Abstract
Current measurements of the biomechanical properties of cells require physical contact with cells or lack sub-cellular resolution. Here, we developed a label-free optical microscopy technique based on Brillouin light scattering capable of measuring intracellular longitudinal modulus with optical resolution. We obtained 3D Brillouin maps of cells in 2D and 3D microenvironments, which reveal mechanical changes due to cytoskeletal modulation and cell volume regulation.
The interaction between the liquid and solid phases within the cytoplasm regulates the rheological behavior of cells1 and has been shown to play a prominent role in how cells deform and move2. In turn, the hydro-mechanical properties of cells, such as viscoelastic moduli and compressibility, influence intracellular water transport, cytoskeletal network modulation and cell volume regulation3, 4. The ability to measure these properties in situ is required to study how cells regulate intracellular mechanics and how the properties change as cells interact with their microenvironment, during aging and injury healing, as well as in the course of disease pathogenesis5. In the past decade, our understanding of cell biomechanics has greatly advanced through rheological studies of cells. Mechanical techniques, such as atomic force microscopy (AFM) and magnetic bead twisting, have been widely used to measure the viscoelastic moduli and deformability of cells6, thus elucidating the different mechanisms by which cells regulate their mechanical properties, i.e. crosslinking, branching and pre-stress in the cytoskeleton (CSK) or variations in solid-liquid volume fraction7. However, these techniques require contact with cells to impose mechanical stress, which limits the applicability to cells cultured on flat 2D substrates8 or to micro-patterned wells that maintain the 3D cell shape9, 10. Optical tweezers or stretchers11, micropipette aspiration12, and, more recently, microfluidic deformation assays13 have been used to assess the elasticity of a cell in suspension, but without sub-cellular resolution. For cells that are confined in 3D hydrogels or microfluidic chips, particle-tracking microrheology (PTM)14 is potentially viable, but it requires the insertion of microbeads into cells or, if tracking sub-cellular components, it is influenced by motor activity15; furthermore, the resolution is limited by the number, size and random location distribution of the beads. Owing to the limitations of these current methods, mapping of mechanical properties of cells within a 3D meshwork of extra-cellular matrix (ECM) proteins has not been performed as of yet16. As a result, there is a dearth of information on the biomechanical properties of cells in 3D environment and how they may differ from those on a flat 2D substrate.
To address this need, here we introduce Brillouin optical cell microscopy for noncontact, label-free, and 3D mapping of the intracellular and extracellular hydro-mechanical properties. This technique is based on the interaction of light with spontaneous acoustic phonons in the gigahertz frequency range17 (Fig. 1a). By measuring the optical frequency shift of the scattered light, local spontaneous pressure waves in the intracellular environments are probed, from which the high frequency longitudinal modulus is determined18. Mechanically, Brillouin interaction can be thought of as a test that measures the ratio of uniaxial stress and strain in confined compression conditions at high frequency19, 20 (Supplementary Note 1 and Supplementary Fig. 1). To enable cellular Brillouin mapping, we have constructed a confocal Brillouin cell microscope by using high numerical aperture (0.6) objective lens and a non-scanning parallel Brillouin spectrometer based on our previous design21 but with an enhanced spectral extinction of 70 dB, spectral resolution of 600 MHz, and low loss (<10 dB) (Fig. 1b, Supplementary Note 2 and Supplementary Fig. 2) as well as a spectrum calibration arm (Supplementary Note 3 and Supplementary Fig. 3). The light source was a 532-nm continuous-wave laser (Online Methods). With the optical power levels of 2–4 mW at the sample and a spectral acquisition time of 100–200 ms, the frequency measurement sensitivity of the instrument was ~10 MHz (Supplementary Note 4 and Supplementary Fig. 4). This is about 0.1% of the Brillouin frequency shift of water of 7.44 GHz at 18 °C.
