Abstract
Joubert syndrome (JBTS) is a primarily autosomal-recessive disorder characterized by a distinctive mid-hindbrain and cerebellar malformation, oculomotor apraxia, irregular breathing, developmental delay, and ataxia. JBTS is a genetically heterogeneous ciliopathy. We sought to characterize the genetic landscape associated with JBTS in the French Canadian (FC) population. We studied 43 FC JBTS subjects from 35 families by combining targeted and exome sequencing. We identified pathogenic (n = 32 families) or possibly pathogenic (n = 2 families) variants in genes previously associated with JBTS in all of these subjects, except for one. In the latter case, we found a homozygous splice-site mutation (c.735+2T>C) in CEP104. Interestingly, we identified two additional non-FC JBTS subjects with mutations in CEP104; one of these subjects harbors a maternally inherited nonsense mutation (c.496C>T [p.Arg166∗]) and a de novo splice-site mutation (c.2572−2A>G), whereas the other bears a homozygous frameshift mutation (c.1328_1329insT [p.Tyr444fs∗3]) in CEP104. Previous studies have shown that CEP104 moves from the mother centriole to the tip of the primary cilium during ciliogenesis. Knockdown of CEP104 in retinal pigment epithelial (RPE1) cells resulted in severe defects in ciliogenesis. These observations suggest that CEP104 acts early during cilia formation by regulating the conversion of the mother centriole into the cilia basal body. We conclude that disruption of CEP104 causes JBTS. Our study also reveals that the cause of JBTS has been elucidated in the great majority of our FC subjects (33/35 [94%] families), even though JBTS shows substantial locus and allelic heterogeneity in this population.
Main Text
Joubert syndrome (JBTS [MIM: 213300]) is a predominantly autosomal-recessive disorder characterized by oculomotor apraxia, hypotonia, neonatal breathing abnormalities, ataxia, and variable developmental delay. The hallmark of JBTS is a malformation involving the brainstem and cerebellum and consisting of cerebellar vermis hypoplasia or aplasia, horizontal elongated cerebellar peduncles, and a deep interpeduncular fossa; together, these take on the pathognomonic appearance of a “molar tooth.”1 A subset of individuals with JBTS also have extraneural manifestations such as polydactyly, retinopathy, cystic kidneys, and liver fibrosis (reviewed by Romani et al.2).
JBTS is a ciliopathy, given that the majority of the known genes associated with JBTS have been shown to play a role in the development and/or function of the non-motile cilia. The cilium is a compartmentalized extension of the extracellular membrane and functions as an antenna by sensing extracellular signals and transducing them intracellularly. The cilium is composed of a microtubule-based cytoskeleton called the axoneme, which nucleates from the basal body, a modified centriolar structure. At the base of the cilium, Y-shaped structures connect the basal body to the cell membrane, forming the transition zone, which constitutes a diffusion barrier between the cilium and the remainder of the plasma membrane (for a review, see Valente et al.3). The majority of genes associated with JBTS encode proteins that localize to the basal body or ciliary transition zone. Many of these proteins physically interact with one another to form large complexes. The most important complex in the pathogenesis of JBTS is the B9 complex (also known as the tectonic complex), in which 9 of its known 15 members are associated with JBTS, Meckel syndrome (MKS [MIM: 249000]), and/or oral-facial-digital syndrome (OFD [MIM: 311200]).4, 5, 6, 7 MKS and OFD are related ciliopathies whose features overlap those of JBTS.
Although JBTS was first described in a French Canadian (FC) family in 1969,8 little is known about its molecular etiology in this population. We thus sought to characterize the genetic landscape associated with JBTS in the FC population by studying a large number of unrelated families. Using a stepwise approach of targeted and whole-exome sequencing (WES), we were able to explain most cases and show that mutations in CEP104 cause JBTS.
This study was approved by our institutional ethics committee. Informed consent was obtained from all participants or their legal guardians. We identified 43 FC individuals with JBTS (from 35 families). All individuals are of FC ancestry and originate from various regions throughout Quebec. The diagnosis of JBTS was based on the presence of (1) at least one JBTS classical neurological manifestation (oculomotor apraxia, ataxia, or history of breathing abnormalities) and (2) the molar tooth sign (MTS) on brain imaging in at least one affected family member (Figure S1). In addition, four fetuses were included in the study. On prenatal imaging, all fetal subjects showed cerebellar vermis hypoplasia or aplasia and elongated cerebellar peduncles, suggestive of JBTS.
