Abstract
An enhanced capacity for de novo lipid synthesis is a metabolic feature of most cancer cells that distinguishes them from their cells of origin. However, the mechanisms through which oncogenes alter lipid metabolism are poorly understood. We find that expression of oncogenic PI3K (H1047R) or K-Ras (G12V) in breast epithelial cells is sufficient to induce de novo lipogenesis, and this occurs through the convergent activation of the mechanistic target of rapamycin complex 1 (mTORC1) downstream of these common oncogenes. Oncogenic stimulation of mTORC1 signaling in this isogenic setting or a panel of eight breast cancer cell lines leads to activation of the sterol regulatory element-binding proteins (SREBP1 and SREBP2), which are required for oncogene-induced lipid synthesis. The SREBPs are also required for the growth factor-independent growth and proliferation of oncogene-expressing cells. Finally, we find that elevated mTORC1 signaling is associated with increased mRNA and protein levels of canonical SREBP targets in primary human breast cancer samples. These data suggest that the mTORC1-SREBP pathway is a major mechanism through which common oncogenic signaling events induce de novo lipid synthesis to promote aberrant growth and proliferation of cancer cells.
Keywords: Akt, Erk, FASN, mTOR, PIK3CA, SCD
Introduction
The genetic events underlying cancer development are accompanied by the induction of a metabolic program, distinct from most normal cells, that facilitates the uncontrolled growth of cancer cells. However, the key molecular connections between the most commonly activated oncogenic pathways in human cancers and this metabolic reprogramming are poorly defined. One long-known, but poorly understood, alteration in cellular metabolism frequently observed in cancer is the activation of de novo lipid synthesis1, a process that only minimally contributes to the lipid content of normal non-proliferating cells. While normal cells generally rely on the uptake of lipids from the circulation, cancer cells often acquire the ability to make their own, which is believed to be required to meet an increased demand for membrane biogenesis during cell proliferation2,3.
The expression of genes encoding lipogenic enzymes, including acetyl-CoA carboxylase (ACACA), fatty acid synthase (FASN) and steroyl-CoA desaturase (SCD), has been found to be elevated in a variety of cancers2,4,5. In normal, lipid-producing tissues, such as the liver, these and most other enzymes involved in de novo sterol and fatty acid synthesis are induced by the sterol regulatory element (SRE) binding protein (SREBP) family of transcription factors, SREBP1 and 26. The SREBPs are produced as inactive precursors, which reside as transmembrane proteins in the endoplasmic reticulum (ER)7–11. When sterols or unsaturated fatty acids become depleted, the membrane-bound SREBP traffics to the Golgi, where it is sequentially cleaved by two site-specific proteases. The N-terminal fragment of SREBP, representing the active transcription factor (referred to as the mature form), is released and can enter the nucleus to activate target genes with SREs in their promoters. Through transcriptional activation of its lipogenic target genes, SREBP is able to induce the de novo synthesis of sterols, fatty acids, and their neutral lipid derivatives.
In addition to its regulation by lipids, SREBP isoform processing and activation have been found to be stimulated by insulin and growth factor signaling through mTORC1 (ref. 12). Activation of mTORC1 signaling induces SREBP activation in cell culture models and in the liver, leading to the accumulation of mature processed SREBP, expression of SREBP target genes, and increased de novo lipid synthesis13–16. The molecular mechanism by which mTORC1 activates SREBP remains unknown but likely involves multiple direct downstream targets. Independent groups have shown that mTORC1 can promote SREBP processing through the mTORC1-regulated protein kinase S6K1 in various settings13,17–20. 4E-BP1, an inhibitor of cap-dependent translation that is blocked by mTORC1 signaling, has also been implicated in the regulation of SREBP downstream of mTORC1 (ref. 18,21). In addition, phosphorylation of the phosphatidic acid phosphatase Lipin1 by mTORC1 has been shown to promote accumulation of mature SREBP in the nucleus through an unknown mechanism22. An important feature of mTORC1 signaling that influences studies on its regulation of SREBP is that the downstream targets of mTORC1 are differentially sensitive to mTOR inhibitors. S6K1 phosphorylation and activation is completely inhibited by rapamycin, while 4E-BP1 and Lipin1 phosphorylation and inhibition are only partially sensitive to rapamycin22–24. As such, it is useful to use both rapamycin, an allosteric inhibitor of mTORC1, and mTOR kinase inhibitors, which completely inhibit both mTORC1 and mTORC2, in such studies.
In normal cells and tissues, mTORC1 activity is tightly controlled by growth factors through the convergence of multiple upstream signaling pathways on a protein complex comprised of the tuberous sclerosis complex (TSC) tumor suppressors, TSC1 and TSC2, and the TBC1D7 protein (the TSC complex)25,26. The TSC complex acts as a GTPase-activating protein (GAP) for Rheb, a Ras-related small G-protein that potently activates mTORC1 when it is GTP-bound27. While loss of function mutations affecting the TSC complex lead to growth factor-independent activation of mTORC1 and are the genetic cause of the tumor syndromes TSC and lymphangioleiomyomatosis (LAM)28, mutations in the complex components are more rare in sporadic cancers. Nonetheless, aberrant activation of mTORC1 is a frequent event in human cancers, across nearly all lineages29. Two of the most commonly activated pathways in cancer, the PI3K-Akt and the Ras-Erk pathways, converge on the TSC complex to activate mTORC130–34.
Here, we find that expression of oncogenic PI3K or K-Ras in normal cells induces de novo lipogenesis and that inhibition of mTORC1 or depletion of the SREBPs blocks this induction. We also find that this is a primary mechanism driving lipid synthesis in a panel of genetically-defined breast cancer lines. We find that depletion of the SREBPs hinders the viability and growth of cells with oncogenic activation of mTORC1 signaling. Lastly, we show an association between mTORC1 activation and expression of lipogenic targets of SREBP in primary human breast cancer samples. These findings identify the mTORC1-SREBP pathway as a major molecular link between oncogenic signaling events and the common increase in de novo lipid synthesis observed in human cancers.