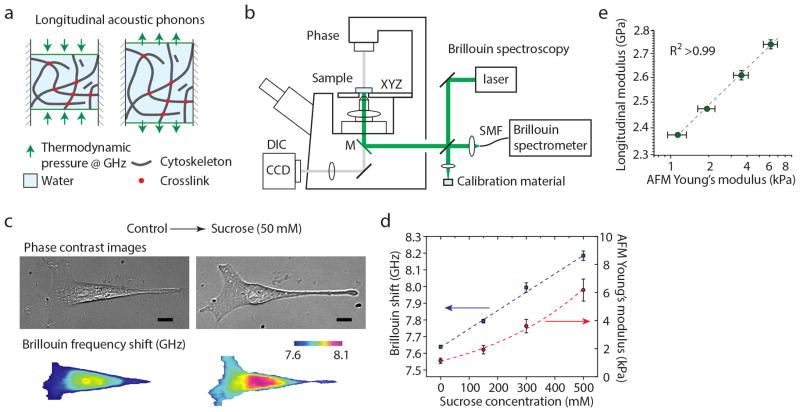
Figure 1. Principle and validation of Brillouin Microscopy.
(a) Schematic of the mechanical interaction probed by Brillouin scattering. Spontaneous fluctuations in density and pressure behave as microscopic mechanical perturbations from which the longitudinal modulus can be extracted with high-resolution optical spectroscopy. The longitudinal modulus depends on intrinsic properties such as the modulus of individual cytoskeletal components, network crosslinking, compressibility of the local microenvironment, and solid-liquid volume fraction. (b) Schematic of the instrument. A standard Olympus IX71 fluorescence microscope is modified to introduce the Brillouin excitation beam through the right-side port; the Brillouin scattered light is collected through the same port and coupled into a fiber, which serves as confocal pinhole. The fiber delivers light into the Brillouin spectrometer (Supplementary Note 2 and Supplementary Fig. 2). (c) Representative Brillouin images (and co-registered phase contrast images) of a cell before and after hyperosmotic shock. A strong stiffening effect is observed. The nucleus is stiffer than the cytoplasm in both conditions. Scale bars, 10 μm. (d) The stiffening effect depends on the osmolarity of the environment as measured by both Brillouin and AFM-based micro-indentation. (e) Validation of Brillouin stiffness measurement against AFM-based micro-indentation tests for cells exposed to different levels of hyperosmotic shocks. The high-correlation observed in log-log-linear fit is consistent with what was previously observed in tissue and biomaterials. Each data point is the average of ~10 measurements on different cells; error bars are s.e.m.
First, we investigated if Brillouin microscopy is sensitive to liquid-solid regulation under different osmotic conditions. Adding sucrose to the cell medium generates osmotic pressure, which has been shown to induce significant increase in cell elastic modulus22. Representative Brillouin confocal sections (Online Methods) of an NIH 3T3 cell before and after hyperosmotic shock (50 mM of sucrose) display a significant increase in Brillouin shift throughout the cell (Fig. 1c). To compute mean longitudinal modulus, we averaged the Brillouin shifts over the cell volume and used an estimated constant value for cell density and refractive index23. This is an approximation as refractive index and density are not uniform throughout the cells and will change in different osmotic conditions; however, we have estimated that the changes in refractive index and density do not substantially affect the modulus estimation (Supplementary Note 5 and Supplementary Fig. 5). The measured Brillouin shift varied linearly with the concentration of sucrose (Fig. 1d).
To compare with the current gold standard for measurements of the Young’s modulus of cells, we performed micro-indentation by using an AFM equipped with a 5 μm diameter bead (Supplementary Note 6 and Supplementary Figs. 6–8). The Young’s modulus derived from the thin-layer Hertzian contact model of AFM indentation showed nearly quadratic increase with sucrose concentration (Fig. 1d and Supplementary Fig. 6)1, 22. The typical values of the high frequency longitudinal modulus are on the order of gigapascals, contributed by the low compressibility of the intracellular microenvironment, and should not be interpreted as the Young’s or shear moduli of the cell that are typically on the order of kilopascals when measured at low frequencies8. However, our data revealed a remarkably high correlation (R2 > 0.99) between the variations of these two types of moduli with the same environmental conditions (Fig. 1e). This result indicates that the underlying biochemical, physical, and structural changes within the cell in response to the osmotic pressure change affect both longitudinal and Young’s moduli in the same direction. We have consistently observed such correlation in biological tissues24 as well as for synthetic hydrogels (Supplementary Note 7 and Supplementary Fig. 9). Furthermore, in hydrogels, we have directly demonstrated that Brillouin signatures are sensitive to mechanical changes due to both polymer concentration and polymer crosslinking (Supplementary Fig. 10). The log-log linear relationship between longitudinal modulus M′ and Young’s modulus E′ is expressed as: log(M′) = a log(E′) + b, where a, and b are material-dependent coefficients24. Our instrument was able to detect a 0.05% change in longitudinal modulus, which corresponds to 2% change in Young’s modulus. This sensitivity is comparable to, if not better than, contact-based mechanical measurements.