We previously explained JBTS in 21 of these individuals (15 families), who showed pathogenic mutations in C5orf42 (MIM: 614571), TMEM231 (MIM: 614949), or CC2D2A (MIM: 612013)9, 10 (see Table S1). In JBTS-affected individuals from the FC population, these studies established the presence of a complex founder effect involving three recurrent mutations in C5orf42 (c.4006C>T [p.Arg1336Trp], c.7400+1G>A, and Ensembl transcript ENST00000509849, c.4690G>A [p.Ala1564Thr] [GenBank: NM_023073.3]), two recurrent mutations in CC2D2A (c.3376G>A [p.Glu1126Lys] and c.4667A>T [p.Asp1556Val] [GenBank: NM_001080522.2]), and two recurrent mutations in TMEM231 (c.12T>A [p.Tyr4∗] and c.625G>A [p.Asp209Asn] [GenBank: NM_001077418.1]), each of which was previously shown to be present on a common haplotype (see Table S1).9, 10 Thus, we used Sanger sequencing to screen all unexplained JBTS-affected families (n = 19) for the seven known recurrent FC mutations. Five individuals (fetus 474, 1673.590, 1951.677, HSJ-JBTS-3, and HSJ-JBTS-4) were found to have homozygous or compound-heterozygous mutations in C5orf42, and two individuals (1342.488 and 1343.488) showed compound-heterozygous mutations in CC2D2A (Table 1). In addition, screening identified a single heterozygous mutation in C5orf42 in one individual (2049.708) and single heterozygous mutations in CC2D2A in two individuals (1610.572 and 1123.415). Sequencing the remaining exons of C5orf42 in individual 2049.708 revealed a heterozygous truncating variant (c.6354dupT [p.Ile2119Tyrfs∗2] [GenBank: NM_023073.3]), whereas sequencing the remaining exons of CC2D2A in individuals 1610.572 and 1123.415 identified a heterozygous canonical splice-site variant (c.2181+1G>A [GenBank: NM_001080522.2]) in the former and a heterozygous missense variant (c.3544T>C [p.Trp1182Arg]) in the latter (Table 1). The c.3544T>C (p.Trp1182Arg) variant has been previously reported as pathogenic in an individual with nephronophthisis.11 These three individuals are compound heterozygous for their respective mutations.
Table 1.
Clinical Characteristics of Previously Unpublished JBTS-Affected Individuals Included in This Study
| Individual | Gene | Mutations | Gender | Age | MTS | OMA | Retinal Involvement | Renal Involvement | Liver Involvement | Limb Anomalies | Developmental Delay | Cognition | Respiratory Abnormality | Hypotonia | Ataxia | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fetus 474 | C5orf42 | c.[4006C>T];[6407del], p.[Arg1336Trp];[Pro2136Hisfs∗31]a | F | 22 weeks | + | NA | − | − | − | − | NA | NA | NA | NA | NA | − |
| HSJ-JBTS-3 | C5orf42 | homozygous c.4006C>T (p.Arg1336Trp) | F | 28 years | + | + | −f | −h | −h | − | − | borderline | − | + | + | strabismus |
| HSJ-JBTS-4 | C5orf42 | homozygous c.4006C>T (p.Arg1336Trp) | F | 3 years | + | + | +f | −u | −u | + | + | NA | − | NA | NA | bifid epiglottis, strabismus, central polydactyly on the right, bilateral preaxial poldaytyly in feet |
| 1712.604 | NPHP1 | homozygous deletion of NPHP1 | F | 2 years | + | + | −e | −u | −u | − | − | NA | − | − | + | − |
| 1915.669 | NPHP1 | homozygous c.555delA (p.Lys185Asnfs∗7) | F | 3 years | + | + | −f | −u | −u | − | + | NA | − | + | NA | − |
| HSJ-JBTS-1 | TMEM67 | homozygous c.2132A>C (p.Asp711Ala) | F | 3.5 years | + | + | −f | −u | −u | − | + | mild ID | − | + | + | − |
