Abstract
Advances in next generation sequencing (NGS) technology have provided the tools to comprehensively and accurately characterize the microbial community in the respiratory tract in health and disease. The presence of commensal and pathogenic bacteria has been found to have important effects on the lung immune system. Until relatively recently, the lung has received less attention compared to other body sites in terms of microbiome characterization, and its study carries special technological difficulties related to obtaining reliable samples as compared to other body niches. Additionally, the complexity of the alveolar immune system, and its interactions with the lung microbiome, are only just beginning to be understood. Amidst this complexity sits Mycobacterium tuberculosis (Mtb), one of humanity’s oldest nemeses and a significant public health concern, with millions of individuals infected with Mtb worldwide. The intricate interactions between Mtb, the lung microbiome, and the alveolar immune system are beginning to be understood, and it is increasingly apparent that improved treatment of Mtb will only come through deep understanding of the interplay between these three forces. In this review, we summarize our current understanding of the lung microbiome, alveolar immunity, and the interaction of each with Mtb.
Keywords: Lung microbiome, pulmonary alveoli, innate immune, mycobacterium tuberculosis, alveolar macrophages, phagosome
1. INTRODUCTION
Tuberculosis (TB) is one of the oldest illnesses in history and still remains a major health problem worldwide, with more than 8 million new cases and more than 1 million deaths occurring every year. Exposure to Mycobacterium tuberculosis (Mtb) causes active disease in nearly 10% of exposed people, while in the remaining 90% the immune response inhibits Mtb multiplication (Zumla, Raviglione, Hafner, & von Reyn, 2013). In a portion of the exposed population, some Mtb bacilli are not killed but instead remain in a non-replicating state dubbed latent tuberculosis infection. In latent TB, the inactive bacilli can regain replication capacity and cause active tuberculosis if the immune response is disrupted. This constitutes an issue of significant global health concern, as these represent potential cases of reactivation and transmission, which could hamper control of the disease (Chapman & Lauzardo, 2014). People at increased risk for tuberculosis reactivation include recent immigrants from countries with a high incidence of tuberculosis, children younger than age 5, individuals who have been infected with Mtb within the past 2 years, and anyone in an immune-compromised state.
The human body is the habitat of microbial symbionts ten-times more in number than Homo sapiens cells.(Cox, Cookson, & Moffatt, 2013) The term “microbiota” refers to the microbes that live in a specific location (e.g. the human body, the gut, the soil, etc.). The collective genomes of the complete microbiota present in the human body is referred to as the “human microbiome” (Cox, et al., 2013). Advances in next generation sequencing (NGS) technology now allow us to comprehensively profile the microbial community in the various niches, including the respiratory tract, in both physiological and pathological conditions (Hui, Lau, Chan, & Tsui, 2013). NGS technology circumvents the need to cultivate organisms by analyzing nucleic acids directly extracted from samples and has led to an exponential growth in our understanding of the role of the microbiome in almost every field (Blaser, 2014). Several lines of research have demonstrated that commensal microbiota is in constant contact with the host immune system and thus influence immune function (Spasova & Surh, 2014). Recent studies have analyzed the bacterial composition of the microbiota in the respiratory tract of chronic obstructive pulmonary disease (COPD), influenza and TB patients, providing clues to the pathogenesis of these conditions(Hui, et al., 2013).
2. The lung microbiome: historically neglected
The presence of commensal microorganisms within the human body has been recognized for well over a century, dating back to Escherich’s 1886 description of Enterobacteria in the digestive tracts of infants (Escherich, 1886). The importance of the microbiome and our ability to shape it has likewise been recognized for many decades, including early work describing shifts in our intestinal bacteria with dietary changes (Herter & Kendall, 1910; Torrey, 1919) and later work on the protective potential of the microbiota against pathogenic group A Streptococcal infection (Crowe, Sanders, & Longley, 1973). In recent years, an explosion of work has documented the detailed mechanisms of many host-microbiome interactions, including pathways that influence systemic immunity (Belkaid & Hand, 2014), cardiovascular health (Wang et al., 2011), and neurological development (Diaz Heijtz et al., 2011). Today, the notions that 90% of “our” cells are microbial (Savage, 1977) and that the genomes of these microorganisms may contain 100-fold more genes than our own genome (Bäckhed, Ley, Sonnenburg, Peterson, & Gordon, 2005) are practically common knowledge.
However, the growth of knowledge of the microbiome and its importance has not been equally applied. In-depth study of many body sites, particularly the gut but also the vagina, oral cavity, and skin, has created a sturdy, albeit incomplete, picture of the importance of the microbiome at each of these sites. In sharp contrast is the lung, which remained poorly studied and poorly understood until very recently and continues to suffer from a relative dearth of research when placed against other sites (Dickson, Erb-Downward, & Huffnagle, 2014; Segal & Blaser, 2014). This deficit is in large part historical. One can look as far back as Escherich (Escherich, 1886), Metchnikoff (Metchnikoff, 1908), and Döderlein (Döderlein, 1892), to find mention of the importance of microorganisms in sites such as the gut and vagina. By contrast, medical science had long assumed that the lung was a sterile environment unless perturbed by respiratory pathogens. Indeed, many culture-based investigations reported no cultivatable organisms in the healthy lung (Baughman, Thorpe, Staneck, Rashkin, & Frame, 1987; Chastre et al., 1988; Higuchi, Coalson, & Johanson, 1982; Johanson, Pierce, & Sanford, 1969; Thorpe, Baughman, Frame, Wesseler, & Staneck, 1987). The assumption of sterility was so firmly entrenched that the original iteration of the Human Microbiome Project (HMP) omitted the lung from its list of sampling sites (Peterson et al., 2009).
It was only with new appreciation for the unseen diversity in already well-established body sites that the scientific community cast a new eye at the lung. Genomic studies of 16S ribosomal RNA diversity in the gut quickly proved that as few as a quarter of all organisms present may be able to be cultured by present methods (Suau et al., 1999). The soil, perhaps an even more complex system, may hold upwards of 99% of its total microbial diversity in the category of unculturables (Torsvik, Goksøyr, & Daae, 1990). “Negative by culture” was clearly no longer sufficient to rule a body site sterile.
Facing the clear evidence of the inadequacy of culture-based studies to exclude the presence of microorganisms, a number of investigations have conclusively demonstrated that the lung harbors a community of microorganisms even in the healthy state (Borewicz et al., 2013; Charlson et al., 2012; Charlson et al., 2011; Dickson, Erb-Downward, Freeman, et al., 2014; Dickson, Erb-Downward, et al., 2014a, 2014b; Erb-Downward et al., 2011; Hilty et al., 2010; Huang et al., 2011; Pragman, Kim, Reilly, Wendt, & Isaacson, 2012; Segal et al., 2013; Sze et al., 2012). The recently-launched second round of the HMP includes the lung (Integrative, 2014), and the NHLBI-sponsored Lung HIV Microbiome Project has already produced a series of important contributions (Lozupone et al., 2013; Morris et al., 2013; Twigg et al., 2013). In addition to bacteria, resident viruses (Lysholm et al., 2012; Willner et al., 2009; Willner et al., 2012; Young et al., 2015), and fungi (Delhaes et al., 2012; Huffnagle & Noverr, 2013; Willger et al., 2014) in the lung microbiome are beginning to be appreciated, although the contributions of each to health and disease remain unclear, and precisely which fungal and viral species are present in the healthy lung continues to be very poorly understood.
3. Niches and sampling controversies in the study of the lung microbiome
Despite advances in our understanding of the lung microbiome in recent years, important controversies remain unsettled. First among these is the blurred line between the upper respiratory tract (URT), which includes the nose and oropharynx, and the lower respiratory tract (LRT), which includes the bronchi and alveoli. Decades of research have not settled the argument of how significant contamination from the URT is when evaluating the lower. Studies during the culture-based era confronted this issue, with multiple groups commenting on the difficulty of obtaining samples from the LRT, whether the trachea or a deeper bronchus, without contamination by the URT (Davidson, Tempest, & Palmer, 1976; Matthew, Holstrom, & Kaspar, 1977).
Decades after these initial studies, the issue of contamination continues to vex researchers. One effort to resolve the controversy has been to sample both the URT and the LRT and simply subtract the former from the latter (Charlson, et al., 2012; Charlson, et al., 2011), although this strategy has not yet seen wide acceptance or validation (Dickson, Erb-Downward, & Huffnagle, 2013). Another proposal has been to apply what is known as the “neutral theory” of community ecology. In this strategy, the basic assumption is that all species in the community (the lung microbiota, in this case) are functionally equivalent and “do not differ on a per capita basis” in key features such as birth, death, and dispersal (Hubbell, 2005). Thus, the observed distribution of species can be said to not be the result of active environmental selection, and dispersal from a source community together with random birth and death of microorganisms should explain any community structure. The null hypothesis then becomes that simple dispersal from the URT explains any variation between the LRT and URT, and any bacteria found to significantly differ between the two sites could be said to have some advantage in the lung (Morris, et al., 2013). However, much like the proposal to subtract URT from LRT, this strategy has yet to come into wide use.