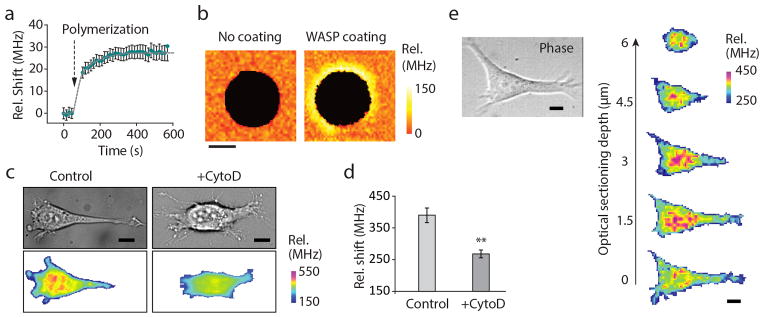
Cytoskeleton modifications are important in the regulation of cell mechanical properties. Using reconstituted actin gels in vitro, we verified that the Brillouin frequency shift was sensitive to two major mechanisms of cytoskeleton stiffening: actin polymerization (Fig. 2a) and branching of actin fibers with high spatial and temporal resolution (Fig. 2b). The Brillouin shifts increase as cells spread on a 2D substrate (Supplementary Note 8 and Supplementary Fig. 11) and decrease when actin polymerization is inhibited with Cytochalasin D25 (Fig. 2c and d). The confocal sectioning of Brillouin microscopy allowed us to analyze longitudinal modulus with 3D resolution. The Brillouin images of a NIH 3T3 fibroblast cultured on a polyacrylamide gel substrate revealed a stiffer nucleus and softer cytoplasm26 (Fig. 2e).
Figure 2. Brillouin microscopy measurements of relevant mechanical changes within a cell.
(a) The Brillouin frequency shift is sensitive to the mechanical changes occurring as actin polymerizes into a gel. In this plot, the frequency shift is relative to the Brillouin shift of 7.44 GHz for pure water at room temperature. (b) In reconstituted actin gels, Brillouin microscopy shows the ability to map stiffer regions of the gels where actin branching is promoted. Scale bar, 5 μm. (c) Representative images of two different NIH 3T3 cells, treated with Cytochalasin D vs untreated control. Scale bars, 10 μm (d) A highly statistically significant (p-value <0.0001, unpaired two-tailed t-test) reduction in Brillouin shift is observed when averaging over the whole cell volume between Cytochalasin D-treated cells (N=14) and controls (N=13). (e) Representative three-dimensional confocal reconstruction of the intracellular stiffness within a NIH 3T3 fibroblast cultured on a stiff polyacrylamide gel substrate. Scale bars, 10 μm. Error bars are s.e.m.
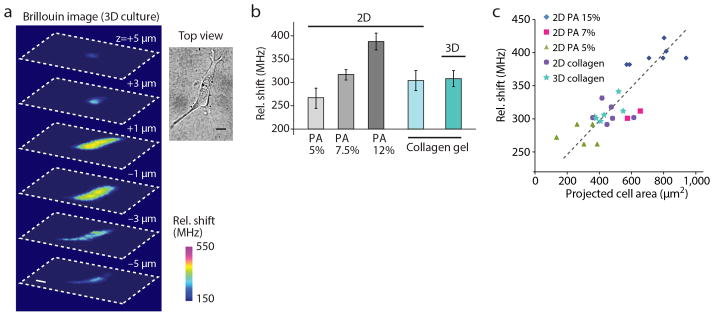
The non-contact nature of the Brillouin technique enables the mechanical measurement of cells in physiologic 3D collagen matrices (Fig. 3a). For direct comparison with cells in 2D, we measured cells grown on flat polyacrylamide substrates with shear moduli of 1, 2.5, and 15 kPa (Fig. 3b). Consistent with previous studies27, we observed an increase in Brillouin shifts with increasing substrate rigidity. The Brillouin shifts from cells cultured within 3D collagen gels with a shear modulus of ~300 Pa was nearly identical to cells grown on top of the same collagen gels and comparable to those of cells grown on stiffer 2.5 kPa polyacrylamide gels (Fig. 3b). Cells grown on polyacrylamide substrates with low modulus (~1 kPa) were somewhat rounded and not fully spread; on the other hand, cells on 2D collagen gels and within 3D collagen gels (~300 Pa) appeared well spread. This morphological behavior has been reported in soft fibrin and collagen gels28, 29, and has been attributed to the higher density of adhesion sites or the nonlinear elasticity of collagen gels. We found a strong correlation (p<0.001) between cell stiffness and projected cell area (Fig. 3c). All of the data obtained on 2D substrates and in 3D hydrogels in various conditions collapse on a single trend of cell stiffness vs. maximum projected cell area (Fig. 3c). This finding, together with the identical intracellular modulus in 2D and 3D collagen gels, may indicate that cellular stiffness is similarly regulated in both 2D and 3D.