| 1123.415 | CC2D2A | c.[3544T>C];[4667A>T], p.[Trp1182Arg];[Asp1556Val] | F | 4 years | + | + | −f | −u | −u | − | + | NA | − | + | + | − |
| 1673.590 | C5orf42 | homozygous c.4006C>T (p.Arg1336Trp) | F | 12 years | + | + | −f | −u | −u | − | + | mild ID | − | + | + | ADHD, motor apraxia |
| 1951.677 | C5orf42 | c.[4006C>T];[7400+1G>A], p.[Arg1336Trp];[?] | F | 1 years | + | + | +e | −u | −u | − | + | NA | + | + | NA | swallowing difficulties |
| 1342.488 | CC2D2A | c.[4667A>T];[3376G>A], p.[Asp1556Val];[Glu1126Lys] | M | 28 years | + | + | −f | −h | −h | − | + | moderate ID | − | + | + | autism, ADHD |
| 1343.488 | M | 31 years | + | + | −f | −h | −h | − | + | normal (university) |
− | + | + | − | ||
| 2049.708 | C5orf42 | c.[4690G>A];[6354dupT], p.[Ala1564Thr];[Ile2119Tyrfs∗2] | F | 1.5 years | + | − | −f | −u | −u | + | + | NA | + | + | + | oromotor apraxia, swallowing difficulties, tongue and hypothalamic hamartomas, neonatal seizures |
| 1610.572 | CC2D2A | c.[4667A>T];[2181+1G>A], p.[Asp1556Val];[?] | F | 2 years | + | + | −e | −u | −u | − | + | NA | − | + | + | − |
| 1310.476 | NPHP1 | hemizygous c.555delA (p.Lys185Asnfs∗7), NPHP1 deletion |
M | 18 years | + | + | −f | +u | −u | − | + | mild ID | + | + | + | renal failure, renal transplant, severe dysphasia |
| HSJ-JBTS-2 | TCTN1 | c.[342−2A>C];[898C>T], p.[?];[Arg300∗] | M | 27 weeks | + | NA | NA | −u | −u | + | NA | NA | NA | NA | NA | abnormal gyration of the frontal lobes |
| 1639.581 | B9D1 | c.[493C>T];[151T>C], p.[Gln165∗];[Ser51Pro] | M | 22 years | + | + | −f | −h | −h | − | − | normal | − | + | + | congenital club feet, dysphasia |
| 1767.621 | CEP290 | c.[6401T>C];[4195−1G>A], p.[Ile2134Thr];[?] | F | 21 weeks | + | NA | NA | + u | −u | + | NA | NA | NA | NA | NA | occipital encephalocele, olfactory bulb agenesis, bifid uterus, renal cysts |
| Fetus2 621 | F | 20 weeks | + | NA | NA | + u | −u | − | NA | NA | NA | NA | NA | renal cysts | ||
| 1686.595 | OFD1 | hemizygous c.920T>A (p.Val307Asp) | M | 15 years | + | + | +fb | +u | −u | − | + | severe ID | + | + | + | autism, end-stage renal failure, swallowing difficulties, abdominal situs inversus |
| 1299.472 | C2CD3 | c.[5929C>T];[5227G>T], p.[Arg1977∗];[Gly1743Cys] | M | 7 years | + | − | −f | −u | −u | − | + | NA | − | + | + | − |
| 1294.472 | M | 4 years | + | + | −e | −u | −u | − | + | NA | + | + | + | swallowing difficulties, oromotor apraxia, gastrostomy, fundoplication | ||
| 1763.618 | CEP104 | homozygous c.735+2T>C | F | 2 years | + | + | +e | −u | −u | − | + | NA | + | + | + | − |
| GeneDx01 | CEP104 | c.[2572−2A>G];[496C>T], p.[?];[Arg166∗] | F | 2.5 years | + | + | −f | −u | −u | − | + | NA | − | − | − | − |
| 842629 | CEP104 | homozygous c.1328_1329insT (p.Tyr444fs∗3) | M | 3.5 years | + | + | − | − | −u | − | + | severe ID | − | + | + | self-mutilation |
All individuals, except for GeneDx01 and 842629, are of French Canadian ancestry. Individuals 1342.488 and 1343.288 are siblings, as are 1299.472 and 1294.47. The following abbreviations are used: ADHD, attention deficit and hyperactivity disorder; e, electroretinogram; F, female; f, fundoscopy; h, history; ID, intellectual disability; M, male; MTS, molar tooth sign; NA, not available or not applicable; OMA, oculomotor apraxia; u, ultrasound.
