Summary
Macrophages are essential for systemic iron recycling, and also control iron availability to pathogens. Iron metabolism in mammalian cells is orchestrated post-transcriptionally by iron-regulatory proteins (IRP)-1 and -2. Here, we generated mice with selective and combined ablation of both IRPs in macrophages to investigate the role of IRPs in controlling iron availability. These animals are hyperferritinemic but otherwise display normal clinical iron parameters. However, mutant mice rapidly succumb to systemic infections with Salmonella Typhimurium, a pathogenic bacterium that multiplies within macrophages, with increased bacterial burdens in liver and spleen. Ex vivo infection experiments indicate that IRP function restricts bacterial access to iron via the EntC and Feo bacterial iron-acquisition systems. Further, IRPs contain Salmonella by promoting the induction of lipocalin 2, a host antimicrobial factor that inhibits bacterial uptake of iron-laden siderophores, and by suppressing the ferritin iron pool. This work reveals the importance of the IRPs in innate immunity.
Graphical abstract

Introduction
Iron supply for the hemoglobinization of new red blood cells in the erythroid marrow depends largely on reutilization of the metal by the liver and spleen monocyte-macrophage system (MPS), which clears old erythrocytes, frees iron from hemoglobin, and exports the metal back into the circulation through the iron exporter ferroportin (FPN, a.k.a. SLC40A1) (Ganz, 2013). Iron recycling by the MPS diminishes in response to infection, which is viewed as an innate defense mechanism to reduce the iron concentration in the circulation and thereby withhold the metal from invaders (Drakesmith and Prentice, 2012, Nairz et al., 2014). Macrophage iron metabolism is thus critical both for securing body iron sufficiency and immunity.
Systemic iron fluxes are controlled in part by the hormone hepcidin (a.k.a. HAMP), which inhibits iron export from macrophages by binding to and triggering the degradation of FPN (Nemeth et al., 2004). In addition to humoral control of systemic iron metabolism by hepcidin, iron metabolism is also regulated cellularly by the iron regulatory proteins (IRP)-1 and -2 (a.k.a. ACO1 and IREB2, respectively) (Kühn, 2014). IRPs respond to changes in cellular iron levels and in turn enact posttranscriptional regulation of key iron metabolism genes via their interaction with cis-regulatory iron responsive elements (IRE) present on target mRNAs, including those encoding the transferrin receptor (TFR1), the ferritin-H (FTH1) and –L (FTL1) iron storage proteins, and the iron exporter FPN. The role of macrophage IRPs in body iron recycling and immunity is not known.
Here we use Cre/Lox technology to generate mice with cell-type selective, complete loss of IRP expression in macrophages. Earlier work investigating mice with complete IRP deficiency in hepatocytes or duodenal enterocytes, respectively, had shown early post-natal death in both cases, reflecting essential functions of the IRPs for organismal survival (Galy et al., 2008, 2010). This study reveals important molecular functions of the IRPs in the control of macrophage iron metabolism, and uncovers that at least this mammalian cell type is viable without IRPs; it also unveils the critical importance of the IRP/IRE system for macrophage-mediated immunity and host resistance to infection with intracellular bacteria.
Results
Role of IRPs in macrophage and body iron homeostasis
Animals homozygous for floxed Irp alleles (Aco1flox/flox,Ireb2flox/flox) (Galy et al., 2005) were bred to a mouse line with targeted insertion of Cre into the Lysozyme2 (Lyz2) locus enabling selective expression of CRE recombinase in monocytes/macrophages and neutrophils (Clausen et al., 1999). Aco1flox/flox,Ireb2flox/flox,Lyz2+/Cre mice (designated IrpLyzCre(+)) are born at Mendelian ratio and do not overtly differ from Aco1flox/flox,Ireb2flox/flox,Lyz2+/+control littermates (IrpLyzCre(−)). To assess the consequences of IRP ablation for macrophage iron metabolism, we analyzed bone marrow-derived macrophages (BMDMs) from IrpLyzCre(+) versus IrpLyzCre(−) animals; for comparison, we also analyzed BMDMs deficient for either of the two IRPs. Western blotting shows efficient ablation of IRP1 and/or IRP2 in BMDMs, with residual amounts of IRP (Figure 1A, left) possibly reflecting mosaic activity of the Lyz2-Cre strain (Tuckerman et al., 2007). IRP-target genes are not significantly affected by single IRP1 deficiency, and IRP2 disruption alone causes a minor increase in FTL1 (Figure 1A, left). Contrasting with this, the simultaneous ablation of both IRPs causes a marked increase in FPN, FTH1, and FTL1 expression (Figure 1A, left), most likely due to translational derepression of the corresponding mRNAs (Galy et al., 2013) and to a minor extent to increased mRNA expression (Figure 1A, right); double IRP deficiency expectedly decreases TFR1 protein and mRNA levels (Figure 1A). Similar to BMDMs, elicited peritoneal macrophages (PMΦ) from IrpLyzCre(+) mice display high ferritin and low TFR1 expression (Figure 1B); FPN protein and mRNA could not be reliably detected in our PMΦ cultures (Figure 1B, see also Van Zandt et al., 2008). These results confirm that IRP1 and IRP2 control key iron-handling molecules in macrophages. They also show that each IRP can largely compensate for the lack of function in the other, hence the rest our study focuses on double IRP deficiency.