Figure 3. Extra-cellular matrix effect on cell stiffness.
(a) Representative 3D Brillouin image reconstruction of a NIH 3T3 cell cultured within a collagen gel matrix and related top-view obtained with bright-field microscopy. Scale bars, 10 μm. (b) Average Brillouin shift of NIH 3T3 fibroblasts cultured on top of polyacrylamide gel substrates of ~1 kPa (N=8), 2.5 kPa (N=8) and 15 kPa (N=12), respectively, and collagen gels (N=5), compared to the average Brillouin shift of cells cultured within collagen gels (N=5). Error bars are s.e.m. (c) Average Brillouin shift of NIH 3T3 fibroblasts plotted against projected cell area in the different 2D and 3D microenvironments.
In conclusion, Brillouin optical microscopy enables mapping the longitudinal modulus of live cells both on 2D substrates and in 3D matrices. Longitudinal modulus may not be directly linked to the tension within the cytoskeletal network or the cortical contractility30. However, we have demonstrated that Brillouin technology is sensitive to the biomechanical changes within a cell due to several important mechanisms such as biopolymer content, polymerization, branching, and liquid-solid volume fraction. Brillouin microscopy opens up new research avenues for the biomechanical investigation of cells and their microenvironment in 3D at subcellular resolution.
Online Methods
Brillouin scattering
Spontaneous Brillouin scattering arises from the interaction of light with acoustic phonons inside the material. The Stokes frequency downshift occurs when light scattering generates an acoustic phonon. Anti-Stokes frequency upshift results when a photon gains energy from a phase-matched thermally-generated acoustic phonon. In the approximation of an isotropic material within the probed voxel, the phase-matching acoustic frequency is given by , with K the photon wavenumber, θ the angle between incident and scattered photons, M′ the real part of the material longitudinal modulus and ρ the mass density of the sample. In backward epi detection, the frequency shift at the peak of the Brillouin spectrum is given at θ = π so that the real part of the longitudinal modulus is computed to be M′ =ρλ2Ω2/(4n2), where λ is the wavelength of the incident radiation and n is the index of refraction (Supplementary Note 1).
Confocal microscope
The light source of the Brillouin confocal microscope is a frequency-doubled Nd-YAG laser (Torus, Laser Quantum, Inc.) emitting a single longitudinal mode at 532 nm. The laser beam was cleaned with a spatial filter, expanded to overfill the back-aperture of the objective lens and inserted into an IX-71 microscope (Olympus). The laser beam entered the microscope through the right-side port, and was reflected to the sample by a 100% reflection mirror sitting below the filter-cube turret of the microscope. Thus, to operate the microscope in Brillouin modality, we moved the filter turret to an empty slot so that all scattered light was reflected out of the microscope through the same right port. Light was focused onto the cell samples via a microscope objective lens of NA greater than 0.6 in epi-illumination, with spatial resolution of at least ~0.5 × 0.5 × 2 μm3. For calibration, we used a pair of automated shutters and reference materials (Supplementary Fig. 3). For 3D imaging, Brillouin shift were acquired from each location within the sample, and then samples were translated stepwise using 3-axis motorized stages (Prior). Scattered light from the samples was collected by a single-mode optical fiber (Thorlabs) serving as a confocal pinhole, and delivered to the apodized VIPA spectrometer (Supplementary Fig. 2). With 2 to 4 mW illumination power at the sample, it generally took 2 to 5 minutes to acquire a 2D frame depending on the number of pixels. In these conditions cell morphology was not affected during repeated measurements, and no apparent difference in mechanical properties was observed between fresh cells and cells that were imaged multiple times. At higher powers exceeding 10 mW, cells remained viable but they tended to change their shape after the imaging session.