The genotype of the fetus was not tested but was assumed on the basis of the genotype of similarly affected sibling 1304.474 (Table S1).
Hypertensive retinopathy.
In parallel, disease in three individuals was elucidated on a clinical basis. Two affected individuals were found to have mutations in NPHP1 (MIM: 607100): individual 1712.604 showed a 0.152 Mb homozygous deletion encompassing NPHP1 on chromosomal microarray in region 2q13 (110,826,262–110,978,224; UCSC Genome Browser hg19 assembly), and individual 1915.669 showed a homozygous frameshift (c.555delA [p.Lys185Asnfs∗7] [GenBank: NM_207181.2]) in NPHP1. Parents were confirmed to be heterozygous carriers of the mutations. Individual HSJ-JBTS-1 showed a homozygous mutation (c.2132A>C [p.Asp711Ala] [GenBank: NM_153704.5]) in TMEM67 (MIM: 609884). This mutation has been previously identified in a French individual with JBTS.12
We then performed WES in the remaining eight individuals (from seven families) in whom disease had not been explained. Genomic DNA from each sample was captured with the Agilent SureSelect 50 Mb exome capture oligonucleotide library and sequenced with paired-end 100 bp reads on an Illumina HiSeq 2000, resulting in an average of 12.8 Gb of raw sequence for each sample. Exome capture and sequencing were performed at the McGill University and Génome Québec Innovation Center (MUGQIC). Data were analyzed as previously described.13 We aligned reads to human genome assembly hg19 by using the Burrows-Wheeler Aligner (BWA v.0.5.9) and then used Picard (v.1.48) to remove putative PCR-generated duplicate reads. The median read depth of bases in consensus coding sequence (CCDS) exons was 102× (determined with Genome Analysis Toolkit v.1.6.7).14 On average, 91% (±3%) of bases in CCDS exons were covered by at least 20 reads. We used SAMtools (v.0.1.17) mpileup and bcftools to call sequence variants and required at least three variant reads and ≥20% variant reads for each called position, as well as Phred-like quality scores of at least 20 for single-nucleotide variants (SNVs) and at least 50 for small insertions or deletions (indels). We used ANNOVAR15 and custom scripts to annotate variants according to the type of mutation, occurrence in dbSNP, 1000 Genomes minor allele frequencies, and NHLBI Exome Sequencing Project (ESP) Exome Variant Server (EVS) minor allele frequencies. To identify potentially pathogenic variants, we filtered out (1) synonymous variants or intronic variants other than those affecting the consensus splice sites, (2) variants seen in more than 5 of 1,128 exomes (sequenced at the MUGQIC) from individuals with rare, monogenic diseases unrelated to JBTS, and (3) variants with a minor allele frequency greater than 0.5% in either the 1000 Genomes or the NHLBI ESP exome datasets.
We first looked at the exome datasets for rare variants in the 23 genes already associated with JBTS (INPP5E [MIM: 613037], TMEM216 [MIM: 613277], AHI1 [MIM: 608894], NPHP1, CEP290 [MIM: 610142], TMEM67, RPGRIP1L [MIM: 610937], ARL13B [MIM: 608922], CC2D2A, CXORF5 [OFD1; MIM: 300170], KIF7 [MIM: 611254], TCTN1 [MIM: 609863], TCTN2 [MIM: 613885], TMEM237 [MIM: 614424], CEP41 [MIM: 610523], TMEM138 [MIM: 614459], C5orf42, TMEM231, TCTN3 [MIM: 613847], ZNF423 [MIM: 604557], CSPP1 [MIM: 611654], B9D1 [MIM: 614209], KIAA0586 [MIM: 610178], MKS1 [MIM: 609883], B9D2 [MIM: 611951] and C2CD3 [MIM: 615944]9, 10, 12, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39), as well as the JBTS candidate gene TTC21B (MIM: 612014).40
In one individual (1310.476), we identified homozygous frameshift mutation c.555delA (p.Lys185Asnfs∗7) (GenBank: NM_207181.2) in NPHP1; this is the same mutation that was identified in individual 1915.669. However, array genomic hybridization showed that this individual is also heterozygous for a 116 kb deletion encompassing NPHP1 in chromosomal region 2q13 (110,862,369–110,978,000; hg19 genome assembly). This deletion is similar to that found in subject 1712.604. The mother is heterozygous for the frameshift mutation, but paternal DNA was unavailable. It thus appears that the proband in this family is compound heterozygous for mutations in NPHP1. The presence of the same nonsense mutation and deletion in NPHP1 in unrelated FC individuals represents another example of a complex founder effect in this JBTS population.