Results
Oncogenic PI3K and K-Ras are sufficient to induce de novo lipid synthesis and do so in an mTORC1-dependent manner
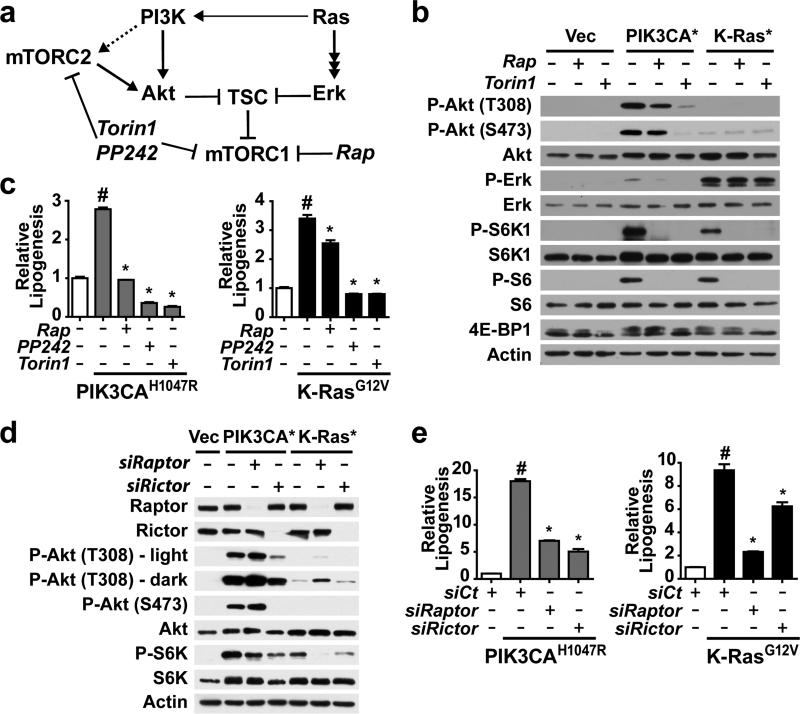
Since both the PI3K and Ras pathways are frequently activated in cancer and converge to activate mTORC1 (Figure 1a), we asked whether activating mutants commonly found in human cancer (PIK3CAH1047R and K-RasG12V) were sufficient to stimulate lipogenesis. We generated an isogenic set of cell lines stably expressing either empty vector or one of these two oncogenic mutants in MCF10a cells, a non-transformed human breast epithelium cell line. The oncogene-expressing cells exhibited growth-factor independent activation of mTORC1, as detected by phosphorylation of its downstream targets, S6K1 and 4E-BP1, and the S6K target ribosomal S6, which were sensitive to rapamycin and the mTOR kinase inhibitor Torin1 (Figure 1b). Consistent with these oncogenes activating mTORC1 primarily through distinct pathways, the PIK3CAH1047R cells displayed activated Akt but not Erk, whereas the K-RasG12V cells had activated Erk with minimal activation of Akt. To measure specific effects on de novo lipid synthesis, cells were labeled with 14C-acetate in order to avoid established effects of these oncogenes on glucose uptake. Importantly, both oncogenic PI3K and K-Ras stimulated an increase in the incorporation of acetate into lipid (Figure 1c). Rapamycin significantly reduced this oncogene-induced lipogenesis, and treatment with two structurally distinct mTOR kinase inhibitors, PP242 (ref. 23) or Torin1 (ref. 24) led to a further reduction.
Figure 1.
Oncogenic PI3K and K-Ras promote de novo lipogenesis through mTORC1 activation. (a) Model of the convergent regulation of mTORC1 through the PI3K and K-Ras pathways and the action of different classes of mTOR inhibitors. (b) Growth-factor independent activation of mTORC1 signaling by oncogenes. MCF10a cells stably expressing empty vector, PIK3CAH1047R, or K-RasG12V were serum starved for 16 h in the presence of vehicle, rapamycin (20 nM), or Torin1 (250 nM). Immunoblots are of proteins and phosphorylated (P) proteins in the cytosolic fraction, with phosphorylation of 4EBP1 detected by mobility shift. (c) Oncogene and mTORC1-dependent induction of de novo lipid synthesis in MCF10a cells. Incorporation of 1-[14C]-acetate into the lipid fraction was measured in the cells from a in the presence of vehicle, rapamycin (20 nM), PP242 (2.5 μM), or Torin1 (250 nM). Representative data are shown as mean ± s.e.m. relative to vector-expressing cells (white bar), n=4. (d,e) Effects of Raptor and Rictor depletion on signaling and de novo lipogenesis. The cells in a were transfected with siRNAs targeting Raptor or Rictor. Cells were lysed 72 h post-transfection following 16 h serum starvation, to analyze signaling (d) or lipid synthesis, as measured and presented in b (e). Representative data are shown as mean ± s.e.m. relative to vector-expressing cells (white bar), n=3. (c,e) #P-value < 0.05 compared to vector-expressing cells; *P-value < 0.05 compared to vehicle-treated cells expressing the same oncogene.
To determine whether the effects of mTOR inhibitors on the induction of lipid synthesis by oncogenic PI3K and K-Ras were through inhibition of mTORC1 or mTORC2 (Figure 1a), siRNAs targeting either mTORC1, through Raptor knockdown, or mTORC2, through Rictor knockdown, were introduced into these cells (Figure 1d). In both oncogene-expressing lines, Raptor knockdown decreased S6K1 phosphorylation. Rictor knockdown had the strongest effect on S6K1 phosphorylation in the PIK3CAH1047R cells, where the mTORC2 target Akt lies upstream of mTORC1. Importantly, de novo lipogenesis in the oncogene-expressing cells mirrored effects of these siRNAs on mTORC1 signaling, with Raptor knockdown inhibiting in both lines and Rictor knockdown having more pronounced effects in the PIK3CAH1047R cells (Figure 1e). Therefore, oncogenic PI3K and K-Ras are sufficient to stimulate de novo lipid synthesis and do so through the common downstream activation of mTORC1.
Activation of SREBP downstream of mTORC1 is required for oncogene-induced lipid synthesis
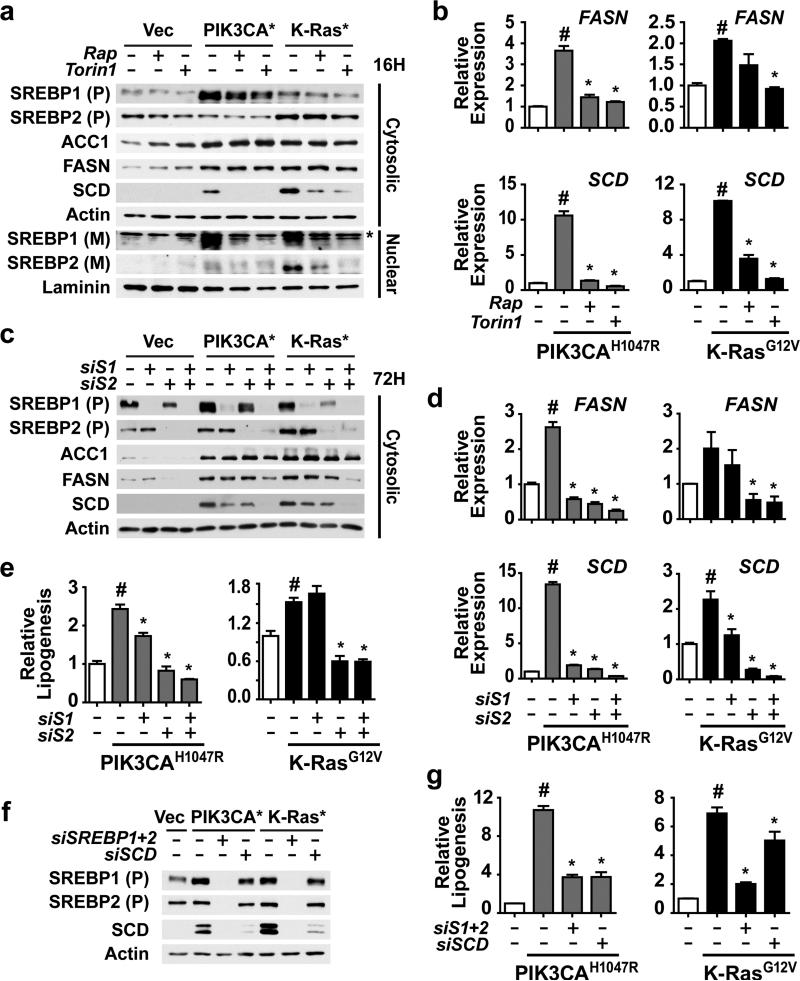
Given that mTORC1 has been found previously to stimulate de novo lipid synthesis through activation of the SREBP transcription factors in other settings13,14, we assessed the effects of oncogenic PI3K and K-Ras on SREBP isoforms and canonical gene targets. Following cellular fractionation to detect the cytosolic, inactive precursor (P) and nuclear, active mature (M) forms of SREBP1 and 2, we found that levels of the SREBP1 precursor were modestly elevated in the oncogene-expressing lines, whereas the SREBP2 precursor was increased only in the oncogenic K-Ras cells (Figure 2a). These increases matched closely with differential changes in the transcript levels of the genes encoding these proteins, SREBF1 and SREBF2 (Figure S1a). Consistent with previous findings suggesting that mTORC1 signaling promotes the processing of SREBP13,20, the nuclear mature forms of both SREBP1 and 2 were elevated in the oncogene-expressing cells, and treatment with either rapamycin or Torin1 blocked this increase (Figure 2a). The oncogene-mediated activation of SREBP was reflected in elevated transcript (Figures 2b and S1a) and protein (Figure 2a) levels of three canonical SREBP targets, ACC1, FASN and SCD. While the transcript levels of ACC1, FASN and SCD were all sensitive to mTORC1 inhibitors, only SCD was decreased at the protein level following overnight treatment, reflecting the long-lived nature of the ACC1 and FASN proteins35,36.