Figure 1. IRP function in macrophage iron metabolism and body iron recycling.
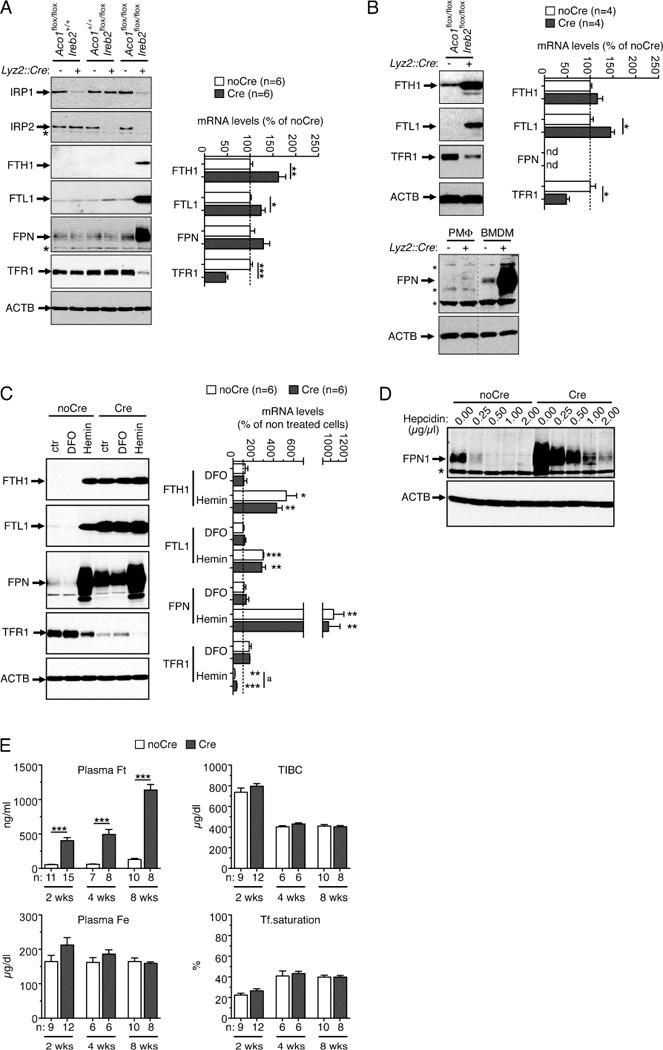
A) Left panels, western-blot analysis (left panels) of IRPs and IRP-target genes in BMDMs from mice carrying either of the two or both floxed Irp1 (Aco1) and Irp2 (Ireb2) alleles together or not with the Lyz2-Cre gene, as indicated, and the corresponding control cells. Histograms: qPCR analysis of IRP-target mRNA expression in doubly deficient (Cre) versus control (noCre) BMDMs. B) Western-blot (left) and qPCR (right) analysis of IRP-target genes in doubly deficient (Cre) versus control (noCre) peritoneal macrophages (PMΦ); lysates from BMDMs (A) were used as a positive control for FPN expression (bottom panels). The qPCR data in (A) and (B) display transcript levels as percentage of control (i.e. noCre cells) after calibration to β-actin. FPN mRNA expression in PMΦ is close to background and is indicated as not detected (nd). C) Western-blot (left) and qPCR (right) analysis of IRP-target genes in Cre versus noCre BMDMs exposed to the iron chelator DFO, the iron donor hemin, or left untreated (ctr). For each gene, mRNA levels are expressed as percentage of control (i.e. untreated noCre BMDMs) after calibration to β-actin. Statistical differences were assessed between treated and untreated cells for each genotype (asterisks) or between genotypes for each treatment (upper case letter). D) Western-blot analysis of FPN decay in BMDMs exposed to increasing amounts of synthetic hepcidin; the asterisk indicates a cross-reacting protein. For western blotting (A–D), β-actin (ACTB) was used to ascertain equal loading. E) Consequences of IRP ablation in macrophages on blood iron parameters in 2, 4 and 10 week-old mice. TIBC, total iron binding capacity. Histograms display averages ± SEM. The number of mice (n) is indicated. p: Student’s t-test (*,a: p<0.05; **: p<0.01; ***: p<0.001). See also Figure S1 and Table S1.
