heterozygotes each having both a DHS and high sequencing coverage (>30 reads per sample). Examining the pairwise correlations in allelic ratios between samples revealed increased similarity among those from the same individual or cell type (Fig. 3a).
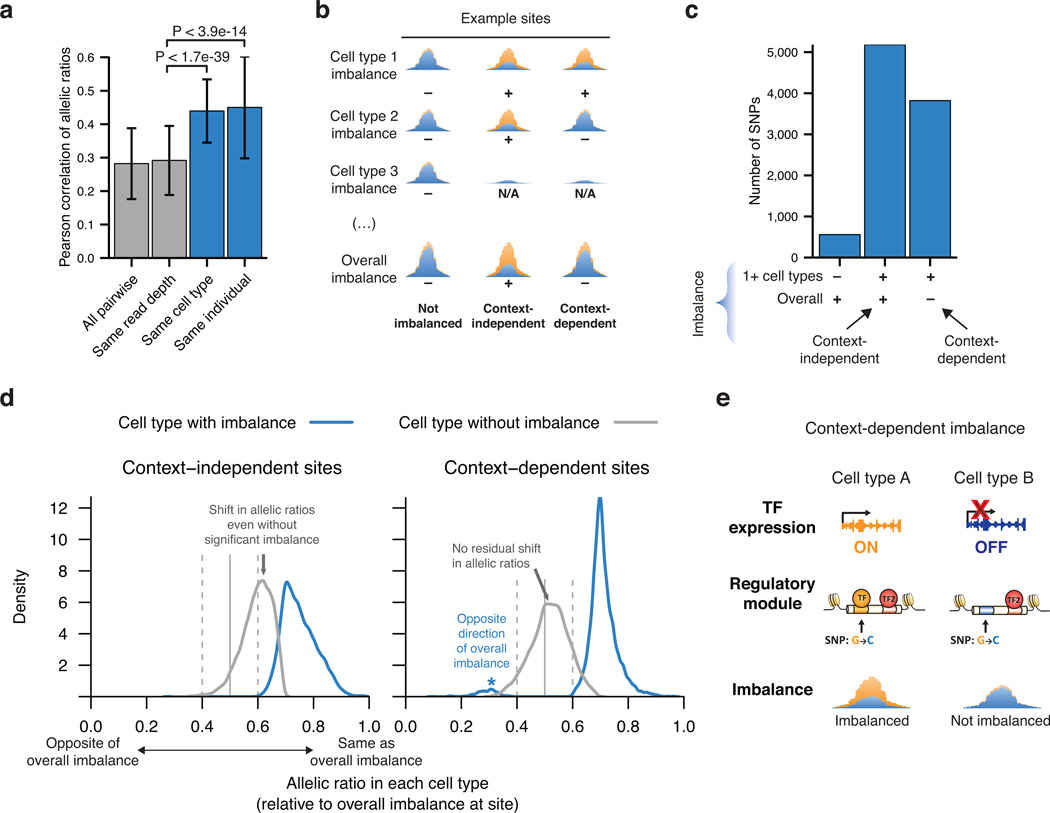
Figure 3. Cross-cell type analysis of imbalance.
(a) Pairwise Pearson correlations of allelic ratios between samples. Note increased correlation among samples from the same individual or cell type compared to all pairwise samples. Error bars represent standard deviation. P-values derived from the Mann-Whitney U test. (b) Sites were classified as context-independent or -dependent by presence or absence (+ or −) of cell-type specific and/or overall imbalance; N/A indicates absence of DHS. (c) Analysis of the relationship between imbalance in one or more cell types and overall imbalance at the same site. 29,889 sites without any imbalance not shown. (d) Allelic ratios per cell type, oriented such that 1.0 represents the direction of overall imbalance at each site. Allelic ratios deviate from 0.5 at context-independent sites even in cell types without significant imbalance (gray arrows). In contrast, context-dependent sites are characterized by strong imbalance only in a subset of cell types. A minority of context-dependent sites display discordant imbalance between samples (blue asterisk). Sites without overall imbalance are shown in Supplementary Fig. 8d. (e) Model of context-dependent imbalance at a composite regulatory element bound by both cell-type-specific and constitutive TFs.
To examine imbalance across cell types at high resolution, we then summed reads from all samples for a given cell type and analyzed each cell type for imbalance (Supplementary Table 10 and Fig. 3b). To avoid confounding cell-type selectivity with variable sensitivity, we required at least 50 reads at each site, subsampled each site to 3 cell types, and then further downsampled the allele counts to the lowest of the three (Supplementary Fig. 8a). Focusing on sites with imbalance detectable in one or more cell types, we defined two classes of sites: those also with imbalance manifest across all cell types (‘context-independent’ sites) and those without imbalance across all cell types (‘context-dependent’ sites) (Fig. 3b–c and Supplementary Fig. 8b–c). Allelic ratios at context-independent sites were shifted towards the direction of overall imbalance, even in cell types without significant imbalance themselves (Fig. 3d and Supplementary Fig. 8d). This high concordance of allelic ratios across cell types suggests that imbalance at these sites occurs consistently across all cell types despite varied detection power. In contrast, at context-dependent sites allelic ratios exhibited a clear bifurcation between cell types with and without imbalance, reflective of a binary presence or absence of imbalance at the same site in different cell types. The direction of imbalance at these context-dependent sites was largely consistent across samples and cell types (Fig. 3d), suggesting that context sensitivity represents a consistent genetic effect reflecting a feature of the cellular environment such as TF levels rather than epigenetic propagation of altered TF occupancy, (Fig. 3e).
