Abstract
Epigenome editing with the CRISPR/Cas9 platform is a promising technology to modulate gene expression to direct cell phenotype and to dissect the causal epigenetic mechanisms of gene regulation. Fusions of the nuclease-inactive dCas9 to the KRAB repressor (dCas9-KRAB) can silence target gene expression, but the genome-wide specificity and the extent of heterochromatin formation catalyzed by dCas9-KRAB is not known. We targeted dCas9-KRAB to the HS2 enhancer, a distal regulatory element that orchestrates expression of multiple globin genes. Genome-wide analyses demonstrated that localization of dCas9-KRAB to HS2 specifically induced H3K9 tri-methylation (H3K9me3) at the enhancer and reduced the chromatin accessibility of both the enhancer and its promoter targets. Targeted epigenetic modification of HS2 silenced the expression of multiple globin genes, with minimal off-target changes in gene expression. These results demonstrate that repression mediated by dCas9-KRAB is sufficiently specific to disrupt the activity of individual enhancers via local modification of the epigenome.
Introduction
Custom control over epigenetic regulation is becoming increasingly attainable with the expansion of genome engineering technologies that combine epigenetic modulators with programmable DNA-targeting platforms. Engineered zinc finger proteins, transcription activator-like effectors, and the clustered regularly interspersed short palindromic repeat (CRISPR)/dCas9 system can localize effector domains to target genomic regions to control epigenetic state1–12. In the CRISPR/dCas9 system adapted from Streptococcus pyogenes, co-delivery of a single guide RNA (sgRNA) directs dCas9 binding through an 18–20 nucleotide protospacer region complementary to the target genomic sequence13–15. The ease and versatility of this RNA-guided epigenome editing platform has enabled its rapid application for designing gene regulatory networks, screening for cellular phenotypes, and directing cell fate9, 16–21. Site-specific epigenome engineering with the CRISPR/dCas9 system is also a promising technology to investigate the function of distal regulatory elements.9, 10 Large-scale efforts to map the human epigenome have revealed more than one million DNase I hypersensitive sites (DHS), many of which likely act as cell-type specific enhancers22–24. Epigenome editing proteins can be targeted to candidate regulatory elements in order to modify local chromatin structure and determine the role these distal elements have in influencing endogenous gene expression.
For targeted gene repression, the most commonly employed effector is the Kruppel-associated box (KRAB) domain1, 3, 6. When localized to DNA, KRAB recruits a heterochromatin-forming complex that causes histone methylation and deacetylation25–28. dCas9-KRAB fusions have effectively silenced non-coding RNAs and single genes when targeted to promoter regions, 5’ untranslated regions, and proximal enhancer elements6, 9, 16, 18. Libraries of sgRNAs targeting dCas9-KRAB to genomic regions proximal to transcription start sites have also been used for high-throughput gene silencing screens16.
Unmodified dCas9 is known to bind at off-target loci containing a 5 bp seed sequence followed by a protospacer adjacent motif (PAM)29, 30. The potential for binding and subsequent epigenome editing by dCas9-KRAB at off-target sites has not been evaluated. Off-target activity is a particular concern given recent evidence that the addition of a KRAB domain to a targeted zinc finger protein can dramatically increase off-target interactions31. Other studies also suggest that targeting the KRAB domain to genomic loci with other DNA-binding proteins can lead to long-range, stable H3K9me3 and chromatin condensation2, 25–27, 32, 33. If similar issues occur with the dCas9-KRAB platform, long-range epigenetic effects would convolute loss-of-function screens and the annotation of gene regulatory elements. Therefore we sought to evaluate the genome-wide specificity of gene regulation, DNA-binding, and chromatin remodeling catalyzed by dCas9-KRAB targeted to an endogenous distal regulatory element.
We targeted dCas9-KRAB to the HS2 enhancer in the globin locus control region (LCR)34. The LCR contains five DHS enhancer regions (HS1–5) that orchestrate expression of hemoglobin subunit genes in erythroid cells during development. Studies of HS2 have revealed fundamental mechanisms of enhancer activity including transcription factor binding, maintenance of chromatin accessibility, and chromatin looping to the globin promoters35–38. Here, we demonstrate that targeting dCas9-KRAB to the HS2 enhancer disrupted the expression of multiple globin genes. Genome-wide analysis of dCas9-KRAB binding and repression activity established that RNA-guided synthetic repressors are highly specific for endogenous target loci. Deposition of H3K9me3 was limited to the intended HS2 region, leading to decreased chromatin accessibility at both the targeted enhancer and its associated promoters. These results demonstrate that dCas9-KRAB can alter the local epigenome of individual enhancers, and support the use of dCas9-KRAB as a highly specific epigenome editing tool for directing cell phenotype and revealing connections between regulatory elements and gene expression.
Results
dCas9-KRAB silences globin genes from a distal enhancer
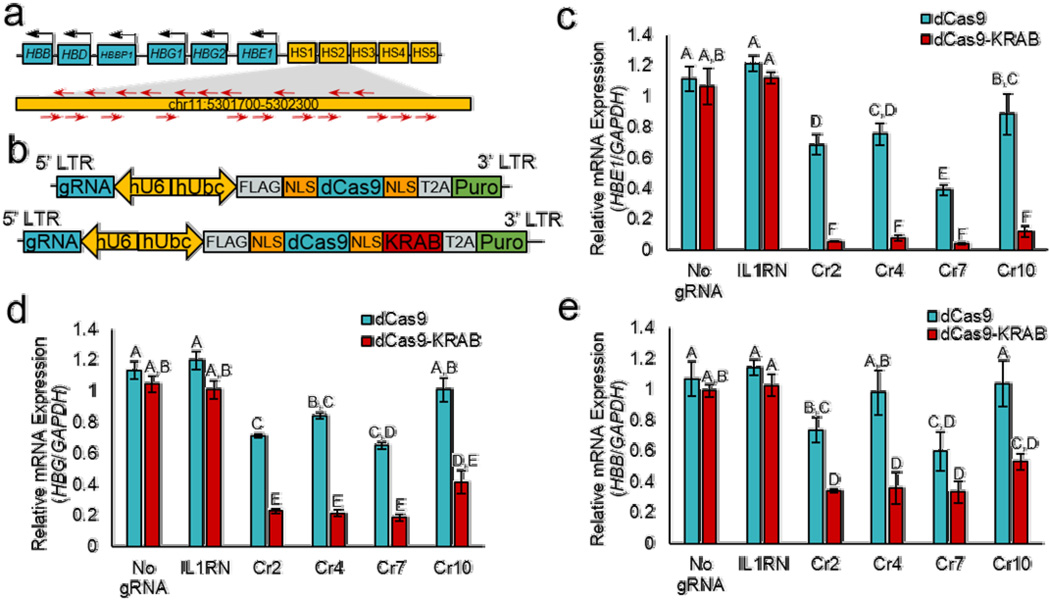
To find optimal sgRNAs for repression of the HS2 enhancer by dCas9-KRAB, we designed a panel of 21 sgRNAs (Cr1 – Cr21) to cover the 400 base pair core of the HS2 enhancer (Fig. 1a, Supplementary Table 1). Each sgRNA was transiently transfected into human K562 erythroid leukemia cells that were modified to stably express dCas9 or dCas9-KRAB (Supplementary Fig. 1a). The activity of each sgRNA was screened three days after transfection by qRT-PCR for HBE1, HBG1 and HBG2 (HBG1/2), and HBB mRNA expression (Supplementary Fig. 1b–d). Due to the sequence similarity between HBG1 and HBG2, PCR primers do not distinguish between the two transcripts. When transfected into K562 cells expressing dCas9-KRAB, most of the 21 sgRNAs caused decreased HBE1 and HBG1/2 expression. The sgRNAs had no effect on globin gene expression in unmodified K562 cells or when designed to target the IL1RN promoter as a negative control7, demonstrating that the effects were both HS2- and dCas9-specific. Reduced globin gene expression likely resulted from a combination of steric blocking and epigenome modification because dCas9-KRAB was more effective than dCas9 in silencing HBE1 and HBG1/2. HBB is not highly expressed in K562 cells39, and transient delivery of sgRNAs did not impact HBB expression. The repressive effects did not persist at six days after transient transfection of sgRNA expression plasmids (Supplementary Fig. 2), likely due to time-dependent reduction of the transfected DNA.
