Abstract
Acquired therapeutic resistance is the major drawback to effective systemic therapies for cancers. Aggressive triple-negative breast cancers (TNBC) develop resistance to chemotherapies rapidly, whereas the underlying mechanisms are not completely understood. Here we show that genotoxic treatments significantly increased the expression of miR-181a in TNBC cells, which enhanced TNBC cell survival and metastasis upon Doxorubicin treatment. Consistently, high miR-181a level associated with poor disease free survival and overall survival after treatments in breast cancer patients. The up-regulation of miR-181a was orchestrated by transcription factor STAT3 whose activation depended on NF-κB-mediated IL-6 induction in TNBC cells upon genotoxic treatment. Intriguingly, activated STAT3 not only directly bound to MIR181A1 promoter to drive transcription, it also facilitated the recruitment of MSK1 to the same region where MSK1 promoted a local active chromatin state by phosphorylating histone H3. We further identified BAX as a direct functional target of miR-181a, whose suppression decreased apoptosis and increased invasion of TNBC cells upon Dox treatment. These results were further confirmed by evidence that suppression of miR-181a significantly enhanced therapeutic response and reduced lung metastasis in a TNBC orthotopic model. Collectively, our data suggested that miR-181a induction played a critical role in promoting therapeutic resistance and aggressive behavior of TNBC cells upon genotoxic treatment. Antagonizing miR-181a may serve as a promising strategy to sensitize TNBC cells to chemotherapy and mitigate metastasis.
Keywords: miR-181a, breast cancer, DNA damage, therapeutic resistance, metastasis
INTRODUCTION
As triple-negative breast cancers (TNBC) lack molecular targets, chemotherapy is the only currently available systemic treatment for TNBC, and prognosis remains poor 1. Although patients with TNBC initially are sensitive to chemotherapy, they rapidly develop resistance to the treatment, which leads to a high incidence of early relapse. Moreover, TNBC is characterized by aggressive metastases, which may further contribute to the rapidly arising therapeutic resistance 2. Therefore, understanding the pathogenesis of therapeutic resistance and metastasis of TNBC is of great importance to develop novel targeted strategies to improve the prognosis in TNBC patients. Nevertheless, the molecular mechanisms underlying therapeutic resistance and their association with metastasis are still ill-defined.
Previous studies demonstrated that chemotherapeutic drugs and radiation used for cancer treatment effectively activate the transcription factor NF-κB, which is a key modulator of inflammation, immune response and cell survival 3. The pro-survival function of NF-κB has been linked with therapeutic resistance in a variety of cancers, which is associated with up-regulation of anti-apoptotic genes driven by NF-κB 4. In addition, NF-κB also regulates expression of noncoding RNAs, such as microRNAs 5. MicroRNAs negatively regulate target gene expression at the post-transcriptional level by recognizing specific sequence primarily within the 3′- UTR (untranslated region) and, to a lesser extent, in coding sequence of mRNA 6. Most miRNA genes are transcribed into primary-miRNAs by RNA polymerase II, which can be sequentially processed into mature miRNAs. The transcription of miRNA can be regulated by upstream genetic regulatory elements, such as conserved transcription factor binding sites, as well as by epigenetic modifications 7. The expression of miRNAs is frequently dysregulated in human malignancies, and miRNAs can function as either oncogenes or tumor suppressors depending on the genes they target in specific cancers 8, 9. Moreover, alteration of miRNA expression in varying cancers has been shown to promote therapeutic resistance to both conventional chemotherapy and targeted therapies 10. Therefore, targeting miRNA may serve as a promising strategy in combination with targeted therapy or chemotherapy to improve anticancer efficacy 10, 11.
Stimulation with genotoxic agents such as ionizing radiation (IR) and chemotherapeutic drugs was found to alter miRNA expression in cancer cells at the transcriptional and post-transcriptional levels 12-15. We recently showed that DNA damaging agents can transcriptionally upregulate miR-21 and miR-125b by activating NF-κB in cultured cancer cells, which promoted cancer cell survival upon genotoxic stress 16, 17. Whether additional miRNAs can be induced by genotoxic agents in cancer cells and clinical relevance of the pro-survival function mediated by chemodrug-induced miRNAs in cancer therapeutic response remain to be further elucidated.
In this study, we identified miR-181a as a major upregulated miRNA in TNBC cells in response to chemotherapeutic drugs. The induction of miR-181a correlated with enhanced cancer cell survival and aggressiveness in response to doxorubicin (Dox) treatment. Consistently, MIR181A1 amplification was found in approximately 12% of breast cancer samples in TCGA database. In addition, high miR-181a level significantly associated with poor distant metastasis free survival (DMFS) in breast cancer patients. We further showed that STAT3 (signal transducer and activator of transcription 3), which was activated by genotoxic treatment in a NF-κB-dependent manner, orchestrated transcriptional activation of miR-181a both as a transcription factor and a regulator of epigenetic modification. Furthermore, we identified the pro-apoptotic gene BAX as a novel functional target of miR-181a, whose repression supported increased cell survival and metastasis in TNBC cells exposed to Dox. Accordingly, miR-181a inhibition significantly reduced TNBC cell resistance to Dox treatment as well as mitigated lung metastasis in vivo. Overall, our data support a critical role of miR-181a induction in promoting acquired resistance to chemotherapeutic agents and metastasis in TNBC cells.
RESULTS
miR-181a induction by DNA damaging agents promotes breast cancer cell survival and metastasis
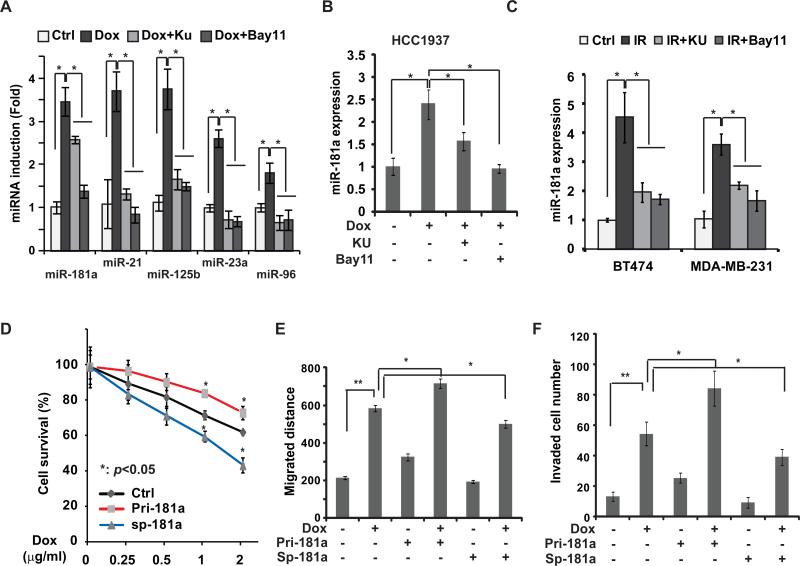
In an effort to identify miRNAs whose expression was altered in TNBC cells by Dox treatment, we treated MDA-MB-231 cells with Dox (2 μg/ml) or vehicle control and quantified miRNA profile using NanoString nCounter analysis. To enrich those miRNAs whose expression may be regulated by genotoxic NF-κB activation, we also included two additional groups in which cells were treated with Dox in combination with inhibitor of ATM or IKK, two essential kinases required for NF-κB activation by DNA damage (supplementary Fig. S1A) 18. Consistent with our previous studies 16, 17, nCounter quantification and qPCR analyses indicated that miR-21 and miR-125b were among the most upregulated miRNAs whose induction depended on both ATM and IKK activity (Fig. 1A and Supplementary Fig. S1B). We also found miR-181a expression was significantly increased in MDA-MB-231 cells treated by Dox, whose induction was attenuated by ATM and IKK inhibitors. In parallel, Dox-induced miR-181a upregulation was abrogated in MDA-MB-231 cells in which RELA/p65 was depleted by siRNA as well as in p65-deficient MEFs, suggesting a critical role of NF-κB/p65 in regulating miR-181a induction by DNA damage (supplementary Fig. S1C, S1D). Similar results were observed in another TNBC cell line HCC1937 cells (Fig. 1B). Moreover, we detected significant induction of miR-181a in MDA-MB-231 cells and HER2+ breast cancer BT474 cells upon IR, which was sensitive to inhibition of ATM or IKK, suggesting that genotoxic agent-induced miR-181a up-regulation may not limited to TNBC cells (Fig. 1C and supplementary Fig. S2C). In addition to mature miR-181a, we also detected significant induction of primary transcripts of MIR181A1 in MDA-MB-231 and BT474 cells in response to genotoxic treatment, which was attenuated by inhibition of ATM or IKK (supplementary Fig. S1E, 1F). These data suggest that genotoxic agents may induce miR-181a up-regulation at the transcriptional level.