Chromatin features at imbalanced variants
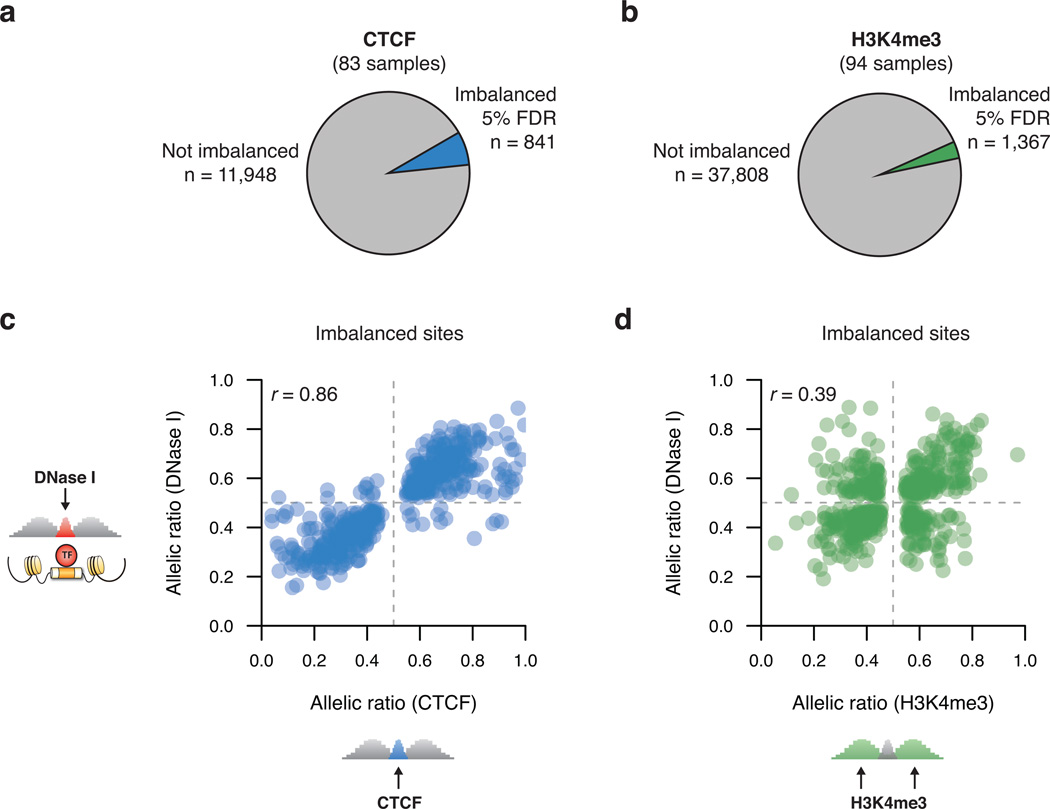
DHSs mark sites of TF binding in place of a canonical nucleosome, and are flanked by histones bearing characteristic covalent modifications1. To investigate the independent responses of these structural features to sequence variation, we surveyed chromatin immunoprecipitation (ChIP-seq) profiles of trimethylation of histone 3 lysine 4 (H3K4me3) and of occupancy of the master genome regulator and transcription factor CTCF (Supplementary Fig. 9 and Supplementary Table 2). We identified vastly fewer imbalanced variants for H3K4me3 and CTCF than for DNase I (Fig. 4a–b and Table 1)17,18. The majority of variants imbalanced in CTCF occupancy also exhibited imbalance in DNase I, consistent with prior work4. However, most imbalanced H3K4me3 variants exhibited no imbalance in DNase I (Supplementary Table 11). Moreover, while the direction of imbalance was highly consistent between DNase I and CTCF, allelic ratios for H3K4me3 showed low correlation with DNase I (Fig. 4c–d). Thus, these results confirm the reliability of DNA accessibility as an indicator of allelic TF occupancy, and suggest that at many sites H3K4me3 patterns vary independently of TF activity26.
Figure 4. Imbalance in CTCF occupancy and H3K4me3.
(a–b) Extent of imbalance (5% FDR) in CTCF (a) and H3K4me3 (b). (c–d) Allelic consistency between DNase I and CTCF (c) and H3K4me3 (d); shown are sites imbalanced for both. r, Pearson correlation of allelic ratios. DNase I SNPs imbalanced at 5% FDR were used.
Table 1.
Summary of experimental data and imbalanced variants identified.
| Assay | # Samples | # Individuals | # Cell types | # Sequencing reads (* 109) |
# Sequencing reads per sample (* 106) |
# peaks per sample |
# SNPs tested |
# Imbalanced SNPs (*) |
|---|---|---|---|---|---|---|---|---|
| DNase I | 493 | 166 | 114 | 26.2 | 53.2 | 173,032 | 362,291 | 64,599 |
| CTCF | 83 | 39 | 28 | 1.0 | 12.4 | 71,998 | 12,490 | 841 |
| H3K4me3 | 94 | 45 | 49 | 1.7 | 17.9 | 61,991 | 39,175 | 1,367 |
| all | 670 | 182 | 120 | 28.9 | - | - | 372,440 | 66,609 |
SNPs were considered significantly imbalanced at 5% FDR. Read counts represent non-redundant reads used for analysis (see Online Methods, Supplementary Tables 1–4).
TF-centric profiles of sequence variation
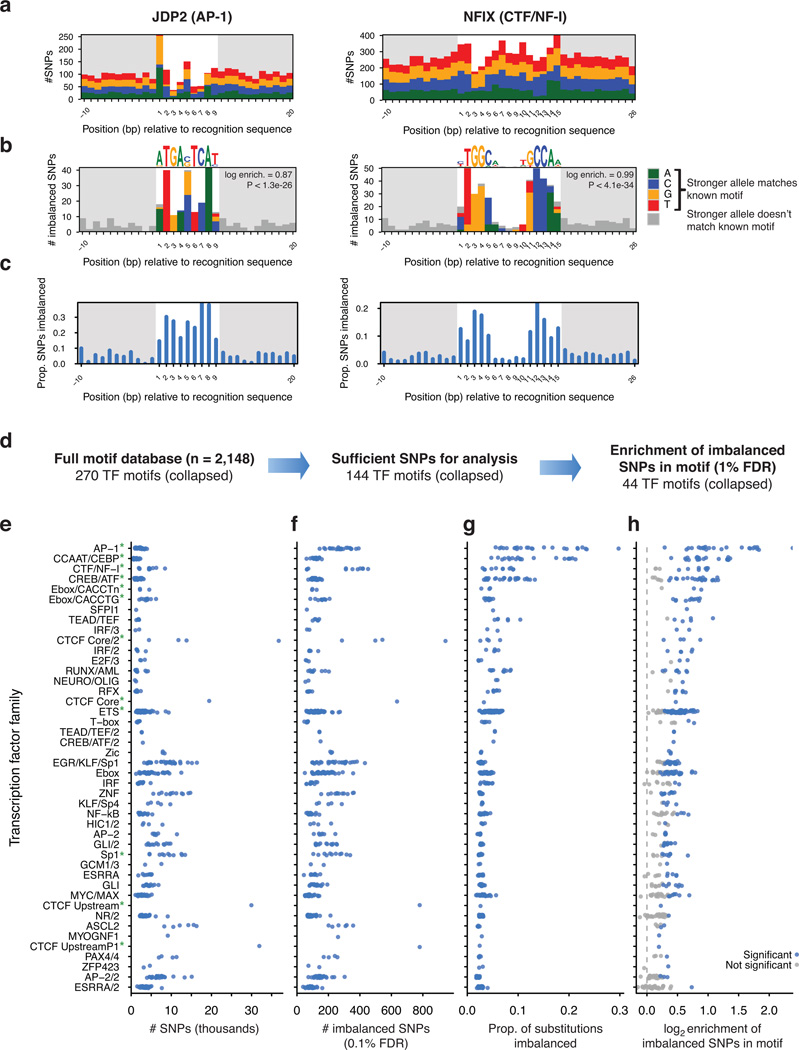
To ascribe imbalanced variants to an effect on the activity of individual TFs, we aligned SNPs to recognition sequences matching 2,203 TF motifs. These TF motifs collectively cover the majority of mammalian TFs, and comprise 825 distinct TF genes and 270 distinct families of nonredundant binding specificities (Supplementary Fig. 10 and Supplementary Tables 12–14). This showed that heterozygosity was uniform around JDP2 and NFIX recognition sequences except for a slight reduction in diversity at positions in the motif with high information content, attributable to purifying selection22,27,28 (Fig. 5a). In contrast, imbalanced variants were strikingly concentrated at key positions within each recognition sequence, with the higher-accessibility allele qualitatively matching the consensus sequence (Fig. 5b). Accounting for the uneven heterozygosity, the frequency of imbalance at each position was strongly reflective of information content at that position in the TF binding motif (Fig. 5c and Supplementary Fig. 11).