VIPA spectrometer
The spectrometer consists of two apodized cross-axis VIPA stages with a relay telescope and square-hole spatial filter in between (Supplementary Note 2). The two VIPA etalons have identical specifications (R1=99.9%, R2=95%, 1.6° internal tilt, Light Machinery). Linearly variable intensity filters (Rugate and Newport 50FS04DV) were used for apodization. The diffraction pattern after the final VIPA stage was detected by using an EM-CCD camera (Ixon Du197, Andor) with a dispersion slope of 0.3 GHz/pixel.
Data acquisition and analysis
We used LABVIEW for instrument automation, using modified manufacturer sample codes (Prior translational stages, Andor camera) and home-written codes to operate shutters. We used MATLAB for spectral analysis. Our algorithm (provided as a Supplementary Software) extracts the optical spectrum, and measures the Brillouin shift and magnitude by curve-fitting with Lorentzian profiles (Supplementary Fig. 3). Brillouin images were produced in MATLAB typically using ‘jet’ colormap and replacing the red with magenta. To quantify the average Brillouin shift of a cell, we typically acquired several en face sections of the cell under examination at different height (similarly to Fig. 2e). After the measurement, we identified the voxels to average (i.e. belonging to the cell rather than the surrounding medium) using a threshold on Brillouin shift and linewidth.
Cell Culture on two-dimensional substrates
NIH 3T3 fibroblast cell line tested for mycoplasma contamination was purchased from American Type Culture Collection (ATCC); after purchasing, cells were frozen at early passage and a fresh cell culture was routinely started from frozen stock. Cells were maintained and inspected daily under inverted phase microscope to compare morphology at different cell densities against reference images. Cells were grown on custom-made 2D substrates consisting of a collagen-coated polyacrylamide gel layer attached to 20 mm glass bottom dishes. Glass-bottom dishes were first treated with 200 μl (3-Aminopropyl)trimethoxysilane (APTMS) for 3 min and 400 μl 0.5% gluteraldehyde solution for 30 min to condition the glass for attachment to polyacrylamide. 15–30 μl of polyacrylamide solution with 5–12% acrylamide and 0.04–0.4% bis-acrylamide was then deposited in each glass well to create 50–100 μm tall gels. Circular coverslips treated with Rain-X were placed face-down on the gel to create a flat, even surface. After solidifying, the coverslips were discarded and gels were treated with 1 ml hydrazine hydrate for 4 hours, followed by 2 ml 5% glacial acetic acid for 1 hour to allow attachment of collagen (type I from rat-tail) to the polyacrylamide gel. Gels were then washed with PBS 3x for 30 min each, sterilized under UV light and left overnight under a layer of 1.5 mg/ml oxidized collagen31. Gels were then washed in serum-free DMEM 3x for 30 min each and left in DMEM for at least 24 hours before plating cells. Cells were plated on the gel substrate in DMEM supplemented with 10% FBS and allowed to spread for at least 24 hours before imaging.
Osmotic pressure experiments
Hyperosmotic environments of varying strength were imposed on cells by adding 2x strength sucrose solutions of 0, 150, 300 or 500 mOsm in equal volume to existing cell media. Sucrose solutions were made by dissolving sucrose in DMEM supplemented with 1% FBS and sterilizing for 30 min under UV light. Cells were grown on polyacrylamide gel and cell media was switched to DMEM supplemented with 1% FBS at least 6 hours prior or overnight. Cell stiffness was measured before and immediately after sucrose addition with Brillouin microscopy and AFM as a comparison. To supplement AFM measurements, average cell dimensions were obtained using confocal microscopy (Supplementary Note 6).