A third effort has considered the lung and its compartments as akin to islands within a large sea. One group has proposed an “island model” of biological communities as applied to the lung (Dickson, Erb-Downward, & Huffnagle, 2014). Here, the composition of the lung microbiome depends on extinction and migration and may shift with time and external forces. A condition that would tend to predispose to bacterial entry (e.g. aspiration, oral infection) might tend to push the lung microbiome towards a more oral or oro-pharyngeal composition. Similarly, a condition that impaired lung immunity (e.g. immunodeficiency) or mucus clearance (e.g. cystic fibrosis) might tend to increase the bacterial burden in the lung and could result in the dominance of different species than in the healthy lung. This theory also suggests that an “island” that lies close to a source population (e.g. the proximity of the larynx to the epiglottis and URT) may adopt a composition more similar to that of the source population than more distant sites (Dickson, Erb-Downward, & Huffnagle, 2014). Two very recent reports have brought new interesting evidence on the lung microbiome composition and possible origin. The first one described that bacterial communities of healthy lungs shared significant similarity with the mouth, but not the nose, suggesting that microbial migration from the oral cavity might be the source of the lung microbiome during health, (Bassis CM, 2015). The second one demonstrates that the lung microbiome in health is more influenced by microbial immigration and elimination, i.e. the “adapted island model of lung biogeography”, than by the effects of local growth conditions on bacterial reproduction rates (Dickson RP, 2015).
Further confusion arises when considering the proper method to sample the lung microbiome. The ideal method is perhaps to take a sample of lung tissue and digest the entire sample, isolating microbial DNA from amidst the host DNA. However, while a small number of studies involving lung transplantation have been able to utilize this method (Erb-Downward, et al., 2011; Goddard et al., 2012; Rudkjøbing et al., 2011), for obvious reasons it cannot be applied routinely or to every disease condition.
As a result, most studies utilize sputum or broncho-alveolar lavage (BAL) samples. Sputum sampling has the distinct advantage of being non-invasive, facilitating collection and potentially enabling longitudinal study designs. However, sputum may only represent higher levels of the LRT, leaving deeper airways out of subsequent analyses. Furthermore, sputum inevitably will contact the URT and oral cavity during the expectoration and collection process, and both classic culture-based (Davidson, et al., 1976; Matthew, et al., 1977) and modern NGS-based studies (Cabrera-Rubio et al., 2012; Goddard, et al., 2012) have suggested that sputum does not accurately recapitulate the LRT, although there continues to be debate on this issue.
BAL was proposed by early investigators of the lung microbial community as a means to acquire samples without contamination by the URT, particularly in conjunction with protected brush or dual BAL tube sampling (Chastre, et al., 1988; Davidson, et al., 1976). Many recent studies have utilized BAL (Borewicz, et al., 2013; Cabrera-Rubio, et al., 2012; Dickson, Erb-Downward, et al., 2014a; Erb-Downward, et al., 2011; Hilty, et al., 2010; Segal, et al., 2013), but it is not a complete solution. Similarly to sputum, BAL cannot completely sample the deepest airways of the lung, potentially biasing any analyses in favor of the microbiome of the more proximal lower airways. Additionally, BAL is an invasive procedure, which both magnifies the risk of performing it, particularly in a lung that may already be weakened by disease, and may make subjects more reluctant to participate in research studies.
4. Composition of the lung microbiome
The composition of the healthy lung microbiome remains an area of active debate. As discussed earlier, the line between the URT and LRT is not clear, and just how much the URT contributes to the diversity of the LRT has and continues to be an area of active debate (Dickson, et al., 2013). One early study comparing oral and oropharyngeal wash and swab samples with bronchoscopic and lower airway protected brush sampling reported an equivalent microbiome between the URT and LRT, with no lung-specific taxa shared between individuals (Charlson, et al., 2011).
In contrast, many other studies have reported distinct and unique lung microbiomes. While the exact composition has differed slightly between studies, many have reported high proportions of Firmicutes (including members of the family Veillonellaceae), Bacteroidetes (including Prevotella spp.), Proteobacteria (including Pseudomonas spp. and other Gammaproteobacteria), and Fusobacteria (Borewicz, et al., 2013; Erb-Downward, et al., 2011; Hilty, et al., 2010). However, even in these commonalities there remain controversies. One study has reported more Bacteroidetes than Proteobacteria in controls when compared to individuals with COPD or asthma (Hilty, et al., 2010), while others have seen abundant Proteobacteria in healthy lungs (Borewicz, et al., 2013). There remains much more controversy surrounding the role of organisms including Neisseriaceae, Haemophilus spp., Streptococcaceae, and Actinobacteria, with varying reports finding these in diseased (Cabrera-Rubio, et al., 2012; Goddard, et al., 2012; Hilty, et al., 2010; Rudkjøbing, et al., 2011) and healthy lungs (Erb-Downward, et al., 2011).
A major limitation of efforts to determine the “normal” lung microbiome remains the small number of healthy individuals sampled and limited geographical diversity of sampling sites. Insight may be gained from examining the history of gut microbiome research. A similar debate of gut microbiome composition occurred upon the discovery of “enterotypes”, or distinct gut microbiomes that differ by region and diet (Arumugam et al., 2011). Initially hailed as a major explanation for microbiome diversity worldwide, subsequent re-analysis and re-examination in other studies with more sampled individuals demonstrated that the distinct regional and dietary clusters noted may be more accurately said to resemble positions on a spectrum (Knights et al., 2014). It is likely that the precise composition of the healthy lung microbiome will remain unsettled for some time.
One commonality between studies has been that the lung contains a lower bacterial burden than other body sites by at least several fold (Charlson, et al., 2011; Hilty, et al., 2010). One investigation reported roughly 2000 bacterial genomes per cm2 in the lung, a population tenfold less than that of the skin and more akin to the population found in the upper two-thirds of the small intestine (Hilty, et al., 2010). It is important to note that this microbial burden appears to be highly plastic and subject to external forces, for that same study reported that the lungs of smokers contain ten-fold more bacterial genomes per cm2, a value akin to the skin.
The diversity of a microbiome, which relates to the number of distinct taxa present, has been shown to impact human health and disease. It has long been known that altering microbiome diversity can predispose individuals to certain diseases (Legatzki, Rösler, & von Mutius, 2014), with reduced diversity of the gut microbiome and its association with increased incidence of allergic disease representing perhaps the strongest link (Bisgaard et al., 2011). However, the effect of diversity within the lung is much less clear. One study of asthmatics and moderate COPD suggested that higher lung microbiome diversity was associated with increased airways hyperreactivity and asthma severity, while lower diversity was found in the lungs of patients with severe COPD (Huang, et al., 2011). Contrastingly, a separate study of moderate COPD sufferers found no dearth of diversity (Cabrera-Rubio, et al., 2012), suggesting that the traditional assumptions on beneficial diversity may not apply in the lung.
A final important consideration centers on the geography of the lung. Much like the gut, the LRT and lung are not a monotonous, consistent tube; rather, the physiology and anatomy of the airways vary greatly as one descends into the deeper portions of the LRT. Oxygen tension, pH, temperature, and mucus production shift with descent into the deeper airways. Temperature, for example, rises with distance from the main stem bronchi (Dickson, Erb-Downward, & Huffnagle, 2014). Thus, it is not surprising that several groups have reported that distinct microbial communities reside at different levels of the LRT(Cabrera-Rubio, et al., 2012; Erb-Downward, et al., 2011). Unfortunately, evaluation of these differences is complicated to achieve with BAL and particularly with sputum samples, where controlling for the source depth is difficult if not impossible. One of the aforementioned studies avoided this challenge by utilizing lung explant samples (Erb-Downward, et al., 2011), but this is unlikely to be widely applicable, particularly in healthy lungs.
5. Lung microbiome in pulmonary TB
The relative dearth of information concerning the lung microbiome is a challenge for researchers in the field of pulmonary TB. Whereas a student of inflammatory bowel disease can draw upon a huge body of gut microbiome study (Hold et al., 2014), no pulmonary disease is so similarly blessed. The microbiome in the context of TB remains an understudied area, despite the heavy burden of the disease worldwide (Jassal & Bishai, 2010). Many existing studies have focused on microbiomes outside of the lungs, including reports of increased Candida spp. in patients being treated for TB infection (Querido et al., 2011) and a loss of diversity in the gut microbiota as a result of TB treatment (Dubourg et al., 2013). In animal models, experimental Mtb infection is associated with a loss of gastrointestinal microbiome diversity, perhaps arising from competition with the newly-introduced Mtb bacteria (Winglee et al., 2014). One intriguing study reported that patients with latent TB infections and seropositivity for Helicobacter pylori exhibited more robust immune response (including increased IFN-gamma induced by Mtb antigen) than individuals with no evidence of H. pylori exposure. Furthermore, experimental work in rhesus macaques revealed that H. pylori seemed to provide protection against Mtb infection (Perry et al., 2010). This finding is well in line with other published work demonstrating that H. pylori exposure protects against allergic asthma (Arnold et al., 2011; Engler et al., 2014; Oertli et al., 2012), and it may be that the H. pylori organism induces broad changes to systemic immunity that promote containment of TB.