Figure 2.
Activation of SREBP1 and SREBP2 by mTORC1 is required for PI3K- and K-Ras-induced de novo lipogenesis. (a) Regulation of SREBP isoforms by oncogenes and mTORC1. MCF10a cells stably expressing empty vector, PIK3CAH1047R,or K-RasG12V were serum starved for 16 h in the presence of vehicle, rapamycin (20 nM), or Torin1 (250 nM). Cytosolic and nuclear fractions were collected to detect the cytosolic precursor (P) and the nuclear mature (M) forms of SREBP1 and 2. * denotes a cross-reacting band. (b) Oncogene and mTORC1-dependent induction of FASN and SCD expression. RNA was isolated from cells treated as in a for analysis by qRT-PCR. Representative data are shown as mean ± s.e.m. relative to vector-expressing cells (white bar), n=2. (c-e) Effects of SREBP1 and SREBP2 knockdown on SREBP targets and de novo lipogenesis. The cells in a were transfected with siRNAs targeting SREBP1, SREBP2, or both. Cells were lysed 72 h post-transfection following 20 h serum starvation for immunoblotting of the cytosolic fraction (c) or RNA extraction for qRT-PCR analysis (d). Representative data are shown as mean ± s.e.m. relative to vector-expressing cells (white bar), n=3. (e) Incorporation of 1-[14C]-acetate into the lipid fraction was measured in these cells, with data shown as mean ± s.e.m. relative to vector-expressing cells (white bar), n=2. (f,g) Effects of SREBP and SCD knockdown on lipogenesis. The cells in a were transfected with siRNAs targeting SREBP1 and SREBP2, or SCD. Cells were lysed as in c for immunoblotting (f). Incorporation of 1-[14C]-acetate into the lipid fraction was measured in these cells (g). Data are shown as mean ± s.e.m. relative to vector-expressing cells (white bar), n=2. (b,d,e) # P-value < 0.05 compared to vector-expressing cells; * P-value < 0.05 compared to vehicle-treated cells expressing the same oncogene.
To determine whether the oncogene-induced increase in expression of lipogenic genes and stimulation of de novo lipid synthesis were through this mTORC1-dependent activation of SREBP isoforms, we used siRNAs to knock down SREBP1, SREBP2, or both. Consistent with redundant regulation of lipogenic gene targets by SREBP1 and 2, the double knockdown resulted in the strongest decrease in FASN and SCD protein (Figure 2c) and transcript levels (Figure 2d). While the transcript levels of ACC1 were affected by SREBP knockdown, especially in the PIK3CAH1047R cells (Figure S1b), ACC1 protein levels were largely unaffected (Figure 2c). As observed in previous studies13,37,38, we found that SREBP2 knockdown, with siRNA sequences that do not directly target SREBP1, decreases SREBF1 transcript levels (Figure S1b), and this is also reflected in SREBP1 protein levels (Figure 2c). This cross-regulation likely reflects the presence of functional SREs in the SREBF1 gene promoter39. Importantly, as with mTORC1 inhibitors, oncogene-induced de novo lipogenesis was eliminated with siRNA-mediated knockdown of SREBP isoforms, with SREBP2 playing the dominant role in these cells (Figure 2e).
Given that treatment with mTOR inhibitors for 16 h is sufficient to block oncogene-driven lipogenesis (Figure 1c), but only SCD protein levels, not ACC1 or FASN, are decreased in this time frame (Figure 2a), we determined whether inhibition of SCD could explain these inhibitory effects on lipogenesis. Indeed, siRNA-mediated knockdown of SCD significantly reduced de novo lipogenesis, with a more pronounced effect in the PI3KH1047R cells, where the decrease mimicked SREBP1/2 knockdown (Figures 2f and 2g). Therefore, oncogenic PI3K and K-Ras can stimulate lipogenesis through the mTORC1-mediated activation of SREBP and its subsequent induction of SCD, the enzyme responsible for generating the mono-unsaturated fatty acids (MUFAs) prevalent in membrane phospholipids.
mTORC1 and SREBP drive de novo lipid synthesis in breast cancer cells
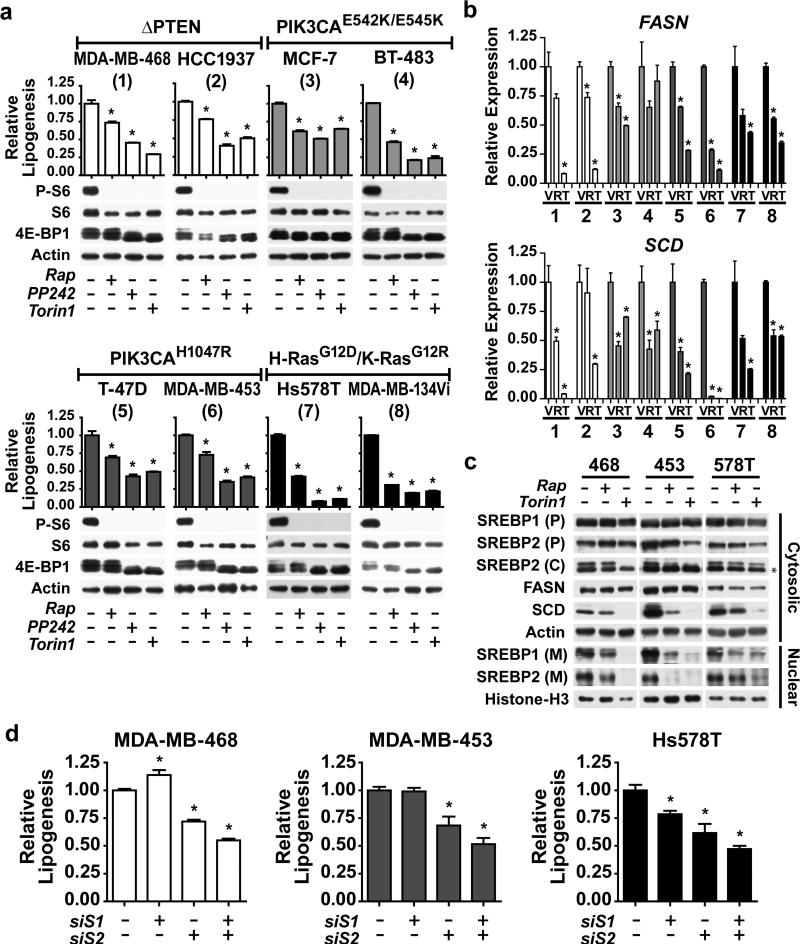
To determine whether mTORC1 signaling also promotes de novo lipid synthesis in the more complex genetic setting of cancer cells, we profiled a panel of eight genetically-defined breast cancer cell lines of luminal and basal subtype that, among other mutations, have oncogenic activation of the PI3K or Ras signaling pathways (Table 1). Consistent with the presence of these mutations leading to activation of the upstream pathways, all eight cell lines displayed growth-factor independent activation of mTORC1 signaling, which was sensitive to rapamycin, PP242 or Torin1 (Figure 3a). De novo lipogenesis was also constitutively active in these cells, and was significantly reduced by rapamycin in all cases, with mTOR kinase inhibitors leading to a further reduction in some lines (Figure 3a). Inhibition of mTOR decreased the transcript levels of SREBF1, SREBF2, FASN, and SCD in these lines (Figures 3b and S2a). Similar to the oncogene-expressing MCF10a cells, mTOR inhibitors had little effect on levels of SREBP precursors, but the mature forms of both SREBP1 and 2 were reduced by rapamycin and Torin1 in three representative lines from this panel (Figure 3c). As a control, we confirmed that the SREBP precursor and mature forms detected by these antibodies are reduced by siRNA-mediated knockdown of SREBP1 and 2 in these cells (Figure S2b). SCD protein levels were also reduced by mTOR inhibitors (Figure 3c), whereas a substantial decrease in FASN levels were not observed until 96 h of treatment (Figure S2c), despite effects on its transcript levels at 18 h (Figure 3b).