In another individual (1639.581), we identified compound-heterozygous mutations in B9D1: a nonsense mutation (c.493C>T [p.Gln165∗] [GenBank: NM_001243473.1]) and a missense mutation (c.151T>C [p.Ser51Pro] [GenBank: NM_015681.3]) predicted to be damaging (Table 1; Table S1). These variants are both extremely rare given that they are absent in the EVS. In a third individual (1686.595), we identified a hemizygous c.920T>A (p.Val307Asp) missense variant (GenBank: NM_003611.2) in OFD1 on chromosome X. This missense change, which is located in a coiled-coil domain, is predicted to be damaging by PolyPhen-2 (score = 0.804), SIFT (score = 0.003), and CADD (C score = 26.4) and is absent in the EVS. Segregation studies were not performed because parental DNA was unavailable. It remains unclear whether this mutation is pathogenic.
In the two affected siblings of family 472, we identified compound-heterozygous mutations (GenBank: NM_001286577.1) in C2CD3: a nonsense mutation (c.5929C>T [p.Arg1977∗]) and a missense mutation (c.5227G>T [p.Gly1743Cys]) predicted to be damaging (Table 1; Table S1). This missense mutation affects a conserved amino acid (including in zebrafish) located in the fifth C2 domain (Figure S2). The two siblings with C2CD3 mutations have a classical form of JBTS with severe global developmental delay but without any extraneural manifestations. Mutations in C2CD3 have previously been reported in only five individuals, who all showed OFD-like phenotypes.41
In one fetus (HSJ-JBTS-2), we identified compound-heterozygous mutations (GenBank: NM_001082538.2) in TCTN1: c.342−2A>C and c.898C>T (p.Arg300∗). The splicing mutation, c.342−2A>C, had previously been reported to be causative for JBTS.42 In another fetus (1767.621), we identified two rare compound-heterozygous variants (GenBank: NM_025114.3) in CEP290: missense mutation c.6401T>C (p.Ile2134Thr) and canonical splice-site mutation c.4195−1G>A. This acceptor splice-site mutation has been previously reported in a subject with a JBTS-related ciliopathy, Senior-Loken syndrome (MIM: 610189), which is characterized by the presence of nephrophthiasis and congenital Leber amaurosis.43 The c.6401T>C (p.Ile2134Thr) missense mutation is predicted to be damaging by PolyPhen-2 (score = 0.964), SIFT (score = 0.03), and CADD (C score = 29). It affects an amino acid that is perfectly conserved down to the lower vertebrates. However, it is found at a minor allele frequency of 0.55% (64/11,600) in the EVS and at a relatively high frequency of 2.2% in in-house FC samples, suggesting that it is unlikely to be pathogenic. Targeted sequencing did not show any other rare variant in CEP290 or in the other known genes previously associated with JBTS. Moreover, multiplex ligation-dependent probe amplification did not identify a deletion or duplication in CEP290. Subsequently, the mother became pregnant, and the second fetus showed the same phenotype and genotype as the proband. It is unclear whether the c.6401T>C (p.Ile2134Thr) missense mutation is pathogenic, whether these fetuses carry another yet unidentified mutation in CEP290, or whether JBTS in this family is explained by mutations in another gene.