AFM-based micro-indentation
An Asylum MFP3D AFM (Asylum Research, Santa Barbara, CA) was used to perform the nanoindentation on single fibroblasts that were adhered to the PDMS coated substrate. The cells were indented via gold-coated polystyrene colloidal probe tips (end radius, R ~ 2.5 μm) attached to cantilivers with nominal spring constant k ~ 0.06 N/m, Novascan, Ames, IA). The thermal noise oscillation method was applied to determine the cantilever spring constant for each probe tip32. The indentation was performed under force control scheme, with maximum force ~2.5 nN. The resulting indentation depths were in the range of 0.2–1 μm. The axial (z direction) displacement of the tip is calculated as the z-piezo subtracted from the vertical deflection of the cantilever. Typical force-displacement curves for cells in different sucrose concentrations are shown in Supplementary Fig. 7. A typical force-displacement curve and the best-fit curve from the thin-layer Hertz model are shown in Supplementary Fig. 8. Different locations on cells were indented to obtain an average for the Young’s modulus of each cell. An indentation speed of 0.1 μm/s was used to probe the Young’s modulus under close-to-equilibrium conditions.
Reconstituted actin gel polymerization and branching
Branching F-actin was polymerized on VCA-coated polystyrene spheres according to the protocol outlined in Pujol et al, 201233. An actin polymerization kit, VCA-Domain WASP protein, ARP2/3 protein, cofilin and gelsolin were all obtained from Cytoskeleton and handled according to provided instructions. 4.5 μm carboxylated polystyrene spheres (Spherotech) were coated in VCA and added to actin buffer solution to polymerize the actin at 6.5 μM actin, 6.5 μM cofilin, 90 nM gelsolin, 180 nM Arp2/3 and 0.01% VCA. To stop polymerization, the actin solution was diluted 1:10 with 6.6 μM phalloidin. Beads were allowed to settle on a gel substrate to reduce motion and imaged immediately. Beads were also imaged with confocal microscopy by modifying the dilution step with 10% Alexa Fluor 594 phalloidin (Life Technologies).
Actin disruption via Cytochalasin D
Actin filament disruption was achieved in NIH3T3 cells by submerging cells in 5 μg/ml cytochalasin D (Sigma Aldrich) in DMEM supplemented with 1% FBS. Cells were plated in DMEM supplemented with 10% FBS at least 24 hours before imaging and cell media was switched to DMEM supplemented with 1% FBS at least 6 hours prior to serum starve the cells and reduce cell movement during imaging. Cells were imaged 30 min to 1 hour after addition of a stock solution of cytochalasin made in DMEM.
Cell culture within 3D collagen gels
Rat tail collagen type I in acetic acid (BD Biosciences) was buffered with 10× PBS with phenol red, titrated to a pH of 8.0 with 0.1 M sodium hydroxide, and brought to a final concentration of 2 mg/mL with water. Cells were lifted from culture with Trypsin-EDTA and centrifuged at 200× g for 5 min, then resuspended in growth medium and mixed with collagen I solution for a final concentration of 4 × 105 cells per mL of total collagen solution. Collagen was polymerized in humidified chambers at 37 °C and 5% CO2 for 20 min prior to adding growth medium to hydrate the gel.
Dependence of cell stiffness on spread area
To quantitatively assess the cell stiffness-morphology relationship, we measured the average cell stiffness vs. the projected cell area for several conditions. For each cell we estimated the Brillouin shift by averaging the frequency shift at each pixel within the cell; we estimated the projected cell area by determining the cell contour from the phase-contrast image and computing the enclosed area with ImageJ. Within the 3D cultures, only cells with extended morphology were analyzed, while round cells were discarded. In the plot, each data point corresponds to a different cell. The points were fit to the same linear regression curve (R>0.8, p-value <0.001).
Supplementary Material
Acknowledgments
We thank AC Martin, FM Mason, E. Moeendarbary, MC Gather and K. Franze for helpful discussions as well as H. Oda, K. Sawicki and K. Berghaus for help with hydrogel measurements. This work was supported in part by the National Institutes of Health through grants K25EB015885 (GS), P41-EB015903 (SHY), R33 CA174550-01 (RDK), National Science of Foundation through grant CBET-0853773 (SHY) and Human Frontier Science Program Young Investigator grant (GS).
Footnotes
Author contributions
GS, SHY conceived the project. GS, WP, RK, SHY devised research plan. GS developed the instrument and performed the experiments. WP, KP developed cell protocols and performed cell-related control measurements. HT, WP, AG designed and performed indentation experiments. GS, WP, SHY wrote the manuscript with input from all authors.
Competing financial interests
The authors declare no competing financial interests.
Code Availability. The MATLAB code used to analyze data is provided as Supplementary Software along with representative Supplementary Data to run the code.