As of early 2015, just a handful of studies exist that have evaluated the respiratory microbiome in the context of TB. The first, in 2012, found that healthy and Mtb-infected lungs shared many organisms, including Bacteroidetes, Proteobacteria, and Actinobacteria, with Firmicutes and Bacteroidetes dominating (Cui et al., 2012). This is consistent with other reports from healthy lungs (Borewicz, et al., 2013; Erb-Downward, et al., 2011; Hilty, et al., 2010). They reported an association between Mtb and the presence of many traditionally “pathogenic” organisms, including Pseudomonas spp. A more recent examination found a similar abundance of Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes in TB lungs. However, they also reported finding Fusobacteria and determined that Proteobacteria and Bacteroidetes were the dominant organisms in TB, with Firmicutes predominantly in control lungs (Cheung et al., 2013). A study published last year which unfortunately did not compare TB sputum to healthy sputum nonetheless noted that Bacteroidetes, Fusobacteria, and Actinobacteria were dominant in TB (Botero et al., 2014). This study also demonstrated that “nasal” clusters separate from “Sputum” and “Oropharynx”, a finding that has been confirmed very recently by another group (Bassis CM, 2015). This group additionally reported on the presence of fungi in the TB lung, with members of the phylum Ascomycota dominating (Botero, et al., 2014). When comparing the sputum to the oropharynx, this group did not note a difference, a finding likely due to contamination during sputum expectoration. The final study found a very different slate of TB-associated organisms, including Streptococcus, Gramulicatella, and Pseudomonas, with healthy lungs possessing more Treponema, Catonella, and Coprococcus (Wu et al., 2013). A compelling aspect of this study is its comparison of recurring and new-onset TB, where an association between Pseudomonas infection and recurrent TB was noted, suggesting that this pathogen may interfere with the clearance of Mtb. Considering the presence of Pseudomonas in several of these surveys, this line of investigation is worthy of additional study and could provide insight into clearance of stubborn, recurrent Mtb infection. Unfortunately, there is little agreement between these surveys and other published lung microbiome analyses. Several assessments of the healthy lung have reported Pseudomonas in “healthy” individuals, and several TB-associated organisms (including Actinobacteria and Streptococcaceae) have been found in healthy as well as diseased lungs (Borewicz, et al., 2013; Erb-Downward, et al., 2011; Hilty, et al., 2010).
In the area of diversity, there is little agreement. One study found increased diversity in the TB lung as compared to the healthy lung (Cui, et al., 2012), while another found no differences in diversity between the two(Cheung, et al., 2013). In this, the TB lung appears to mirror studies of the healthy lung, where reports of increased diversity being associated with disease (Huang, et al., 2011) are conflicted by other reports finding no disease-associated diversity differences (Cabrera-Rubio, et al., 2012).
The lack of consensus between these studies and between these studies and the broader lung microbiome field likely arises from the small sample size. Taken together, the four studies of the lung microbiome in TB through early 2015 include only 134 TB patients and 64 controls, with all but one site located within China (Botero, et al., 2014; Cheung, et al., 2013; Cui, et al., 2012; Wu, et al., 2013). Early attempts to determine the composition of the gut microbiome were hampered by a lack of sample diversity, and further study, including sampling of sites in other regions of the world where Mtb is endemic (e.g. Africa, Southeast Asia, Eastern Europe), will be required before the TB lung microbiome can begin to be firmly characterized. Future studies will also need to complement sputum sampling with BAL to determine if the communities seen in TB sputum are truly representative of the lung microbiome or if they represent contamination from the URT. Unfortunately, this is far easier said than done, particularly when many areas of endemic Mtb have poor health infrastructure and may lack access to BAL equipment and knowledgeable practitioners. Lastly, the importance of an accurate characterization of the lung microbiome is critical when considering certain opportunistic pathogens, such as Rhodococcus, that can complicate diagnosis of Mtb infection. Rhodococci can be mistaken for Mtb infections in a routine AFB stain, and members of the genus can also occur as mixed infections with Mtb (Mistry et al., 2006).
6. Mtb infection of alveolar macrophages and alveolar epithelial cells
The pulmonary alveolus is surrounded by a thin lining of epithelial type I and type II cells. Type I cells are thin, flat, and cover more than 90% of the alveolar surface, allowing for gas exchange. Type II epithelial cells are cuboidal with apical microvilli and are involved in the production and secretion of pulmonary surfactant lipids and proteins (Guillot et al., 2013). Type II epithelial cell also release other soluble substances that have innate immune activity, and these cells are important in initiating and shaping immune responses in the lung (Chuquimia, Petursdottir, Periolo, & Fernandez, 2013; Chuquimia et al., 2012). Alveolar epithelial cells influence the microbicidal capacity of pulmonary macrophages against mycobacteria through a series of secreted factors that surprisingly do not involve IFN-γ-induced proinflammatory cytokines (Petursdottir, Chuquimia, Freidl, & Fernandez, 2014). The airway epithelium is also capable of stimulating Mtb-reactive CD8+ T cells to produce IFN-γ (Harriff et al., 2014), and constitutes the main source of CXCL5 upon Mtb infection (Nouailles et al., 2014), a cytokine which drives PMN recruitment leading to Mtb clearance in mice in a TLR2-dependent manner (Nouailles, et al., 2014).
Infection with Mtb is initiated by inhalation of aerosol droplets carrying a small number of mycobacteria (Gengenbacher & Kaufmann, 2012), the historical Flugge droplets (Pistacchio, 1999). Once Mtb gains access to the pulmonary alveoli, it is internalized and replicates within type II alveolar epithelial cells (Harriff, et al., 2014), alveolar macrophages (AMs), and other phagocytes, including dendritic cells (DCs). The latter transport bacteria to local, draining lymph nodes, interacting with T cells to prime and initiate adaptive immune responses. Infected macrophages migrate and transport Mtb from the airway into pulmonary tissue sites. An inflammatory focal lesion develops, comprised of infected macrophages and monocytes attracted to the site. This primary lesion matures into a granuloma, which is the hallmark of TB (Gengenbacher & Kaufmann, 2012).
Macrophages are the major reservoir of Mtb during the early stage of infection (Bruns & Stenger, 2014). After Mtb is inhaled and transmitted to the lungs, it is phagocytized by alveolar macrophages and eliminated by various mechanisms (Yuk & Jo, 2014). The ability of Mtb to survive within macrophages is an important determinant of the outcome of infection. If the host fails in eradicating the bacteria, the disease is preserved in a latent condition, which can either be terminated with an appropriate host innate immune response or lead to Mtb replication in macrophages and dissemination to other tissues (Yuk & Jo, 2014). Mtb primarily escapes eradication by impairing phagosome maturation (Gengenbacher & Kaufmann, 2012). However, Mtb is capable of interfering with other host mechanisms that attempt to overcome the phagosome maturation block. Various molecules derived from Mtb inhibit apoptosis, efferocytosis, autophagy, and ROS generation (Behar, Divangahi, & Remold, 2010; Bruns & Stenger, 2014). These mechanisms do not act independently but instead in a cooperative way. For example, besides its direct microbicidal effect on Mtb, ROS induces autophagy and apoptosis in human macrophages and may act synergistically with antimicrobial peptides (AMPs). An alternative strategy is for Mtb to egress from the phagosome into the cytosol of macrophages (Behar, et al., 2010; van der Wel et al., 2007)
Tissue destruction and pathogenesis during TB infection is not mediated by pathogens alone but induced by an immunopathological inflammatory response of the host. AM activation, which allows them to act against invading organisms and danger signals, is tightly regulated to limit inflammation and minimize lung injury thus preserving alveolar function (Rajaram, Ni, Dodd, & Schlesinger, 2014). Anti-inflammatory signaling occurs through IL-10 and TGF-β, the mannose receptor (MR, CD206) (Hussell & Bell, 2014), and micro-RNA (Ma et al., 2014). Cell-to-cell interactions, via epithelial cell ligation of AM receptors, also contributes to limiting the progression of inflammation (Rajaram, et al., 2014). AMs are ineffective as antigen presenting cells (APCs) (Fels & Cohn, 1986), which limits the initiation of an effective T cell inflammatory response (Hussell & Bell, 2014), and are considered to be “immunoregulatory macrophages”(Rajaram, et al., 2014).
Several genetic polymorphisms have been associated with susceptibility to Mtb infection in monocytes/macrophages, including the promoter regions of IL10 (Liang, Guo, Li, & Kong, 2014; Moraes et al., 2004), TNFRI, and TNFRII (Stein et al., 2007). Polymorphisms in the Vitamin D (Vit D) Receptor (VDR) gene are also associated with susceptibility to Mtb (Wilkinson et al., 2000). Historically, the association of vitamin D on infectious diseases began with observations on the beneficial action of sunlight in patients with TB (Weinzirl, 1906). The protective effect of vitamin D is mostly due to its modulatory activity on macrophage function (P. T. Liu et al., 2006). TLR1/2 activation induces the expression of VDR (Bruns & Stenger, 2014). Vitamin D upregulates the expression of AMPs like human beta defensin 2 and cathelicidin and restricts the growth of Mtb in human macrophages (Afsal et al., 2014; P. T. Liu, et al., 2006).
7. Phagosomal innate immune recognition of Mtb
The phagosome is the platform where several host defense mechanisms initiate. These include processes such as endosomal TLR activation, endosomal acidification, ROS and RNS production, hydrolytic enzyme activity, and AMP microbicidal effects.