We next determined how IRP deficiency affects the response of macrophages to iron challenge (Figure 1C). In IrpLyzCre(−) BMDMs, iron chelation by deferoxamine (DFO) augments TFR1 mRNA and protein levels and slightly decreases FPN protein expression; the expected suppression of ferritin could not be observed because of already low basal expression. Opposite to DFO, the iron source hemin suppresses TFR1 and stimulates FTL1, FTH1, and FPN at the protein and less so at the mRNA level. IRP-deficient BMDMs still respond to iron fluctuation with further reduction of TFR1 upon iron loading (TFR1 mRNA ratio hemin/non-treated: IrpLyzCre(−), 0.18±0.04; IrpLyzCre(+), 0.34 ±0.08) and a slight increase upon iron chelation. These effects could possibly reflect IRP-independent regulation of TFR1 mRNA turnover and/or changes in Tfr1 transcription. FTL1 protein expression remains unresponsive to DFO in IrpLyzCre(+) cells, and is slightly increased by hemin (Figure 1C, left) accompanied by an increase of the FTL1 mRNA (Figure 1C, right, the same is observed for FTH1); FPN mRNA stimulation by hemin (Figure 1C, right) is accompanied by a marked FPN upregulation at the protein level (Figure 1C, left). These data indicate that ferritin regulation in macrophages is dominated by IRP-dependent control of ferritin translation, whereas FPN levels are set through both IRP-dependent and IRP-independent regulatory pathways. Importantly, IRP deficiency does not interfere with hepcidin-mediated degradation of FPN (Figure 1D). However, because basal FPN expression is elevated in IRP deficiency, higher doses of hepcidin are required to suppress FPN. This shows that both IRPs and hepcidin are important to set FPN levels and hence the iron export capacity of macrophages.
Similar to primary macrophage cultures (Figure 1), IRP ablation increases ferritin expression posttranscriptionally in vivo, both in liver and spleen macrophages (Supplementary figure 1 available online); FPN expression is augmented in liver (Figure S1A) but not in spleen (Figure S1B). Of note, IrpLyzCre(+) mice display a marked increase in plasma ferritin concentration (Figure 1E), as predicted for animals with augmented macrophage ferritin expression (Cohen et al., 2010). Although ferritin and FPN mediate, respectively, macrophage iron retention and export, misregulation of these critical iron management molecules in MPS cells has no detectable impact on plasma iron levels and transferrin saturation values (Figure 1E); hematological parameters and tissue iron stores are also unaffected (Table S1). Hence, IRPs control key iron handling molecules in MPS macrophages in vivo but are dispensable for maintaining the body iron balance under standard laboratory conditions.
The macrophage IRP/IRE system protects mice against Salmonella.
Hypoferremia is a common response to infection (Nairz et al., 2014). Part of this response involves Hepcidin stimulation and subsequent inhibition of FPN-mediated iron efflux from macrophages. Although IRP deficiency antagonizes hepcidin-mediated suppression of FPN (Figure 1D), it does not significantly mitigate the hypoferremia induced by aseptic inflammatory stimuli in mice (Figure S2). This shows that macrophage iron retention in response to acute inflammation relies predominantly on IRP-independent mechanisms. It also suggests that macrophage IRP deficiency would unlikely alter iron availability to microbes present in the circulation. We thus investigated the impact on iron availability towards intracellular pathogens, and infected mice with Salmonella enterica serovar Typhimurium (designated S.Tm). This Gram-negative bacterium persists within MPS cells, including spleen and liver macrophages, it causes systemic disease in mice (Vazquez-Torres et al., 1999, 2004), and its dependence on iron is well established (Nairz et al., 2007). Remarkably, IrpLyzCre(+) mice succumb faster than IrpLyzCre(−) littermates when exposed to a lethal dose of S.Tm (Figure 2A, top). This effect is unrelated to the insertion of Cre into the Lyz2 locus since Lyz2+/Cre animals display the same survival as wild-type (Figure 2A, bottom). The higher vulnerability of IrpLyzCre(+) animals to S.Tm is associated with higher bacterial multiplication in liver and spleen (Figure 2B). Importantly, IrpLyzCre(+) mice are unable to mount an adequate immune response against S.Tm, as evidenced by the blunted induction of several immune response genes in liver and spleen (Figure 2C). These results reveal that the macrophage IRP/IRE system is critically important for the host defense against Salmonella (note that IRP depletion in IrpLyzCre(+) neutrophils might also impact on mouse survival).
Figure 2. Macrophage IRPs protect mice against infection with Salmonella Typhimurium.

A) Top, IrpLyzCre(+) mice (Cre) and control littermates (noCre) were subjected to systemic infection with 500 CFU of Salmonella Typhimurium (S.Tm). The representative Kaplan-Meyer curve displays mouse survival over time. Bottom, same experiment with Lyz2+/Cre versus wild-type mice. B) IrpLyzCre(+) and IrpLyzCre(−) mice were injected with a sublethal dose (200 CFU) of S.Tm and the bacterial load of liver (top) and spleen (bottom) tissues was determined at different time points after infection. The data expressed as CFU per gram of tissue were log transformed. C) The induction of immune response genes in liver and spleen was assayed by qPCR 6 hr after infection, i.e. before differences in bacterial loads (B) are manifest. The data are expressed as percentage of control (i.e. noCre animals injected with vehicle) after calibration to tubulin (TUBB5) mRNA. B and C) The data are displayed as box and whiskers with 10 to 90 percentiles (outliers are represented as dots). (n) indicates the number of mice. p: A, long-Rank test; B and C, Student’s t-test (*: p<0.05; **: p<0.01; ***: p<0.001, ns: not significant). See also Figure S2.