References
- 1.Moeendarbary E, et al. Nat Mat. 2013;12:253–261. doi: 10.1038/nmat3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stroka KM, et al. Cell. 2014;157:611–623. doi: 10.1016/j.cell.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charras GT, Mitchison TJ, Mahadevan L. J Cell Sci. 2009;122:3233–3241. doi: 10.1242/jcs.049262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart MP, et al. Nature. 2011;469:226–230. doi: 10.1038/nature09642. [DOI] [PubMed] [Google Scholar]
- 5.Ingber D. Ann Med. 2003;35:564577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 6.Bao G, Suresh S. Nat Mat. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 7.Ingber DE. J Cell Sci. 2003;116:1157–1173. doi: 10.1242/jcs.00359. [DOI] [PubMed] [Google Scholar]
- 8.Mofrad MRK, Kamm RD. Cytoskeletal Mechanics. Cambridge Univ Press; 2006. [Google Scholar]
- 9.Ng L, et al. J Biomech. 2007;40:1011–1023. doi: 10.1016/j.jbiomech.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Lam W, Rosenbluth M, Fletcher D. Blood. 2007;109:3505–3508. doi: 10.1182/blood-2006-08-043570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guck J, et al. Biophys J. 2001;81:767–784. doi: 10.1016/S0006-3495(01)75740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans E, Yeung A. Biophys J. 1989;56:151–160. doi: 10.1016/S0006-3495(89)82660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto O, et al. Nat Meth. 2015;12:199–202. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- 14.Mason TG, Ganesan K, vanZanten JH, Wirtz D, Kuo SC. Phys Rev Lett. 1997;79:3282–3285. [Google Scholar]
- 15.Yap B, Kamm R. J Appl Physiol. 2005;98:1930–1939. doi: 10.1152/japplphysiol.01226.2004. [DOI] [PubMed] [Google Scholar]
- 16.Panorchan P, Lee J, Kole T, Tseng Y, Wirtz D. Biophys J. 2006;91:3499–4006. doi: 10.1529/biophysj.106.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dil JG. Rep Progr Phys. 1982;45:285–334. [Google Scholar]
- 18.Scarcelli G, Yun SH. Nat Phot. 2008;2:39–43. doi: 10.1038/nphoton.2007.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenberg SR, Grodzinsky AJ. J Orth Res. 1985;3:148–159. doi: 10.1002/jor.1100030204. [DOI] [PubMed] [Google Scholar]
- 20.Koski KJ, Akhenblit P, McKiernan K, Yarger JL. Nat Mat. 2013;12:262–267. doi: 10.1038/nmat3549. [DOI] [PubMed] [Google Scholar]
- 21.Scarcelli G, Yun SH. Optics Express. 2011;19:10913–10922. doi: 10.1364/OE.19.010913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou E, et al. Proc Natl Acad Sci USA. 2009;106:10632–10637. doi: 10.1073/pnas.0901462106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barer R, Joseph S. Quart J Micr Sci. 1954;95:399–423. [Google Scholar]
- 24.Scarcelli G, Kim P, Yun SH. Biophys J. 2011;101:1539–1545. doi: 10.1016/j.bpj.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakatsuki T, Schwab B, Thompson N, Elson E. J Cell Sci. 2001;114:1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- 26.Tseng Y, Lee JSH, Kole TP, Jiang I, Wirtz D. J Cell Sci. 2004;117:2159–2167. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 27.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Biophys J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali MY, Chuang CY, Saif MTA. Soft Matt. 2014;10:8829–8837. doi: 10.1039/c4sm01602e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winer JP, Oake S, Janmey PA. Plos One. 2009;4:e6382. doi: 10.1371/journal.pone.0006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nijenhuis N, Zhao X, Carisey A, Ballestrem C, Derby B. Biophys J. 2014;107:1502–1512. doi: 10.1016/j.bpj.2014.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damljanovic V, Lagerholm B, Jacobson K. Biotechniques. 2005;39:847–851. doi: 10.2144/000112026. [DOI] [PubMed] [Google Scholar]
- 32.Hutter JL, Bechhoefer J. Rev Sci Instr. 1993;64:1868–1873. [Google Scholar]
- 33.Pujol T, du Roure O, Fermigier M, Heuvingh J. Proc Natl Acad Sci USA. 2012;109:10364–10373. doi: 10.1073/pnas.1121238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.