Mtb invasion starts with host recognition of Mtb outer surface molecules that can bind to the host pathogen-associated receptors, including toll-like receptors (TLRs). In mycobacterial infection, several TLRs are involved in recognition of Mtb components and activation of the innate immune responses. In active disease, Mtb primarily infects professional phagocytes in the lungs where it uses strategies such as prevention of phagolysosome maturation, subversion of host cell death pathways, and the immune system to survive and replicate (Keown, Collings, & Keenan, 2012; Soldati & Neyrolles, 2012). Recent evidence has highlighted the importance of the TLR adaptor molecule MYD88 for confinement of Mtb in the phagosome (Rahman, Sobia, Gupta, Kaer, & Das, 2014) as well as for mycobacterial clearance and an adequate innate and adaptive immune response (Berod et al., 2014; Fremond et al., 2004; Reiling, Ehlers, & Holscher, 2008).
Since Mtb is a bacteria that remains in the phagosome, inhibiting phagosomal acidification and recruitment of lysosomal proteins is crucial for its survival (Keown, et al., 2012; Soldati & Neyrolles, 2012). While escape of Mtb from its vacuole within infected macrophages has been reported (Behar, et al., 2010; van der Wel, et al., 2007), controversy remains as to whether these observations are truly physiological or due to the experimental technique used. An alternative hypothesis of bacteria-mediated pore formation within the phagosomal membrane was later reported (Teitelbaum et al., 1999). Phagosomal signaling occurs after activation of endosomal TLR ligands once their ligands are endosomically released. The phagosome may constitute the platform from which components of Mtb could be recognized by pattern recognition receptors (PRRs) present therein, such as endosomal TLRs. The subcellular localization of endosomal TLRs guarantees release of ligands from the pathogen in a naturally-occurring, immunity-promoting manner (Sander et al., 2011).
Endosomal TLR signaling may play an important role in the outcome of infection, as these receptors are involved in the generation of Type I IFNs upon recognition of pathogen-associated nucleic acids in the phagosome (Gonzalez-Navajas, Lee, David, & Raz, 2012; He, Jia, Jing, & Liu, 2013; Puig et al., 2012). A type I IFN gene signature is associated with active TB (Berry et al., 2010; Mayer-Barber et al., 2014), and Type I IFNs appear to suppress IFN-γ-triggered responses (de Paus et al., 2013; Teles et al., 2013), including Mtb killing by macrophages (McNab et al., 2014).
Several studies have demonstrated the association of specific endosomal TLR polymorphisms with susceptibility to tuberculosis (Basu, Shin, & Jo, 2012; Bharti et al., 2014; Davila et al., 2008; Thada, Valluri, & Gaddam, 2013; Velez et al., 2010). However, not much is known about nucleic acid recognition and signaling in tuberculosis, and its role in susceptibility to the disease remains unclear (Juarez et al., 2010; Kiemer et al., 2009; Yamashiro, Oliveira, & Bafica, 2014). Interestingly, stimulation with DNA from attenuated Mtb is able to generate a greater TNF-α response compared to DNA from the virulent strain in human alveolar macrophages (Kiemer, et al., 2009). A recent report demonstrated that TLR3 recognizes BCG RNA and is necessary for mycobacterial RNA-induced IL-10 production (Bai et al., 2014). RNA from M. leprae is able to elicit a TNF-α and IFN-γ response (Lorenzi et al., 2010). Recognition of Mtb DNA might occur through TLR9, as IFN-β production upon Mtb infection of murine DCs is dependent on TLR9 (Simmons et al., 2010).
The impact of studying TLR-signaling goes beyond innate immunity, as it appears to be important in linking the adaptive immune system to Mtb, and this linkage appears to be crucial in determining the fate of the disease. TLR9 regulates Th1 responses and also cooperates with TLR2 in mediating optimal resistance to Mtb (Bafica et al., 2005). Relatively recent studies have demonstrated that different antigenic proteins from Mtb trigger different responses in peripheral blood mononuclear cells (PBMCs) from patients with active TB (Kassa et al., 2012). Tuberculin purified protein derivative (PPD) elicits inflammatory responses via TLR2-ROS-MAPK signaling pathway (Yang et al., 2008). Early Secreted Antigen ESAT-6, expressed by virulent Mtb strains but not by BCG, promotes protective T Helper 17 cell responses in a TLR-2-dependent manner (Chatterjee et al., 2011). In contrast to TLR2, TLR9 signaling from DCs controls IFN-γ production by CD4+ T cells in Mtb–infected animals (Bafica, et al., 2005). TLR profiling in these cells, as well as their endosomal TLR signaling characterization, would allow us to elaborate a model linking innate recognition with adaptive immunity. The implications of this go beyond merely pathogenetic description, as TLR8-TLR2 and TLR9-TL2 crosstalk could be important in promoting vaccine enhancing responses, particularly in light of increasing evidence of the benefits of using endosomal agonists as adjuvants for TB vaccines (Baldwin et al., 2009; Commandeur et al., 2014).
8. Antimicrobial peptides (AMPs) lung epithelial activation and cellular interaction
AMPs are naturally occurring antimicrobial molecules which are important in host defense against pathogenic microbes. AMPs, including defensins and cathelicidins, not only play an important role in killing various pathogens but additionally act as regulators of the innate immunte system (van der Does, Bergman, Agerberth, & Lindbom, 2012). AMPs also participate at the interface of innate and adaptive immunity as chemoattractants for immune effector cells, modulators of the production of a variety of inflammatory mediators by multiple cell types, and regulators of the differentiation of monocytes into dendritic cells(van der Does, et al., 2012). LL-37, the only human member of the cathelicidin family, enhances opsonic phagocytosis of Gram-negative and Gram-positive bacteria by human monocytes as well nonopsonized Escherichia coli by human macrophages (Wan et al., 2014). Cathelicidin is also required for Mtb-induced ROS release and proinflammatory cytokine production (van der Does et al., 2010). A recent study has shown that increase in macrophage phagocytosis of Mtb and cathelicidin gene upregulation by vitamin D is more prominent in pulmonary TB patients without lung cavitation, suggest the potential use of vitamin D in less severe forms of TB disease (Afsal, et al., 2014). However, Vit D is not the only way to increase the concentration of AMPs. Phenylbutyrate, an HDAC inhibitor, also induces cathelicidin and autophagy in human macrophages (Mily et al., 2013). LL-37 also elevates the expression of FcRs on macrophages (Wan, et al., 2014) and directs macrophage differentiation toward macrophages with a proinflammatory signature (van der Does, et al., 2010).
Furthermore, AMPs may show synergistic effects with antituberculous drugs such as rifampicin (Khara et al., 2014). The use of AMPs in tuberculosis therapy holds immense potential in the future, especially against drug-resistant Mtb (Lan et al., 2014) (Padhi, Sengupta, Sengupta, Roehm, & Sonawane, 2014).
9. Differences in the host response against different M. tuberculosis genotypes and Mycobacteria
Mtb is one member of the group called Mycobacterium tuberculosis Complex (MTBC). The degree of genetic diversity within MTBC created debate in the field until newer and more reliable phylogenetic analysis of MTBC strains through the use of Whole Genome Sequencing (WGS) revealed that there exists more genetic diversity between strains, with genetic distance sometimes comparable to the interspecies distance between Mtb and M. bovis (Gagneux, 2013; Galagan, 2014).
There are functional and clinical consequences of such genetic diversity (Coscolla & Gagneux, 2010). Supporting the pathological relevance of these genetic studies, it has been shown that different isolates of Mtb possess different lung epithelial invasive potentials (Ashiru, Pillay, & Sturm, 2012). Regarding the host-pathogen interaction, differences may alter the balance of an adequate immune response that allows the mycobacteria to establish a granuloma for transmission and inflammation that can lead to bacterial elimination (Galagan, 2014). Evidence of strain variation suggests that specific Mtb proteins may play a role in immune evasion via antigenic variation (H. Liu et al., 2013; Vordermeier et al., 2012). Correlation between phylogenetically-related clinical isolates representative of the global diversity of the human MTBC and inflammatory response has been reported (Portevin, Gagneux, Comas, & Young, 2011). Although the latter study measured several cytokines from infected human peripheral blood monocyte-derived macrophages and found a wide variation in the response to different strains, no evaluation of Type I or Type II IFNs was performed. A very recent publication has reported that virulent, modern Beijing strains induced lower levels of pro-inflammatory cytokines (Chen et al., 2014). The difference in Type I IFN responses elicited by different Mtb has been less studied. More virulent strains may induce higher Type I IFN responses, as new evidence points out the inhibitory effect of these proteins on monocyte and macrophage responsiveness to IFN-γ (de Paus, et al., 2013; McNab, et al., 2014; Teles, et al., 2013).
Mycobacteria are ubiquitous in the environment, but they are not part of the normal human microbial flora (Hernandez-Pando et al., 1997). Non-tuberculous mycobacteria (NTM) differ from Mtb both in their wide environmental distribution and in their limited capacity to cause disease. They seem to play a role in the exacerbation of chronic pulmonary disorders like cystic fibrosis, chronic obstructive pulmonary disease (COPD), and an TB-like syndrome in the immunocompromised (Daley & Griffith, 2010; Winthrop et al., 2010). A recent report indicated that NTM may interfere with important aspects of TB control and management, including new anti-tuberculosis vaccine efficacy, the immuno-diagnostic Tuberculin skin test (TST), QuantiFERON TB Gold In Tube assay (QFTGIT), and immune biomarkers (Dhanasekaran et al., 2014). Carriage of NTM may reduce the specificity of future diagnostic and predictive immune biomarkers relevant to TB management. Of greater importance is the realization that certain types of human exposure to some environmental mycobacteria may lead to increased susceptibility to mycobacterial disease (Hernandez-Pando, et al., 1997).