Figure 5. Profiles of TF sensitivity to sequence variation.
(a–c) Concentration of imbalanced SNPs within recognition sequences for AP-1 and CTF/NF-I transcription factors. Shown are: (a) all SNPs tested for imbalance; (b) significantly imbalanced variants; and (c) the proportion of imbalanced SNPs per position. Color indicates sites where the allele with higher accessibility has higher information content according to the motif. White background denotes width of motif. (d) Survey of TF motifs analyzed for profiles of imbalance. Similar motifs were grouped into nonredundant TF cluster (Supplementary Fig. 10). TFs with insufficient overlapping SNPs were not analyzed (Online Methods). (e–h) TF clusters with enrichment of imbalanced SNPs. Each point represents an individual motif. Shown are: (e) the number of SNPs overlapping recognition sites; (f) number of imbalanced SNPs; (g) frequency of substitutions resulting in imbalance; and (h) the log2 enrichment of proportion of imbalanced SNPs lying in TF recognition sequences relative to non-imbalanced SNPs. Green asterisks in (e) mark TF clusters highlighted in the main text. Significance of enrichment of significant SNPs in motifs in (h) was assessed by permutation.
Our analysis yielded sufficient overlapping SNPs for assessment of the profile of imbalance at 144 TF clusters, likely reflective of the number of genomic matches to each TF consensus sequence and the cellular selectivity of the cognate TF activity (Fig. 5d and Supplementary Tables 15–16). Although most imbalance was found to match sequence preferences predicted by TF motifs, we found that only a minority of variants overlapping TF recognition sequences resulted in allelic imbalance (Fig. 5e–g). Fully 44 nonredundant TF clusters showed statistically significant enrichment of imbalance within the TF motif (Fig. 5h and Supplementary Fig. 11). The TF clusters with significant enrichment of imbalance recruit a variety of tissue-specific or inducible regulators, comprising constitutive factors like CTF/NF-I, CCAAT/CEBP, CTCF, and SP1; resident nuclear factors including the AP-1 complex, CREB/ATF, and ETS families; and multifunctional sequence elements like the E-box. This suggests that these factors are directly responsible for the potentiation of DNA accessibility in a wide variety of cellular contexts, and indeed, many of these factors were previously identified as key determinants of accessible chromatin2,29.
Site-dependent buffering of sequence variation
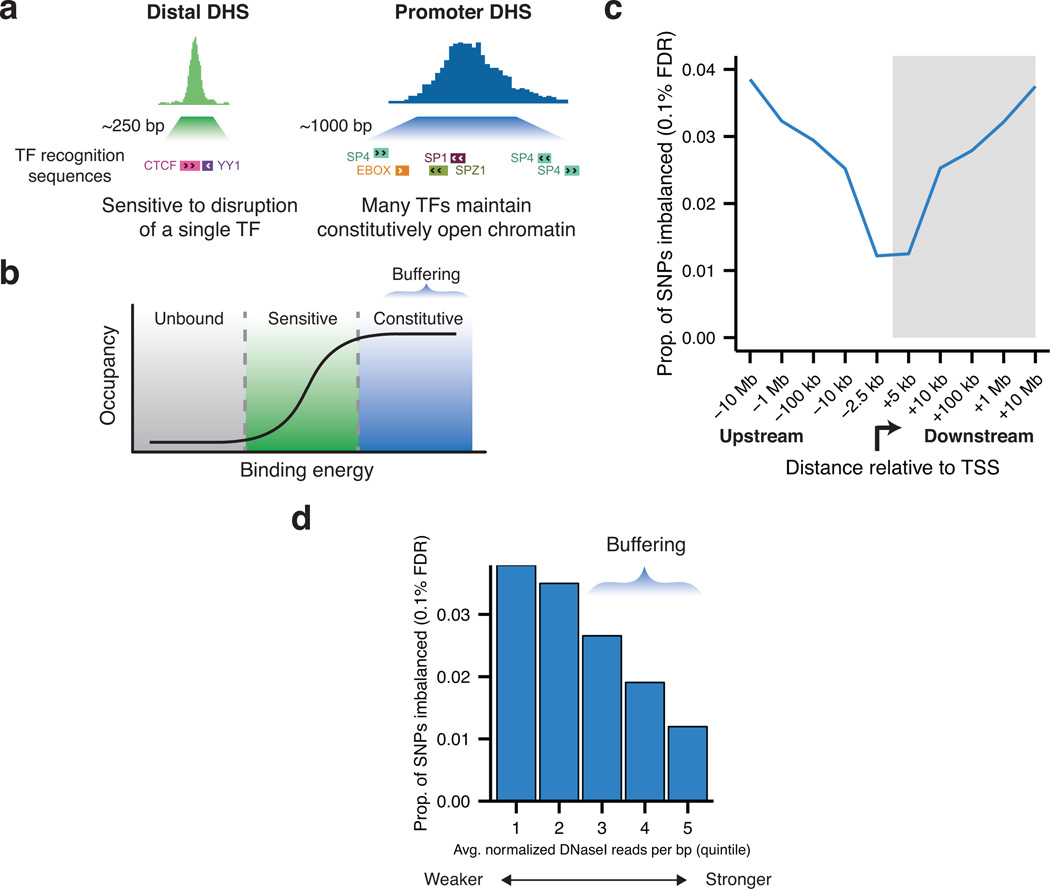
That only a minority of the variants in the present study result in imbalance (though all overlap DHSs) suggests that local features buffer the effect of sequence variation on TF occupancy17. Promoters represent the prototypical transcriptional regulatory element, being distinguished from more distal DHSs by their length and intense accessibility, and are easily identifiable by sequence features and their accessibility across a broad range of cell types (Fig. 6a). We reasoned that the combinatorial binding of numerous TFs at promoters may result in a highly accessible chromatin state that is buffered to perturbation by point variation (Fig. 6b). Indeed, we found that transcription start sites exhibited a reduced frequency of imbalance, despite higher detection power from the increased sequencing depth (Fig. 6c and Supplementary Fig. 12a–c). Indeed, the strength and cell-type activity spectrum of the DHS were both negatively correlated with frequency of imbalance (Fig. 6d and Supplementary Fig. 12d). By measuring the number of independent TF binding sites in the flanking 500 bp revealed by DNase I footprints, we found that additional factor occupancy was itself directly associated with buffering (Supplementary Fig. 12e). These results suggest that the effects of sequence variation on TF activity are buffered by site-dependent features, imparting a regulatory structure on the genome and confirming the need to study regulatory variants at their native loci.