Figure 1.
Genotoxic treatments induce miR-181a upregulation in breast cancer cells. (A) qPCR analysis of miRNA expression in MDA-MB-231 cells treated with Dox (2μg/ml) alone or along with KU55933 (Ku) or Bay11-7085 (Bay11) for 8 h, *: p< 0.05. (B) Similar analysis of miR-181aexpression in HCC1937 cells was performed as in (A), *: p< 0.05. (C) MDA-MB-231 and BT474 cells were treated with IR (10Gy) alone or in the absence or presence of KU55933 or Bay11-7085, miR-181a expression was measured with qPCR. (D) MDA-MB-231 cells were transfected with control, pri-miR-181a, or miR-181a sponge and treated for 24 h with Dox at indicated doses, the live cell population was examined by WST-1 assay. Data from three independent experiments were pooled and shown as mean ± S.D. *, p <0.05. (E) Monolayer MDA-MB-231 cells transfected with control, pri-miR-181a or miR-181a-sponge were wounded and incubated with Dox for 4 h as shown, the migrated distance was determined after 24 h and quantitation from 3 experiments was shown as mean ± S.D., *: p< 0.05, **: p< 0.01. (F) MDA-MB-231 cells transfected with control, pri-miR-181a or miR-181a-sponge were incubated by Dox as shown, cell invasion was measured with modified Boyden Chamber assay. Numbers of invading cells were quantified and data from triplicate experiments were shown as mean ± S.D., *: p< 0.05, **: p< 0.01.
To determine pathological significance of miR-181a induction, we overexpressed miR-181a in MDA-MB-231 cells and found that it significantly enhanced cells survival upon Dox treatment compared with mock transfected cells. In contrast, antagonizing miR-181a by miR-181a-sponge inhibitor substantially increased MDA-MB-231 cell sensitivity to Dox and resulted in reduced cell survival upon treatment (Fig. 1D). Moreover, overexpression of miR-181a increased, while inhibiting miR-181a reduced, MDA-MB-231 cell migration and invasion following Dox treatment (Fig. 1E, F). These results are in line with previous studies indicating a strong association between therapeutic resistance and aggressive metastasis in TNBC 1, and suggesting that miR-181a induction by Dox in TNBC cells may contribute to acquired resistance and promote metastasis.
miR-181a is amplified in breast cancer patients and associates with poor clinical outcomes
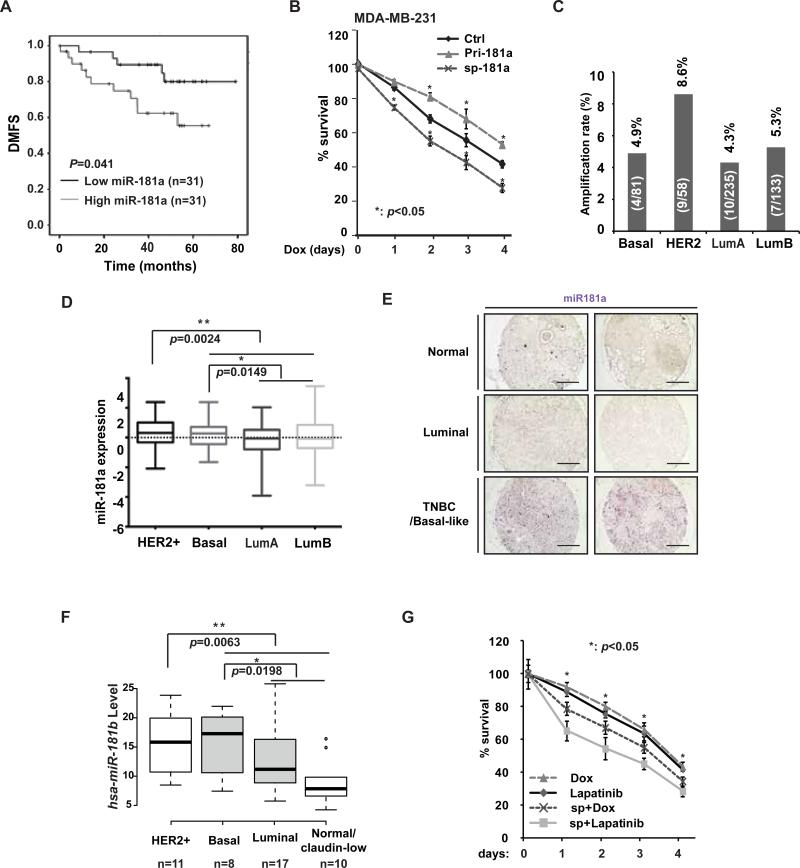
Distinct roles of miR-181a in cancer progression have been reported in different cancer types. miR-181a was shown to promote ovarian cancer progression by promoting epithelial-mesenchymal transition (EMT) 19, while ectopic miR-181a expression inhibited acute myeloid leukemia tumor growth 20. To determine the potential function of miR-181a in breast cancer pathogenesis, we analyzed two independent clinical patient data sets. We collected 62 FFPE samples of TNBC patients (Supplementary Tab. S1) and analyzed miR-181a level by qPCR. When stratified by median miR-181a level, high expression group significantly correlated with poor DMFS among these TNBC patients (Fig. 2A). In another publicly available data set (GSE19536) 21, we found high miR-181a level was associated with poor DFS in breast cancer patients (Supplementary Fig S2A), although it did not reach statistical significance likely due to small cohort numbers. Consistently, MDA-MB-231 cells with increased miR181a level showed significantly enhanced survival upon prolonged Dox treatment, whereas inhibiting miR-181a promoted Dox-induced cell death (Fig. 2B). Furthermore, in patient data collected by TCGA invasive breast cancer study, we found MIR181A1 was amplified in about 12% of breast cancer patients (Supplementary Fig. S2B). Among those patients characterized by molecular subtypes, higher rate of MIR181A amplification was found in HER2+ subtype compared to the other subtypes (Fig. 2C). In accordance, miR-181a expression level was significantly higher in HER2+ and basal subtype breast cancers, which tend to be more aggressive and associate with poor prognosis (Fig. 2D). Consistently, we detect substantially higher miR-181a levels in TNBC/basal-like breast cancer tissues than that in luminal breast cancer and normal breast tissues (Fig. 2E). Subtype-specific enrichment of miR-181 cluster upregulation was also confirmed in a miRNA profiling analysis among 51 human breast cancer cell lines 22. The level of miR-181b, a miR-181 family member co-transcribed with miR-181a in same cluster, was significantly increased in HER2+ and basal-like breast cancer cell lines compared to luminal and normal-like/claudin-low breast cancer cells (Fig. 2F).
Figure 2.
Increased miR-181a level associates with poor clinical outcomes in TNBC patients. (A) Kaplan-Merier curves of 62 TNBC patients after stratification by the median level of miR-181a were used for depicting distant metastasis-free survival (DMFS). (B) MDA-MB-231 cells were transfected with control, pri-miR-181a, or miR-181a sponge and treated with Dox (0.5 μg/ml) for times as indicated, the cell survival was determined by WST-1 assay. Data from three independent experiments were pooled and shown as mean ± S.D. *, p <0.05. (C and D) Amplification rate (amplified cases/total cases of each subtype, C) or expression level (D) of the MIR181A1 gene in different subtypes of human invasive breast cancer (TCGA dataset). (E) The expression of miR-181a was visualized by ISH staining in breast cancer and normal breast samples. Representative images from 2 cases in each group were shown. Scale bar: 400 μM. (F) miR-181b level in 46 human breast cancer cell lines was grouped in 4 subtypes. Data were retrieved from Ref 22. (G) BT474 cells was transfected with control or miR-181a sponge and treated with Dox (0.5 μg/ml) or lapatinib (0.05μM) for the times as indicated, the live cell population data from three independent experiments were pooled and shown as mean ± S.D. *, p <0.05.
The significantly increased level of miR-181a in HER2+ breast cancer cells led us to speculate that miR-181a may also promote HER2+ breast cancer cell survival. Indeed, inhibiting miR-181a substantially increased HER2+ BT474 cell sensitivity to chemotherapeutic drug Dox and HER2-targeting kinase inhibitor Lapatinib, resulting in further decreased cell survival upon treatments (Fig. 2G). All these data indicate that miR-181a functions as an oncomiR in breast cancer development and progression. The miR-181a-mediated therapeutic resistance may be not limited in TNBC cells upon chemotherapy, it also plays a critical role in modulating HER2+ breast cancer cell response to HER2-targeted therapies.