Figure 1. Silencing of downstream globin genes by dCas9-KRAB transcription factors targeted to the distal HS2 enhancer.
(a) A panel of 21 sgRNAs were designed to target dCas9-KRAB to the HS2 enhancer, a distal activator of globin genes. Cr2, Cr4, Cr7, and Cr10 sgRNAs were selected for further study and stably delivered to K562 cells using the lentiviral vector shown in (b). (c–e) Repression of the HS2 enhancer was assayed by qRT-PCR of (c) HBE1, (d) HBG1/2, and e) HBB genes and fold-changes were calculated relative to non-transduced K562 cells (mean ± s.e.m). Within each panel, groups that share the same letter (A – F) are not significantly different by multi-way ANOVA followed by Tukey’s post-hoc test, P < 0.05 (n=3 independent experiments).
To further characterize the mechanisms of targeted repression by dCas9 and dCas9-KRAB, four sgRNAs from the initial screen with the strongest effects on globin gene expression (Cr2, Cr4, Cr7, and Cr10) were transferred into a lentiviral vector and co-expressed with either dCas9 or dCas9-KRAB (Fig. 1b). Stable co-expression of each of the four sgRNAs with dCas9-KRAB substantially repressed HBE1, HBG1/2, and HBB expression seven days after transduction relative to co-delivery of the sgRNA with dCas9 (Fig. 1c–e). Globin gene expression was not altered by dCas9 or dCas9-KRAB delivered alone or co-delivered with an IL1RN-targeted sgRNA. As observed with transient sgRNA delivery, targeting dCas9-KRAB to HS2 reduced globin expression more than targeting dCas9. Moreover, stable expression of HS2-targeted sgRNAs with dCas9-KRAB, but not with dCas9, reduced HBG1/2 protein expression over 21 days (Supplementary Fig. 3). These results demonstrate that dCas9-KRAB repressors targeted by a single sgRNA can interrupt distal enhancer activity and silence expression of multiple genes >10 kb away.
dCas9-KRAB repression is highly specific
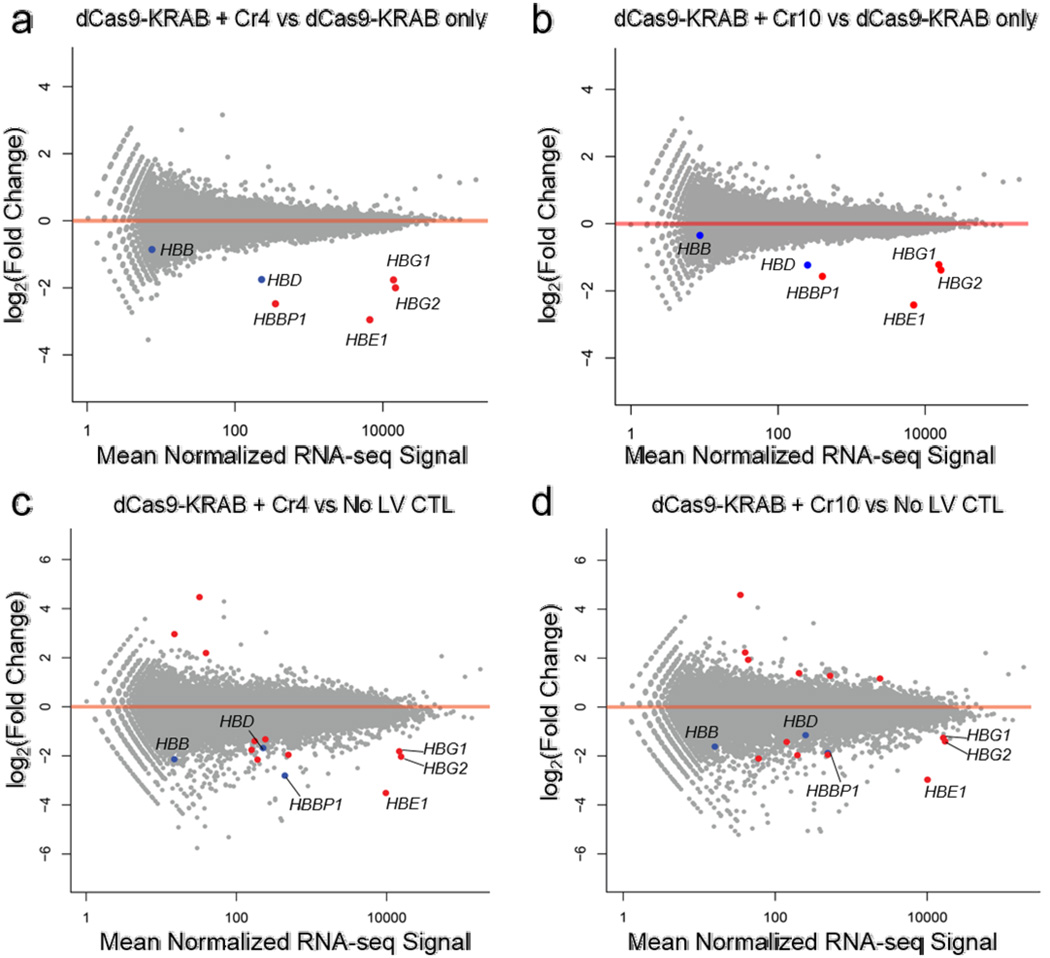
Previous studies using multiplexed sgRNAs with dCas9-VP64 activators have established the specificity of endogenous gene activation7, 40. Additionally, sgRNA-mediated silencing with dCas9-KRAB was highly specific as determined by microarray analyses when targeting an endogenous promoter9 and RNA-seq when targeting a reporter gene6. To determine if that specificity extends to dCas9-KRAB-mediated repression of an endogenous enhancer, we used RNA-seq to measure the transcriptome-wide effects of targeting dCas9-KRAB and dCas9 to HS2 using the Cr4 and Cr10 sgRNAs (Fig. 2a and b, Supplementary Fig. 4a and b).