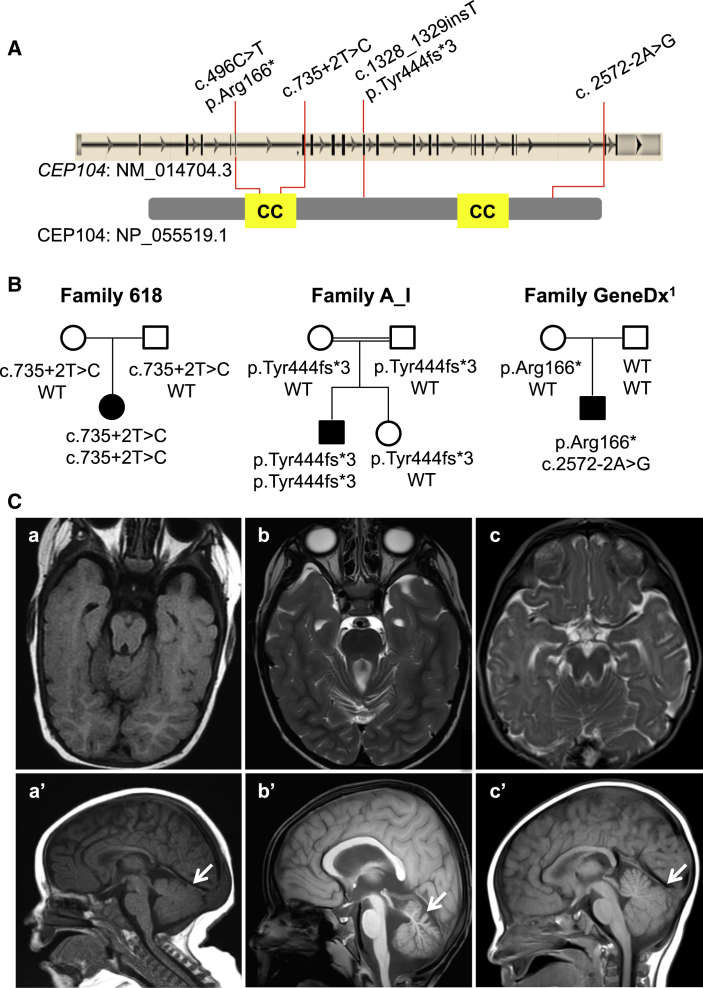
Only one individual (1763.618) remained without causative or candidate variants in known genes previously associated with JBTS, MKS, or OFD. We identified ten genes containing rare homozygous or multiple heterozygous variants in the exome of this subject (Table S2). For each of these genes, we searched PubMed (by using the search term “cilia” and the name of the gene of interest) to determine whether their products localize to the cilia or have a role in cilia function or development. Only one of these genes, CEP104, is known to be implicated in cilia function.44, 45, 46 In subject 1763.618, CEP104 harbors a homozygous splice-site variant (c.735+2T>C [GenBank: NM_014704.3]) affecting the canonical donor splice site following exon 7; this site encodes part of a coiled-coil domain (Figure 1A). This variant, which is absent in public SNP databases (dbSNP, EVS, 1000 Genomes, and the ExAC Browser) and 201 in-house FC control exomes, is predicted by MutationTaster and Human Splicing Finder to abolish the donor splice and thus most likely cause the skipping of this exon. Sanger sequencing confirmed that this mutation is homozygous in the affected individual and heterozygous in the unaffected parents (Figure 1B). This individual has a pure neurologic form of JBTS with a MTS on brain imaging (Figure 1C), oculomotor apraxia, hypotonia, and ataxia. She does not have breathing irregularities, polydactyly, or retinal, renal, or liver involvement. She has significant developmental delay given that she is not yet sitting independently, nor is she saying any words at the current age of 2.5 years (Table 1).
Figure 1.
CEP104 Mutations in JBTS-Affected Families
(A) Localization of the identified mutations in CEP104 (upper panel) and CEP104 (lower panel; 925 amino acids). The coiled-coiled (CC) domains (amino acid positions 209–289 and 677–725 according to UniProt: O60308) are highlighted in yellow.
(B) Segregation of CEP104 mutations within families.
(C) Brain MRI images of the JBTS individuals with CEP104 mutations from family 618 (a and a′), family A_I (b and b′), and GeneDx1 (c and c′). Axial T1-weighted (a) and T2-weighted (b and c) images show the molar tooth sign with deepened interpeduncular fossa and elongated, thickened, and abnormally orientated superior cerebellar peduncles. Sagittal T1-weighted images (a′, b′, and c’) show hypoplasia of the cerebellar vermis (arrow).