NF-κB-mediated STAT3 activation is essential for miR-181a induction by genotoxic drugs
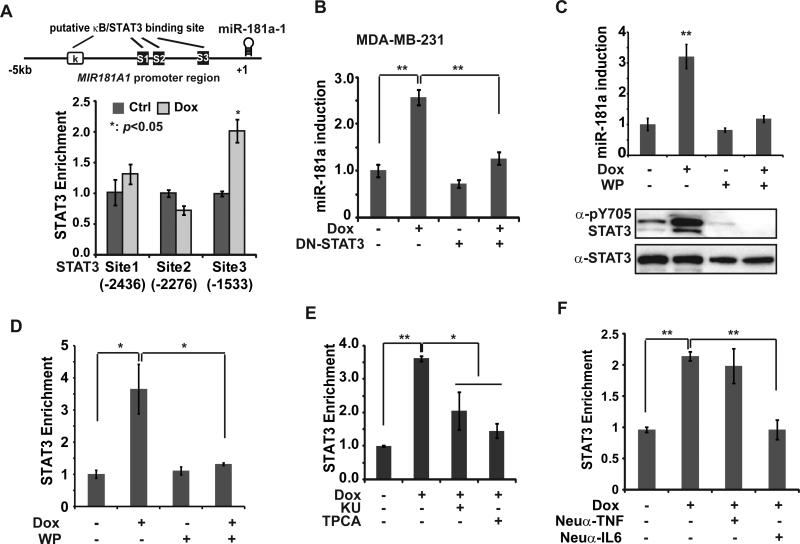
Bioinformatic analysis of MIR181A1 promoter region revealed several binding sites of transcription factors NF-κB and STAT3 which both can be activated by DNA damage. By chromatin IP (ChIP), we detected a significant increase of STAT3 recruitment to the proximal STAT3 site (S3) (Fig. 3A). However, no substantial enrichment of NF-κB/p65 was detected on MIR181A1 promoter (supplementary Fig. S3A). Moreover, luciferase reporter assay confirmed that deletion of STAT3 site3 almost abrogated MIR181A1 promoter-driven transcription in response to Dox treatment, while mutation of NF-κB sites showed little effect (supplementary Fig. S3B). These data indicate that STAT3, but not NF-κB, is a direct transcription regulator of miR-181a induction upon genotoxic treatment.
Figure 3.
IL-6-dependent STAT3 activation is essential for miR-181a induction upon genotoxic stress. (A) ChIP analyses of STAT3 recruitment to the miR-181a promoter region was carried out in MDA-MB-231 cells upon Dox treatment. Schematic representation of putative NF-κB-binding and STAT3-binding sites within human MIR181A1 promoter was shown. (B) MDA-MB-231 cells were transfected with vector or STAT3-Y705F mutant, and treated with Dox (2 μg/ml) for 8 h; miR-181a expression was determined by qPCR, **: p< 0.01. (C) qPCR analysis of miR-181a expression in MDA-MB-231 cells treated with Dox (2 μg/ml) alone or along with WP1066 (10 μM) for 8 h, **: p< 0.01. Total cell extracts from parallel treated cells were probed with indicated antibodies. (D) ChIP analyses of STAT3 recruitment to the miR-181a promoter region (S3) was carried out in MDA-MB-231 cells treated as in (C), *: p< 0.05. (E) STAT3 binding to miR-181a promoter region (S3) was analyzed by ChIP in MDA-MB-231 cells treated by Dox alone or in the absence or presence of KU55933 or TPCA1 (1 μM) *: p< 0.05; **: p< 0.01. (F) ChIP analyses of STAT3 recruitment to the miR-181a promoter region was performed in MDA-MB-231 cells treated with Dox alone or along with neutralizing anti-TNFα or anti-IL-6 antibody, **: p< 0.01.
To further confirm the critical role of STAT3 in inducing miR-181a expression in TNBC cells, we transfected a dominant-negative STAT3 mutant (DN-STAT3 Y705F) into MDA-MB-231 cells. Dox treatment induced robust up-regulation of miR-181a, which was significantly attenuated by transfection of DN-STAT3 (Fig. 3B). Consistently, a STAT3 inhibitor WP1066 blocked STAT3 phosphorylation at Tyr705 and abrogated miR-181a induction in Dox-treated MDA-MB-231 cells (Fig. 3C). Moreover, inhibiting STAT3 activation by WP1066 also blocked STAT3 recruitment to MIR181A promoter in MDA-MB-231 cells upon Dox treatment (Fig. 3D). Taken together, this evidence supports that DNA damage-induced STAT3 activation is essential for miR-181a induction in TNBC cells.
As our original screen showed that miR-181a induction by Dox was sensitive to inhibition of ATM or IKK (Fig. 1A), and our ChIP analyses did not support a direct role of NF-κB in regulating miR-181a transcription, we reasoned that genotoxic NF-κB activation may indirectly modulate miR-181a expression by activating STAT3 in TNBC cells upon DNA damage. In agreement with this notion, treatment with ATM or IKK inhibitor, KU55933 or TPCA-1 respectively, substantially decreased STAT3 enrichment on MIR181A promoter in Dox-treated MDA-MB-231 cells (Fig. 3E). Our previous studies showed that genotoxic stress-induced NF-κB activation can induce up-regulation and secretion of proinflammatory cytokines, such as IL-6, IL-8 and TNFα 16, 23. Accordingly, we found Dox-induced increase in IL-6 transcription and secretion was diminished in cells with p65 deficiency (supplementary Fig S3D-F). Moreover, p65 deficiency also attenuated STAT3 activation upon Dox treatment (supplementary Fig. S3C). Consistently, blocking IL-6, but not TNFα, with neutralizing antibody in culture media remarkably reduced STAT3 recruitment to MIR181A promoter in response to Dox (Fig. 3F). In parallel, treatment with recombinant IL6 was able to restore the miR-181a induction by Dox attenuated by IKK inhibitor TPCA1 (supplementary Fig. S3G). All these data support a critical role of NF-κB-IL6-STAT3 axis in up-regulating miR-181a transcription in TNBC cells upon genotoxic stimulation.
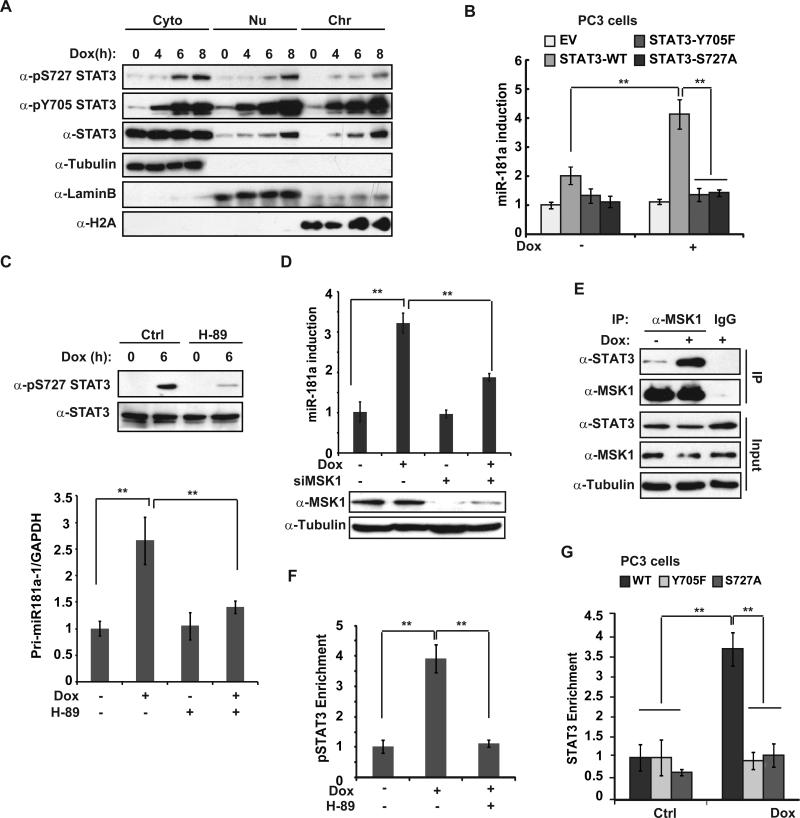
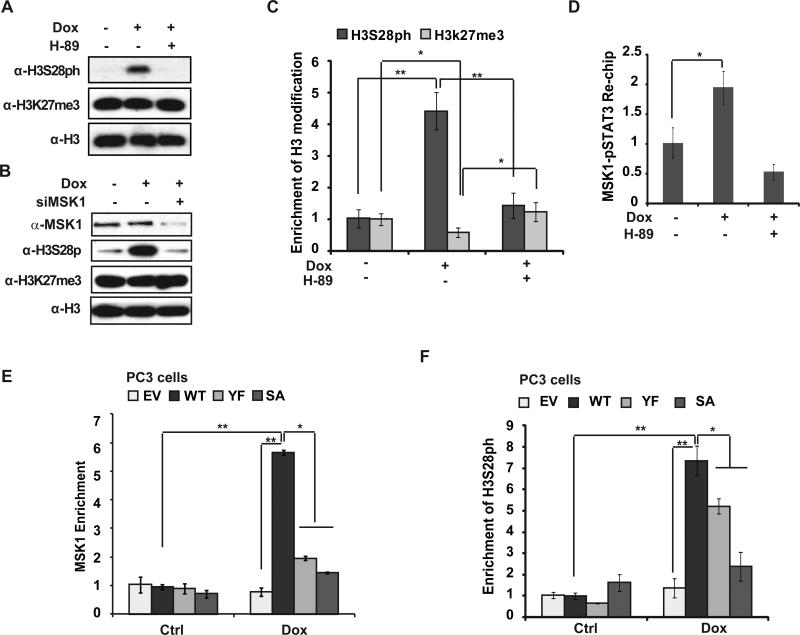
IL-6-induced Tyr705 phosphorylation is essential for STAT3 activation. Meanwhile, phosphorylation at Ser727 was also implicated to play critical role in modulating STAT3 transactivity 24. We found STAT3 was phosphorylated at Ser727 with a slightly delayed kinetics compared to Tyr705 phosphorylation in MDA-MB-231 cells upon Dox treatment (Fig. 4A and Supplementary Fig. S4A). Increased phosphorylation of STAT3 upon genotoxic treatment was detected both in cytoplasm and nuclei of MDA-MB-231 cells. Moreover, chromatin-bound STAT3 was phosphorylated at both Tyr705 and Ser727, supporting the critical role of phosphorylation on both residues in mediating STAT3-depedent gene transactivation (Fig. 4A). Accordingly, miR-181a induction by Dox in prostate cancer PC3 cells, which is STAT3 deficient25, was significantly enhanced by reconstitution of wild type STAT3, but neither STAT3-Y705F nor -S727A mutant (Fig. 4B).