IRPs limit Salmonella proliferation by controlling iron bioavailability
To better define the mechanism(s) through which IRPs protect mice against Salmonella, we infected primary macrophages ex vivo. Consistent with the data obtained in vivo (Figure 2A), PMΦ lacking IRP expression fail to restrict intracellular S.Tm proliferation over time (Figure 3A); the same result was obtained when infecting BMDMs (Figure 3B). Of note, IRP ablation does not alter the capacity of macrophages to produce reactive oxygen species upon infection with S.Tm (Figure S3A), nor the engulfment of fluorescently-labeled heat-killed S.Tm particles (Figure S3B). Hence, a simple defect in the oxidative and phagocytic activities of IrpLyzCre(+) macrophages cannot account for their reduced capacity to contain S.Tm. However, IRP deficiency dampens the induction of several cytokines in macrophages infected ex vivo (Figure 3C), mirroring the blunted immune response observed in mice (Figure 2C); IrpLyzCre(+) cells apparently also produce less nitric oxide than IrpLyzCre(−) (Figure 3D) but this unlikely explains their anti-microbial deficit towards S.Tm (Figure S3C). IRP deficiency thus seems to broadly weaken the macrophage innate immune response to S.Tm infection. To determine whether the anti-microbial deficit of IrpLyzCre(+) macrophages is related to iron metabolism misregulation or reflects potential iron-independent functions of the IRP/IRE system, we added the iron chelator deferasirox (DFX) to macrophages during infection. Importantly, iron chelation alone abrogates the disadvantage conferred by IRP deficiency on macrophage antibacterial activity (Figure 3E), showing that IRPs control intracellular Salmonella proliferation at least in part through influencing iron management.
Figure 3. IRPs promote macrophage immunity and inhibit Salmonella proliferation.

A) PMΦ from IrpLyzCre(+) (Cre) versus IrpLyzCre(−) (noCre) mice were infected ex vivo with S.Tm and intracellular bacterial proliferation (averages ± SEM) was recorded over time. IRP-deficient PMΦ exhibit a tendency for reduced anti-bacterial activity 12 hr after infection reaching statistical significance at the 24hr time point. B) IrpLyzCre(+) BMDMs display the same inability to contain S.Tm proliferation 24 hr post-infection. C) The concentration of cytokines in the culture medium of PMΦ was determined 24 hr after infection with S.Tm. The data are displayed as box and whiskers with 10 to 90 percentiles. D) PMΦ were infected as in (A) in the presence of the iron chelator Deferasirox (DFX) versus vehicle (ctr.) and the intracellular bacterial load (averages ± SEM) was determined 24h later. The sample size (n) is indicated. p: Student’s t-test (*: p<0.05; **: p<0.01; ***: p<0.001). See also Figure S3.
IRPs control Salmonella iron uptake through LCN2 and ferritin
To compare the effect of iron chelation on macrophage antimicrobial activity with the effects of genetic microbial iron limitation, we infected macrophages with a triple sit feo entC mutant strain of S.Tm defective for the three main iron acquisition pathways (Crouch et al, 2008). Proliferation of this mutant is expectedly reduced compared to wild-type bacteria (Figure 4A). Importantly, the sit feo entC S.Tm mutant proliferates almost identically in IrpLyzCre(+) as in IrpLyzCre(−) (Figure 4A). Likewise, while microbial 59Fe uptake is ~3-fold higher when bacteria infect IRP-deficient versus control macrophages (Figure 4B), iron acquisition by the sit feo entC S.Tm mutant is diminished and unaffected by the IRP status of the host cell (Figure 4B). These results demonstrate that the macrophage IRP/IRE system limits bacterial iron assimilation and growth.
Figure 4. IRPs control bacterial iron uptake through LCN2 and ferritin.

A) PMΦ from IrpLyzCre(+) (Cre) versus IrpLyzCre(−) (noCre) animals were infected, respectively, with a wild-type (S.Tm WT) or a triple mutant strain (S.Tm triple) of S.Tm deficient for the three main bacterial iron uptake pathways; intracellular bacterial proliferation was assayed 24 hr later. B) PMΦ were infected as in (A) in the presence of a 59Fe tracer and microbial 59Fe uptake was determined 24 hr post infection. C) PMΦ were infected with S.Tm mutant strains deficient for either of the three main iron uptake pathways, as indicated below the histogram; bacterial proliferation was measured 24 hr later. Numbers above the bars indicate the Cre/noCre ratios. D) Cre versus noCre BMDMs were infected, respectively, in the presence of recombinant LCN2 versus vehicle (left) or an anti-LCN2 antibody versus isotype control (right). E) BMDMs were labeled with a 59Fe tracer prior infection and samples were collected at different time points to determine respectively, bacterial number, bacterial iron uptake, and the amount of 59Fe in ferritin immunoprecipitates. Bacterial 59Fe uptake (black lines, left axis) is indicated as percentage of noCre BMDMs 12 hr after infection; ferritin-associated 59Fe (red lines, right axis) is given as percentage of noCre BMDMs at time 0 hr. Note that the inverse correlation between ferritin-bound iron levels and bacterial iron uptake is qualitative and does not necessarily imply the uptake of all the iron associated with ferritin by S.Tm. Data are presented as averages ± SEM. The sample size (n) is indicated. p: Student’s t-test (*: p<0.05; **: p<0.01; ***: p<0.001). See also Figure S4.