Figure 6. Buffering of regulatory variation.
(a) Schematic of chromatin environment at promoter DHSs indicating increased size, accessibility, and density of TF binding relative to distal DHSs. (b) Threshold model of TF occupancy explaining buffering of point changes at strong sites. (c) Frequency of imbalance relative to transcription start sites (TSS) demonstrates buffering within the promoter region. Buffering is strongest between −2.5 kbp and +5.0 kbp of TSS. Bins are labeled by endpoint furthest from TSS. (d) Frequency of imbalance, broken down by site strength as measured by DNase I accessibility across all cell types having a DHS.
TF-centric prediction of variants affecting DNA accessibility
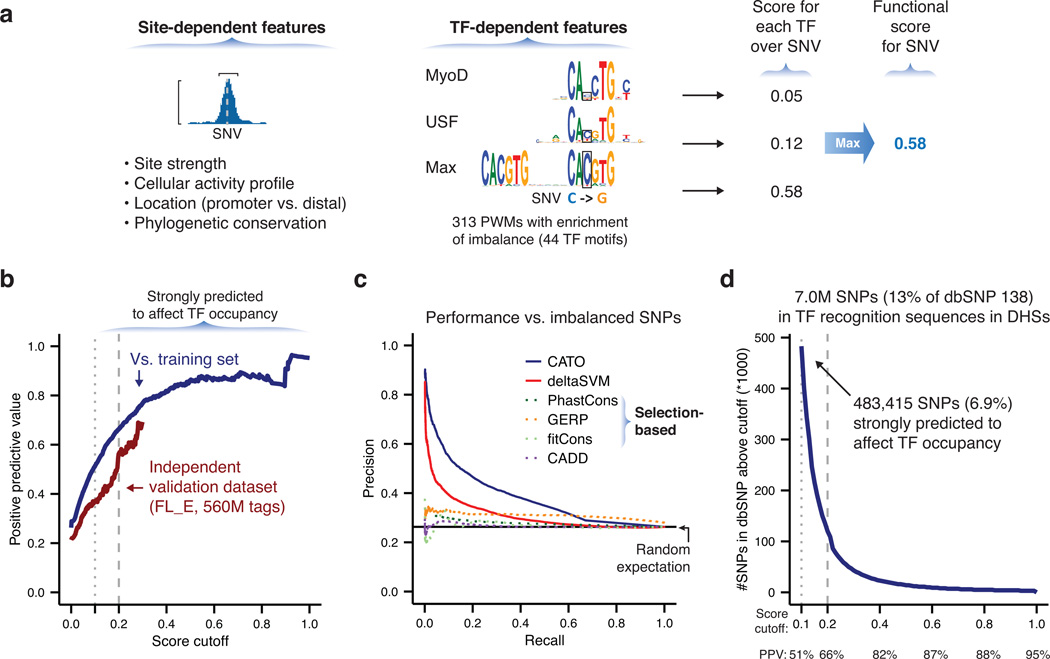
Given the challenges to studying functional sequence variation at endogenous loci, existing methods for prediction of functional regulatory variation limit their consideration to overlap with TF binding sites. Moreover, site strength and broad cell-type activity are often interpreted as positive factors indicative of reproducibility rather than indication of reduced penetrance. To overcome these deficiencies, we used the experimentally determined sensitivity profiles delineated by the SNPs overlapping each motif to train logistic models for the genome-wide prediction of variation affecting TF occupancy. We quantified the effect of single nucleotide variants on the energy of TF binding as the difference in information content between the two alleles and the specific position in the recognition sequence disrupted. We also incorporated features associated with TF occupancy at a specific recognition sequence, including the occupancy measured by DNase I footprinting, the score of the match to the motif, and phylogenetic conservation. To account for variation in detection power across our experimental data set, we included the read depth and number of heterozygous samples as covariates. We trained a separate model for each of 314 motifs enriched for imbalanced SNPs (Supplementary Fig. 13a and Supplementary Table 17). Cell-type activity spectrum (MCV), the position of the SNP relative to the TF motif, and the score of the match to the TF motif all had strong coefficients in the model. While other factors had individually small effects, their combined contribution was substantial. Finally, we recalibrated the raw regression scores in terms of the empirical rate of significant variants to provide a standardized score on an intuitive scale. As a given single nucleotide variant (SNV) generally overlaps multiple degenerate TF recognition sequences, we assigned an overall score as the maximum score for any individual TF (Fig. 7a). This resulted in a simple scoring scheme providing recalibrated probability of affecting the binding of any TF, as well as a quantitatively ranked list of TF families whose binding might be altered.
Figure 7. Recognition of variation affecting TF occupancy genome-wide.
(a) Scores for noncoding variants in DHS were calculated from the maximum of scores for all overlapping TF-specific models. (b–c) Measurement of model performance versus experimentally determined imbalanced variants (Online Methods). (b) Positive predictive value (PPV; the proportion of predicted variants that are true positives, also known as precision) is plotted for increasing score cutoffs. At a score cutoff of 0.1 (dotted line), 51% of predictions are true positives. Red line measures performance on held-out validation FL_E data set. (c) Precision (as in b) versus recall (the overall proportion of imbalanced SNPs that are correctly predicted). A higher area under the curve represents higher model performance. (d) Identification of common human sequence variants affecting TF occupancy. Cumulative distribution showing the number of SNPs exceeding a given score cutoff. PPV at selected cutoffs is transcribed from data in (b).
For a cutoff of 0.1, the model demonstrated a positive predictive value of 51%, with increased accuracy at more stringent cutoffs, and demonstrated nearly the same positive predictive values on a separate erythroblast DNase I validation data set (Fig. 7b). Precision-recall analysis shows that the TF-based model outperforms other approaches on both the training set of imbalanced SNPs and an independent set of dsQTLs4, and that inference of natural selection from phylogenetic constraint or population diversity offers poor predictive power for variation in regulatory regions (Fig. 7c and Supplementary Fig. 13b–c).
To illustrate the genome-wide recognition of variants affecting TF occupancy using our experimental models of sensitivity to sequence variation for 313 motifs, we scored 50M variants in dbSNP 138, a large collection of human sequence variation30. Although 7.0M of these variants lie in a DHS and alter a TF recognition sequence (simply requiring a log odds difference between alleles > 2), it is unclear how many of these affect binding in vivo. We identified 483,415 SNVs with a score of 0.1 or higher, illustrating the potential of our method to focus global analyses on a minority of noncoding variants likely to affect TF occupancy (Fig. 7d and Supplementary Data 3). Thus our approach provides a scalable method for high-throughput identification of regulatory variants and will likely prove broadly applicable to the study of human disease and the interpretation of personal genomes.