Figure 4.
Phosphorylation of STAT3 S727 is required for STAT3-mediated miR-181a transactivation in response to DNA damage. (A) MDA-MB-231 cells were treated with Dox for the times as indicated. Cytoplasmic, nuclear soluble and chromatin binding proteins were fractionated and immunoblotted using antibodies as shown. (B) PC3 cells stably expressing vector, STAT3-WT, STAT3-Y705F or STAT3-S727A mutant were treated with Dox as shown, miR-181a expression was analyzed with qPCR, **: p< 0.01. (C) qPCR analysis of Pri-miR-181aexpression in MDA-MB-231 cells treated with Dox (2 μg/ml) alone or along with H-89 (10 μM) for 6 h, **: p< 0.01. Parallel total protein extract samples were analyzed by Western blot. (D) qPCR quantification of miR-181a was carried out in control or MSK1-depleted MDA-MB-231 cells exposed to Dox as in (C). **: p< 0.01. (E) MDA-MB-231 cells were treated with Dox or left untreated. Whole cell extracts were IPed with anti-MSK1 and analyzed by immunoblotting. (F) ChIP analyses of STAT3 recruitment to the miR-181a promoter region was carried out in MDA-MB-231 cells upon Dox treatment in the presence or absence of H-89. (G) STAT3 binding to miR-181a promoter region was analyzed by ChIP in PC3 cells stably expressing STAT3-WT, STAT3-Y705F or STAT3-S727A mutant treated with Dox (2 μg/ml) for 6 h. *: p< 0.05; **: p< 0.01.
The phosphorylation of STAT3 on Ser727 and Tyr705 by DNA damage is regulated by distinct mechanisms, as reconstitution with STAT3-Y705F in PC3 cells did not block Ser727 phosphorylation by Dox, and vice versa (Supplementary Fig. S4B). A number of kinases, such as mTOR, MAPK and MSK1, have been shown to phosphorylate STAT3 at Ser727, depending on the stimulus and cell type 26. Interestingly, we previously showed that MSK1 was activated in TNBC cells by genotoxic agents and MSK1-dependent histone H3 phosphorylation was required for miR-21 induction upon Dox treatment 16. We found that MSK1 inhibitor H-89 abolished STAT3 Ser727 phosphorylation in MDA-MB-231 cells exposed to Dox, as well as attenuated the upregulation of miR-181a transcription by Dox (Fig. 4C). The decrease of miR-181a induction and cell survival upon Dox treatment was also observed in MSK1-depleted MDA-MB-231 cells (Fig. 4D and supplementary Fig. S4C). Moreover, genotoxic treatment enhanced interaction between MSK1 and STAT3 in MDA-MB-231 cells (Fig. 4E), which was independent of NF-κB or IL-6 (supplementary Fig. S4D), suggesting a direct response to DNA damage. Consistently, blocking MSK1 activation by H-89 or MSK1 knockdown also significantly reduced STAT3 recruitment at MIR181A1 promoter (Fig. 4F and supplementary Fig. S4E, S4F). In line with this observation, mutation at either Tyr705 or Ser727 dramatically reduced STAT3 enrichment at MIR181A1 promoter in PC3 cells treated by Dox (Fig. 4G). All these data suggest that phosphorylation on both Tyr705 and Ser727 are required for MIR181A1 promoter recruitment thereby driving miR-181a transactivation in cancer cells upon genotoxic treatment.
Activated STAT3 recruits/stabilizes MSK1 binding to MIR181A promoter upon genotoxic treatment
Since MSK1 can phosphorylate histone H3 at Ser10 and Ser28, which both mark an active chromatin state 27, we reasoned that MSK1 may also have a role in regulating miR-181a induction in TNBC cells by modifying chromatin structure. We detected a robust induction of histone H3S28 phosphorylation in MDA-MB-231 cells in response to Dox treatment which was abolished by H-89 or MSK1 knockdown, indicating a critical role of MSK1 in regulating H3S28 phosphorylation upon DNA damage (Fig. 5A, 5B). Global H3K27 methylation level appeared unaffected by Dox and/or MSK1 inhibition. However, ChIP analyses revealed that Dox treatment induced a significant decrease of H3K27 methylation (H3K27me3) concurrent with increased H3S28 phosphorylation at the MIR181A promoter region (Fig. 5C). This observation is consistent with previous reports showing that phosphorylation of H3S28 leads to decreased methylation at neighboring H3K27, thereby activating transcription of a subset of genes 28, 29.
Figure 5.
DNA Damage-induced epigenetic activation at MIR181A1 promoter depends on STAT3-meidated MSK1 enrichment. (A) MDA-MB-231 cells were treated with Dox, acid-extracted histone proteins were analyzed by the indicated antibodies. (B) Similar immunoblotting analyses as in (A) were carried out in control or MSK1-depleted MDA-MB-231 cells. (C) ChIP analyses of histone marker H3S28ph and H3K27me3 enrichment around the STAT3-site (S3) within the MIR-181A promoter region was carried out in MDA-MB-231 cells treated by Dox in the presence or absence of H-89 (10 μM). Histone H3 ChIP was used for normalization. (D) Re-ChIP analyses of activated STAT3 (pS727)-MSK1 co-recruitment to the MIR-181A promoter region was carried out in MDA-MB-231 cells treated as in (C). (E and F) MSK1 (E) or histone H3S28ph (F) enrichment at MIR-181A promoter region was analyzed by ChIP in PC3 cells stably expressing STAT3-WT, STAT3-Y705F or STAT3-S727A mutant treated with Dox (2 μg/ml) for 6 h. *: p< 0.05; **: p< 0.01.
In agreement with the local change of histone H3 modifications, we also detected a significant increase of MSK1 binding at MIR181A promoter. Interestingly, the chromatin region with enriched MSK1 binding overlapped with STAT3 binding site (S3) within MIR181A promoter (supplementary Fig. S4G), suggesting MSK1 and STAT3 may form a complex on MIR181A promoter. In accordance, ChIP-reChIP assay using sequential IP with MSK1 and pS727-STAT3 antibodies confirmed the co-recruitment of MSK1 and activated STAT3 to the S3 site within MIR181A promoter in Dox-treated MDA-MB-231 cells (Fig. 5D).
To explore the interrelationship between STAT3 and MSK1 recruitment to MIR181A promoter upon DNA damage, we analyzed MSK1 recruitment in STAT3-deficient PC3 cells and PC3 cells reconstituted with STAT3-WT, -Y705F, or -S727A mutant. Interestingly, we found MSK1 enrichment at MIR181A promoter depended on STAT3 (Fig. 5E). Moreover, Y705F or S727A mutation, which blocked STAT3 binding to MIR181A promoter (Fig. 4G), also abolished MSK1 recruitment to MIR181A promoter in Dox-treated PC3 cells (Fig. 5E). Consistently, in PC3 cells expressing STAT3-Y705F or -S727A mutant, Dox-induced H3S28ph was also remarkably reduced compared to that in STAT3-WT reconstituted cells (Fig. 5F). Collectively, these results indicate that STAT3 activation by phosphorylation at Y705 and S727 is required for genotoxic stimulation-induced MSK1 enrichment at MIR181A promoter, where stabilized MSK1 binding further phosphorylates H3S28 and promotes a local active chromatin state, facilitating MIR181A transactivation.