Figure 2. Specificity of gene regulation by dCas9-KRAB repressors targeted to the HS2 enhancer.
RNA-seq was performed for genome-wide analysis of HS2 sgRNA silencing specificity. (a–d) Differential expression analyses demonstrate specific silencing of globin genes when comparing dCas9-KRAB targeted by (a) Cr4 and (b) Cr10 versus dCas9-KRAB without sgRNA and when comparing dCas9-KRAB guided by c) Cr4 and d) Cr10 to no lentivirus control (No LV CTL) K562s. Red data points indicate FDR < 0.01 by differential expression analysis compared to dCas9-KRAB controls without sgRNA (n = 3 biological replicates). Points labeled in blue indicate other globin genes.
When dCas9-KRAB was targeted to the HS2 enhancer by either Cr4 or Cr10, the only significantly repressed genes were HBG1/2, HBE1, and HBBP1 (false discovery rate, FDR < 0.01), compared to dCas9-KRAB alone (Fig. 2a and b). A similar result was observed when comparing the effect of targeting dCas9-KRAB versus dCas9, both co-delivered with sgRNAs (Supplementary Fig. 4a and b). HBBP1 is a pseudogene that does not encode functional globin protein but may still be regulated by HS2. HBD expression was also reduced by targeting dCas9-KRAB to HS2 with either Cr4 or Cr10, but the change in expression was not significant after multiple hypothesis testing. The only significant off-target effect observed was an increase in expression of the PCSK1N gene when dCas9-KRAB + Cr10 was compared to dCas9 + Cr10 (Supplementary Fig. 4b). PCSK1N regulates processing of neuroendocrine pathway proteins, and is not known to be involved in KRAB-mediated effects. No off-target gene expression differences were observed after treatment with Cr4.
When we compared samples treated with dCas9/dCas9-KRAB and sgRNA to untransduced controls, we detected a larger number of differentially expressed genes, varying from six to ten genes depending on the comparison (Fig. 3c and d, Supplementary Fig. 4c and d, Supplementary Tables 2–5). When we compared dCas9-KRAB without gRNA to un-transduced controls, we observed 29 differentially expressed genes (Supplementary Fig. 4e, Supplementary Table 6). Three genes were differentially expressed in all comparisons to untreated controls, with or without presence of gRNA or the KRAB domain. These modest off-target changes in gene expression may be the result of lentiviral transduction, puromycin selection, or dCas9 expression. Overall, these results indicate that RNA-guided KRAB-mediated gene repression is highly specific and underscore that inclusion of proper controls such as dCas9-KRAB treatment without a functional sgRNA are critical to proper experimental design and interpretation.
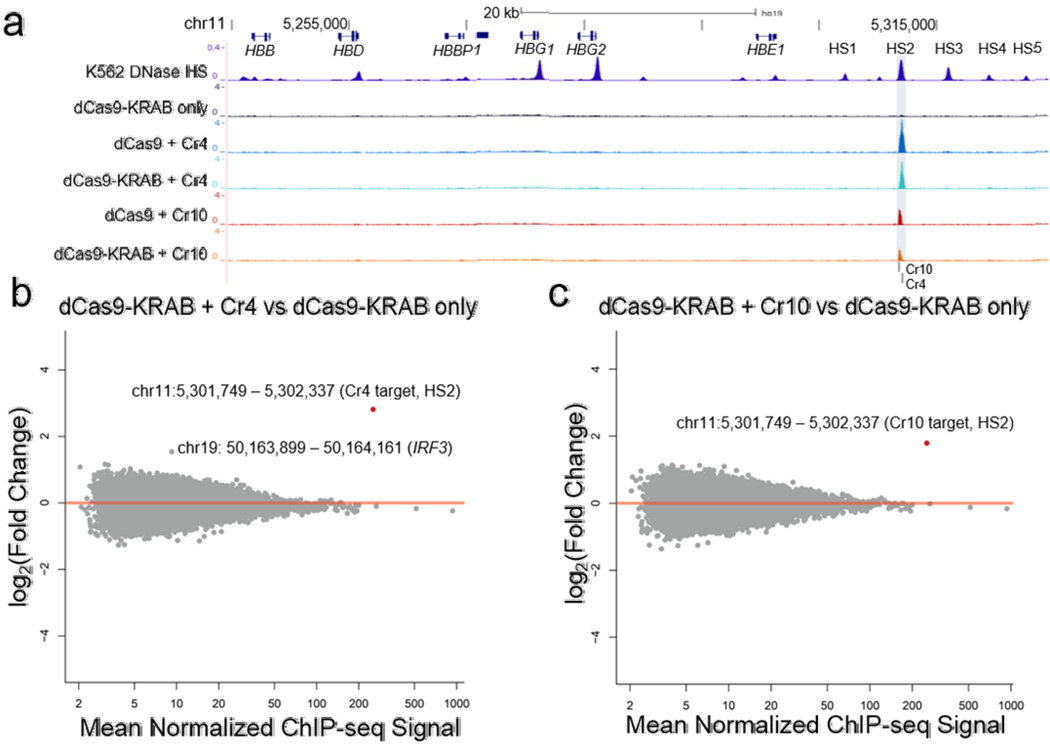
Figure 3. Genome-wide binding activity of dCas9 repressors targeted to the HS2 enhancer.
(a) ChIP-seq tracks demonstrate highly specific binding of FLAG-tagged dCas9 and dCas9-KRAB to the HS2 enhancer (shaded region) of the globin locus (chr11: 5244651 – 5314450), compared to dCas9-KRAB without sgRNA. An ENCODE K562 DNase I hypersensitivity DNase-seq track is included to highlight the globin LCR22. (b,c) Differential analyses of global binding activity include comparisons of dCas9-KRAB targeted by (b) Cr4 and (c) Cr10 versus dCas9-KRAB without sgRNA. Points labeled in red indicate FDR < 0.05 by differential DESeq analysis (n = 3 biological replicates).
dCas9-KRAB binding is highly specific
Previous reports have found evidence that dCas9 promiscuously binds the genome outside of the target site29, 30, 41. However, recent studies have demonstrated that dCas9 and dCas9-KRAB effectors can bind specifically when targeting endogenous gene promoters41. Evaluating off-target localization and potential downstream effects of targeting endogenous enhancers is critical for implementing dCas9-KRAB to control cell phenotype and to investigate epigenome function. To evaluate the genome-wide DNA-binding of dCas9-KRAB, we performed ChIP-seq using a FLAG epitope expressed on the N-terminus of dCas9 or dCas9-KRAB (Fig. 1b). Recruitment of dCas9 and dCas9-KRAB by Cr4 or Cr10 was highly localized to the HS2 enhancer (Fig. 3a), and there were no other significant dCas9-KRAB binding sites in the human genome other than the sgRNA target site (Fig. 3b and c, Supplementary Table 7 and 8).