The intracellular survival of mycobacteria and the immune response that they induce in macrophages could be related to their growth rate. Slow growing mycobacteria such as M. abscessus, M. fortuitum, M. celatum, and M. tuberculosis can gain entrance into human macrophages with much more efficiency than fast-growing mycobacteria (Helguera-Repetto et al., 2014). Viable, slow-growing Mycobacteria persist inside macrophages without causing cell damage or inducing reactive oxygen species (ROS). In contrast, fast-growing mycobacteria, such as M. tuberculosis, destroy the cells and induce high levels of ROS. The modulation of macrophage cytokine production caused by NTM may constitute an immune-evasion strategy used to survive inside macrophages that is different from the one reported for M. tuberculosis (Helguera-Repetto, et al., 2014).
CONCLUSIONS
The rapid development of DNA sequencing technologies has had an enormous impact on our understanding of the relationship between the microbiota and the host immune system. However, despite many advances and a flood of research, our understanding of the changes in the lung microbiome in the context of an old yet still widespread disease such as pulmonary tuberculosis is only starting to emerge. Sampling obstacles and divergent findings between published studies will need to be overcomes to truly dissect the relationships between the host immune system, the microbiome, and Mtb in pulmonary TB. Although the data so far is limited, ongoing and future studies will hopefully provide useful information that will shape our approaches to prevention, immune augmentation and regulation, and therapy of TB. Such advances will open the doors for novel interventions aimed at influencing the lung microbiome and the alveolar immune system to help rid humanity of one of its most pugnacious and persistent foes.
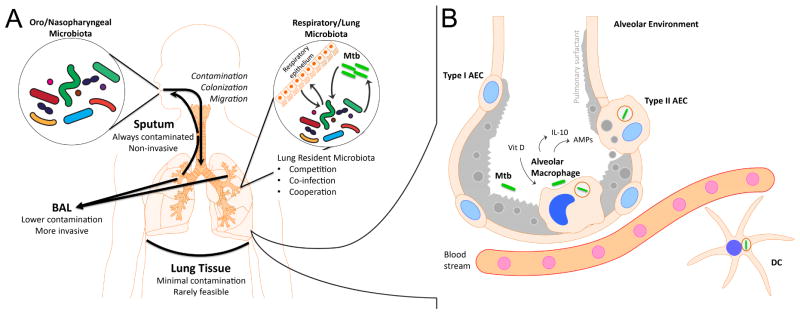
Figure 1.
A. The interaction of the microbiome with Mycobacterium tuberculosis (Mtb) is characterized by complexity and the influence of external forces. Within the lung, the resident microbiota (Firmicutes, Bacteroidetes, Proteobacteria, etc) may cooperate or compete with Mtb. Existing organisms or newly-acquired organisms may also produce co-infections and additional pathology atop of existing pulmonary tuberculosis (TB). In addition, Mtb and the resident microbiota can interact with the respiratory epithelium and adjacent alveolar immune cells, and these interactions can influence the alveolar immune response (Fig 1B). Throughout this process, the microbiome of the Oropharyngeal/Nasopharyngeal spaces may contaminate, colonize, or migrate to the lung, contributing new organisms, including potential pathogens.
To study the lung microbiome, sampling is performed using three primary techniques: induced sputum, broncho-alveolar lavage, and tissue sampling. Induced sputum is simple to obtain but always contaminated by upper respiratory tract and oro/nasopharyngeal microorganisms. Broncho-alveolar lavage (BAL) is less likely to be contaminated by upper respiratory flora but requires an invasive bronchoscopic procedure. Direct extraction from lung tissue has minimal risk of contamination but is only rarely feasible (i.e. lung transplantation).
B. A thin lining of epithelial type I and type II cells surrounds the pulmonary alveolus. Alveolar Epithelial Cell (AEC) Type I cover most of the alveolar surface, allowing efficient gas exchange between the capillaries and the alveolar space. AEC Type II cells produce and secrete pulmonary surfactant as well as cytokines, opsonins and antimicrobial peptides.
Alveolar macrophages (AMs) reside in the luminal side of the alveolus and constitute the first line of immune cell defense in the alveolar environment. AMs also serve to limit inflammation and minimize lung injury to preserve alveolar function. AM activation is tightly regulated, through processes that involve a complex balance between activating signals like TLR-dependent inflammatory responses and repressing signals through IL-10 and TGF-beta which limit the progression of inflammation. The function of AM as antigen presenting cell is limited, and they serve more as immunoregulatory macrophages. Negative regulation of inflammation is also achieved by cell-to-cell interaction with AEC. Mtb infection of AECs and AMs occurs after the bacteria reach the alveoli. Mtb is able to inhibit phagosome maturation, and when a lack of robust oxidative response is present in the host cell, the bacteria is allowed to persist in the phagosome of these cells. Other innate immune cells, including dendritic cells (DCs) are involved in transporting Mtb to draining lymph nodes.
Acknowledgments
FINANCIAL SUPPORT
Supported by NHLBI (F30 HL-126324 to AJA)
Footnotes
CONFLICT OF INTEREST
Authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afsal K, Harishankar M, Banurekha VV, Meenakshi N, Parthasarathy RT, Selvaraj P. Effect of 1,25-dihydroxy vitamin D on cathelicidin expression in patients with and without cavitary tuberculosis. Tuberculosis (Edinb) 2014;94(6):599–605. doi: 10.1016/j.tube.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C, et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. The Journal of Clinical Investigation. 2011;121(8):3088–3093. doi: 10.1172/JCI45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiru OT, Pillay M, Sturm AW. Mycobacterium tuberculosis isolates grown under oxygen deprivation invade pulmonary epithelial cells. Anaerobe. 2012;18(4):471–474. doi: 10.1016/j.anaerobe.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (New York, NY) 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202(12):1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai W, Liu H, Ji Q, Zhou Y, Liang L, Zheng R, et al. TLR3 regulates mycobacterial RNA-induced IL-10 production through the PI3K/AKT signaling pathway. Cell Signal. 2014;26(5):942–950. doi: 10.1016/j.cellsig.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Baldwin SL, Bertholet S, Kahn M, Zharkikh I, Ireton GC, Vedvick TS, et al. Intradermal immunization improves protective efficacy of a novel TB vaccine candidate. Vaccine. 2009;27(23):3063–3071. doi: 10.1016/j.vaccine.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassis CME-DJ, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, Huffnagle GB. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6(2) doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J, Shin DM, Jo EK. Mycobacterial signaling through toll-like receptors. Front Cell Infect Microbiol. 2012;2:145. doi: 10.3389/fcimb.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman RP, Thorpe JE, Staneck J, Rashkin M, Frame PT. Use of the protected specimen brush in patients with endotracheal or tracheostomy tubes. Chest. 1987;91(2):233–236. doi: 10.1378/chest.91.2.233. [DOI] [PubMed] [Google Scholar]
- Behar SM, Divangahi M, Remold HG. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat Rev Microbiol. 2010;8(9):668–674. doi: 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the Microbiota in Immunity and Inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L, Stuve P, Swallow M, Arnold-Schrauf C, Kruse F, Gentilini MV, et al. MyD88 signalling in myeloid cells is sufficient to prevent chronic mycobacterial infection. Eur J Immunol. 2014;44(5):1399–1409. doi: 10.1002/eji.201344039. [DOI] [PubMed] [Google Scholar]
- Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti D, Kumar A, Mahla RS, Kumar S, Ingle H, Shankar H, et al. The role of TLR9 polymorphism in susceptibility to pulmonary tuberculosis. Immunogenetics. 2014;66(12):675–681. doi: 10.1007/s00251-014-0806-1. [DOI] [PubMed] [Google Scholar]
- Bisgaard H, Li N, Bonnelykke K, Chawes BLK, Skov T, Paludan-Müller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. The Journal of allergy and clinical immunology. 2011;128(3):646–652. e641–645. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- Blaser MJ. The microbiome revolution. J Clin Invest. 2014;124(10):4162–4165. doi: 10.1172/JCI78366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borewicz K, Pragman AA, Kim HB, Hertz M, Wendt C, Isaacson RE. Longitudinal analysis of the lung microbiome in lung transplantation. FEMS microbiology letters. 2013;339(1):57–65. doi: 10.1111/1574-6968.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botero LE, Delgado-Serrano L, Cepeda ML, Bustos JR, Anzola JM, Del Portillo P, et al. Respiratory tract clinical sample selection for microbiota analysis in patients with pulmonary tuberculosis. Microbiome. 2014;2 doi: 10.1186/2049-2618-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns H, Stenger S. New insights into the interaction of Mycobacterium tuberculosis and human macrophages. Future Microbiol. 2014;9(3):327–341. doi: 10.2217/fmb.13.164. [DOI] [PubMed] [Google Scholar]
- Cabrera-Rubio R, Garcia-Núñez M, Setó L, Antó JM, Moya A, Monsó E, et al. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. Journal of Clinical Microbiology. 2012;50(11):3562–3568. doi: 10.1128/JCM.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman HJ, Lauzardo M. Advances in diagnosis and treatment of latent tuberculosis infection. J Am Board Fam Med. 2014;27(5):704–712. doi: 10.3122/jabfm.2014.05.140062. [DOI] [PubMed] [Google Scholar]
- Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, et al. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0042786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. American Journal of Respiratory and Critical Care Medicine. 2011;184(8):957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastre J, Fagon JY, Soler P, Bornet M, Domart Y, Trouillet JL, et al. Diagnosis of nosocomial bacterial pneumonia in intubated patients undergoing ventilation: comparison of the usefulness of bronchoalveolar lavage and the protected specimen brush. The American Journal of Medicine. 1988;85(4):499–506. doi: 10.1016/s0002-9343(88)80085-8. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Dwivedi VP, Singh Y, Siddiqui I, Sharma P, Van Kaer L, et al. Early secreted antigen ESAT-6 of Mycobacterium tuberculosis promotes protective T helper 17 cell responses in a toll-like receptor-2-dependent manner. PLoS Pathog. 2011;7(11):e1002378. doi: 10.1371/journal.ppat.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Chang JR, Huang WF, Hsu SC, Kuo SC, Sun JR, et al. The pattern of cytokine production in vitro induced by ancient and modern Beijing Mycobacterium tuberculosis strains. PLoS One. 2014;9(4):e94296. doi: 10.1371/journal.pone.0094296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MK, Lam WY, Fung WY, Law PT, Au CH, Nong W, et al. Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing. PLoS One. 2013;8(1):e54574. doi: 10.1371/journal.pone.0054574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuquimia OD, Petursdottir DH, Periolo N, Fernandez C. Alveolar epithelial cells are critical in protection of the respiratory tract by secretion of factors able to modulate the activity of pulmonary macrophages and directly control bacterial growth. Infect Immun. 2013;81(1):381–389. doi: 10.1128/IAI.00950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuquimia OD, Petursdottir DH, Rahman MJ, Hartl K, Singh M, Fernandez C. The role of alveolar epithelial cells in initiating and shaping pulmonary immune responses: communication between innate and adaptive immune systems. PLoS One. 2012;7(2):e32125. doi: 10.1371/journal.pone.0032125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commandeur S, van den Eeden SJ, Dijkman K, Clark SO, van Meijgaarden KE, Wilson L, et al. The in vivo expressed Mycobacterium tuberculosis (IVE-TB) antigen Rv2034 induces CD4(+) T-cells that protect against pulmonary infection in HLA-DR transgenic mice and guinea pigs. Vaccine. 2014;32(29):3580–3588. doi: 10.1016/j.vaccine.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Coscolla M, Gagneux S. Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech. 2010;7(1):e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MJ, Cookson WO, Moffatt MF. Sequencing the human microbiome in health and disease. Hum Mol Genet. 2013;22(R1):R88–94. doi: 10.1093/hmg/ddt398. [DOI] [PubMed] [Google Scholar]
- Crowe CC, Sanders WE, Longley S. Bacterial interference. II. Role of the normal throat flora in prevention of colonization by group A Streptococcus. The Journal of Infectious Diseases. 1973;128(4):527–532. doi: 10.1093/infdis/128.4.527. [DOI] [PubMed] [Google Scholar]
- Cui Z, Zhou Y, Li H, Zhang Y, Zhang S, Tang S, et al. Complex sputum microbial composition in patients with pulmonary tuberculosis. BMC microbiology. 2012;12 doi: 10.1186/1471-2180-12-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley CL, Griffith DE. Pulmonary non-tuberculous mycobacterial infections. Int J Tuberc Lung Dis. 2010;14(6):665–671. [PubMed] [Google Scholar]
- Davidson M, Tempest B, Palmer DL. Bacteriologic diagnosis of acute pneumonia. Comparison of sputum, transtracheal aspirates, and lung aspirates. JAMA. 1976;235(2):158–163. [PubMed] [Google Scholar]
- Davila S, Hibberd ML, Hari Dass R, Wong HE, Sahiratmadja E, Bonnard C, et al. Genetic association and expression studies indicate a role of toll-like receptor 8 in pulmonary tuberculosis. PLoS Genet. 2008;4(10):e1000218. doi: 10.1371/journal.pgen.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paus RA, van Wengen A, Schmidt I, Visser M, Verdegaal EM, van Dissel JT, et al. Inhibition of the type I immune responses of human monocytes by IFN-alpha and IFN-beta. Cytokine. 2013;61(2):645–655. doi: 10.1016/j.cyto.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, et al. The Airway Microbiota in Cystic Fibrosis: A Complex Fungal and Bacterial Community—Implications for Therapeutic Management. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran S, Jenum S, Stavrum R, Wiker HG, Kenneth J, Vaz M, et al. Effect of non645 tuberculous Mycobacteria on host biomarkers potentially relevant for tuberculosis management. PLoS Negl Trop Dis. 2014;8(10):e3243. doi: 10.1371/journal.pntd.0003243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(7):3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, E-DJ, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, Curtis JL. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Annals of the American Thoracic Society. 2015 doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Freeman CM, Walker N, Scales BS, Beck JM, et al. Changes in the lung microbiome following lung transplantation include the emergence of two distinct pseudomonas species with distinct clinical associations. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0097214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Huffnagle GB. The Role of the Bacterial Microbiome in Lung Disease. Expert review of respiratory medicine. 2013;7(3):245–257. doi: 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an Ecology of the Lung: New Conceptual Models of Pulmonary Microbiology and Pneumonia Pathogenesis. The lancet Respiratory medicine. 2014;2(3):238–246. doi: 10.1016/S2213-2600(14)70028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, et al. Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. Journal of Clinical Microbiology. 2014a;52(10):3605–3613. doi: 10.1128/JCM.01028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, et al. Cell-associated bacteria in the human lung microbiome. Microbiome. 2014b;2 doi: 10.1186/2049-2618-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döderlein A. Das Scheidensekret und seine Bedeutung für das Puerperalfieber. Besold; 1892. [Google Scholar]
- Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P, et al. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur J Clin Microbiol Infect Dis. 2013;32(5):637–645. doi: 10.1007/s10096-012-1787-3. [DOI] [PubMed] [Google Scholar]
- Engler DB, Reuter S, van Wijck Y, Urban S, Kyburz A, Maxeiner J, et al. Effective treatment of allergic airway inflammation with Helicobacter pylori immunomodulators requires BATF3-dependent dendritic cells and IL-10. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(32):11810–11815. doi: 10.1073/pnas.1410579111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escherich T. Die Darmbakterien des Säuglings und ihre Beziehungen zur Physiologie der Verdauung (Enterobacteria of infants and their relation to digestion physiology) Stuttgart: F. Enke; 1886. [Google Scholar]
- Fels AO, Cohn ZA. The alveolar macrophage. J Appl Physiol (1985) 1986;60(2):353–369. doi: 10.1152/jappl.1986.60.2.353. [DOI] [PubMed] [Google Scholar]
- Fremond CM, Yeremeev V, Nicolle DM, Jacobs M, Quesniaux VF, Ryffel B. Fatal Mycobacterium tuberculosis infection despite adaptive immune response in the absence of MyD88. J Clin Invest. 2004;114(12):1790–1799. doi: 10.1172/JCI21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S. Genetic diversity in Mycobacterium tuberculosis. Curr Top Microbiol Immunol. 2013;374:1–25. doi: 10.1007/82_2013_329. [DOI] [PubMed] [Google Scholar]
- Galagan JE. Genomic insights into tuberculosis. Nat Rev Genet. 2014;15(5):307–320. doi: 10.1038/nrg3664. [DOI] [PubMed] [Google Scholar]
- Gengenbacher M, Kaufmann SH. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev. 2012;36(3):514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML, et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(34):13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12(2):125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot L, Nathan N, Tabary O, Thouvenin G, Le Rouzic P, Corvol H, et al. Alveolar epithelial cells: master regulators of lung homeostasis. Int J Biochem Cell Biol. 2013;45(11):2568–2573. doi: 10.1016/j.biocel.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Harriff MJ, Cansler ME, Toren KG, Canfield ET, Kwak S, Gold MC, et al. Human lung epithelial cells contain Mycobacterium tuberculosis in a late endosomal vacuole and are efficiently recognized by CD8(+) T cells. PLoS One. 2014;9(5):e97515. doi: 10.1371/journal.pone.0097515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Jia H, Jing Z, Liu D. Recognition of pathogen-associated nucleic acids by endosomal nucleic acid-sensing toll-like receptors. Acta Biochim Biophys Sin (Shanghai) 2013;45(4):241–258. doi: 10.1093/abbs/gms122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helguera-Repetto AC, Chacon-Salinas R, Cerna-Cortes JF, Rivera-Gutierrez S, Ortiz-Navarrete V, Estrada-Garcia I, et al. Differential macrophage response to slow- and fast-growing pathogenic mycobacteria. Biomed Res Int. 2014;2014:916521. doi: 10.1155/2014/916521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Pando R, Pavon L, Arriaga K, Orozco H, Madrid-Marina V, Rook G. Pathogenesis of tuberculosis in mice exposed to low and high doses of an environmental mycobacterial saprophyte before infection. Infect Immun. 1997;65(8):3317–3327. doi: 10.1128/iai.65.8.3317-3327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herter CA, Kendall AI. The Influence of Dietary Alternations on the Types of Intestinal Flora. Journal of Biological Chemistry. 1910;7(3):203–236. [Google Scholar]