DISCUSSION
We have presented an expansive survey of regulatory variation impacting transcription factor occupancy in vivo. Our approach leverages the focally high coverage provided by DNase-seq reads to efficiently assess regulatory variants in their native genomic and cellular contexts, and the results highlight the fact that genetic variation in regulatory DNA is chiefly interpreted in a cell type-specific fashion. As the power to detect the impact of variation on TF occupancy is determined by the amplitude and cell type activity spectrum of the harboring DHS together with population diversity (Fig 2 and Supplementary Fig. 12a), survey of additional cell types and individuals will uncover further functional variation, and power at weaker DHSs can be boosted using targeted footprinting.
TF-centric models connecting variation at specific recognition sequence positions to specific quantitative effects on occupancy should be of immediate utility in decoding the wealth of regulatory variation manifest in personal genomes. Our modeling approach could readily be extended to incorporate a variety of more granular features, such as cellular context sensitivity, biophysical models of protein-DNA interaction31,32 or DNA shape33, and also offers a novel means of calibrating models of TF-DNA recognition. Our modeling approach indirectly incorporates the baseline effect(s) of nearby TF binding through consideration of chromatin accessibility, but variants are scored independently of nearby recognition sequences or other variants in close linkage. Additional information such as sequence preferences indicative of dimerization or allosteric effects on TF activity34,35 will likely have important utility for connecting altered TF binding within regulatory regions with consequent alterations in gene expression.
Because accessibility is a prerequisite for regulatory DNA function, the cellular spectrum of activity of a given regulatory variant will be governed by the accessibility of its harboring regulatory region. It is presently unclear to what extent the biological consequences of variation within a given TF recognition sequence might be further restricted to specific cellular contexts by differential expression of its cognate TF (Fig. 3e), or that of co-occupying TFs. This issue has major practical implications, as highly prevalent context sensitivity would require surveys of functional variation to be performed separately in every relevant cellular context. Alternatively, less prevalent context sensitivity might allow the supplementation of tissue-specific regulatory maps with eQTL mapping in a proxy tissue. Past eQTL studies have disagreed on the degree of cell-type selectivity36–40, likely because of the conflation of cell-type selectivity with incomplete detection power, a limited range of pure cell populations38, and a bias towards promoters41. The unprecedented range of cell types surveyed herein has revealed two prominent compartments: context-dependent and context-independent regulatory variation. Within these compartments, both the potential for imbalance at a site and its direction of effect are genetically controlled, but the ultimate presence of imbalance can depend on epigenetic context. The sites of context-dependent imbalance reported here can be incorporated into assessments of regulatory variant activity, and future work offering increased resolution will provide insight into the sequence determinants of cellular context-specific functional variation.
The fact that regulatory variants are extensively buffered suggests that most single nucleotide variants in regulatory DNA regions have very modest effects (or little to no effect), on TF occupancy and hence downstream function. An important implication of the dominance of context-sensitive features is the implication that studies employing synthetic constructs – either non-integrating or integrating at exogenous sites – will have limited relevance for interpreting the function of individual sequence variants in vivo. Rather, future work will require high-throughput methods for the study of regulatory activity that do not sacrifice critical features of the endogenous locus.
Connecting the biological impact of sequence variants on TF occupancy with downstream function – such as on gene expression or other molecular phenotypes – remains a challenge, chiefly because we lack both the ability to measure very small effect sizes at the molecular (e.g., expression) level, and an understanding of how effect sizes relate to phenotype. For example, a minute change in transcript expression compounded over weeks or months of developmental time may in fact comprise a substantial biological effect size. Given, however, both the frequency of regulatory variation and the degree of buffering we observe, it seems likely that only a small minority of variants impacting TF occupancy will individually result in a visible phenotype. Yet, this vast landscape of noncoding variation harbors the majority of variants associated with common disease3. Much as the recognition of the triplet code enabled the distinction of synonymous from nonsynonymous coding variants, identification and categorization of variation that affects site-specific TF activity is foundational to our ability to cull meaning from the vast expanse of human noncoding variation.
ONLINE METHODS
DNase I and ChIP-seq profiling
We utilized both novel and published samples produced by the Roadmap Epigenomics and ENCODE projects (Supplementary Tables 1–2) and applied several criteria to ensure data quality. First, we excluded known malignant or transformed cell lines. Second, we excluded samples whose distribution of allelic ratios at heterozygous sites deviated from a mean of 0.5, showed secondary modes, or exhibited excessive variance (all potentially indicating pooling of samples from different individuals). Finally, the signal-to-noise ratio for each sample was computed as the signal portion of tags (SPOT) score, computed using the program Hotspot42. Samples with low enrichment (generally, SPOT score below 0.3) were excluded.
DNase I was performed as described3,43 (Supplementary Table 1). Briefly, nuclei were extracted from cells or tissues and incubated for 3 min at 37 °C with limiting concentrations of the DNA endonuclease, DNase I (Sigma) supplemented with Ca2+ and Mg2+. The digestion was stopped with EDTA and the samples were treated with proteinase K. The small ‘double-hit’ fragments (<500 bp) were recovered by sucrose ultra-centrifugation, end-repaired and ligated with Illumina sequencing adapters. Chromatin immunoprecipitations were performed as described for CTCF44 and H3K4me32 (Supplementary Table 2). Libraries generated from immunoprecipitated or DNase I-treated DNA were sequenced on an Illumina Genome Analyzer IIx or HiSeq 2000/2500 by the High-Throughput Genomics Center (University of Washington) according to a standard protocol.
Short read mapping
We mapped reads to the human genome (GRCh37/hg19) using bowtie45. Single-end (SE) reads were mapped using the command, ‘bowtie --mm -n 3 -v 3 -k 2 --phred64-quals’ (or, '--phred33-quals' for HiSeq). Aligned reads were subsequently processed to retain only unique alignments with 1 fewer mismatch than the next-best alignment, and with no more than 2 mismatches total. Paired-end (PE) reads were mapped using the command, ‘bowtie -n 2 -m 1 -e 70 --best --sam --chunkmbs 256 --phred33-quals --sam --maxins 750’.
Both mates were required to map properly. Reads from several samples with longer read lengths were hard clipped to 36 bp.
Genomic feature overlaps and distance calculations were performed using the BEDOPS suite of software tools46. Duplicate reads were flagged on a per-sample basis using Picard MarkDuplicates and all further analysis considered only non-redundant reads.