S.Tm acquires elemental ferrous iron via the SitABCD and Feo systems and takes up ferric iron with the help of siderophores (Osman and Cavet, 2011). To determine which iron uptake pathway is targeted by the IRPs, we infected macrophages with bacteria deficient for either of the three iron uptake systems. While the antimicrobial deficit of IrpLyzCre(+) macrophages is evident with S.Tm lacking sitABCD, it is abolished when the bacteria lack the siderophore iron uptake pathway (entC) (Figure 4C); disruption of the Feo system has an intermediate effect. IRPs thus restrain siderophore-mediated bacterial iron uptake, and limit elemental iron acquisition via the Feo but not the SitABCD system. In line with those results, we find that mice with macrophage IRP deficiency display the same survival as wild-type littermates when infected with entC or feo mutant bacteria, respectively, but remain significantly more vulnerable to infection with the sitABCD mutant strain (Figure S4). Altogether these data show that macrophage IRPs limit Salmonella siderophore- and Feo-mediated iron assimilation.
Finally, we sought to determine how loss of IRP function promotes bacterial iron assimilation. Lipocalin 2 (LCN2) is a host antimicrobial factor induced upon infection that inhibits bacterial uptake of iron-laden siderophores (Flo et al., 2004, Raffatellu et al., 2009, Deriu et al., 2013). As Lcn2 induction is impaired in IrpLyzCre(+) macrophages (Figure 3C), we hypothesized that these cells fail to limit bacterial iron acquisition due to insufficient LCN2 production. To test this, we either added recombinant LCN2 to the culture medium of infected macrophages, or inhibited endogenous LCN2 with an anti-LCN2 antibody. The anti-bacterial deficit of IrpLyzCre(+) macrophages is fully reverted by exogenous addition of LCN2 (Figure 4D, left), and is only slightly affected by inhibition of endogenous LCN2 (Figure 4D, right); LCN2 inhibition in IrpLyzCre(−) macrophages at least partially phenocopies the anti-bacterial deficit of IRP-null cells (Figure 4D, right). In contrast to LCN2, exogenous addition of TNFA and/or IL6, whose induction is also impaired in IrpLyzCre(+) macrophages (Figure 3D), enhances the bactericidal activity of both IrpLyzCre(+) and IrpLyzCre(−) cells but does not rescue IRP deficiency (Fig. S4B). These results show that the inability of IRP-deficient macrophages to combat S.Tm is at least partly due to LCN2 misregulation.
IrpLyzCre(+) BMDMs also display a tendency towards increased total non-heme iron levels (ferrozine assay: 12.9 ±1.9 versus 7.5 ±1.5 μmol Fe/g total protein in IrpLyzCre(+) versus control BMDMs, n=5 in each group, p= 0.07 Student’s t-test). Iron loading in IrpLyzCre(+) cells occurs in spite of reduction of the TFR1 iron uptake molecule and stimulation of the FPN iron exporter (Figure 1), and could possibly reflect sequestration of the metal into the overabundant ferritin (Galy et al., 2013). We hypothesized that iron bound to ferritin in IrpLyzCre(+) macrophages (Figures 1A and 1B) constitutes a pool of metal exploitable by S.Tm. To test this, IrpLyzCre(−) and IrpLyzCre(+) macrophages were loaded with 59Fe-citrate prior infection, and samples were collected at different time points to determine, respectively, the uptake of 59Fe by S.Tm and the amount of 59Fe in ferritin immunoprecipitates (IP) (Figure 4E). At time zero of infection, ferritin from IrpLyzCre(+) macrophages contains about 2.3-fold more 59Fe compared to IrpLyzCre(−) (Figure 4E, upper histogram). This result confirms that cellular iron accumulates within the excess of ferritin in IRP-deficient cells. Over the course of infection, the amount of 59Fe associated with ferritin decreases both in IrpLyzCre(−) and IrpLyzCre(+) macrophages (Figure 4E), indicating that the pool of iron bound to ferritin is indeed mobilized while S.Tm multiplies. Importantly, the initially enlarged ferritin-bound iron pool of IrpLyzCre(+) macrophages is consumed and becomes nearly identical to IrpLyzCre(−) 12 hr after infection. This experiment shows that the excess of ferritin in IrpLyzCre(+) macrophages provides a surplus of iron contributing to increased bacterial iron uptake.
In conclusion, macrophage IRPs are critical for host resistance to infection with S.Tm in mice. Although additional mechanisms may contribute, the restriction of S.Tm iron acquisition and proliferation by promotion of LCN2 synthesis and suppression of the ferritin iron pool emerge as underlying mechanisms.