- Higuchi JH, Coalson JJ, Johanson WG. Bacteriologic diagnosis of nosocomial pneumonia in primates. Usefulness of the protected specimen brush. The American Review of Respiratory Disease. 1982;125(1):53–57. doi: 10.1164/arrd.1982.125.1.53. [DOI] [PubMed] [Google Scholar]
- Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1) doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World journal of gastroenterology: WJG. 2014;20(5):1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. The Journal of Allergy and Clinical Immunology. 2011;127(2):372–381. e371–373. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell SP. Neutral theory in community ecology and the hypothesis of functional equivalence. Functional Ecology. 2005;19(1):166–172. [Google Scholar]
- Huffnagle GB, Noverr MC. The emerging world of the fungal microbiome. Trends in Microbiology. 2013;21(7):334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui AW, Lau HW, Chan TH, Tsui SK. The human microbiota: a new direction in the investigation of thoracic diseases. J Thorac Dis. 2013;5(Suppl 2):S127–131. doi: 10.3978/j.issn.2072-1439.2013.07.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14(2):81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- Integrative HMPRNC. The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host & Microbe. 2014;16(3):276–289. doi: 10.1016/j.chom.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal Mandeep S, Bishai William R. Epidemiology and Challenges to the Elimination of Global Tuberculosis. Clinical Infectious Diseases. 2010;50(s3):S156–S164. doi: 10.1086/651486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson WG, Pierce AK, Sanford JP. Changing pharyngeal bacterial flora of hospitalized patients. Emergence of gram-negative bacilli. The New England Journal of Medicine. 1969;281(21):1137–1140. doi: 10.1056/NEJM196911202812101. [DOI] [PubMed] [Google Scholar]
- Juarez E, Nunez C, Sada E, Ellner JJ, Schwander SK, Torres M. Differential expression of Toll-like receptors on human alveolar macrophages and autologous peripheral monocytes. Respir Res. 2010;11:2. doi: 10.1186/1465-9921-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassa D, Ran L, Geberemeskel W, Tebeje M, Alemu A, Selase A, et al. Analysis of immune responses against a wide range of Mycobacterium tuberculosis antigens in patients with active pulmonary tuberculosis. Clin Vaccine Immunol. 2012;19(12):1907–1915. doi: 10.1128/CVI.00482-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keown DA, Collings DA, Keenan JI. Uptake and persistence of Mycobacterium avium subsp. paratuberculosis in human monocytes. Infect Immun. 2012;80(11):3768–3775. doi: 10.1128/IAI.00534-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khara JS, Wang Y, Ke XY, Liu S, Newton SM, Langford PR, et al. Anti-mycobacterial activities of synthetic cationic alpha-helical peptides and their synergism with rifampicin. Biomaterials. 2014;35(6):2032–2038. doi: 10.1016/j.biomaterials.2013.11.035. [DOI] [PubMed] [Google Scholar]
- Kiemer AK, Senaratne RH, Hoppstadter J, Diesel B, Riley LW, Tabeta K, et al. Attenuated activation of macrophage TLR9 by DNA from virulent mycobacteria. J Innate Immun. 2009;1(1):29–45. doi: 10.1159/000142731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, et al. Rethinking “enterotypes”. Cell Host & Microbe. 2014;16(4):433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Lam JT, Siu GK, Yam WC, Mason AJ, Lam JK. Cationic amphipathic D-enantiomeric antimicrobial peptides with in vitro and ex vivo activity against drug-resistant Mycobacterium tuberculosis. Tuberculosis (Edinb) 2014;94(6):678–689. doi: 10.1016/j.tube.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Legatzki A, Rösler B, von Mutius E. Microbiome diversity and asthma and allergy risk. Current Allergy and Asthma Reports. 2014;14(10) doi: 10.1007/s11882-014-0466-0. [DOI] [PubMed] [Google Scholar]
- Liang B, Guo Y, Li Y, Kong H. Association between IL-10 gene polymorphisms and susceptibility of tuberculosis: evidence based on a meta-analysis. PLoS One. 2014;9(2):e88448. doi: 10.1371/journal.pone.0088448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jiang Y, Dou X, Wang H, Zhao X, Zhang W, et al. pstS1 polymorphisms of Mycobacterium tuberculosis strains may reflect ongoing immune evasion. Tuberculosis (Edinb) 2013;93(5):475–481. doi: 10.1016/j.tube.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311(5768):1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- Lorenzi JC, Trombone AP, Rocha CD, Almeida LP, Lousada RL, Malardo T, et al. Intranasal vaccination with messenger RNA as a new approach in gene therapy: use against tuberculosis. BMC Biotechnol. 2010;10:77. doi: 10.1186/1472-6750-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Cota-Gomez A, Palmer BE, Linderman DJ, Charlson ES, Sodergren E, et al. Widespread colonization of the lung by Tropheryma whipplei in HIV infection. American Journal of Respiratory and Critical Care Medicine. 2013;187(10):1110–1117. doi: 10.1164/rccm.201211-2145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysholm F, Wetterbom A, Lindau C, Darban H, Bjerkner A, Fahlander K, et al. Characterization of the viral microbiome in patients with severe lower respiratory tract infections, using metagenomic sequencing. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Li Y, Li M, Deng G, Wu X, Zeng J, et al. microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol Immunol. 2014;62(1):150–158. doi: 10.1016/j.molimm.2014.06.014. [DOI] [PubMed] [Google Scholar]
- Matthew EB, Holstrom FM, Kaspar RL. A simple method for diagnosing pneumonia in intubated or tracheostomized patients. Critical Care Medicine. 1977;5(2):76–81. doi: 10.1097/00003246-197703000-00003. [DOI] [PubMed] [Google Scholar]
- Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 2014;511(7507):99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab FW, Ewbank J, Howes A, Moreira-Teixeira L, Martirosyan A, Ghilardi N, et al. Type I IFN induces IL-10 production in an IL-27-independent manner and blocks responsiveness to IFN-gamma for production of IL-12 and bacterial killing in Mycobacterium tuberculosis-infected macrophages. J Immunol. 2014;193(7):3600–3612. doi: 10.4049/jimmunol.1401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metchnikoff E. In: The Prolongation of Life: Optimistic Studies. Mitchell PC, translator. New York & London: G.P. Putnam’s Sons; 1908. [Google Scholar]
- Mily A, Rekha RS, Kamal SM, Akhtar E, Sarker P, Rahim Z, et al. Oral intake of phenylbutyrate with or without vitamin D3 upregulates the cathelicidin LL-37 in human macrophages: a dose finding study for treatment of tuberculosis. BMC Pulm Med. 2013;13:23. doi: 10.1186/1471-2466-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry NF, Dholakia Y, D’Souza DT, Taylor M, Hoffner S, Birdi TJ. Rhodococcus and Mycobacterium Tuberculosis: masquerade or mixed infection. Int J Tuberc Lung Dis. 2006;10(3):351–353. [PubMed] [Google Scholar]
- Moraes MO, Pacheco AG, Schonkeren JJ, Vanderborght PR, Nery JA, Santos AR, et al. Interleukin-10 promoter single-nucleotide polymorphisms as markers for disease susceptibility and disease severity in leprosy. Genes Immun. 2004;5(7):592–595. doi: 10.1038/sj.gene.6364122. [DOI] [PubMed] [Google Scholar]
- Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. American Journal of Respiratory and Critical Care Medicine. 2013;187(10):1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouailles G, Dorhoi A, Koch M, Zerrahn J, Weiner J, 3rd, Fae KC, et al. CXCL5-secreting pulmonary epithelial cells drive destructive neutrophilic inflammation in tuberculosis. J Clin Invest. 2014;124(3):1268–1282. doi: 10.1172/JCI72030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertli M, Sundquist M, Hitzler I, Engler DB, Arnold IC, Reuter S, et al. DC-derived IL-18 drives Treg differentiation, murine Helicobacter pylori-specific immune tolerance, and asthma protection. The Journal of Clinical Investigation. 2012;122(3):1082–1096. doi: 10.1172/JCI61029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi A, Sengupta M, Sengupta S, Roehm KH, Sonawane A. Antimicrobial peptides and proteins in mycobacterial therapy: current status and future prospects. Tuberculosis (Edinb) 2014;94(4):363–373. doi: 10.1016/j.tube.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Perry S, de Jong BC, Solnick JV, de la Luz Sanchez M, Yang S, Lin PL, et al. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One. 2010;5(1) doi: 10.1371/journal.pone.0008804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH Human Microbiome Project. Genome Research. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petursdottir DH, Chuquimia OD, Freidl R, Fernandez C. Macrophage control of phagocytosed mycobacteria is increased by factors secreted by alveolar epithelial cells through nitric oxide independent mechanisms. PLoS One. 2014;9(8):e103411. doi: 10.1371/journal.pone.0103411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistacchio E. Fl gge’s droplets. Infez Med. 1999;7(2):129–132. [PubMed] [Google Scholar]