Genotyping from DNase I and ChIP-seq reads
We identified samples derived from the same genetic background, including biological replicates and multiple tissues sampled from the same donor (Supplementary Table 3). Samples from the same individual were initially verified to match using preliminary per-sample heterozygote genotype calls. In addition, we examined allelic ratios for each sample at final heterozygote calls to identify potential sample mismatches manifest as excessive imbalance. Finally, ‘vcftools --relatedness’47 was used to identify unexpected relatedness suggesting sample swaps. For genotyping, reads from all samples from the same individual were then pooled using ‘samtools merge’.
We called genotypes directly from the combined DNase I, CTCF and H3K4me3 reads using SAMtools21. We merged reads from all samples for a given individual into a single BAM file, adjusted base qualities from Phred+64 to Phred+33 where necessary, removed any reads with more than two mismatches to the reference genome, and corrected SAM tags using ‘samtools calmd’. We called genotypes across all samples using ‘samtools mpileup -Q 20 -I -d 10000 -D -E -g’ and ‘bcftools view --vcg’.
We filtered the resultant genotypes using VCFtools47 to:
-
1.
retain only biallelic autosomal SNPs
-
2.
require a SNP quality ≥500
-
3.
eliminate SNPs with a Hardy-Weinberg equilibrium P < 0.01
-
4.
require ≥30 total reads across all individuals
-
5.
retain only genotypes supported by ≥12 reads
-
6.
retain SNPs with at least 1 heterozygous genotype with genotype quality >50.
We parsed the VCF file using BEDOPS46 to extract heterozygous sites per individual and performed further filtering:
-
7.
exclude SNPs overlapping the ENCODE blacklist22
-
8.
require no other SNP passing above filters within 36 bp
-
9.
require genotype calls to have at least 4 reads for each allele per individual
-
10.
require genotype calls to have a quality score of at least 50
We observed a Ti/Tv ratio of 2.19 for all SNPs, 2.11 for imbalanced SNPs (5% FDR), and 2.02 for the strict imbalanced SNPs (0.1% FDR). The resulting genotypes are summarized per-individual and per-cell type in Supplementary Tables 3 and 4. Linkage disequilibrium was calculated using ‘vcftools --geno-r2’47 on the unphased genotypes.
Short-read mapping bias
We simulated all possible 36-bp SE reads overlapping each SNP, including both the reference and alternate alleles. We then mapped the simulated reads using BWA48 to a hg19all index including hg19 unmapped sequences and alternate haplotypes with the command, ‘bwa aln -l 32 -k 2 hg19all <FASTQ file> | bwa samse hg19all - <FASTQ file>’. Sites with any overlapping read mismapped or mapped with <30 MAPQ were excluded.
Validation of genotypes
We downloaded Illumina 1MDuo genotypes from the ENCODE project for samples matching 23 of the individuals in our study (AG04449_and_AG04450, AG09309, AG09319, AG10803, BJ, GM06990, GM12878, H1, HAEpiC, HCF, HCM, HCPEpiC, HIPEpiC, HMEC, HRCE, HRE, HRPEpiC, IMR90, NH-A_and_NHLF, NHDF-neo, RPTEC, SAEC, SkMC) from the “HAIB Genotype” track in the UCSC Genome Browser22. We computed the sensitivity and specificity of our heterozygote calls relative to each replicate in the HAIB data (Supplementary Table 5). All SNPs in DHSs, CTCF or H3K4me3 peaks of that cell type or on the Illumina design (for HAIB calls or our calls, respectively) were considered for sensitivity calculations. Sensitivity of our genotypes was computed in two distinct senses: (i) raw sensitivity for all heterozygous sites in DHSs on the array design and (ii) sensitivity of pass-filter genotype calls for heterozygous sites.
Identification of allelic imbalance
At each SNP, reads were extracted from all DNase I alignments for each heterozygous individual using SAMtools21, and reads matching each allele were counted. We computed read sums separately for DNase I, H3K4me3, and CTCF data. For DNase I samples, we excluded 3 bp at the 5’ end of the read to exclude any possibility of sequence-specific DNase I cut rate resulting in artificial imbalance49. To correct for potential mapping bias caused by the extra mismatch in reads containing the non-reference allele, a less-stringent mismatch threshold was applied. Reads containing the reference allele were only counted if they contained zero or one base mismatches (over the entire read length) to the reference sequence; reads with the non-reference allele were counted if they had one or two base mismatches (one of which is the SNP). We only counted reads where the SNP position had an Illumina base quality >20. Sites with fewer than 50 reads total across all samples were excluded for lack of power to test for allelic imbalance. PE mate pairs were counted as a single read.
We filtered a small number of SNPs with >5% of reads not matching the two expected SNP alleles across all samples. We required that SNPs must overlap a DNase I hotspot in ≥3 cell types, and required ≥2 heterozygous samples for each SNP. Finally, we excluded SNPs lying within 100 bp of 1000 Genomes indels present at >5% MAF in CEU50.
Sites passing all filters were then tested for imbalance using a two-tailed binomial test. We calculated a false discovery rate using the Benjamini-Hochberg method. We set a loose significance cutoff at 5% FDR; for the more stringent level we additionally required at least 70% imbalance (i.e., a proportion of reads mapping to the reference allele of <30% or >70%) and a 0.1% FDR. Imbalance in ChIP-seq data was established at a 5% FDR and was compared to 5% FDR DNaseI-imbalanced SNPs.
Power to detect imbalance from additional samples
Imbalance was computed considering a subset of samples, starting with the sample with the highest sequencing coverage and re-computing upon adding each successive sample. Coverage was measured as the total number of non-redundant reads overlapping all SNPs. Data for all sites with at least 12 reads were considered. P-value thresholds from FDR analysis of the full data set were used.
Targeted DNase I footprinting
Targeted capture of DNase I libraries was performed as described24. Nuclei from HMF and AG10803 cells were DNase I digested and used to generate Illumina libraries as above. The DNase I libraries were amplified by PCR following the Capture SureSelect protocol recommendations (Agilent) and purified using Agencourt AMPure XP beads (Beckman Coulter Genomics). Five hundred nanograms of each library was hybridized to MethylSeq or Human All Exon kits (Agilent) for 24 h at 65 °C. The biotinylated probe/target hybrids were captured on DynalMyOne Streptavidin T1 (Invitrogen), washed, eluted, and desalted and purified on a MinElute PCR column (Qiagen) as described in the SureSelect protocol. The eluted captured library was amplified by PCR with minimal amount of PCR cycles. Amplified captured libraries were purified using Agencourt AMPure XP beads. The samples were then quantified by Qubit dsDNA assay (Invitrogen). Samples were diluted to a working concentration of 10 nM. Cluster generation was performed for each sample and loaded on to a single lane of an Illumina HiSeq flowcell and sequenced.