Table 1.
Breast cancer cell lines used in this study with known mutations upstream of mTORC1.
| Cell Line | Mutations | Subtypea |
|---|---|---|
| MDA-MB-468 | PTEN p.V85_splice | Basal |
| HCC1937 | PTEN del/del | Basal |
| MCF-7 | PIK3CA E545K | Luminal |
| BT-483 | PIK3CA E542K | Luminal |
| T-47D | PIK3CA H1047R | Luminal |
| MDA-MB-453 | PIK3CA H1047R (PTENb) | Luminal |
| Hs578T | HRAS G12D | Basal |
| MDA-MB-134-VI | KRAS G12R | Luminal |
Subtype is based on the gene expression signature published by Neve et al.6
PTEN mutation with undefined effect.
Figure 3.
Breast cancer lines depend on mTORC1 and its activation of SREBP for de novo lipogenesis. (a) Signaling and de novo lipogenesis in response to mTOR inhibition. Eight breast cancer cell lines were serum starved for 18 h in the presence of vehicle, rapamycin (20 nM), PP242 (2.5 μM), or Torin1 (250 nM). Note: phosphorylation of S6 and, via mobility shifts, 4E-BP1 are shown as markers of mTORC1 activation. Incorporation of 1-[14C]-acetate into the lipid fraction was measured. Representative data are shown as mean ± s.e.m. relative to vehicle-treated cells, n=2. * P-value < 0.05 compared to vehicle-treated cells. (b) mTORC1-dependent FASN and SCD expression in breast cancer cells. The cell lines, numbered as in a, were serum starved for 18 h in the presence of vehicle, rapamycin (20 nM), or Torin1 (250 nM). RNA was isolated for analysis by qRT-PCR, with transcript levels shown as mean ± s.e.m. relative to vehicle-treated cells. * P-value < 0.05 compared to vehicle-treated cells. (c) Dependence of SREBP processing on mTORC1 in breast cancer cells. MDA-MB-468, MDA-MB-453 and Hs578T were treated as b and fractionated into nuclear and cytosolic fractions for immunoblotting. The SREBP full-length precursor (P), processed C-terminus (C) and nuclear mature (M) isoforms were detected. * denotes a cross-reacting band. (d) SREBP knockdown attenuates de novo lipogenesis in breast cancer cells. The cells in c were transfected with siRNAs targeting SREBP1, SREBP2, or both. Cells were lysed 72 h post-transfection following 16 h serum starvation to analyze lipid synthesis as in a. Data are shown as mean ± s.e.m. relative to cells transfected with non-targeting siRNA, n=2. * P-value < 0.05.
To determine the role of SREBP in the induction of lipogenesis in breast cancer cells, SREBP1, SREBP2, or both were knocked-down using siRNAs in these three lines, which we confirmed by measuring SREBF1, SREBF2, and SCD transcript levels (Figure S3). Depletion of SREBP2 alone or in combination with SREBP1 attenuated the ability of these cells to synthesize lipids de novo (Figure 3d). Therefore, aberrant mTORC1 signaling and its activation of SREBP isoforms underlie the lipogenic property of heterogeneous breast cancer cell lines.
The SREBPs support oncogene-induced cell proliferation and growth
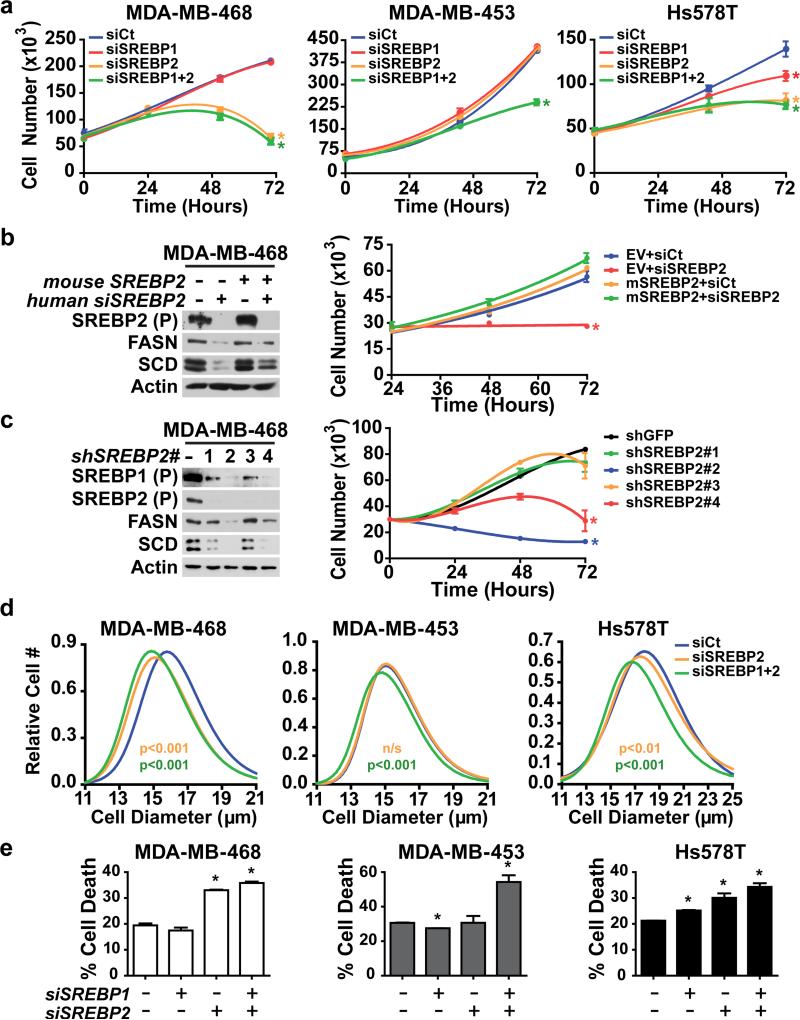
We next determined the importance of downstream activation of SREBP on the growth properties of breast cancer cells with activated mTORC1. The proliferation of MDA-MB-468, MDA-MB-453, and Hs578T grown in full serum was significantly reduced by combined knockdown of SREBP1 and 2, with the Hs578T cells being equally sensitive to SREBP2 knockdown alone (Figure S4). To determine the influence of exogenous lipids on these responses, proliferation was also measured in lipid-depleted serum. While the overall responses to SREBP isoform knockdown were similar, reducing serum lipids greatly sensitized the MDA-MB-468 cells to the knockdown of SREBP2 (Figure 4a). Exogenous expression of mouse SREBP2, which is resistant to the siRNAs targeting human SREBP2 and, thereby, restores FASN and SCD expression, rescued the inhibitory effects of SREBP2 knockdown on the proliferation of these cells (Figure 4b). As further confirmation of the specificity of these effects, four different shRNA sequences targeting SREBP2 were tested. The inhibition of proliferation with these shRNAs closely matched the degree of decrease in FASN and SCD protein levels elicited by the individual shRNAs (Figure 4c). The knockdown of SREBP isoforms also led to a reduction in cell size, with the double knockdown having the strongest effect in all three cell lines tested (Figure 4d). The differential effects of SREBP1 and SREBP2 depletion on the proliferation and growth of these cell lines were also observed for measurements of cell death, with the double knockdown most strongly decreasing viability (Figure 4e). Therefore, SREBP plays a key role in supporting cell growth, proliferation and survival in these breast cancer cells.