BAX is a functional direct target of miR-181a and mediates miR-181a-promoted breast cancer cell drug resistance and aggressive invasion
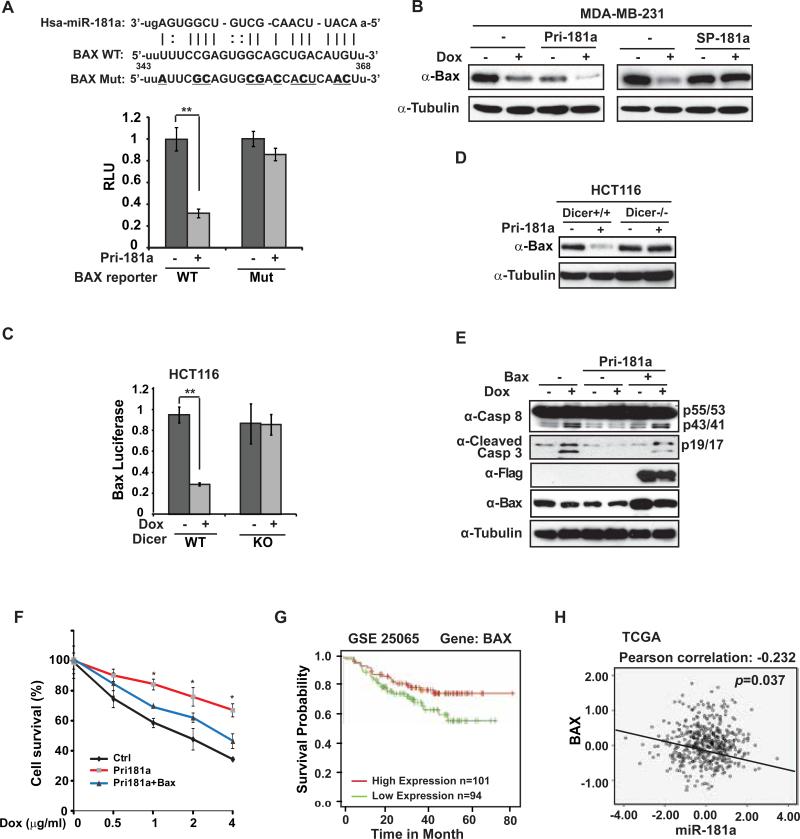
To delineate the underlying mechanism of miR-181a-promoted breast cancer therapeutic resistance and aggressive behavior, we searched potential target gene of miR-181a using several bioinformatics tools such as TargetScan, PicTar, miRanda and Rna22. We particularly focused on apoptosis-related Bcl-2 family genes as several members within this group including BCL2, MCL1 and BCL2L11 have been identified as miR-181a targets in different cells 30. Intriguingly, we found a miR-181a targeting sequence within coding region of BAX, which is highly conserved among different species as well as eight human isoforms (Fig. 6A and supplementary Fig. S5A). Moreover, overexpression of miR-181a significantly down-regulated expression of a luciferase reporter fused to wild type BAX miR-181a-targeting sequence, but not mutated sequence (Fig. 6A). Consistently, BAX expression was substantially decreased in MDA-MB-231 cells upon Dox treatment and/or overexpressing miR-181a. Furthermore, miR-181a sponge inhibitor was able to rescue the decrease of BAX protein level in response to Dox treatment, (Fig. 6B). All these data strongly suggest a critical role of miR-181a in suppressing BAX expression in breast cancer cells upon DNA damage.
Figure 6.
miR-181a promotes breast cancer cell resistance to Dox by repressing BAX. (A) MDA-MB-231 cells were transfected with control or pri-miR-181a plasmid along with WT or mutant BAX luciferase reporter as shown. The activity of both Renilla and Firefly luciferases was assayed using the dual-luciferase reporter assay system and data from triplicate experiments were showed as mean ± S.D. *, p< 0.05. Schematic representation of the putative miR-181a target site within the coding region of BAX gene (n.t.343-368) was shown on the top. Mutations used for abolishing miR-181a recognition sequence were underlined in bold. (B) MDA-MB-231 cells were transfected with pri-miR-181a or miR-181a sponge as shown. BAX expression was determined by Western blot. Tubulin was used as loading control. (C) HCT116 WT and DICER−/− cells were transfected with control or pri-miR-181a plasmid along with WT or mutant BAX luciferase reporter as shown. The luciferase activity were assayed in (A) and data from triplicate experiments were showed as mean ± S.D. *, p< 0.05. (D) BAX expression was determined by Western blot in HCT116 WT and DICER−/− cells transfected with control or pri-miR-181a. (E) MDA-MB-231 cells were transfected with pri-miR-181a and BAX as indicated. Cells were treated with Dox (2 μg/ml) and total cell extracts were analyzed by immunoblotting with indicated antibodies. (F) MDA-MB-231 cells were transfected with control, pri-miR-181a alone or along with BAX. Cell survival upon Dox treatment with indicated doses for 48 h was analyzed with WST-1 assay. Data from three independent experiments were pooled and shown as mean ± S.D. *, p <0.05. (G) Low expression of Bax associated with poor relapse-free survival in breast cancer patients. Patient data obtained from dataset GSE25065 were stratified by median level of BAX expression and analyzed with Kaplan-Merier curves. (H) Pearson analysis of gene expression data from breast cancer patients (TCGA dataset) was used for depicting the correlation between Bax and miR-181a.
To further validate whether miR-181a-dependent suppression of BAX is mediated by miRNA-dependent pathway, we compared the response of BAX-miR-181a reporter to Dox treatment in wild type HCT116 cells and HCT116 cells deficient of Dicer, which is essential for miRNA biogenesis 31. As expected, Dox treatment induced a dramatic decrease in BAX-miR-181a reporter activity in HCT116 cells, but little effect was detected in Dicer-deficient cells (Fig. 6C). In agreement, overexpression of primary-miR-181a was able to decrease BAX expression in HCT116 cells, but not in HCT116 Dicer-deficient cells (Fig. 6D), supporting that Dox-induced BAX downregulation is primarily mediated by miR-181a-dependent post-transcriptional repression.
To determine the functional significance of BAX suppression in miR-181a-mediated chemotherapeutic resistance, we transfected MDA-MB-231 cells with miR-181a alone or along with BAX construct. Dox-induced Caspase 3/8 activation was substantially decreased in miR-181a transfected cells, which was correlated with decreased BAX expression. Moreover, co-transfection of BAX along with miR-181a rescued miR-181a-dependent BAX down-regulation and reinforced Dox-induced Caspase activation in MDA-MB-231 cells (Fig. 6E). Enhanced cell survival by miR-181a overexpression in Dox-treated MDA-MB-231 cells was significantly attenuated by co-transfection of BAX (Fig. 6F). We also found ectopic expression of BAX was able to mitigate the increased migration and invasion in miR-181a-overexpressed MDA-MB-231 cells upon Dox treatment (supplementary Fig. S5B, S5C). These observations were further supported by clear association between low BAX expression and poor overall survival in a breast cancer patient dataset (GSE25065, Fig. 6G), which is also in agreement with the data obtained from KM Plotter 32 showing that high BAX expressing is associated with better RFS in ER−/PR− breast cancer patients (supplementary Fig. S5D). Moreover, we confirmed the negative correlation between miR-181a and BAX expression level in both TCGA breast cancer dataset and our own TNBC patient sample collection (Fig. 6H and supplementary Fig. S5E). Altogether, these data indicate that BAX is a critical functional target gene of miR-181a whose suppression may contribute to acquired therapeutic resistance and increased metastasis in TNBC cells.