The addition of KRAB to dCas9 did not alter binding signal (Supplementary Fig. 5a and b). With a genome-wide FDR < 0.05, there was one genomic window, located on chromosome 2, with a significant decrease in ChIP-seq signal when comparing dCas9-KRAB + Cr4 versus dCas9 + Cr4 (Supplementary Table 9). The region contained a 5 bp protospacer seed sequence matched to Cr429, 30. No genome-wide significant changes in ChIP-seq signal were observed when comparing dCas9-KRAB + Cr10 versus dCas9 + Cr10 (Supplementary Fig. 5a and b). When comparing dCas9-KRAB + Cr4/10 versus dCas9 + Cr4/10, we observed a decrease in ChIP-seq signal at HS2, but it did not reach genome-wide significance (P = 4.5 ×10−4 and P = 1.9 × 10−4 for Cr4 and Cr10, respectively) (Supplementary Fig. 5a and b). The decrease in FLAG ChIP-seq signal at the HS2 enhancer and off-target site was perhaps due to epitope or target site inaccessibility after formation of the repressive complex by KRAB. These findings show that RNA-guided dCas9-KRAB repressors can localize to intended genomic loci with exceptional specificity.
dCas9-KRAB binding disrupts transcription factor binding
We hypothesized that targeting dCas9-KRAB to HS2 would reduce binding of endogenous transcription factors at the enhancer. The GATA2 and AP-1 transcription factors are well known to bind the HS2 enhancer and regulate globin gene expression, as confirmed in K562 cells by data from the ENCODE project24, 42. The binding motifs for both GATA2 and AP-1 are adjacent to the Cr4 and Cr10 sgRNA targets within the enhancer34, 35, 42 (Supplementary Fig. 6a). We used ChIP-qPCR to evaluate disruption of the interactions of GATA2 and the AP-1 subunit FOSL1 with HS2 by dCas9-KRAB or dCas9. Recruiting dCas9-KRAB to HS2 with Cr4 or Cr10 significantly reduced binding of both GATA2 (Supplementary Fig. 4b) and FOSL1 (Supplementary Fig. 6c) compared to dCas9-KRAB only controls. In contrast, localizing dCas9 to the HS2 enhancer did not significantly reduce binding of endogenous transcription factors. These results show that dCas9-KRAB can disrupt interactions between endogenous transcription factors and their DNA binding sites.
dCas9-KRAB induces histone methylation at enhancers
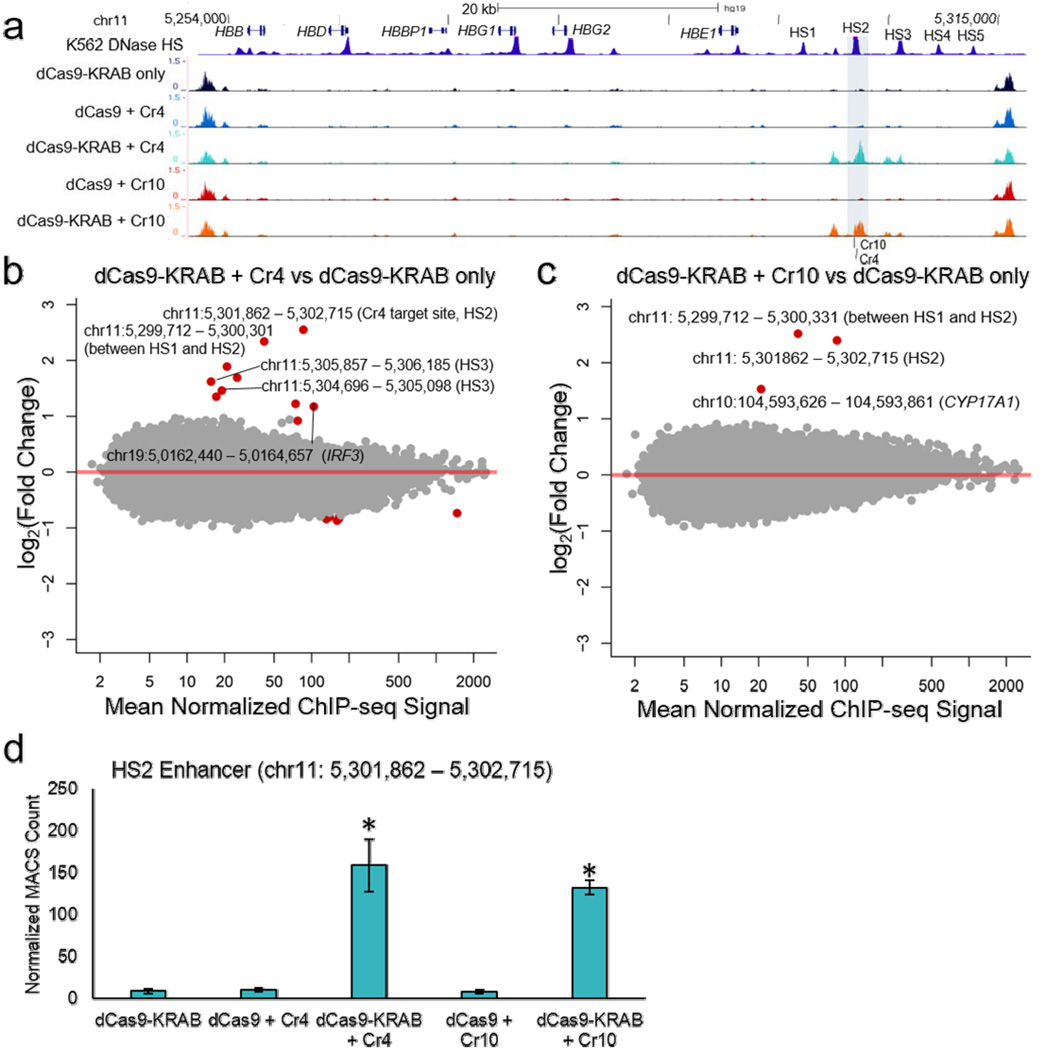
To investigate the epigenetic consequences of KRAB-mediated silencing, global H3K9me3 patterns were assayed by ChIP-seq (Fig. 44). dCas9-KRAB targeted to endogenous promoters can induce repressive histone methylation9. Furthermore SETDB1, a methyltransferase recruited by the KRAB repression complex, catalyzes H3K9me3 when localized to genomic loci27. Co-delivery of dCas9-KRAB + Cr4/Cr10 significantly increased H3K9me3 signal at the target HS2 enhancer relative to dCas9-KRAB without a sgRNA (Fig. 4b–c; Supplementary Fig. 7a and b; Supplementary Fig. 8; Supplementary Tables 10 and 11). The strength of H3K9me3 ChIP-seq signal induced by dCas9-KRAB at the HS2 enhancer was comparable to nearby endogenous H3K9me3 signal at sites that flank the globin locus (Fig. 4a), indicating that dCas9-KRAB induces histone methylation at physiologically relevant levels. Furthermore, the H3K9me3 observed at HS2 did not spread beyond the DHS overlapping the enhancer, covering a span of 853 bp. The effect at HS2 was specific to the KRAB domain, as no changes in H3K9me3 occurred at the enhancer when dCas9 was targeted with Cr4 or Cr10 (Fig. 4d; Supplementary Fig. 7c–f; Supplementary Table 12 and 13). For Cr4, increased H3K9me3 signal was also observed at the immediately adjacent HS3 enhancer, and both Cr4 and Cr10 increased H3K9me3 at the DHS between HS1 and HS2. Together, these results demonstrate that dCas9-KRAB can induce histone modifications associated with heterochromatin when targeted by sgRNAs to active enhancers.