Figure 4.
The SREBPs support proliferation, growth, and survival in breast cancer cell lines. (a) Effect of SREBP1 and SREBP2 depletion on proliferation in breast cancer cells. MDA-MB-468, MDA-MB-453 and Hs578T cells were transfected with siRNAs targeting SREBP1, SREBP2, or both and were switched to lipid-reduced serum 24 h after the knockdown (t = 0 h). For all proliferation graphs, data are shown as mean ± s.e.m., n=3. * P-value < 0.05 compared to control cells at the final time point. (b) Rescue of human SREBP2 knockdown with mouse SREBP2 expression. MDA-MB-468 cells stably expressing mouse SREBP2 were transfected with siRNA targeting human SREBP2. Cells were lysed for immunoblotting 72 h post-transfection, following 16 h serum starvation. To measure proliferation, cells were cultured in lipid-reduced serum and counted every 24 h. (c) Effects of SREBP2 shRNA on SREBP target expression and proliferation. MDA-MB-468 cells stably expressing four different shRNA sequences targeting SREBP2 were either serum starved for 16 h for immunoblot analysis or cultured in lipid-reduced serum to measure proliferation. (d) Effects of SREBP knockdown on cell size. Cell diameter was measured at 48 h in cells from a, in solution. Color-coded P-values, compared to cells with control siRNAs, correspond to the color-coding in the legend (>1000 cells measured for each). (e) Effect of SREBP knockdown on breast cancer cell viability. Percent cell death was determined by counting Annexin-V and/or propidium iodide positive cells treated as in a by flow cytometry 72 h after siRNA transfection. Data are shown as mean ± s.e.m. relative to cells transfected with non-targeting siRNA, n=2. * P-value < 0.05.
To better define the cellular role of SREBP as a downstream effector of oncogenic signaling pathways, we compared the PIK3CAH1047R and K-RasG12V-expressing MCF10a cells to the isogenic vector-expressing cells. When grown in full serum, the proliferation rate of the vector control cells was comparable to the oncogene-expressing cells (Figure 5a), and knocking down SREBP isoforms, particularly SREBP2, only modestly reduced proliferation in the oncogene-expressing lines (Figure 5b). To better distinguish between the control cells, which exhibit growth-factor dependent mTORC1 activation, and the oncogene-expressing lines, with constitutive mTORC1 activation (Figure 1b), proliferation was also measured in low serum and growth factor conditions, where both the PI3K and K-Ras cells exhibit a proliferation advantage over the controls (Figure 5c). Under these conditions, SREBP2 knockdown arrested the proliferation of both oncogene-expressing lines, an effect enhanced when combined with SREBP1 knockdown (Figure 5d). The oncogene-expressing cells also exhibited an increase in cell size relative to the control cells (Figure 5e), and their size was significantly reduced upon knockdown of SREBP2 alone or in combination with SREBP1 (Figure 5f). Collectively, these findings reveal a requirement for SREBP in the aberrant, growth factor-independent growth and proliferation of oncogene-expressing cells.
Figure 5.
Effects of oncogene expression and SREBP depletion on cell growth and proliferation in MCF10a cells. (a,c) Proliferation of PIK3CAH1047R- and K-RasG12V-expressing MCF10a cells compared to vector-expressing cells cultured in full growth medium (a) or in low serum conditions (c) starting 24 h post-knockdown (t = 0 h). For all proliferation graphs, time points are shown as mean ± s.e.m., n=3. * P-value < 0.05 compared to control cells at the final time point. (b,d) Effect of SREBP depletion on proliferation of oncogene-expressing MCF10a cells. Cells from a and c cultured in full serum (b) or low serum (d) were counted every 24 h following the siRNA-mediated knockdown of SREBP1, SREBP2, or both. (e,f) Oncogene- and SREBP-dependent effects on cell growth. The diameters of the cells treated as in c (e) and d (f) were measured at 48 h, following trypsinization. Color-coded P-values, compared to cells with control siRNAs, correspond to the color-coding in the legend (>1000 cells measured for each).
Association of mTORC1 activation with the expression of SREBP targets in human breast cancer
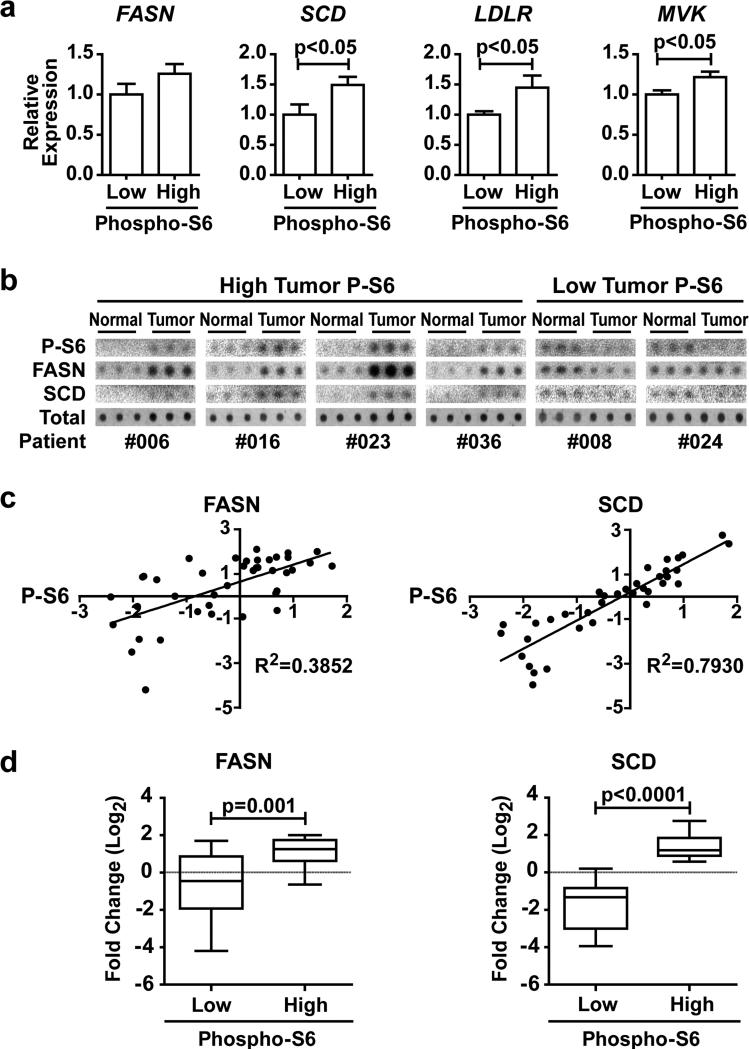
To determine whether the mTORC1-SREBP pathway is activated in human breast cancers, we analyzed the coupled gene expression and reverse phase protein array data from the breast invasive carcinoma dataset of The Cancer Genome Atlas (TCGA) 40–42. Using phosphorylation of ribosomal S6 (P-S6) as an indication of mTORC1 activation, the expression of canonical SREBP targets in cells with low and high mTORC1 signaling was compared. When compared to tumors with low P-S6 (n=116), those with high P-S6 (n=112) displayed increased expression of the SREBP target genes FASN, SCD, LDLR, and MVK (Figure 6a). To further assess a connection between mTORC1 activation and increased protein levels of SREBP targets, we used arrays of protein extracts from primary breast cancer samples comprised of matched pairs of tumor and adjacent normal breast tissue from each patient (n=40), comparing P-S6 levels to that of FASN and SCD. To validate the antibodies for this assay, we used dot blots of MDA-MB-468 cell lysates and confirmed the decreased signal in lysates from Torin1-treated cells (Figure S5). Examples of the array data with triplicate spots of normal and tumor tissue from each of 6 patients are shown in Figure 6b. An association between the fold change in P-S6 levels in the tumor samples relative to their matched normal tissue and the fold change in FASN and, especially, SCD protein levels was evident in this analysis (Figure 6c). FASN and SCD protein levels were significantly increased in tumor samples with elevated P-S6 compared to tumor samples with decreased P-S6 (Figure 6d). Taken together with our isogenic and breast cancer cell data, these findings indicate that the aberrant activation of mTORC1 in cancer promotes a lipogenic program through the activation of SREBP and its gene targets.