Discussion
Iron recycling and storage by macrophages is critical for systemic iron homeostasis and host defense (Nairz et al., 2014). While the role of hepcidin in the control of macrophage iron fluxes and inflammatory hypoferremia is recognized (Ganz, 2013), additional hepcidin-independent modes of iron metabolism regulation are being discovered (Deschemin and Vaulont, 2013, Guida, Altamura et al., 2015). Although it represents a key regulator of cellular iron transport and storage, the role of the IRP/IRE system in macrophage iron metabolism has not yet been understood. Through genetic disruption of the macrophage IRP/IRE system in mice, this work uncovers the important function of the macrophage IRPs, including the importance of the IRP/IRE system for innate immunity.
Disruption of IRP function in macrophages affects their iron storage and export capacity, similar to enterocytes and hepatocytes (Galy et al., 2008, 2010). Surprisingly, IrpLyzCre(+) mice develop normally, are viable and fertile, and show no sign of illness when maintained in a standard laboratory environment. This contrasts with the severe wasting and perinatal lethality of mice with constitutive, cell-specific loss of IRP function in enterocytes or hepatocytes, respectively (Galy et al., 2008, 2010). Furthermore, macrophage IRP function seems dispensable for maintaining systemic iron homeostasis under steady-state conditions. Changes in macrophage iron recycling and storage in IrpLyzCre(+) mice are apparently compensated for by the organism. Mosaic activity of the Lyz2-Cre deletor strain (Tuckerman et al., 2007) might also mitigate the iron phenotype of IrpLyzCre(+) mice. Importantly, IRPs in macrophages strongly influence plasma ferritin concentration, a widely used clinical parameter. This result supports earlier work concluding that macrophages represent a major source of ferritin in the blood (Cohen et al., 2010).
Iron holds a key position at the host-pathogen interface (Nairz et al., 2014). Here we demonstrate that macrophage IRP function is protective against Salmonella. IRP ablation in macrophages broadly hampers the immune response to S.Tm infection. IRPs most likely regulate the immune response in an indirect manner since the transcripts encoding the cytokines assayed in our study are devoid of IRE, as assessed using the sIRES tool (Campillos et al., 2010). The HIF2α (hypoxia-inducible factor 2 alpha, a.k.a. EPAS1) transcription factor could be a possible intermediate linking IRP function to cytokine regulation. Indeed the HIF2α mRNA bears an IRE in its 5’UTR (Sanchez et al., 2007), and HIF2α is required for cytokine induction in macrophages (Imtiyaz et al., 2010). However, IRP ablation leads to HIF2α overexpression (Galy et al., 2013) and would thus be predicted to potentiate the immune response, which is opposite to what we observe. IRPs could influence cytokine production via modulation of cellular iron levels. Iron indeed modulates the immune response in multiple ways. On the one hand the metal can negatively affect immune defense pathways (Nairz et al., 2014), but it also seems to promote toll-like receptor signaling (Wang et al., 2009), and elevation of intracellular iron levels in FPN-deficient BMDMs increases cytokine expression upon LPS stimulation (Zhang et al., 2011). Although IRP-deficiency has pleiotropic effects on the immune response and impaired cytokine production could play an important role, the failure of IRP-deficient macrophages to combat S.Tm can be imputed at least in part to their inability to limit microbial iron acquisition. We identify two molecular mechanisms by which IRPs restrict bacterial iron uptake. First, IRPs are needed for sufficient host LCN2 production and restriction of siderophore-mediated iron uptake; since similar to cytokines the LCN2 mRNA does not bear a recognizable IRE, the exact mechanisms of IRP-dependent LCN2 regulation remains to be defined. It will also be interesting to determine how S.Tm mobilizes the iron bound to ferritin. Induction of ferritin degradation, as observed in Neisseria meningitidis-infected cells, is one possible scenario (Larson et al., 2004). Of note, reduction of macrophage iron levels through enhancement of FPN-mediated iron efflux is viewed as a defense mechanism against intracellular bacteria, including S.Tm (Chlosta et al., 2006, Paradkar et al., 2008, Nairz et al., 2007). S.Tm proliferation in IRP-deficient BMDMs is increased in spite of FPN overexpression. This indicates that high FPN expression alone cannot inhibit intracellular bacterial proliferation when ferritin is hyper induced.
Experimental procedures
Mice
All mouse lines were backcrossed to C57BL/6 animals for at least 10 generations. For all studies of IrpLyzCre(+) mice, control animals consisted of IrpLyzCre(−) littermates. Unless specified, 10 to 12 week-old male mice were used. For infection experiments, mice were injected (i.p.) with the indicated CFU of S.Tm diluted in PBS (control mice received PBS alone). To increase macrophage number in peritoneal exudates, mice received (i.p.) 0.075 ml of 4% water-solubilized thiogylcolate medium (Sigma-Aldrich, Taufkirchen, Germany) per g body weight. Mice were kept under a constant light/dark cycle on a standard diet and had access to food and water ad libitum. They were euthanised by CO2 inhalation to collect tissues, macrophages, or blood. Animal handling was in accordance with approved guidelines from the EMBL, the Medical University of Innsbruck Animal ethics committee, and the Austrian Ministry for Science and Education (approvals: BMWF-66.011/0151-II/3b/2011 and BMWFW-66.011/0111-WF/V/3b/2014).