Targeted capture data were analyzed as in the preceding sections, except we corrected for a slight increase in the proportion of reads matching the reference sequence for SNPs lying directly over a capture probe. We calculated melting temperatures (Tm) for RNA probes (Supplementary Table 8) using the package MELTING with the options ‘-S SEQ -H dnarna -nn sug95 -P 6.15e14 -E Na=1’51. We then empirically determined expected allelic ratios of reads mapping to the reference for each SNP as a function of the Tm of the overlapping probe. We used 0.5 as the expected allelic ratio for SNPs not overlapping probes. We then performed the binomial test for imbalance relative to the expected allelic ratio. We also repeated the identification of imbalanced SNPs in the genomic samples, but including only reads from the genomic HMF and AG10803 samples. For both the genomic and targeted data, we required at least 50 reads across both samples, kept only sites where one or both samples were heterozygous, and required the presence of a hotspot in at least one of the two cell types. Significant imbalance was established at 5% FDR using Benjamini-Hochberg and >60% imbalance.
Cross-cell type analysis of imbalance
To assess imbalance on a per-sample basis, we identified a set of well-sequenced sites in high-depth samples, requiring ≥30 reads per sample, and ≥3 heterozygous samples per site (Supplementary Table 9). We retained only samples with ≥1,000 sites meeting these coverage requirements.
The analysis of context-sensitive sites (Fig. 3b–e) was performed similarly, except samples of the same cell type from different individuals were further collapsed and we required ≥50 reads per cell type (Supplementary Table 10). To avoid confounding cell-type selectivity with variable detection sensitivity, we subsampled each site to 3 cell types, and further downsampled the allele counts to the lowest of the 3 cell types. We applied the same significance criteria as before to counts across all samples, except that samples were called significant at a 5% FDR and an allelic ratio of >60%. P-value thresholds from FDR analysis of the full data set were used as cutoffs.
Genomic identification of TF recognition sequences
Potential sites of transcription factor binding were identified by scanning the entire human genome using position weight matrices curated from four major TF motif collections: TRANSFAC52, JASPAR53, UniPROBE54, and a published SELEX dataset35. To avoid ascertainment bias for motifs better matching the reference allele of common polymorphisms, we created an alternate genome to complement the reference GRCh37/hg19 human genome. This alternate genome incorporates the non-reference allele at the location of each SNP identified in the CEU population of the 1000 Genomes Project50. Both the reference and alternate genomes were then scanned for motif occurrences with a threshold P < 10−4 using the program FIMO55. A 5th order Markov model was generated from 36-bp-mappable human genome sequence and used as the background model.
Clustering TF motifs by similarity
We generated all-vs.-all pairwise similarity scores for each TF motif using TOMTOM56 employing the same 5-order HMM background model:
tomtom -dist kullback -query-pseudo 0.1 -target-pseudo 0.1 -text
-min-overlap 0 -thresh 1
The pairwise scores were then collated into a matrix, and we used Cluster 3.0 to perform hierarchical clustering using Pearson correlation as the distance metric and complete linkage. The resultant tree was cut at height 0.1 using a custom python script. The original TOMTOM alignments were used to assign a relative orientation to motifs in each cluster for uniform visualization of cluster members. Motifs were mapped to gene names as previously described27. Well-known TF clusters were assigned names manually, otherwise a name was generated from the first motif in the cluster. Any redundancy in cluster names was resolved by appending “/2”, “/3”, etc.
TF-centric prediction of variants affecting DNA accessibility
All SNPs tested for imbalance in DNase I accessibility were aligned relative to all database motifs. The proportion of SNPs that were allelically imbalanced at each position relative to the motif was computed using the imbalanced SNPs with a 0.1% FDR and an allelic ratio of ≥70%. We considered motifs with a median of ≥40 SNPs per position in motif and ≥3 positions with ≥7 significant SNPs; positions with <7 SNPs were considered missing data. For SNPs overlapping multiple matches to the same motif, we chose best motif position and orientation per SNP based on footprint occupancy score (FOS; a quantitative measurement of factor occupancy27,57) and FIMO P-value. For each SNP overlapping a TF recognition sequence, we measured the strength of the perturbation as the log odds difference between the two alleles according to the position weight matrix using a 40% G+C background.
For each motif, the enrichment of imbalanced SNPs was computed as log2 of the proportion of imbalanced SNPs lying within the recognition sequence (relative to the flanking 20 bp) divided by the proportion of non-imbalanced SNPs lying within the recognition sequence. To compute the statistical significance of the enrichment of imbalanced SNPs in each motif relative to flanking sequence, we computed the enrichment after permuting the assignments between imbalanced SNPs and their position in the motif or in the flanking regions. We performed 1,000 permutations, and fit a normal distribution to estimate a P-value. To correct for multiple testing, we estimated an FDR using Benjamini-Hochberg. An FDR cutoff of 1% corresponds approximately to a 0.25 log enrichment.
Definition of genomic regions
SNPs were annotated as follows:
Location relative to genes was computed using RefSeq.
CpG islands were downloaded from the UCSC Genome Browser.
Sequence conservation was measured using the PhastCons 100-way alignment from the UCSC genome browser.
Unthresholded hotspots and 1% FDR peaks were called using the program Hotspot42.
The cell-type activity spectrum (termed MCV, for multi-cell verified) was computed using ‘bedmap --count’ with the combined list of all DHSs across all cell types in Supplementary Table 1. An additional 22 malignant or immortalized cell lines were included for prediction.
Normalized DHS strength was computed as the number of reads per 1 million reads sequenced; the mean was taken for all DHS overlapping a given SNP.
The average DHS width was computed as the average width of all overlapping unthresholded hotspots across all cell types.
DNase I footprints were collated from 85 high-depth samples, and the lowest FOS was taken from the overlapping footprints (requiring FOS < 0.95, and 1 bp of overlap with SNPs or ≥3 bp with TF recognition sequences).
The number of factors occupying the 500-bp region surrounding each SNP was computed by counting the all distinct TF clusters overlapping a DNase I footprint by at least 3 bp in at least one cell type.
Prediction of SNPs perturbing TF recognition sequences
We used the glm() function R to fit a logistic model for each motif, considering all SNPs directly overlapping the recognition sequence (using the strict 0.1% FDR set of significant SNPs, as before):
significant ~ log(Read depth) + Num. hets.^2 + MCV^2 + CpG Island
+ 3' UTR + coding + intron + intergenic + Dist. to TSS^2 +
DHS strength^2 + Width of DHS + #nearby binding sites^2 +
PhastCons + Footprint presence + Footprint occupancy +
log(score)^2 + logodds difference + x2 + … + xn
Scores were scaled to an empirical percent significant score using a regression on binned raw regression scores:
pctSig ~ exp(score.bin)
We used to the predict() function to apply the model for each motif, and selected the maximum score from all motifs at a SNP. Performance was plotted against experimentally determined imbalanced variants (5% FDR; only considering SNPs with ≥3 heterozygotes and >100 reads) using ROCR58. The covariate terms Num. hets and log(Read depth) were set to 0 for computation of the empirical percent significant score, plotting of classifier performance, and predictions.