Figure 6.
Expression of SREBP targets is associated with mTORC1 activation in human breast cancer. (a) Comparison of SREBP target gene expression and P-S6 levels in data from primary breast cancer samples. Expression of FASN, SCD, LDLR, and MVK in breast carcinoma samples from the TCGA, grouped by high (n=112) or low (n=116) P-S6-S240/244 levels. Data are shown as mean ± s.e.m. relative to low P-S6 samples. (b,c) Association of FASN and SCD protein levels with P-S6 levels in primary human breast cancers. Dot blots of six different matched pairs of breast cancer and normal tissue are shown, each spotted in triplicate (b). The log2 fold change of P-S6 levels in paired tumor versus normal tissue is graphed with log2 fold change of either FASN (n=40) or SCD (n=37) (c), with the coefficient of determination (R2) provided. (d) The data from c was grouped into high and low fold changes of P-S6 levels in tumor versus normal tissue and graphed for the fold-change in FASN and SCD protein levels.
Discussion
An important metabolic characteristic of cancer cells that distinguishes them from their cells of origin is their enhanced capacity to synthesize, de novo, the major macromolecules needed to make new cells, including proteins, nucleotides, and lipids43. While there has been much progress in defining the unique metabolic properties of cancer cells, how these properties are acquired over the course of oncogenic transformation is less well understood. In this study, we demonstrate that mTORC1 activation downstream of oncogenic PI3K and Ras signaling is a major mechanism by which cancer cells stimulate aberrantly elevated rates of de novo lipid synthesis. Furthermore, we show that the SREBP family of transcription factors, well-established promoters of lipid synthesis in physiological settings, such as the liver6, are the key downstream effectors of mTORC1 promoting oncogene-induced lipid synthesis.
SREBP has emerged as a major effector of mTORC1 signaling. Porstmann et al. were the first to show that aberrant activation of an oncogene (Akt) could activate SREBP44, which was later found to be through downstream activation of mTORC1 (ref. 14). In a bioinformatics search for common cis-regulatory elements in the promoters of mTORC1-induced genes, we identified SREs, which are recognized by SREBP, to be most prevalent, demonstrating the importance of SREBP as a primary downstream transcriptional effector of mTORC1 (ref. 13). How mTORC1 activates SREBP is unknown, although multiple direct downstream targets of mTORC1 have been implicated12. These distinct mechanisms are likely to underlie the differential sensitivity of SREBP1/2 processing and lipid synthesis to rapamycin and TOR kinase inhibitors. It is now clear that the mTORC1-SREBP1c pathway represents a major route by which insulin signaling activates physiological lipid synthesis in the liver15,16,20,22. Recent studies have also implicated an important role for the SREBPs in cancer cell viability37,45–47. We find that activation of the mTORC1-SREBP pathway underlies the elevated lipid synthesis observed in cancer cells, providing a key link between oncogenic signaling and this metabolic process. Relative to SREBP1, SREBP2 appears to have a stronger effect on de novo lipogenesis and proliferation in the cell settings used in this study. It is not clear whether this is due to a specific function of SREBP2, which unlike SREBP1, is embryonic lethal if knocked out in mice48, or whether it is due to the influence of SREBP2 on SREBP1 expression detected here and in previous studies13,37,38, resulting in a functional decrease in both isoforms upon SREBP2 knockdown.
Lipogenic enzymes transcriptionally activated by the SREBPs have emerged as potential therapeutic targets in cancer. Chemical inhibition or genetic knockdown of ATP citrate lyase (ACLY)49,50, FASN2, and SCD51–53 have been found to reduce proliferation and survival in a variety of cancer cells and xenograft tumor models2,50. Recent studies have suggested that proliferating cells need to coordinate protein synthesis with the synthesis of MUFAs, prevalent in membrane phospholipids, to prevent endoplasmic reticulum (ER) stress37,54, suggesting a potential selective pressure for the co-regulation of protein and lipid synthesis by mTORC1. SCD is required for the production of MUFAs and is, therefore, particularly important for the proliferation and survival of cancer cells51–53. Blocking the mevalonate pathway downstream of SREBP, which is responsible for isoprenoid and cholesterol production, has also been found to reduce cancer cell viability55,56.
Despite the apparent importance of de novo lipogenesis in some cancer settings, it is unclear what role lipid uptake plays and whether exogenous lipids become limiting for cancer cells in the tumor microenvironment. In our study, depletion of serum lipids had little effect on the sensitivity of breast cancer cells to knockdown of SREBP isoforms. However, in the oncogene-expressing MCF10a lines, a requirement for SREBP was only revealed under conditions of low serum, where oncogenic PI3K and K-Ras exert a proliferation advantage. Interestingly, cancer cells cultured in hypoxic conditions have been shown to be dependent on the uptake of exogenous lipids for sustained growth and survival, and this has been attributed to the oxygen dependence of SCD for its production of endogenous MUFAs54,57. Consistent with our findings that SCD is essential for the mTORC1- and SREBP-dependent stimulation of de novo lipid synthesis downstream of oncogenes (Figure 2g), Kamphorst et al57 have demonstrated that activated Akt stimulates the de novo production of MUFAs. In contrast, they found that oncogenic Ras actually decreases cellular MUFA content and enhances the percentage of MUFAs acquired through uptake of exogenous lipids. In addition to increasing lipid uptake57, we show in a distinct isogenic setting that oncogenic K-Ras can also enhance de novo lipid synthesis under conditions where exogenous lipids are not readily available. Interestingly, relative to cells expressing oncogenic PI3K, SCD was found to be less critical for Ras-stimulated lipogenesis in our assays, consistent with these previous findings57. It is important that we gain a better understanding of the balance between lipid synthesis and uptake in the tumor microenvironment and how distinct oncogenic lesions influence this in different cancers. It is interesting to note that a major physiological effect of systemic treatment with rapamycin is an increase in circulating lipids58,59, likely resulting from the stimulation of lipolysis in adipose tissue60–62. Such an increase in the availability of exogenous lipids to growing tumors might overcome the suppressive effects of mTORC1 inhibition on de novo synthesis within the tumor.