Macrophages
BMDMs and PMΦ were isolated and grown as described previously (Ferring-Appel et al., 2009). For iron challenges, BMDMs were treated for 12 hr with 100 μM heme arginate (Leiras Oy, Turku, Finland) or 100 μM DFO (Sigma-Aldrich), respectively; for hepcidin treatment, cells were exposed for 5 hr to increasing doses of hepcidn 25 (Peptides international, Louisville, KY). For all treatments, cells received vehicle as control. For infection experiments, macrophages were infected at a MOI of 10 and intracellular bacterial multiplication was determined as described previously (Nairz et al., 2013). To test the role of LCN2, macrophages were infected in the presence of recombinant murine LCN2 (100 ng/ml, R&D Systems GmbH, Wiesbaden-Nordenstadt, Germany) versus vehicle, or with a rat monoclonal anti-mouse LCN2 antibody (25 μg/ml, clone 228418, R&D Systems GmbH) versus isotype control (25 μg/ml, clone 5447, R&D Systems GmbH). To test the influence of iron chelation, infected macrophages were treated with 100 μM DFX (Novartis, Basel, Switzerland) versus vehicle.
Bacterial strains
We used the S. Tm. wild-type strain ATCC 14028s and isogenic mutant derivatives deficient for either (entC::aph, Δsit::bla and Δfeo::Tn10 (Tetr), respectively) or all three iron acquisition pathways (Crouch et al., 2008).
Statistical analyses
Data are presented as mean ±SEM unless otherwise specified. Statistical analyses were performed using the Prism software (GraphPad Software, La Jolla, CA). Differences between two mean values were evaluated by two-tailed Student’s t-test (data were log-transformed for the comparison of bacterial loads and mRNA levels of immune response genes). Multiple groups were compared by one-way ANOVA with Bonferroni’s post hoc analysis. To compare survival curves the log-rank test was used. A p value < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We are grateful to I. Förster (University of Bonn, Germany) for her kind gift of the Lyz2-Cre knockin strain. We wish to thank S. Berger, S. Engl and M. Seifert for excellent technical support, and the staff of the animal, gene core and flow cytometry facilities of the EMBL. This work was supported in part by a grant from the BMBF (HepatoSys and Virtual Liver) to M.W.H., the Austrian Research Fund (FWF; project TRP-188 to G.W.), and the Medical University of Innsbruck (MUI-START project 2012032003 to M.N.). F.C.F. is supported by the National Institutes of Health (AI39557, AI44486).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional and detailed experimental procedures can be found in the supplement.
Author contributions
B. G., M. W. H. and G. W. conceived the study, analyzed the data and wrote the manuscript. B. G. and M. N. designed the experiments. D. C., D. F. A., B. G., D. H., M. N., A. S., T. S. and L. V. performed experiments. F. C. F. contributed crucial reagents and edited the manuscript.
References
- Campillos M, Cases I, Hentze MW, Sanchez M. SIREs: searching for iron-responsive elements. Nucleic Acids Res. 2010;38:W360–367. doi: 10.1093/nar/gkq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlosta S, Fishman DS, Harrington L, Johnson EE, Knutson MD, Wessling-Resnick M, Cherayil BJ. The iron efflux protein ferroportin regulates the intracellular growth of Salmonella enterica. Infect Immun. 2006;74:3065–3067. doi: 10.1128/IAI.74.5.3065-3067.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Cohen LA, Gutierrez L, Weiss A, Leichtmann-Bardoogo Y, Zhang DL, Crooks DR, Sougrat R, Morgenstern A, Galy B, Hentze MW, et al. Serum ferritin is derived primarily from macrophages through a nonclassical secretory pathway. Blood. 2010;116:1574–1584. doi: 10.1182/blood-2009-11-253815. [DOI] [PubMed] [Google Scholar]
- Crouch ML, Castor M, Karlinsey JE, Kalhorn T, Fang FC. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2008;67:971–983. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschemin JC, Vaulont S. Role of hepcidin in the setting of hypoferremia during acute inflammation. PLoS One. 2013;8:e61050. doi: 10.1371/journal.pone.0061050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakesmith H, Prentice AM. Hepcidin and the iron-infection axis. Science. 2012;338:768–772. doi: 10.1126/science.1224577. [DOI] [PubMed] [Google Scholar]