Inhibiting miR-181a sensitizes breast cancer to Dox treatment and mitigating lung metastasis in vivo
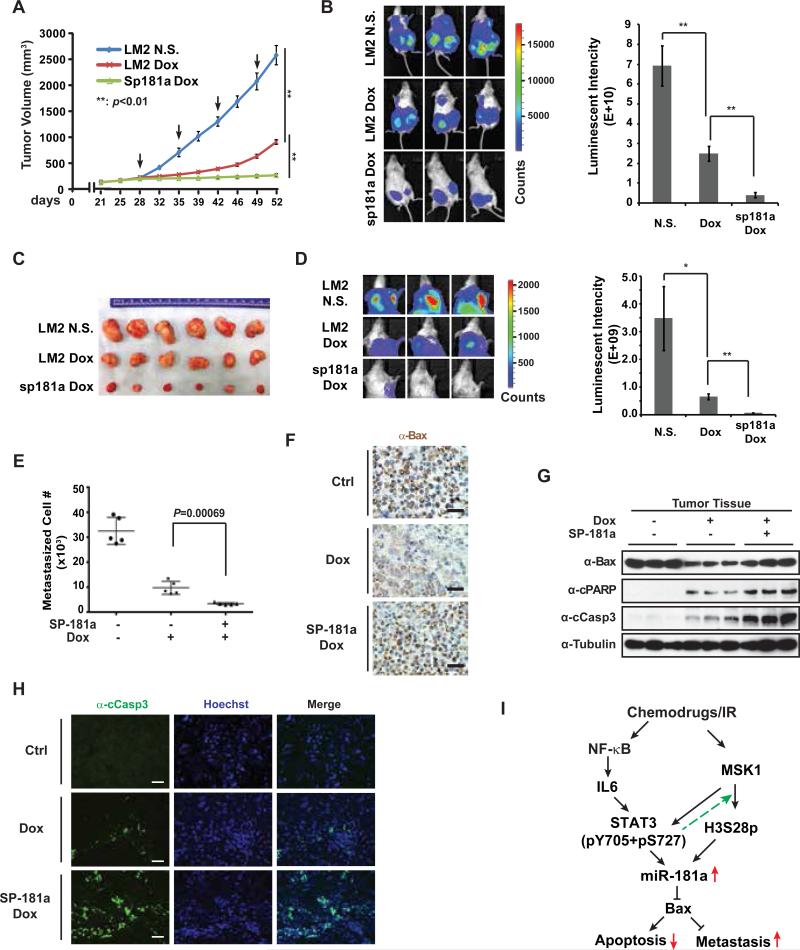
Our data from in vitro cell culture model suggested that miR-181a induction upon genotoxic treatment in TNBC cells may play important roles in mediating therapeutic resistance and cancer metastasis. To validate the potential pathophysiological significance of miR-181a induction in therapeutic response of breast cancer, we generated an orthotopic breast cancer model using MDA-MB-231 LM2 subline which has high lung metastasis tendency 33. As shown in Fig. 7A, Dox treatment remarkably reduced tumor growth compared to saline-treated group. However, the tumor-suppressive efficacy of Dox treatment substantially decreased after the third injection of Dox (around 44 days) as the tumors resumed an accelerated growth, suggesting potential acquired resistance to Dox treatment. In contrast, LM2 cells stably expressing miR-181a sponge inhibitor showed extended growth suppression in response to Dox treatment (Fig. 7A). After 4 cycles of Dox treatment, xenograft tumors from LM2-miR-181a sponge cells showed significantly decreased size and tumor weight compared to LM2 xenograft (Fig. 7A-C and supplementary Fig. S6A). These data were consistent with the observation that antagonizing miR-181a with sponge inhibitor significantly sensitized LM2 cells to Dox treatment in vitro (supplementary Fig. S6B). In addition to reduced tumor growth, we also observed significantly reduced lung metastasis in mice transplanted with LM2-miR181a sponge cells relative to LM2-transplanted mice (Fig. 7D, 7E). Moreover, when normalized to primary tumor weight, lung metastasis of LM2-miR181a sponge cells still showed significant decrease compared to parent LM2 cells, suggesting that the decreased lung metastasis by antagonizing miR-181a was not solely due to reduced primary tumor growth (supplementary Fig. S6C). Altogether, these data strongly support an important role of miR-181a induction in response to Dox treatment in promoting therapeutic resistance and remote metastasis of TNBC cells in vivo.
Figure 7.
Inhibition of miR-181a sensitizes breast cancer to Dox treatment and mitigates lung metastasis in mice. (A) NSG mice transplanted with MDA-MB-231 LM2 /miR-181a sponge stable cells (n=5) or MDA-MB-231 LM2/vector control cells (n=5) were treated with Dox (2 mg/kg) or N.S when xenograft tumor volume reached ~100 mm3 as shown. Xenograft tumor growth was monitored and showed as the tumor volume. Arrow: Dox or N.S. treatment. (B) Bioluminescence imaging was performed at the end point of experiment to visualize tumor size. Quantified bioluminescent signals are shown as mean ± SEM on the right. **: p< 0.01. (C) Representative images of the xenograft tumors isolated from three indicated groups after treatment. (D) Pulmonary tumor outgrowth was monitored by bioluminescent imaging as in (B). *: p< 0.05; **: p< 0.01. (E) Approximate lung metastasized MDA-MB-231 cells were quantified by qPCR determination of human ALU loci in mouse lung tissues. (F) Representative images of IHC staining of BAX in xenograft tumor after treatments. The scale bar represents 20 μm. (G) Protein samples extracted from xenograft tumors were immunoblotted by antibodies against BAX, cleaved PARP, cleaved caspase-3, and tubulin. (H) Cleaved caspase-3 was detected with immunofluorescence staining in tumors from indicated group. Nuclei were visualized by Hoechst staining. The scale bar represents 20 μm. (I) A model depicting the role of miR-181a upregulation in modulating breast cancer cell response to chemotherapy.
To validate the negative correlation between miR-181a and BAX expression in breast cancers and the potential role of BAX in mediating TNBC cells response to chemotherapeutic drugs, we determined the miR-181a level in xenograft tumors with qPCR and BAX expression with immunohistochemistry. In agreement with the BAX-suppressive effect of miR-181a observed in cultured LM2 cells (supplementary Fig. S6D), we found Dox treatment significantly increased miR-181a level in LM2 xenograft tumors, which negatively correlated to BAX expression (Fig. 7F and supplementary Fig. S6E). Moreover, BAX expression was recovered by expression of miR-181a sponge inhibitor in LM2 cells. In line with these results, we found Dox-induced apoptosis was further enhanced in breast cancer cells expressing miR-181a sponge, which is associated with increased BAX expression in LM2 xenograft tumors (Fig. 7G, 7H). Collectively, these data suggest that BAX is a critical mediator of Dox-induced breast cancer cell apoptosis, whose downregulation by miR-181a may lead to tumor resistance to Dox treatment.
DISCUSSION
The potential significance of miRNAs in cancer therapeutic response and spontaneous/acquired resistance has been highlighted by recent progress in mechanistic studies as well as development of miRNA-based therapeutic approaches 11. Here we showed that induction of miR-181a played a critical role in mediating acquired resistance to chemotherapeutic drug Doxorubicin in TNBC cells. Moreover, the Dox-resistance also closely correlated with increased metastasis of TNBC cells, and inhibiting miR-181a sensitized TNBC cells to Dox treatment and reduced metastasis both in vitro and in vivo. Intriguingly, our mechanistic exploration revealed a pivotal role of transcription factor STAT3 in orchestrating miR-181a induction upon Dox treatment. Recruitment of STAT3 on MIR181A promoter in response to genotoxic treatment not only activated transcriptional machinery, it may also facilitate and/or stabilize binding of MSK1 on MIR181A promoter. The MIR181A promoter-bound MSK1 phosphorylated histone H3S28 and led to local depletion of transcription-suppressive marker H3K27me3, likely by displacing polycomb group protein complex in this region 28. The increased H3S28 phosphorylation and decreased H3K27 methylation may alter the chromatin conformation and facilitate STAT3-driven transactivation of miR-181a in breast cancer cells upon DNA damage (Fig. 7I).
Accumulating evidence suggests that binding of transcription factor may change the local chromatin state. It was reported that promoter-bound STAT3 facilitated histone methyl transferase SET9 recruitment to SOCS3 promoter region where it promoted transcription by methylating histone H3 on Lys4 (H3K4me) 34. Moreover, augmented STAT3 and STAT5 recruitment to the IL10 promoter and enhancer increased histone H3K18 acetylation by recruiting p300, which facilitated IL10 transactivation in T cells 35. In parallel, two recent studies showed that NF-κB could facilitate an active chromatin state by recruiting chromatin remodeling complex SWI/SNF to specific gene promoters 36, 37. In this study, we found MSK1 is required for STAT3 phosphorylation at Ser727 upon DNA damage and blocking STAT3-S727 phosphorylation diminished STAT3 enrichment on MIR181A promoter region. Our previous study indicated MSK1 was activated by genotoxic treatments in breast cancer cells in a manner dependent on ATM and MAPK14 16. Moreover, we detected phosphorylated STAT3 (pS727) in cytoplasmic fraction of MDA-MB-231 cells after Dox treatment. Therefore, following IL6R/JAK-dependent STAT3-Y705 phosphorylation, DNA damage-activated MSK1 may enhance STAT3 activation by phosphorylating STAT3-S727, which in turn translocated into the nucleus and bound to specific gene promoters such as MIR181A promoter. Promoter-bound STAT3 further induced epigenetic activation of MIR181A by enhancing MSK1 binding to the promoter and facilitating histone H3S28 phosphorylation. Interestingly, a recent genome-wide analysis revealed that H3S28ph marked around 50% of stress-induced genes, which was mostly enriched at promoters and 5’-UTRs 38. These finding suggest that STAT3-mediated H3S28ph may play a much broader role in regulating gene transcription in cells exposed to genotoxic stress, which warrants further investigation.