In a separate study, a 3.5-year-old Arab Israeli JBTS-affected boy, who was born to consanguineous parents (first cousins), was found to bear a homozygous frameshift variant (c.1328_1329insT [p.Tyr444fs∗3] [GenBank: NM_014704.3]) in CEP104 (Figures 1A and 1B). This variant was identified by exome capture (Agilent SureSelect v.4) and paired-end 100 bp sequencing (HiSeq 2000) initially done in both the affected individual and his unaffected sister. Exome sequence analyses and rare-variant filtering were performed as previously described.47 The mean coverage of the targeted exomes was 62×, and 97% of bases were covered at ≥10×. No rare recessive (homozygous, hemizygous, or potentially compound-heterozygous) variants were identified in genes previously associated with JBTS, OFD, or MKS in the exome of the affected individual. The c.1328_1329insT (p.Tyr444fs∗3) variant was confirmed by Sanger sequencing to be homozygous in the affected subject but heterozygous in his unaffected sister and parents. It is extremely rare in that it is absent from public databases (dbSNP, 1000 Genomes, EVS, and the ExAC Browser) and 350 in-house ethnically matched exomes. Examination of high-quality SNPs in the exome dataset indicated that this variant is located in a homozygous region that spans at least 8 Mb (chr1: 1,653,004–9,777,599; hg19 genome assembly). Like subject 1763.618, this individual shows a pure neurologic from of JBTS with oculomotor apraxia, profound psychomotor delay, self-mutilation, and a MTS on brain imaging (Figure 1C).
In order to identify additional subjects with mutations in CEP104, we sequenced its coding regions in a cohort of 96 individuals with unexplained MKS but did not identify any candidate variants. We next queried CEP104 in GeneMatcher, a freely accessible website that enables the identification of individuals with variants in candidate genes.48 We identified one gene match on the basis of an entry deposited by the diagnostic laboratory GeneDx. WES was performed and analyzed in this subject and his unaffected parents on a clinical basis, as recently described.49 The per-sample mean coverage of the target exome was 114×, and 99.9% of bases were covered at ≥10×. This WES analysis showed two variants in CEP104: c.496C>T (p.Arg166∗) and c.2572−2A>G (Figures 1A and 1B). The c.496C>T variant is extremely rare in that it has been reported at a MAF of 0.00015 (2/13,004) and 0.00002 (3/121,316) in the EVS and ExAC Browser, respectively. The c.2572−2A>G variant, which is not reported in the EVS or ExAC Browser, destroys the canonical splice acceptor site of intron 20 (MutationTaster and Human Splice Finder). This individual inherited the variant c.496C>T (p.Arg166∗) from his mother, whereas the variant c.2572−2A>G occurred de novo. These results were confirmed by Sanger sequencing. As predicted, this individual (referred to herein as GeneDx01), a 28-month-old white non-FC male, has a JBTS phenotype similar to that of the FC and Arab Israeli individuals, including global developmental delay, oculomotor apraxia, and MTS on brain imaging (Figure 1C). He has a pure neurologic phenotype with absence of polydactyly, retinopathy, and nephropathy (Table 1). Analysis of the exome data did not reveal any other de novo variants or rare recessive variants in genes previously associated with JBTS, MKS, or OFD. We were not able to phase the de novo c.2572−2A>G variant by using the trio exome sequencing data because of the absence of informative markers in the region. However, it is most likely that this variant is in trans with the maternally inherited nonsense variant, given that the majority (∼76%) of de novo point mutations arise in the paternal germline.50 Moreover, de novo mutations are rare events in that the average exome contains only zero to three such mutations.51 The likelihood that a JBTS-affected individual who does not show any rare variants in known ciliopathy-related genes carries a de novo mutation in cis with a rare nonsense mutation in a strong candidate gene is extremely low.