- Ferring-Appel D, Hentze MW, Galy B. Cell-autonomous and systemic context-dependent functions of iron regulatory protein 2 in mammalian iron metabolism. Blood. 2009;113:679–687. doi: 10.1182/blood-2008-05-155093. [DOI] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring D, Hentze MW. Generation of conditional alleles of the murine Iron Regulatory Protein (IRP)-1 and -2 genes. Genesis. 2005;43:181–188. doi: 10.1002/gene.20169. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring-Appel D, Kaden S, Gröne HJ, Hentze MW. Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring-Appel D, Sauer SW, Kaden S, Lyoumi S, Puy H, Kölker S, Gröne HJ, Hentze MW. Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metab. 2010;12:194–201. doi: 10.1016/j.cmet.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring-Appel D, Becker C, Gretz N, Gröne HJ, Schümann K, Hentze MW. Iron regulatory proteins control a mucosal block to intestinal iron absorption. Cell Rep. 2013;3:844–857. doi: 10.1016/j.celrep.2013.02.026. [DOI] [PubMed] [Google Scholar]
- Ganz T. Systemic iron homeostasis. Physiol Rev. 2013;93:1721–1741. doi: 10.1152/physrev.00008.2013. [DOI] [PubMed] [Google Scholar]
- Guida C, Altamura A, Klein F, Galy B, Boutros M, Ulmer AJ, Hentze MW, Muckenthaler MU. TLR2/6 control a novel inflammatory pathway for rapid hepcidin-independent hypoferremia. Blood. 2015;125:2265–2275. doi: 10.1182/blood-2014-08-595256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiyaz HZ, Williams EP, Hickey MM, Patel SA, Durham AC, Yuan LJ, Hammond R, Gimotty PA, Keith B, Simon MC. Hypoxia-inducible factor-2alpha regulates macrophage function in mouse models of acute and tumor inflammation. J Clin Invest. 2010;120:2699–2714. doi: 10.1172/JCI39506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn LC. Iron regulatory proteins and their role in controlling iron metabolism. Metallomics. 2015;7:232–243. doi: 10.1039/c4mt00164h. [DOI] [PubMed] [Google Scholar]
- Larson AJ, Howie HL, SO M. Neisseria meningitidis accelerates ferritin degradation in host epithelial cells to yield an essential iron source. Mol Microbiol. 2004;53:807–820. doi: 10.1111/j.1365-2958.2004.04169.x. [DOI] [PubMed] [Google Scholar]
- Nairz M, Theurl I, Ludwiczek S, Theurl M, Mair SM, Fritsche G, Weiss G. The coordinated regulation of iron homeostasis in murine macrophages limits the availability of iron for intracellular Salmonella Typhimurium. Cell Microbiol. 2007;9:2126–2140. doi: 10.1111/j.1462-5822.2007.00942.x. [DOI] [PubMed] [Google Scholar]
- Nairz M, Schleicher U, Schroll A, Sonnweber T, Theurl I, Ludwiczek S, Talasz H, Brandacher G, Moser PL, Muckenthaler MU, et al. Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection. J Exp Med. 2013;210:855–873. doi: 10.1084/jem.20121946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz M, Haschka D, Demetz E, Weiss G. Iron at the interface of immunity and infection. Front Pharmacol. 2014 doi: 10.3389/fphar.2014.00152. Published online 2014 Jul 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- Osman D, Cavet JS. Metal sensing in Salmonella: implications for pathogenesis. Adv Microb Physiol. 2011;58:175–232. doi: 10.1016/B978-0-12-381043-4.00005-2. [DOI] [PubMed] [Google Scholar]
- Paradkar PN, De Domenico I, Durchfort N, Zohn I, Kaplan J, Ward DM. Iron depletion limits intracellular bacterial growth in macrophages. Blood. 2008;112:866–874. doi: 10.1182/blood-2007-12-126854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- Tuckermann JP, Kleiman A, Moriggl R, Spanbroek R, Neumann A, Illing A, Clausen BE, Stride B, Förster I, Habenicht AJ, et al. Macrophages and neutrophils are the targets for immune suppression by glucocorticoids in contact allergy. J Clin Invest. 2007;117:1381–1390. doi: 10.1172/JCI28034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zandt KE, Sow FB, Florence WC, Zwilling BS, Satoskar AR, Schlesinger LS, Lafuse WP. The iron export protein ferroportin 1 is differentially expressed in mouse macrophage populations and is present in the mycobacterial-containing phagosome. J Leukoc Biol. 2008;84:689–700. doi: 10.1189/jlb.1107781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A, Jones-Carson J, Bäumler AJ, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks WT, Fang FC. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres A, Vallance BA, Bergman MA, Finlay BB, Cookson BT, Jones-Carson J, Fang FC. Toll-like receptor 4 dependence of innate and adaptive immunity to Salmonella: importance of the Kupffer cell network. J Immunol. 2004;172:6202–6208. doi: 10.4049/jimmunol.172.10.6202. [DOI] [PubMed] [Google Scholar]
- Wang L, Harrington L, Trebicka E, Shi HN, Kagan JC, Hong CC, Lin HY, Babitt JL, Cherayil BJ. Selective modulation of TLR4-activated inflammatory responses by altered iron homeostasis in mice. J Clin Invest. 2009;119:3322–3328. doi: 10.1172/JCI39939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang F, An P, Guo X, Shen Y, Tao Y, Wu Q, Zhang Y, Yu Y, Ning B, et al. Ferroportin1 deficiency in mouse macrophages impairs iron homeostasis and inflammatory responses. Blood. 2011;118:912–922. doi: 10.1182/blood-2011-01-330324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