Our data here suggest that oncogenic activation of mTORC1 and its downstream induction of an SREBP-driven lipogenic program are key elements to the transforming capacity of PI3K and K-Ras signaling. Together with established downstream effectors of these pathways that promote cancer cell survival and proliferation, activation of the mTORC1-SREBP pathway serves to enhance cell autonomous growth.
Materials and methods
Cell culture
All cell lines were obtained from ATCC (Manassas, VA, USA) and maintained in RPMI-1640 with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. Lipid-reduced FBS was made by mixing with fumed silica (20 mg/ml) (S5130, Sigma) for 3 hours. Pools of MCF10A cells stably expressing pBabe-Puro-vector, -PI3KCAH1047R (Addgene #12524)63, or K-RasG12V (Addgene #9052), via retroviral transduction, were selected and cultured in DMEM-F12 with 5% horse serum, EGF (20 ng/ml), hydrocortisone (0.5 mg/ml), cholera toxin (100 ng/ml), Insulin (10 μg/ml), and puromycin (1 μg/ml) at 37°C and 5% CO2. Rapamycin (553210, Calbiochem, San Diego, CA, USA), PP242 (4257, Tocris, Bristol, UK), and Torin1 (4247, R&D Systems, Minneapolis, MN, USA) were used to inhibit mTOR.
Plasmids and siRNAs
The pLKO mouse SREBP2 plasmid (Addgene #32018,) was a gift from David Sabatini22. SREBP2 shRNAs from the RNAi consortium were used: shSREBP2#1 (TRCN0000020665), shSREBP2#2 (TRCN0000020667), shSREBP2#3 (TRCN0000020666), shSREBP2#4 (TRCN0000020668). All siRNA experiments used ON-TARGET-plus SMARTpool siRNAs (30 nM; Dharmacon/GE, Lafayette, CO, USA) transfected using Lipofectamine RNAiMAX (Invitrogen, Grand Island, NY, USA), according to the manufacturer's instructions for reverse transfection. For Raptor and Rictor knockdowns, siRNAs were transfected on two consecutive days to achieve efficient knockdown. Each siRNA and shRNA targeting SREBP2 recognized a unique sequence in SREBF2, with at least 5 mismatches toward any sequence in SREBF1, and vice versa.
Immunoblotting
Cells were lysed in NP-40 buffer (40 mM HEPES, pH 7.4; 400 mM NaCl; 1 mM EDTA, pH 8.0; 1% NP-40 (CA-630, Sigma); 5% glycerol; 10 mM pyrophosphate; 10 mM β-glycerophosphate; 50 mM NaF; 0.5 mM orthovanadate) containing Protease Inhibitor Cocktail (Sigma) and 1 mM DTT. Nuclear isolation was performed with a Nuclear Extract Kit (40010, Active Motif, Carlsbad, CA, USA), with 10 μg/ml ALLN (208719, Millipore, Bedford, MA, USA) treatment 20 min prior to isolation, and ALLN added to the hypotonic and lysis buffers. The nuclear fraction was washed with hypotonic buffer prior to lysis.
Antibodies used for immunoblots recognized SREBP1 (sc-8984, Santa Cruz, Santa Cruz, CA, USA), SREBP2 precursor and processed C-terminus (557037, BD, Franklin Lakes, NJ, USA), SREBP2 mature N-terminus (30682, Abcam, Cambridge, MA, USA), Actin (A5316, Sigma), and from Cell Signaling Technologies (Danvers, MA, USA): ACC1 (3676), FASN (3180), SCD (2438), HA (2367), P-Akt-T308 (9275), P-Akt-S473 (4051), Total-Akt (4691), P-S6K1-T389 (9234), Total-S6K1 (2708), P-S6-S240/S244 (2215), Total-S6 (2217), 4E-BP1 (9644), Ras (3965), P-Erk1/2-T202/Y204 (9106), Total-Erk1/2 (9102), Lamin A/C (2032), Histone H3 (4499).
De novo lipid synthesis
Cells grown in 6-well plates were serum starved 16-18 h, with 5 μCi/mL 1-14C-acetate (NEC084H001MC, Perkin Elmer, Waltham, MA, USA) added to the media for the final 4 h. Cells were washed twice with PBS prior to lysis in 0.5% Triton X-100. Lipids were extracted with 2:1 (v/v) chloroform/methanol (500 μl) followed by low-speed centrifugation (1000 rpm, 20 min). 14C-labeled lipid in the denser fraction was quantified in duplicate samples using a LS6500 scintillation counter (Beckman Coulter), and normalized to protein concentration for MCF10a and cell number for breast cancer cells.
mRNA expression analysis
RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). cDNA was synthesized using the Superscript III First Strand Synthesis System (Invitrogen) and quantified using SYBR-Green for qRT-PCR (Applied Biosystems 7300 Real Time PCR System, Carlsbad, CA, USA). Each condition was run in triplicate and normalized to RPLP0 (F-cagattggctacccaactgtt, R-gggaaggtgtaatccgtctcc) mRNA levels. Primer sequences: SREBF1 (F-tgcattttctgacacgcttc; R-ccaagctgtacaggctctcc), SREBF2 (F-tggcttctctccctactcca, R-gagaggcacaggaaggtgag), ACACA (F-atgtctggcttgcacctagta, R-ccccaaagcgagtaacaaattct), FASN (F-aaggacctgtctaggtttgatgc, R-tggcttcataggtgacttcca), SCD (F-cccagctgtcaaagagaagg, R-caagaaagtggcaacgaaca).
Cell proliferation, size and death
Cell number and size were measured in solution, following trypsinization, using a Z2 Coulter Counter (Beckman Coulter, Danvers, MA). Medium was replaced every 24 h. For cell death, adherent and non-adherent cells were combined, washed in PBS, and resuspended in annexin buffer (10 mM HEPES; 140 mM NaCl; 2.5 mM CaCl2, pH 7.4) prior to incubation with Annexin-V/FITC conjugate (A13199, Invitrogen) for 15 min. Cells for each sample were resuspended in annexin buffer containing propidium iodide (PI; P4170, Sigma) before analyzing by flow cytometry (FACS Calibur, BD). Percent cell death was calculated by dividing the sum of the cells positive for PI, Annexin-V, or both by the total number of cells.
Analysis of TCGA data
Reverse-phase protein array and gene expression data were downloaded from cBioPortal40–42. Samples with P-S6-S240 levels greater than one standard deviation above average were classified as “High Phospho-S6”, whereas those greater than one standard deviation below average were classified as “Low Phospho-S6”.
Breast cancer patient lysate arrays
Breast cancer patient lysate arrays (PMA2-001-L, Protein Biotechnologies, Ramona, CA, USA) were blotted following the manufacturer's instructions. Colloidal gold staining (170-6527, BioRad, Waltham, MA, USA) was used to determine total protein content. Signal from 40 of 55 samples was detectable for FASN and 37 of 55 samples for SCD. Dot intensity was measured with ImageJ64. “High Phospho-S6” samples had a fold change in P-S6 levels between the tumor and normal tissues greater than log2(0.5), whereas “Low Phospho-S6” were those lower than log2(-0.5).
Statistical analysis
All data were analyzed with GraphPad Prism (La Jolla, CA, USA). P-values were calculated by an unpaired two-tailed Student's t-test, where appropriate.