Figure 4. Genome-wide H3K9me3 signal in K562 cells treated with dCas9-KRAB targeted to the HS2 enhancer.
(a) ChIP-seq tracks show increased H3K9me3 signal at the HS2 enhancer (shaded area) and flanking DHS sites in the globin LCR (chr11: chr11:5241410 – 5317466). An ENCODE K562 DNase I hypersensitivity DNase-seq track is included to highlight the globin LCR22. (b,c) Global analysis of H3K9me3 patterns was performed by ChIP-seq for (b) Cr4 or (c) Cr10, comparing dCas9-KRAB with sgRNA versus dCas9-KRAB without sgRNA. Points labeled red indicate FDR < 0.05 by differential expression analysis compared to dCas9-KRAB without sgRNA (n = 3 biological replicates). (d) Counts for the HS2 enhancer (chr11: 5301862–5302715) from MACS-based peak calls were normalized to total counts (mean ± s.e.m). * indicated significance (p<0.05) by Student’s t-test compared to dCas9-KRAB only control.
To identify off-target H3K9me3 sites, we first compared dCas9-KRAB + Cr4/Cr10 to dCas9-KRAB without a sgRNA (Supplementary Table 10 and 11). For Cr4, ten off-target changes in H3K9me3 signal were detected, and eight of these contained a protospacer seed sequence match for Cr4 (Fig. 4b). Only one off-target change in H3K9me3, found in the IRF3 gene, coincided with dCas9-KRAB binding by anti-FLAG ChIP-seq with an FDR < 0.10 (FDR = 0.0785; Fig. 3b; Supplementary Table 7). For Cr10, one off-target H3K9me3 site was observed nearby the CYP17A1 gene, and this region did not contain a match for the protospacer target seed (Fig. 4c). Although we expected KRAB to only cause increases in H3K9me3, five of the eleven total off-target regions (45%) had decreased H3K9me3 signal. All changes in H3K9me3 signal outside the globin LCR were smaller in magnitude and less statistically significant than at sites within the LCR (Supplementary Fig. 7a and b).
We performed a similar analysis to identify differences in H3K9me3 that occur when adding KRAB to dCas9. For the Cr4 gRNA, we identified 44 off-target changes in H3K9me3 (Supplementary Fig. 7c). Of these changes, 39 (89%) had a decrease in H3K9me3 ChIP-seq signal that we do not expect to be directly due to KRAB domain activity. The off-target changes were also low in magnitude compared to changes in the LCR (Supplementary Fig. 7c and e). We further analyzed the 20 off-target regions with the strongest statistical significance, and found 14 to contain Cr4 seed sequences (Supplementary Tables 12 and 13). For dCas9-KRAB + Cr10 compared to dCas9 + Cr10, no off-target H3K9me3 modifications were identified (Supplementary Fig. 7d and f; Supplementary Table 13). Overall, these results show that dCas9-KRAB generated H3K9me3 at the globin LCR with nearly perfect specificity, and off-target histone modification events were typically subtle and varied depending on sgRNA target sequence. Since many of the off-target H3K9me3 changes were decreases in H3K9me3 signal, it may be that these are false positives at this threshold of statistical significance. Furthermore, we did not observe any downstream effects on nearby gene expression associated with off-target H3K9me3 (Fig. 2a–d).
dCas9-KRAB reduces chromatin accessibility at enhancers
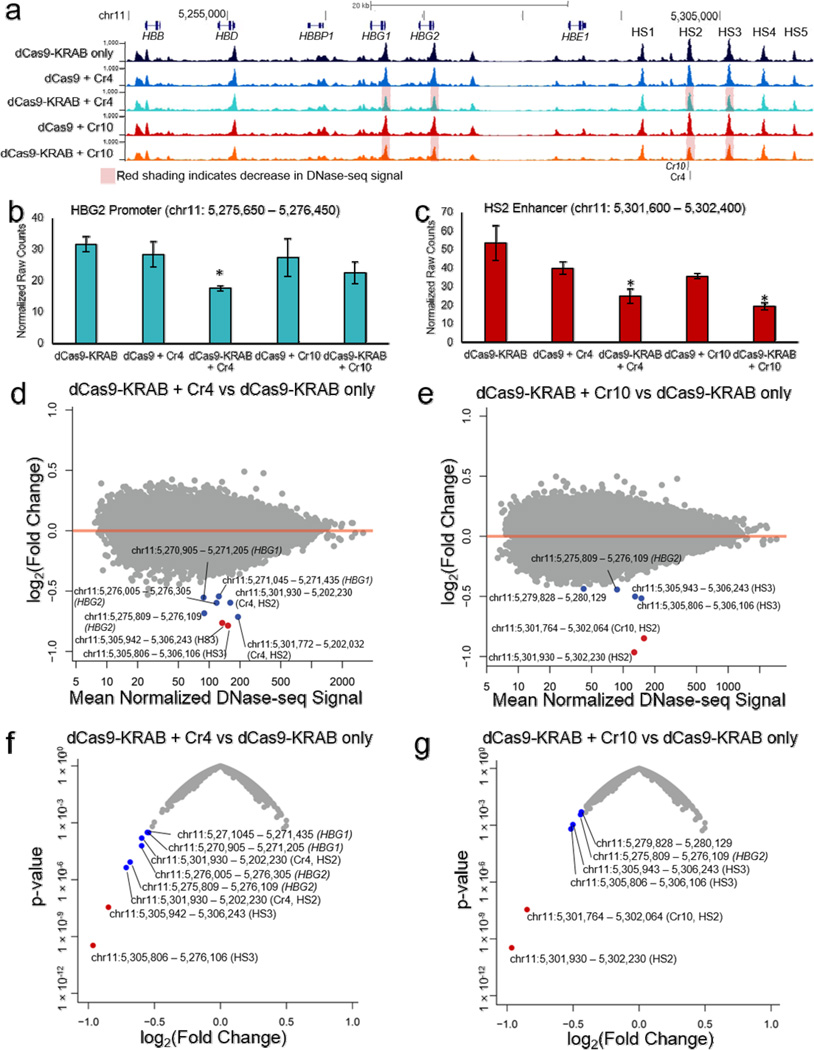
As an orthogonal measure of the specificity of epigenome editing by dCas9-KRAB, we used DNase I hypersensitive sequencing (DNase-seq) to identify genome-wide changes in chromatin accessibility concurrent with KRAB-mediated gene silencing. Targeting dCas9-KRAB to the HS2 enhancer decreased chromatin accessibility at HS2, HS3, and the DHS between HS1 and HS2 (Fig. 5a–e; Supplementary Fig. 9), which were the same regions that displayed increases in H3K9me3 (Fig. 4a). In addition, we also observed decreased chromatin accessibility at the HBG1 and HBG2 promoters (Fig. 5b; Supplementary Fig. 9e–h). These promoters are up to 25 kb from the targeted enhancer region, indicating that dCas9-KRAB-mediated H3K9me3 of the LCR directly affects chromatin structure at the target promoters.