Distinct roles of miR-181a have been reported in regulating chemotherapeutic resistance depending on cancer types and drugs investigated. Induction of miR-181a upon cisplatin treatment was shown to enhance apoptosis in non-small cell lung cancer A549 cells 39. In breast cancer MCF7 cells, miR-181a was proposed to enhance therapeutic efficacy in mitoxantone-resistant cells by repressing breast cancer resistance protein (BCRP/ABCG2) 40. In contrast, miR-181a promoted chemoresistance to cisplatin in both cervical cancer cells and tongue squamous carcinoma cells, and PRKCD and Twist1, respectively, were identified as miR-181a targets responsible for cisplatin resistance 41, 42. In a recent study aiming to identify miRNAs correlating with resistance to anthracyclines or taxanes-based adjuvant chemotherapy in TNBC patients, both miR-181a and miR-181b were found to be upregulated in TNBC tumors and associated with chemoresistance 43. In agreement with these findings, we found miR-181a induction in TNBC MDA-MB-231 cells repressed apoptosis in response to Dox treatment, suggesting a resistance-promoting effect. The pro-apoptotic gene BAX was identified as a functional direct target of miR-181a in MDA-MB-231 cells whose downregulation may lead to increased resistance to Dox in TNBC cells. BAX and BAK are the effectors of Bcl-2 family to induce mitochondrial outer membrane permeabilization (MOMP) and subsequent cytochrome C release, which depends on their translocation and oligomerization on outer mitochondrial membrane upon stress stimulation. A recent study showed that inhibiting effectors, BAX/BAK per se, is more effective to suppress MOMP-induced apoptosis 44. These findings agree with our results that miR-181a-dependent suppression of BAX effectively reduced, whereas stabilizing BAX with miR-181a-sponge inhibitor enhanced, apoptosis in Dox-treated breast cancer cells. Although BAX suppression appeared to play a major role in mediating Dox-resistance, we cannot exclude that other miR-181a targets may also contribute to the acquired resistance to Dox in TNBC cells. For instance, BCL2L11 has been identified as a miR-181a target gene in mouse breast cancer cells treated with transforming growth factor (TGF)-β, whose downregulation promoted metastasis by inhibiting anoikis 45. Moreover, TGF-β-induced miR-181a was shown to suppress DNA damage apical kinase ATM in breast cancer cells thereby promoting the increase of breast cancer stem-like cell population 46, which has been linked with higher chemotherapeutic resistance.
Besides inhibiting pro-apoptotic genes such as BCL2L11 and BAX, miR-181a may also promote cancer metastasis by enhancing cancer cell motility and altering cancer microenvironment. In colorectal tumors from patients with liver metastases, miR-181a was found to be the most elevated miRNA compared to those patients without metastases 47. WIF1 was identified as a direct target gene of miR-181a which promoted colorectal cancer cell motility and invasion. Moreover, miR-181a was enriched in recurrent ovarian tumors compared to the matched-primary tumors, which may promote EMT by repressing its functional target Smad7 19. In addition, the effect of upregulated miR-181a may not be limited to cancer cells themselves, as miR-181a can be secreted into tumor microenvironment and blood/lymphatic circulation which may facilitate breast cancer cell local invasion as well as metastasis to remote sites. Accordingly, a recent study showed that miR-181a is one of predominant miRNAs present in exosomes isolated from serum from stage II and III breast cancer patients 48. Therefore, genotoxic therapy-induced upregulation of miR-181a may have profound impact on breast cancer migration and invasion via both autonomous and non-autonomous mechanisms, resulting in aggressive metastasis.
Our studies indicate that miR-181a upregulation in response to genotoxic treatment in breast cancer cells may serve as an important mechanism promoting acquired therapeutic resistance and aggressive metastasis. The findings that HER2+ breast cancer patients have a higher rate of miR-181a amplification and inhibiting miR-181a also sensitized breast cancer cells to HER2-targeted therapy imply that blocking miR-181a may not only improve the TNBC response to genotoxic chemotherapy, it may also enhance the efficacy of targeted therapy in HER2+ breast cancer patients. Therefore, developing miR-181a-targeting therapeutic approaches may hold high promise to sensitize breast cancers to both conventional chemotherapy and targeted therapy by simultaneously affecting multiple cellular processes involved in therapeutic resistance and metastasis.
MATERIALS AND METHODS
Cell Culture
MDA-MB-231, HCC1937, BT474, PC3 and HEK293 cell lines were obtained from ATCC. Cells resuscitated from original passage and passaged within 6 months were used in all experiments. ATCC cell lines were characterized by Short Tandem Repeat (STR) profiling. HCT 116 and HCT 116 Dicer−/− cells were a generous gift from Dr. Bert Vogelstein (Johns Hopkins University). PC3 cells stably expressing STAT3-WT or -Y705F mutant has been reported previously 25. PC3 cells stably expressing STAT3-S727A was generated by transfecting PC3 cells with respective construct followed with G418 selection. All the cells were cultured with their recommended conditions.
Patient cohort
A series of clinical data from 62 TNBC patients was collected at FUSCC (Supplemental Table 1). Tumors with immunohistochemical (IHC) phenotypes of ER-, PR- and HER2- were included. The protocol was reviewed and approved by an independent institutional review board at Fudan University, and all patients gave their written informed consent before inclusion in this study. All eligible patients proceeded to surgery between January 1, 2009 and December 31, 2010. Post-operative chemotherapy and radiation therapy were suggested according to breast cancer guidelines of FUSCC. Follow-up for these patients was completed on Jun 30, 2014. Tumor tissue was obtained from patients undergoing surgery and immediately stored at −80°C.
Chromatin immunoprecipitation (ChIP)
ChIP assays were carried out as previously described 16. In brief, cells were cross-linked with 1% formaldehyde, sonicated and then immunoprecipitated with antibodies against STAT3, MSK1, H3, H3K27me3, H3S28ph, or pS727-STAT3 suspended in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1). The ChIP-PCR primers were designed to amplify the promoter regions containing respective binding sites in the miR-181a promoter. For ChIP-reChIP experiments, reChIP buffer (Dilution Buffer, 10mM DTT) was added to beads after 1st IP following washes and incubated at 37°C for 50 minutes. The sample was then diluted 40 times in dilution buffer, and subjected to 2nd IP.
Animal Studies
Female NOD scid gamma (NSG) mice (age 6 wk) were maintained in the UTHSC animal facility. All animal studies were conducted in accordance with NIH animal use guidelines and a protocol approved by UTHSC IACUC. Power analysis was performed with G*Power online analysis freeware, using repeated-measures ANOVA with between-subjects factors. 1 × 106 MDA-MB-231 LM2 parental or sponge-miR181a stable cells were injected into mice mammary fat pad (2/each mice). 10 mice injected with each cell line were randomly divided into 2 groups (5/group) with total of 4 groups. Group assignment and tumor monitoring were carried out double-blinded. Tumor volume was assessed by caliper measurement using the formula (width2× length/2) (mm3) and mice were imaged by Xenogen IVIS system (Caliper Life Sciences; Alameda, CA). When palpable tumors reached ~100 mm3, the mice were treated with intraperitoneal injection of Doxorubicin (2mg/kg) or Saline once a week. After four cycles, the mice were sacrificed and primary tumors were isolated for further experiments. The right lobes of the lungs were digested to prepare DNA using QIAamp genomic DNA kit (QIAGEN) according to the manufacturer's instruction. Purified DNA was subjected to qPCR analysis using primers specific for the human Alu sequences, as described previously 49.
Statistical Analysis
The results were presented as Mean ± SD, and analyzed with one-way ANOVA and Student's t-test. Disease free survival analysis was estimated by the Kaplan-Meier method. The Spearman rank test was used for correlation analysis. All statistical analyses were performed using SPSS 22.0 software (SPSS Inc). P<0.05 was denoted as statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Bert Vogelstein (Johns Hopkins University) for wild type and DICER−/− HCT116 cells. The BAX expression construct was a generous gift from Dr. Douglas Green (St. Jude Children's Research Hospital). We also thank Dr. R. Laribee (UTHSC) for stimulating discussion and critical reading of the manuscript. This work was supported in part by grants from National Cancer Institute (R01 CA149251 to ZW), American Cancer Society (RSG-13-186-01-CSM to ZW), and National Natural Science Foundation of China (NSFC) (81272924 to JW).