Supplementary Material
Acknowledgements
This work was supported in part by NSF predoctoral fellowship DGE-1144152 (S.J.H.R.), a postdoctoral fellowship from the LAM Foundation (I.B.S.), a Sanofi Innovation Award (B.D.M.), and NIH grants R01-CA181390 and P01-CA120964 (B.D.M.).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Medes G, Thomas A, Weinhouse S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 1953;13:27–29. [PubMed] [Google Scholar]
- 2.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 3.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 4.Kuhajda FP, Jennert K, Wood FD, Hennigart RA, Jacobs LB, Dick JD, et al. Fatty acid synthesis: A potential selective target for antineoplastic therapy. Proc Natl Acad Sci USA. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Ding S, Habib N, Fermor B, Wood C, Gilmour R. Partial characterization of a cDNA for human stearoyl-CoA desaturase and changes in its mRNA expression in some normal and malignant tissues. Int J Cancer. 1994;57:348–352. doi: 10.1002/ijc.2910570310. [DOI] [PubMed] [Google Scholar]
- 6.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Sato R, Brown MS, Hua X, Goldsteln JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 8.Hua X, Sakai J, Brown MS, Goldstein JL. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J Biol Chem. 1996;271:10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- 9.Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 11.Jeon T, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2011;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ricoult SJH, Manning BD. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 2013;14:242–251. doi: 10.1038/embor.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci USA. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Ogawa W, Emi A, Hayashi K, Senga Y, Nomura K, et al. Role of S6K1 in regulation of SREBP1c expression in the liver. Biochem Biophys Res Commun. 2011;412:197–202. doi: 10.1016/j.bbrc.2011.07.038. [DOI] [PubMed] [Google Scholar]
- 18.Wang BT, Ducker GS, Barczak AJ, Barbeau R, Erle DJ, Shokat KM. The mammalian target of rapamycin regulates cholesterol biosynthetic gene expression and exhibits a rapamycin-resistant transcriptional profile. Proc Natl Acad Sci USA. 2011;108:15201–15206. doi: 10.1073/pnas.1103746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Yuan H, Niu Y, Niu W, Fu L. The role of AMPK/mTOR/S6K1 signaling axis in mediating the physiological process of exercise-induced insulin sensitization in skeletal muscle of C57BL/6 mice. Biochim Biophys Acta. 2012;1822:1716–1726. doi: 10.1016/j.bbadis.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Owen JL, Zhang Y, Bae S-H, Farooqi MS, Liang G, Hammer RE, et al. Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc Natl Acad Sci USA. 2012;109:16184–16189. doi: 10.1073/pnas.1213343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luyimbazi D, Akcakanat A, McAuliffe PF, Zhang L, Singh G, Gonzalez-Angulo AM, et al. Rapamycin regulates stearoyl CoA desaturase 1 expression in breast cancer. Mol Cancer Ther. 2010;9:2770–2784. doi: 10.1158/1535-7163.MCT-09-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e1000038. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Manning BD. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 29.Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008;27:S43–51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 31.Inoki K, Li Y, Zhu T, Wu J, Guan K. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Bio. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 32.Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci USA. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 34.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci USA. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tweto J, Liberati M, Larrabee A. Protein turnover and 4’-phosphopantetheine exchange in rat liver fatty acid synthetase. J Biol Chem. 1971;246:2468–2471. [PubMed] [Google Scholar]
- 36.Nakanishi S, Numa S. Purification of rat liver acetyl coenzyme A carboxylase and immunochemical studies on its synthesis and degradation. Eur J Biochem. 1970;16:161–173. doi: 10.1111/j.1432-1033.1970.tb01068.x. [DOI] [PubMed] [Google Scholar]
- 37.Griffiths B, Lewis CA, Bensaad K, Ros S, Zhang Q, Ferber EC, et al. Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab. 2013;1:3. doi: 10.1186/2049-3002-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, et al. Sterol regulatory element–binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howell JJ, Ricoult SJH, Ben-Sahra I, Manning BD. A growing role for mTOR in promoting anabolic metabolism. Biochem Soc Trans. 2013;41:906–912. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- 44.Porstmann T, Griffiths B, Chung Y-L, Delpuech O, Griffiths JR, Downward J, et al. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene. 2005;24:6465–6481. doi: 10.1038/sj.onc.1208802. [DOI] [PubMed] [Google Scholar]
- 45.Guo D, Prins RRM, Dang J, Kuga D, Iwanami A, Soto H, et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal. 2009;2:ra82. doi: 10.1126/scisignal.2000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krycer JRJ, Phan L, Brown AJA. A key regulator of cholesterol homeostasis, SREBP-2, can be targeted in prostate cancer cells with natural products. Biochem J. 2012;446:191–201. doi: 10.1042/BJ20120545. [DOI] [PubMed] [Google Scholar]
- 47.Williams KJ, Argus JP, Zhu Y, Wilks MQ, Marbois BN, York AG, et al. An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer Res. 2013;73:2850–2862. doi: 10.1158/0008-5472.CAN-13-0382-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Migita T, Narita T, Nomura K, Miyagi E, Inazuka F, Matsuura M, et al. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–8554. doi: 10.1158/0008-5472.CAN-08-1235. [DOI] [PubMed] [Google Scholar]
- 51.Fritz V, Benfodda Z, Rodier G, Henriquet C, Iborra F, Avancès C, et al. Abrogation of de novo lipogenesis by stearoyl-CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol Cancer Ther. 2010;9:1740–1754. doi: 10.1158/1535-7163.MCT-09-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roongta U V, Pabalan JG, Wang X, Ryseck R-P, Fargnoli J, Henley BJ, et al. Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol Cancer Res. 2011;9:1551–1561. doi: 10.1158/1541-7786.MCR-11-0126. [DOI] [PubMed] [Google Scholar]
- 53.Minville-Walz M, Pierre A-S, Pichon L, Bellenger S, Fèvre C, Bellenger J, et al. Inhibition of stearoyl-CoA desaturase 1 expression induces CHOP-dependent cell death in human cancer cells. PLoS One. 2010;5:e14363. doi: 10.1371/journal.pone.0014363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young RM, Ackerman D, Quinn ZL, Mancuso A, Gruber M, Liu L, et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 2013;27:1115–1131. doi: 10.1101/gad.198630.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freed-Pastor WA, Mizuno H, Zhao X, Langerød A, Moon S-H, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clendening JW, Penn LZ. Targeting tumor cell metabolism with statins. Oncogene. 2012;31:4967–4978. doi: 10.1038/onc.2012.6. [DOI] [PubMed] [Google Scholar]
- 57.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci USA. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morrisett JD, Abdel-Fattah G, Hoogeveen R, Mitchell E, Ballantyne CM, Pownall HJ, et al. Effects of sirolimus on plasma lipids, lipoprotein levels, and fatty acid metabolism in renal transplant patients. J Lipid Res. 2002;43:1170–1180. [PubMed] [Google Scholar]
- 59.Kasiske BL, de Mattos A, Flechner SM, Gallon L, Meier-Kriesche HU, Weir MR, et al. Mammalian target of rapamycin inhibitor dyslipidemia in kidney transplant recipients. Am J Transpl. 2008;8:1384–1392. doi: 10.1111/j.1600-6143.2008.02272.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhang C, Yoon M-S, Chen J. Amino acid-sensing mTOR signaling is involved in modulation of lipolysis by chronic insulin treatment in adipocytes. Am J Physiol Endocrinol Metab. 2009;296:E862–868. doi: 10.1152/ajpendo.90651.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakrabarti P, English T, Shi J, Smas CM, Kandror K V. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soliman GA, Acosta-Jaquez HA, Fingar DC. mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids. 2010;45:1089–1100. doi: 10.1007/s11745-010-3488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neve RM, Chin K, Fridlyand J, Yeh J, Frederick L, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.