Figure 5. Changes in global chromatin landscape with dCas9-KRAB localized to the HS2 distal enhancer.
(a) Genome browser tracks of DNase-seq alignments at the globin locus (chr11: 5244651 – 5314450) show reduced DHS peaks at the HS2 and HS3 enhancers, as well as HBG1 and HBG2 promoter regions, in conditions containing dCas9-KRAB with sgRNA compared to dCas9-KRAB without sgRNA. Red shading labels the HBG1 promoter, HBG2 promoter, HS2 enhancer and HS3 enhancer regions for dCas9-KRAB + Cr4/10, which demonstrated decreased chromatin accessibility when compared to dCas9-KRAB with no sgRNA or dCas9 + Cr4/10. (b,c) Normalized DNase-seq cut counts within 800 bp window surrounding the (b) HBG2 promoter and (c) HS2 enhancer are shown (mean ± s.e.m, n = 3 biological replicates. * indicates p <0.05 compared to the dCas9-KRAB only sample (Student’s t-test). (d,e) Differential genome-wide analysis of changes in chromatin accessibility induced by dCas9-KRAB targeted by (d) Cr4 and (e) Cr10 compared to dCas9-KRAB without sgRNA in K562 cells. (f,g) Volcano plots of significance (p-value) versus fold-change for differential DESeq expression analysis of dCas9-KRAB guided by (f) Cr4 or (g) Cr10 compared to dCas9-KRAB without sgRNAs. Points labeled red indicate FDR < 0.05 by DESeq analysis. Points labeled in blue indicate other regions in the globin promoters or globin LCR.
The most significant and highest magnitude changes for both Cr4 and Cr10 occurred at the HS2 and HS3 enhancers and the HBG1 and HBG2 promoters (Fig. 5d–e). The effects on chromatin state were largely dependent on the addition of the KRAB domain to dCas9, as supported by comparing dCas9-KRAB to dCas9 with the same gRNAs (Fig. 5b and c; Supplementary Fig. 9a–h; Supplementary Fig. 10a–d). No significant effects on chromatin accessibility were found outside the globin locus, providing further evidence that dCas9-KRAB targeting has highly specific effects on the epigenome. Collectively these findings suggest substantial and highly specific changes to chromatin accessibility concomitant with gene repression and the introduction of repressive histone modifications by the targeted dCas9-KRAB.
Discussion
The simplicity and efficacy of the RNA-guided CRISPR/dCas9 targeting system has the potential to transform epigenome editing into a widely used research tool, but the success of these applications relies on precise editing of epigenetic state. Programmable KRAB fusion proteins have previously been employed to repress enhancers up to 20 kb from a single target gene9, 18. This study expands the capability of synthetic gene regulation by targeting dCas9-KRAB to an enhancer located 10 to 50 kb away from target genes and by comprehensively characterizing the remarkable specificity of this technology.
A recent study showed that targeting dCas9-KRAB to a putative distal enhancer silenced downstream gene expression but did not induce repressive histone marks at the target enhancer9. Here, we demonstrate that dCas9-KRAB can deposit H3K9me3 at endogenous enhancers, indicating that dCas9-KRAB-mediated histone methylation may be locus or cell type-dependent. We also did not find evidence that addition of the KRAB domain to dCas9 increased off-target binding, in agreement with other recent dCas9-KRAB genome-wide binding studies41. This is in contrast to another study of engineered zinc finger-KRAB fusion proteins31. That difference may be due to the distinct mechanism of protein-DNA binding versus RNA-DNA interactions mediated by dCas9 or locus- or cell type-specific effects. Overall, our findings advocate the use of dCas9-KRAB repressors to achieve precise silencing of endogenous genomic targets.
Histone methylation induced by KRAB or HP1 has been estimated to spread up to 20 kb from the repressor binding site using synthetic reporter assays with other DNA targeting proteins26, 32, 33. In contrast, with dCas9-KRAB, increases in H3K9me3 were found up to 4.5 kb away from the sgRNA target site. Furthermore, our H3K9me3 ChIP-seq data show that epigenome modification occurred only at flanking DHS sites, and did not span the region between those sites. That observation suggests that heterochromatin spreading induced by KRAB may not occur linearly along the genome, and may instead spread through three-dimensional interactions between discrete nearby open chromatin sites. Spreading of histone methylation over long distances may therefore be site-specific and dependent on three-dimensional chromatin structure at those sites.
The DNase I hypersensitivity profiles of enhancers correlate with associated promoters22, and chromatin conformation analyses have shown that the globin LCR physically interacts with HBG1, HBG2, and HBE1 in K562 cells38. Furthermore, forcing DNA looping interactions with the globin LCR activates developmentally silenced embryonic and fetal globin expression43. When dCas9-KRAB was targeted to HS2, promoter regions of HBG1 and HBG2 also had reduced DNase-seq signal, while the HS1 region between HS2 and the target promoters was not affected. Additionally, HBG1 and HBG2 promoters did not have increased H3K9me3 signal. The reduced hypersensitivity observed at HBG1 and HBG2 is therefore unlikely to be caused by heterochromatin spreading from the HS2 enhancer, but rather may be the result of disruption of long-range interactions between the promoters and the HS2 enhancer. Potential mechanisms for the disruption of DNA looping include interference with endogenous transcription factor binding and alteration of the chromatin state of the enhancer region. Overall, these results suggest that the accessible chromatin structure and active state of some promoters requires continuous input from enhancer elements and support the use of dCas9-KRAB as a potential method to identify and disrupt these enhancer-promoter interactions.
The KRAB domain is distinctive from epigenome editing effectors because, rather than catalyzing a single type of histone modification, it draws a diverse group of histone modifiers that cooperate to form heterochromatin25, 27, 28, 32. This broad efficacy likely contributes to the effectiveness of KRAB at silencing different types of genomic elements, including transcribed genes and proximal and distal regulatory elements6, 9, 16, 18. This versatility, along with the exceptional specificity demonstrated in this study, distinguishes dCas9-KRAB as a promising platform for applications such as gene therapy, cellular reprogramming, and high throughput screens of regulatory element function and modulators of cell phenotype16.
Online Methods
Plasmids
The sgRNA plasmid and lentiviral plasmid encoding dCas9 are available on Addgene44 (Addgene #53188 and #53191). The KRAB domain was cloned in-frame with the dCas9 ORF at the C-terminus using NheI sites. For sgRNA screening, the oligonucleotides containing HS2 protospacer sequences were synthesized (IDT-DNA), hybridized, phosphorylated, and inserted into phU6-gRNA plasmids using BbsI sites. The protospacer target sequences for the panel of 21 HS2 enhancer sgRNAs are provided in Supplementary Table 1. U6-sgRNA expression cassettes were transferred in reverse orientation upstream of the hUbC promoter at the PacI sites. For sgRNA and dCas9 transduction experiments, a puromycin resistance cassette was linked to the dCas9 and dCas9-KRAB effectors using a T2A ribosome skipping peptide.