- Portevin D, Gagneux S, Comas I, Young D. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7(3):e1001307. doi: 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0047305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Tosh KW, Schramm LM, Grajkowska LT, Kirschman KD, Tami C, et al. TLR9 and TLR7 agonists mediate distinct type I IFN responses in humans and nonhuman primates in vitro and in vivo. J Leukoc Biol. 2012;91(1):147–158. doi: 10.1189/jlb.0711371. [DOI] [PubMed] [Google Scholar]
- Querido SM, Back-Brito GN, Dos Santos SS, Leao MV, Koga-Ito CY, Jorge AO. Opportunistic microorganisms in patients undergoing antibiotic therapy for pulmonary tuberculosis. Braz J Microbiol. 2011;42(4):1321–1328. doi: 10.1590/S1517-838220110004000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Sobia P, Gupta N, Kaer LV, Das G. Mycobacterium tuberculosis subverts the TLR-2-MyD88 pathway to facilitate its translocation into the cytosol. PLoS One. 2014;9(1):e86886. doi: 10.1371/journal.pone.0086886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram MV, Ni B, Dodd CE, Schlesinger LS. Macrophage immunoregulatory pathways in tuberculosis. Semin Immunol. 2014;26(6):471–485. doi: 10.1016/j.smim.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling N, Ehlers S, Holscher C. MyDths and un-TOLLed truths: sensor, instructive and effector immunity to tuberculosis. Immunol Lett. 2008;116(1):15–23. doi: 10.1016/j.imlet.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Rudkjøbing VB, Thomsen TR, Alhede M, Kragh KN, Nielsen PH, Johansen UR, et al. True microbiota involved in chronic lung infection of cystic fibrosis patients found by culturing and 16S rRNA gene analysis. Journal of Clinical Microbiology. 2011;49(12):4352–4355. doi: 10.1128/JCM.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474(7351):385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annual review of microbiology. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1(1) doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal LN, Blaser MJ. A Brave New World: The Lung Microbiota in an Era of Change. Annals of the American Thoracic Society. 2014;11(Suppl 1):S21–S27. doi: 10.1513/AnnalsATS.201306-189MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DP, Canaday DH, Liu Y, Li Q, Huang A, Boom WH, et al. Mycobacterium tuberculosis and TLR2 agonists inhibit induction of type I IFN and class I MHC antigen cross processing by TLR9. J Immunol. 2010;185(4):2405–2415. doi: 10.4049/jimmunol.0904005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T, Neyrolles O. Mycobacteria and the intraphagosomal environment: take it with a pinch of salt(s)! Traffic. 2012;13(8):1042–1052. doi: 10.1111/j.1600-0854.2012.01358.x. [DOI] [PubMed] [Google Scholar]
- Spasova DS, Surh CD. Blowing on embers: commensal microbiota and our immune system. Front Immunol. 2014;5:318. doi: 10.3389/fimmu.2014.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CM, Zalwango S, Chiunda AB, Millard C, Leontiev DV, Horvath AL, et al. Linkage and association analysis of candidate genes for TB and TNFalpha cytokine expression: evidence for association with IFNGR1, IL-10, and TNF receptor 1 genes. Hum Genet. 2007;121(6):663–673. doi: 10.1007/s00439-007-0357-8. [DOI] [PubMed] [Google Scholar]
- Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Applied and Environmental Microbiology. 1999;65(11):4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. American Journal of Respiratory and Critical Care Medicine. 2012;185(10):1073–1080. doi: 10.1164/rccm.201111-2075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum R, Cammer M, Maitland ML, Freitag NE, Condeelis J, Bloom BR. Mycobacterial infection of macrophages results in membrane-permeable phagosomes. Proc Natl Acad Sci U S A. 1999;96(26):15190–15195. doi: 10.1073/pnas.96.26.15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RM, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 2013;339(6126):1448–1453. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thada S, Valluri VL, Gaddam SL. Influence of Toll-like receptor gene polymorphisms to tuberculosis susceptibility in humans. Scand J Immunol. 2013;78(3):221–229. doi: 10.1111/sji.12066. [DOI] [PubMed] [Google Scholar]
- Thorpe JE, Baughman RP, Frame PT, Wesseler TA, Staneck JL. Bronchoalveolar lavage for diagnosing acute bacterial pneumonia. The Journal of Infectious Diseases. 1987;155(5):855–861. doi: 10.1093/infdis/155.5.855. [DOI] [PubMed] [Google Scholar]
- Torrey JC. The Regulation of the Intestinal Flora of Dogs through Diet. The Journal of Medical Research. 1919;39(3):415–447. [PMC free article] [PubMed] [Google Scholar]
- Torsvik V, Goksøyr J, Daae FL. High diversity in DNA of soil bacteria. Applied and Environmental Microbiology. 1990;56(3):782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg HL, Morris A, Ghedin E, Curtis JL, Huffnagle GB, Crothers K, et al. Use of bronchoalveolar lavage to assess the respiratory microbiome: signal in the noise. The Lancet Respiratory Medicine. 2013;1(5):354–356. doi: 10.1016/S2213-2600(13)70117-6. [DOI] [PubMed] [Google Scholar]
- van der Does AM, Beekhuizen H, Ravensbergen B, Vos T, Ottenhoff TH, van Dissel JT, et al. LL-37 directs macrophage differentiation toward macrophages with a proinflammatory signature. J Immunol. 2010;185(3):1442–1449. doi: 10.4049/jimmunol.1000376. [DOI] [PubMed] [Google Scholar]
- van der Does AM, Bergman P, Agerberth B, Lindbom L. Induction of the human cathelicidin LL-37 as a novel treatment against bacterial infections. J Leukoc Biol. 2012;92(4):735–742. doi: 10.1189/jlb.0412178. [DOI] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129(7):1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Velez DR, Wejse C, Stryjewski ME, Abbate E, Hulme WF, Myers JL, et al. Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and West Africans. Hum Genet. 2010;127(1):65–73. doi: 10.1007/s00439-009-0741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vordermeier HM, Hewinson RG, Wilkinson RJ, Wilkinson KA, Gideon HP, Young DB, et al. Conserved immune recognition hierarchy of mycobacterial PE/PPE proteins during infection in natural hosts. PLoS One. 2012;7(8):e40890. doi: 10.1371/journal.pone.0040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, van der Does AM, Tang X, Lindbom L, Agerberth B, Haeggstrom JZ. Antimicrobial peptide LL-37 promotes bacterial phagocytosis by human macrophages. J Leukoc Biol. 2014;95(6):971–981. doi: 10.1189/jlb.0513304. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzirl J. The Action of Sunlight upon Bacteria with Special Reference to B. tuberculosis. Public Health Pap Rep. 1906;32(Pt 2):128–153. doi: 10.1093/infdis/4.supplement_3.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson RJ, Llewelyn M, Toossi Z, Patel P, Pasvol G, Lalvani A, et al. Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case-control study. Lancet. 2000;355(9204):618–621. doi: 10.1016/S0140-6736(99)02301-6. [DOI] [PubMed] [Google Scholar]
- Willger SD, Grim SL, Dolben EL, Shipunova A, Hampton TH, Morrison HG, et al. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome. 2014;2(1) doi: 10.1186/2049-2618-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One. 2009;4(10) doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Haynes MR, Furlan M, Hanson N, Kirby B, Lim YW, et al. Case Studies of the Spatial Heterogeneity of DNA Viruses in the Cystic Fibrosis Lung. American Journal of Respiratory Cell and Molecular Biology. 2012;46(2):127–131. doi: 10.1165/rcmb.2011-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winglee K, Eloe-Fadrosh E, Gupta S, Guo H, Fraser C, Bishai W. Aerosol Mycobacterium tuberculosis infection causes rapid loss of diversity in gut microbiota. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winthrop KL, McNelley E, Kendall B, Marshall-Olson A, Morris C, Cassidy M, et al. Pulmonary nontuberculous mycobacterial disease prevalence and clinical features: an emerging public health disease. Am J Respir Crit Care Med. 2010;182(7):977–982. doi: 10.1164/rccm.201003-0503OC. [DOI] [PubMed] [Google Scholar]
- Wu J, Liu W, He L, Huang F, Chen J, Cui P, et al. Sputum microbiota associated with new, recurrent and treatment failure tuberculosis. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0083445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro LH, Oliveira SC, Bafica A. Innate immune sensing of nucleic acids from mycobacteria. Microbes Infect. 2014 doi: 10.1016/j.micinf.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Yang CS, Shin DM, Lee HM, Son JW, Lee SJ, Akira S, et al. ASK1-p38 MAPK-p47phox activation is essential for inflammatory responses during tuberculosis via TLR2-ROS signalling. Cell Microbiol. 2008;10(3):741–754. doi: 10.1111/j.1462-5822.2007.01081.x. [DOI] [PubMed] [Google Scholar]
- Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2015;15(1):200–209. doi: 10.1111/ajt.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuk JM, Jo EK. Host immune responses to mycobacterial antigens and their implications for the development of a vaccine to control tuberculosis. Clin Exp Vaccine Res. 2014;3(2):155–167. doi: 10.7774/cevr.2014.3.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368(8):745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]