Financial support: This work was supported in part by grants from National Cancer Institute (R01CA149251 to ZW), American Cancer Society (RSG-13-186-01-CSM to ZW), and National Natural Science Foundation of China (NSFC) (81272924 to JW),
Footnotes
Conflicts of interest: The authors disclose no potential conflicts of interest.
REFERENCES
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-Negative Breast Cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 3.Wu ZH, Miyamoto S. Many faces of NF-kappaB signaling induced by genotoxic stress. J Mol Med. 2007;85:1187–1202. doi: 10.1007/s00109-007-0227-9. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin AS. Regulation of cell death and autophagy by IKK and NF-κB: critical mechanisms in immune function and cancer. Immunol Rev. 2012;246:327–345. doi: 10.1111/j.1600-065X.2012.01095.x. [DOI] [PubMed] [Google Scholar]
- 5.Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-κB. Immunol Rev. 2012;246:205–220. doi: 10.1111/j.1600-065X.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garofalo M, Croce CM. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat. 2013;16:47–59. doi: 10.1016/j.drup.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato M, Paranjape T, Muller RU, Nallur S, Gillespie E, Keane K, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhry MA, Sachdeva H, Omaruddin RA. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010;29:553–561. doi: 10.1089/dna.2009.0978. [DOI] [PubMed] [Google Scholar]
- 14.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Wan G, Berger FG, He X, Lu X. The ATM Kinase Induces MicroRNA Biogenesis in the DNA Damage Response. Mol Cell. 2011;41:371–383. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu J, Shi Y, Tan G, Yang CH, Fan M, Pfeffer LM, et al. DNA damage induces NF-kappaB-dependent microRNA-21 upregulation and promotes breast cancer cell invasion. J Biol Chem. 2012;287:21783–21795. doi: 10.1074/jbc.M112.355495. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Tan G, Niu J, Shi Y, Ouyang H, Wu Z-H. NF-κB-dependent microRNA-125b upregulation promotes cell survival by targeting p38α upon UV radiation. J Biol Chem. 2012;287:33036–33047. doi: 10.1074/jbc.M112.383273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCool KW, Miyamoto S. DNA damage-dependent NF-kappaB activation: NEMO turns nuclear signaling inside out. Immunol Rev. 2012;246:311–326. doi: 10.1111/j.1600-065X.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parikh A, Lee C, Joseph P, Marchini S, Baccarini A, Kolev V, et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial–mesenchymal transition. Nat Commun. 2014;5:2977. doi: 10.1038/ncomms3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickey CJ, Schwind S, Radomska HS, Dorrance AM, Santhanam R, Mishra A, et al. Lenalidomide-mediated enhanced translation of C/EBPα-p30 protein up-regulates expression of the antileukemic microRNA-181a in acute myeloid leukemia. Blood. 2013;121:159–169. doi: 10.1182/blood-2012-05-428573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enerly E, Steinfeld I, Kleivi K, Leivonen S-K, Aure MR, Russnes HG, et al. miRNA-mRNA Integrated Analysis Reveals Roles for miRNAs in Primary Breast Tumors. PLoS ONE. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riaz M, van Jaarsveld MT, Hollestelle A, Prager-van der Smissen WJ, Heine AA, Boersma AW, et al. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res. 2013;15:R33. doi: 10.1186/bcr3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu J, Shi Y, Xue J, Miao R, Huang S, Wang T, et al. USP10 inhibits genotoxic NF-kappaB activation by MCPIP1-facilitated deubiquitination of NEMO. EMBO J. 2013;32:3206–3219. doi: 10.1038/emboj.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sansone P, Bromberg J. Targeting the Interleukin-6/Jak/Stat Pathway in Human Malignancies. J Clin Oncol. 2012;30:1005–1014. doi: 10.1200/JCO.2010.31.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang CH, Yue J, Fan M, Pfeffer LM. IFN Induces miR-21 through a Signal Transducer and Activator of Transcription 3–Dependent Pathway as a Suppressive Negative Feedback on IFN-Induced Apoptosis. Cancer Research. 2010;70:8108–8116. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakahara R, Kunimoto H, Tanino K, Kojima H, Inoue A, Shintaku H, et al. Phospho-Ser727 of STAT3 regulates STAT3 activity by enhancing dephosphorylation of phospho-Tyr705 largely through TC45. Genes Cells. 2012;17:132–145. doi: 10.1111/j.1365-2443.2011.01575.x. [DOI] [PubMed] [Google Scholar]
- 27.Baek Sung H. When Signaling Kinases Meet Histones and Histone Modifiers in the Nucleus. Mol Cell. 2011;42:274–284. doi: 10.1016/j.molcel.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 28.Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb Group Protein Displacement and Gene Activation through MSK-Dependent H3K27me3S28 Phosphorylation. Mol Cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 29.Lau PN, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci U S A. 2011;108:2801–2806. doi: 10.1073/pnas.1012798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang Y-B, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion. 2012;12:213–219. doi: 10.1016/j.mito.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr., Sjoblom T, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Györffy B, Lanczky A, Eklund A, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Research and Treatment. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 33.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedrich CM, Rauen T, Apostolidis SA, Grammatikos AP, Rodriguez Rodriguez N, Ioannidis C, et al. Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc Natl Acad Sci U S A. 2014;111:13457–13462. doi: 10.1073/pnas.1408023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonnay F, Nguyen XH, Cohen - Berros E, Troxler L, Batsche E, Camonis J, et al. Akirin specifies NF-κB selectivity of Drosophila innate immune response via chromatin remodeling. EMBO J. 2014 doi: 10.15252/embj.201488456. 10.15252/embj.201488456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tartey S, Matsushita K, Vandenbon A, Ori D, Imamura T, Mino T, et al. Akirin2 is critical for inducing inflammatory genes by bridging I κ B - ζ and the SWI/SNF complex. EMBO J. 2014 doi: 10.15252/embj.201488447. 10.15252/embj.201488447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawicka A, Hartl D, Goiser M, Pusch O, Stocsits RR, Tamir I, et al. H3S28 phosphorylation is a hallmark of the transcriptional response to cellular stress. Genome Res. 2014 doi: 10.1101/gr.176255.114. 10.1101/gr.176255.176114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, et al. miR-181a and miR-630 Regulate Cisplatin-Induced Cancer Cell Death. Cancer Research. 2010;70:1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 40.Jiao X, Zhao L, Ma M, Bai X, He M, Yan Y, et al. MiR-181a enhances drug sensitivity in mitoxantone-resistant breast cancer cells by targeting breast cancer resistance protein (BCRP/ABCG2). Breast Cancer Res Treat. 2013;139:717–730. doi: 10.1007/s10549-013-2607-x. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Ke G, Han D, Liang S, Yang G, Wu X. MicroRNA-181a enhances the chemoresistance of human cervical squamous cell carcinoma to cisplatin by targeting PRKCD. Exp Cell Res. 2014;320:12–20. doi: 10.1016/j.yexcr.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 42.Liu M, Wang J, Huang H, Hou J, Zhang B, Wang A. miR-181a–Twist1 pathway in the chemoresistance of tongue squamous cell carcinoma. Biochem Biophys Res Commun. 2013;441:364–370. doi: 10.1016/j.bbrc.2013.10.051. [DOI] [PubMed] [Google Scholar]
- 43.Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, et al. MicroRNA Profiling Implies New Markers of Chemoresistance of Triple-Negative Breast Cancer. PLoS ONE. 2014;9:e96228. doi: 10.1371/journal.pone.0096228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Llambi F, Moldoveanu T, Tait Stephen WG, Bouchier-Hayes L, Temirov J, McCormick Laura L, et al. A Unified Model of Mammalian BCL-2 Protein Family Interactions at the Mitochondria. Mol Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123:150–163. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Yu Y, Tsuyada A, Ren X, Wu X, Stubblefield K, et al. Transforming growth factor-[beta] regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30:1470–1480. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji D, Chen Z, Li M, Zhan T, Yao Y, Zhang Z, et al. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer. 2014;13:86. doi: 10.1186/1476-4598-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou W, Fong Miranda Y, Min Y, Somlo G, Liu L, Palomares Melanie R, et al. Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan M, Krutilina R, Sun J, Sethuraman A, Yang CH, Wu Z-h, et al. Comprehensive Analysis of MicroRNA (miRNA) Targets in Breast Cancer Cells. J Biol Chem. 2013;288:27480–27493. doi: 10.1074/jbc.M113.491803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.