Cell culture
K562 cells and HEK293T cells were obtained from the American Tissue Collection Center (ATCC) through the Duke University Cancer Center Facilities. Cell lines were verified via morphological inspection. K562 cells were maintained in RPMI1640 supplemented with 10% FBS and 1% penicillin-streptomycin. HEK293T cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. All cell lines were cultured at 37C with 5% CO2.
Lentiviral transduction
K562s were transduced with lentivirus to stably express dCas9 or dCas9-KRAB. To produce VSV-G pseudotyped lentivirus, HEK293T cells were plated at a density of 5.1e3 cells/cm2 in high glucose DMEM (GIBCO, #11995) supplemented with 10% FBS and 1% pencillin-streptomycin. The next day after seeding, cells in 10-cm plates were co-transfected with the appropriate dCas9/dCas9-KRAB lentiviral expression plasmid (20 µg), the second-generation packaging plasmid psPAX2 (Addgene #12260, 15 µg), and the envelope plasmid pMD2.G (Addgene #12259, 6 µg) by calcium phosphate precipitation45. After 14–20 hours, transfection medium was exchanged for 10 mL of fresh 293T medium. Conditioned medium containing lentivirus was collected 24 and 48 hours after the first media exchange. Residual producer cells were cleared from the lentiviral supernatant by filtration through 0.45 µm cellulose acetate filters and concentrated 20-fold by centrifugation through a 100 kDa molecular weight cutoff filter (Millipore). Concentrated viral supernatant was snap-frozen in liquid nitrogen and stored at −80 °C for future use. For transduction, concentrated viral supernatant was diluted 1:10 with K562 media. To facilitate transduction, the cationic polymer polybrene was added at a concentration of 4 µg/mL to the viral media. Non-transduced (NT) cells did not receive virus but were treated with polybrene as a control. The day after transduction, the medium was exchanged to remove the virus. For cells that were transduced with lentivirus containing a puromycin resistance gene, 1 µg/ml puromycin was used to initiate selection for transduced cells approximately 96 hours after transduction.
Transient transfection
Six to eight days after transduction with dCas9 or dCas9-KRAB lentivirus, K562s were transiently transfected with plasmid encoding HS2 sgRNAs via electroporation. 2e6 cells were transfected with 5 µg of plasmid DNA in 200 uL of Opti-MEM (Gibco) within 2 mm cuvettes at 160V, 950 µF, and infinite resistance. Transfection efficiencies of greater than 70% were routinely achieved, as assayed by flow cytometry after delivery of a control eGFP expression plasmid (unpublished data, P. Thakore).
Western blot
Cells were lysed in RIPA buffer (Sigma), and the BCA assay (Pierce) was performed to quantify total protein. Lysates were mixed with LDS sample buffer (Invitrogen) and boiled for 5 min; equal amounts of total protein were run in NuPAGE Novex 4–12% Bis-Tris polyacrylamide gels (Life Technologies) and transferred to nitrocellulose membranes. Nonspecific antibody binding was blocked with 5% nonfat milk in TBS-T (50 mM Tris, 150 mM NaCl and 0.1% Tween-20) for 30 min. The membranes were then incubated with primary antibody in 5% milk in TBS-T: mouse anti-γ hemoglobin subunit (Santa Cruz, clone 51–7) diluted 1:1000 overnight at 4°C, anti-FLAG (Sigma, F7425) diluted 1:1000 for 60 min at room temperature, or rabbit anti-GAPDH (Cell Signaling, clone 14C10) diluted 1:5000 for 60 min at room temperature. Membranes labeled with primary antibodies were incubated with anti-mouse (Santa Cruz, SC-2005) or anti-rabbit HRP-conjugated antibody (Sigma-Aldrich, A6154) diluted 1:5000 for 60 min and washed with TBS-T for 60 min. Membranes were visualized using the Immun-Star WesternC Chemiluminescence Kit (Bio-Rad) and images were captured using a ChemiDoc XRS+ system and processed using ImageLab software (Bio-Rad).
Quantitative reverse transcription-PCR
Cells were harvested for total RNA isolation using the RNeasy Plus RNA isolation kit (Qiagen). cDNA synthesis was performed using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) using QuantIT Perfecta Supermix was performed with the CFX96 Real-Time PCR Detection System (Bio-Rad) with the oligonucleotide primers reported in Supplementary Table 2. The results are expressed as fold-increase mRNA expression of the gene of interest normalized to Gapdh expression by the ΔΔCt method.
RNA-sequencing
RNA-seq libraries were constructed as previously described46. Briefly, seven days after transduction, cells were harvested and mRNA was purified from total RNA using oligo(dT) Dynabeads (Invitrogen). First-strand cDNA was synthesized using the SuperScript VILO cDNA Synthesis Kit (Invitrogen) and second-strand cDNA was synthesized using DNA polymerase I (New England Biolabs). cDNA was purified using Agencourt AMPure XP beads (Beckman Coulter). Purified cDNA was treated with Nextera transposase (Illumina) for 5 min at 55 °C to simultaneously fragment and insert sequencing primers into the double-stranded cDNA. Transposase activity was halted using QG buffer (Qiagen) and fragmented cDNA was purified on AMPure XP beads. Indexed sequencing libraries were PCR-amplified and sequenced for 50-bp paired-end reads on an Illumina HiSeq 2000 instrument at the Duke Genome Sequencing Shared Resource and for 75 paired-end reads on an Illumina MiSeq. Reads were trimmed to 50 bp and aligned to the delivered lentiviral vector were removed from analysis using Bowtie247. Filtered reads were then aligned to human RefSeq transcripts using Bowtie2. Statistical analysis, including multiple hypothesis testing, on three independent biological replicates was performed using DESeq48.
Chromatin immunoprecipitation
ChIP experiments were performed in in biological triplicate, starting from independent cell transductions and harvested seven days after transduction. For each replicate, 2 × 107 nuclei were re-suspended in 1 mL of RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS in PBS at pH 7.4). Samples were sonicated using a Diagenode Bioruptor XL sonicator at 4°C to fragment chromatin to 200–500 bp segments. Insoluble components were removed by centrifugation for 15 min at 15000 rpm. We conjugated 5 µg of anti-FOSL1 (Santa Cruz Biotechnology, sc-183), anti-GATA-2 (Santa Cruz Biotechnology, sc-9008), anti-FLAG (Sigma-Aldrich, F1804), or anti-H3K9me3 (Abcam, ab8898) to 200 µl of either sheep anti-rabbit or sheep anti-mouse IgG magnetic beads (Life Technologies, 11203D/11201D). Sheared chromatin in RIPA was then added to the antibody-conjugated beads and incubated on a rotator overnight at 4°C. After incubation, beads were washed five times with a LiCl wash buffer (100 mM Tris at pH 7.5, 500 mM LiCl, 1% NP-40, 1% sodium deoxycholate), and remaining ions were removed with a single wash with 1 mL of TE (10 mM Tris-HCl at pH 7.5, 0.1 mM Na2EDTA) at 4°C. Chromatin and antibodies were eluted from beads by incubating for 1 h at 65°C in IP elution buffer (1% SDS, 0.1 M NaHCO3), followed by incubating overnight at 65°C to reverse formaldehyde cross-links. DNA was purified using MinElute DNA purification columns (Qiagen).