We downloaded dbSNP 13830 from the UCSC Genome Browser and scored each SNP on assembled chromosomes (i.e., autosomes, chrX and chrY). We conservatively considered only variants overlapping 1% FDR DNase hotspot peaks (considering the cell types in Supplementary Table 1 plus 22 malignancy-derived samples).
Validation of TF-centric models
Fetal-liver derived erythroblast DNase I data (FL_E; see Supplementary Table 1) were analyzed as before. We tested for imbalance at 9,846 SNPs passing all filters, and 1,613 imbalanced variants were identified at a 5% FDR cutoff. Variants were then scored using the TF models generated on the primary data sets, and the PPV computed as before using ROCR.
To assess the significance of the enrichment of predicted SNPs in dsQTLs4 while accounting for possible confounding factors, noncoding SNPs (those not in CCDS) from dbSNP with matching MAF, genic location and distance to TSS were sampled to generate a background distribution. Sets of SNPs from 500 permutations were scored with a significance cutoff at 0.10. To estimate a P-value, the background distribution was fit with a normal distribution.
Model performance was also compared relative to GERP59, PhastCons60, CADD61, fitCons62 (i6 scores across 3 cell types), and deltaSVM63 (maximum score across all cell types in common with this study). For comparison against dsQTLs, the background set from Lee et al.63 was intersected with GM12878 DNaseI peaks. Precision-recall curves were computed using ROCR.
Code availability
We used publically available software tools including the BEDOPS suite46. Analysis was performed using bash, awk and R. Additional code is available on request.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants U54HG004592, U54HG007010, U01ES01156, 1S10RR026770, and 1S10OD017999 to J.A.S. and NIMH fellowship F31MH094073 to M.T.M.. J.V. was supported by a National Science Foundation Graduate Research Fellowship under grant no. DGE-071824.
Footnotes
Accession codes
Data have been deposited in the Gene Expression Omnibus under accessions GSE18927, GSE26328, GSE29692, and GSE55579 for DNase I (Supplementary Table 1) and GSE30263 for ChIP-seq (Supplementary Table 2).
AUTHOR CONTRIBUTIONS
M.T.M., E.H. and J.A.S. conceived and designed the experiments. M.T.M. and E.H. analyzed the data. J.V. and M.T.M. performed TF cluster analysis. R.S. provided bioinformatics support. A.S. generated targeted footprinting data. R.K. assisted with data collection. M.T.M. and J.A.S. wrote the paper. M.T.M. and J.A.S. jointly supervised research.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 2.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmiter RD, Brinster RL. Germ-line transformation of mice. Annu. Rev. Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson KR, Stamatoyannopoulos G. Role of gene order in developmental control of human gamma- and beta-globin gene expression. Mol. Cell. Biol. 1993;13:4836–4843. doi: 10.1128/mcb.13.8.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 9.Archer TK, Lefebvre P, Wolford RG, Hager GL. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 10.Mendenhall EM, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aalfs JD, Kingston RE. What does ‘chromatin remodeling’ mean? Trends Biochem. Sci. 2000;25:548–555. doi: 10.1016/s0968-0004(00)01689-3. [DOI] [PubMed] [Google Scholar]
- 12.Ronald J, et al. Simultaneous genotyping, gene-expression measurement, and detection of allele-specific expression with oligonucleotide arrays. Genome Res. 2005;15:284–291. doi: 10.1101/gr.2850605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni Y, Hall AW, Battenhouse A, Iyer VR. Simultaneous SNP identification and assessment of allele-specific bias from ChIP-seq data. BMC Genet. 2012;13:46. doi: 10.1186/1471-2156-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat. Genet. 2003;33:469–475. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 15.McDaniell R, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurano MT, Wang H, Kutyavin T, Stamatoyannopoulos JA. Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genet. 2012;8:e1002599. doi: 10.1371/journal.pgen.1002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilpinen H, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy TE, et al. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res. 2012;22:860–869. doi: 10.1101/gr.131201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McVicker G, et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heap GA, et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum. Mol. Gen. 2009;19:122–134. doi: 10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stergachis AB, et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science. 2013;342:1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang K, et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat. Methods. 2009;6:613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spivakov M, et al. Analysis of variation at transcription factor binding sites in Drosophila and humans. Genome Biol. 2012;13:R49. doi: 10.1186/gb-2012-13-9-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biddie SC, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Stormo GD. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat Biotechnol. 2011;29:480–483. doi: 10.1038/nbt.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jolma A, et al. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 36.Lee J-H, et al. A robust approach to identifying tissue-specific gene expression regulatory variants using personalized human induced pluripotent stem cells. PLoS Genet. 2009;5:e1000718. doi: 10.1371/journal.pgen.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding J, et al. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am. J. Hum. Gen. 2010;87:779–789. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price AL, et al. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 2011;7:e1001317. doi: 10.1371/journal.pgen.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flutre T, Wen X, Pritchard J, Stephens M. A Statistical Framework for Joint eQTL Analysis in Multiple Tissues. PLoS Genet. 2013;9:e1003486. doi: 10.1371/journal.pgen.1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veyrieras J-B, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
ONLINE REFERENCES
- 42.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.John S, et al. Genome-scale mapping of DNase I hypersensitivity. Curr Protoc Mol Biol. 2013;Chapter 27(Unit 21.27) doi: 10.1002/0471142727.mb2127s103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neph S, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazarovici A, et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6376–6381. doi: 10.1073/pnas.1216822110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Novère N. MELTING, computing the melting temperature of nucleic acid duplex. Bioinformatics. 2001;17:1226–1227. doi: 10.1093/bioinformatics/17.12.1226. [DOI] [PubMed] [Google Scholar]
- 52.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Portales-Casamar E, et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 59.Cooper GM, et al. Characterization of evolutionary rates and constraints in three Mammalian genomes. Genome Res. 2004;14:539–548. doi: 10.1101/gr.2034704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gulko B, Hubisz MJ, Gronau I, Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015;47:276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee D, et al. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 2015;47:955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stergachis AB, et al. Developmental Fate and Cellular Maturity Encoded in Human Regulatory DNA Landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vierstra J, Wang H, John S, Sandstrom R, Stamatoyannopoulos JA. Coupling transcription factor occupancy to nucleosome architecture with DNase-FLASH. Nat. Methods. 2014;11:66–72. doi: 10.1038/nmeth.2713. [DOI] [PubMed] [Google Scholar]
- 66.Kellis M, et al. Reply to Brunet and Doolittle: Both selected effect and causal role elements can influence human biology and disease. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3366. doi: 10.1073/pnas.1410434111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vierstra J, et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maurano MT, et al. Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell Rep. 2015;12:1184–1195. doi: 10.1016/j.celrep.2015.07.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.