ChIP-qPCR
Quantitative real-time PCR (qRT-PCR) using QuantIT Perfecta Supermix was performed with the CFX96 Real-Time PCR Detection System (Bio-Rad) and the oligonucleotide primers reported in Supplementary Table 2. 100 pg of ChIP DNA was loaded into each reaction. The results are expressed as a fold-increase of signal at the HS2 enhancer normalized to signal at the GAPDH promoter by the ΔΔCt method.
ChIP-sequencing
Illumina TruSeq adapted libraries were constructed using an Apollo 324 NGS Library Prep System with a PrepX Complete ILMN DNA Library Kit (WaferGen Biosystems Inc). ChIP products were amplified with 15 cycles of PCR, and fragments 150–700 bp in length were selected using an AxyPrep MAG PCR Clean-Up Kit (Axygen MAG-PCR-CL-50). Libraries were sequenced using single end 50 bp reads on an Illumina HiSeq at the Duke Genome Sequencing Shared Resource. ChIP-seq analyses were executed independently for each epitope to find regions of differential FLAG or H3K9me3 enrichment, as compared to cells treated with lentivirus encoding dCas9-KRAB without sgRNA or dCas9 with sgRNA. To account for background levels of integrated lentiviral DNA, reads aligning to the delivered lentiviral sequences were filtered from analyses using Bowtie247. Next we used Bowtie2 to align the remaining reads to the hg19 reference genome and removed PCR duplicates using the rmdup tool from SAMtools49. Reads from each triplicate for each condition were combined, and peaks were called using MACS50. Resulting peaks from each condition with a q-value ≤ 0.05 were merged using the mergeBed tool from BEDTools51. The number of reads overlapping each of these peaks for each triplicate was determined using the intersectBed tool from BEDTools. Differential enrichment for each peak was evaluated from these read counts using DESeq252 with an FDR cutoff of ≤ 0.05.
DNase I hypersensitivity-sequencing
All experiments were performed in biological triplicate, starting from independent cell transductions. Library preparation and analysis were performed as previously described with the one exception of adding a 5´ phosphate group to oligo 1b to increase ligation efficiency53. For each replicate, seven days after transduction, approximately 2.5 × 107 nuclei were extracted and then digested with different amounts of DNase I for 16 min at 37 °C. Reactions were terminated by the addition of 50 mM EDTA. Libraries were constructed from pooled digests as described and sequenced on the Illumina HiSeq2000 platform with 50-bp single-end reads at the Duke Genome Sequencing Shared Resource. Resulting reads were filtered for delivered vector sequences and aligned by Bowtie54. Peaks were called by MACS version 250, with a cut-off line at FDR < 0.01. Differential DHS sites were determined using DESeq252.
Supplementary Material
Acknowledgments
We thank the Duke Genome Sequencing & Analysis Core for sequencing the RNA-seq, ChIP-seq, and DNase-seq libraries. This work was supported by US National Institutes of Health (NIH) grants R01DA036865, U01HG007900, R21AR065956, P30AR066527, and a NIH Director’s New Innovator Award (DP2OD008586), US National Science Foundation (NSF) Faculty Early Career Development (CAREER) Award (CBET-1151035), and American Heart Association Scientist Development Grant (10SDG3060033) to C.A.G. P.I.T. was supported by a National Science Foundation Graduate Research Fellowship and an American Heart Association Mid-Atlantic Affiliate Predoctoral Fellowship.
Footnotes
Accession Codes
Raw RNA-seq (GSE71557), ChIP-seq (GSE70671), and DNase-seq (GSE70973) data are available on Gene Expression Omnibus.
Competing Interest Statement
C.A.G., G.E.C., T.E.R., P.I.T., and A.M.K. are inventors on patent applications related to genome engineering with the CRISPR/Cas9 system. C.A.G. is a scientific advisor to Editas Medicine, a company engaged in therapeutic development of genome engineering technologies.
Author Contributions
P.I.T, G.E.C., T.E.R. and C.A.G. designed experiments. P.I.T., A.D., A.S., N.K.S., A.M.K., and P.B. performed the experiments. P.I.T., A.D., L.S., A.M.K., G.E.C., T.E.R. and C.A.G. analyzed the data. P.I.T., G.E.C., T.E.R., and C.A.G., wrote the manuscript with editing by all authors.
References
- 1.Beerli RR, Dreier B, Barbas CF., 3rd Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci U S A. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- 3.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendenhall EM, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearns NA, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015 doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015 doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeder ML, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivenbark AG, et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics. 2012;7:350–360. doi: 10.4161/epi.19507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, et al. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res. 2014;42:e155. doi: 10.1093/nar/gku836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakraborty S, et al. A CRISPR/Cas9-Based System for Reprogramming Cell Lineage Specification. Stem Cell Reports. 2014;3:940–947. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and Programmable Regulation of Gene Networks with an Integrated RNA and CRISPR/Cas Toolkit in Human Cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groner AC, et al. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds N, et al. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014 doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 31.Grimmer MR, et al. Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation. Nucleic Acids Res. 2014;42:10856–10868. doi: 10.1093/nar/gku708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayyanathan K, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardison R, et al. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 35.Tuan D, Kong S, Hu K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc Natl Acad Sci U S A. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDowell JC, Dean A. Structural and functional cross-talk between a distant enhancer and the epsilon-globin gene promoter shows interdependence of the two elements in chromatin. Mol Cell Biol. 1999;19:7600–7609. doi: 10.1128/mcb.19.11.7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vakoc CR, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 38.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dean A, Ley TJ, Humphries RK, Fordis M, Schechter AN. Inducible transcription of five globin genes in K562 human leukemia cells. Proc Natl Acad Sci U S A. 1983;80:5515–5519. doi: 10.1073/pnas.80.18.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polstein L, et al. Genome-wide specificity of DNA-binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015 doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJA. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015;43:3389–3404. doi: 10.1093/nar/gkv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikonomi P, et al. Levels of GATA-1/GATA-2 transcription factors modulate expression of embryonic and fetal hemoglobins. Gene. 2000;261:277–287. doi: 10.1016/s0378-1119(00)00510-2. [DOI] [PubMed] [Google Scholar]
- 43.Deng W, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 44.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic acids research. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salmon P, Trono D. Production and titration of lentiviral vectors. Current protocols in neuroscience / editorial board, Jacqueline N. Crawley … [et al] 2006;Chapter 4(Unit 4 21) doi: 10.1002/0471142301.ns0421s37. [DOI] [PubMed] [Google Scholar]
- 46.Gertz J, et al. Transposase mediated construction of RNA-seq libraries. Genome research. 2012;22:134–141. doi: 10.1101/gr.127373.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harbor protocols. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.