CEP104 localizes on both mother and daughter centrioles in non-ciliated retinal pigment epithelial (RPE1) cells but only on the daughter centriole in ciliated cells.44, 45, 46 The mother centriole functions as a basal body by nucleating cilia when the cells are in the G0 stage of the cell cycle; upon re-entry to the division phase, cells resorb their cilia, allowing the centrioles to form spindle poles. Interestingly, CEP104 moves from the mother centriole to the tip of the primary cilium during ciliogenesis.46 Moreover, knockdown of CEP104 in RPE1 cells impairs ciliogenesis.45, 46 Together, these observations suggest that loss of CEP104 function affects ciliogenesis by disrupting the conversion of the mother centriole into the ciliary basal body. CEP104 is known to interact with the centriolar protein complex CEP97-CP110, which inhibits ciliary assembly.52 Depletion of either CEP97 or CP110 promotes primary cilia formation. It has been suggested that CEP104 might be involved in the removal of the inhibitory complex CP110-CEP97 from the mother centriole and thereby allow its conversion into the basal body and the nucleation of the cilia.46 Interestingly, CP110 suppresses the ability of CEP290, a product of a gene associated with JBTS, to cooperate with RAB8A to enable cilia assembly.53, 54 CP110 has also been shown to interact with C2CD3 and KIAA0586, other products of genes associated with JBTS.38, 55
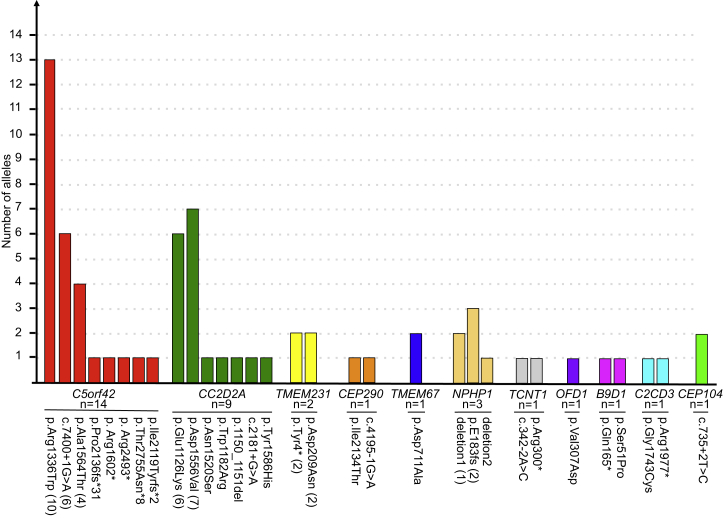
In summary, we identified rare and deleterious mutations in CEP104, a gene involved in ciliogenesis, in three individuals with JBTS. The occurrence of such mutations in unrelated individuals strongly suggests that disruption of CEP104 causes JBTS. In addition, our survey of the genetic landscape associated with JBTS in the FC population revealed great genetic heterogeneity with the possible involvement of as many as 11 genes (Figures S3 and S4). Of a total of 35 families, 14 are affected by causal mutations in C5orf42, nine by mutations in CC2D2A, three by mutations in NPHP1, two by mutations in TMEM231, and one each by mutations in TCTN1, TMEM67, B9D1, C2CD3, or CEP104 (Figure 2). We identified possible pathogenic mutations in CEP290 in one family and OFD1 mutations in another. There is also significant allelic heterogeneity given that a total of eight different mutations were identified in C5orf42 and seven were identified in CC2D2A (Figure 2). Finally, in FC JBTS-affected individuals, we observed the presence of a complex founder effect whereby four genes contained a total of nine recurrent mutations (Figure S4). Previous studies have identified the underlying genetic etiology in 41%–62% of affected individuals by screening known genes associated with JBTS in large heterogeneous JBTS populations.41, 56 In the current study, disease in 94% of the affected FC individuals (in 33/35 families) has been explained. This greater diagnostic yield is possibly related to the fact that we systematically studied an ethnically homogeneous population by using WES. Although we cannot rule out that JBTS in additional FC individuals who have not yet been examined is explained by mutations in other genes, the genetic landscape of JBTS appears to be largely elucidated in this population.
Figure 2.
Locus and Allelic Heterogeneity of JBTS in FC Individuals
The graph shows the number of different alleles associated with JBTS in FC individuals. The total number (n) of JBTS-affected families with mutations in the indicated genes is shown beneath the gene name. The number of families bearing the recurrent alleles is noted in parentheses following the allele name on the x axis. Deletion 1 refers to chr2: 110,826,262–110,978,224, and deletion 2 refers to chr2: 110,862,369–110,978,000 (hg19 genome assembly).
Acknowledgments
Foremost, we thank the families who generously contributed their time and materials to this research study. This work was selected for study by the Care4Rare (Enhanced Care for Rare Genetic Diseases in Canada) Canada Consortium Gene Discovery Steering Committee (Kym Boycott [lead; University of Ottawa], Alex MacKenzie [co-lead; University of Ottawa], Jacek Majewski [McGill University], Michael Brudno [University of Toronto], Dennis Bulman [University of Ottawa], and David Dyment [University of Ottawa]) and was funded in part by Genome Canada, the Canadian Institutes of Health Research (CIHR), the Ontario Genomics Institute, the Ontario Research Fund, Genome Quebec, and the Children’s Hospital of Eastern Ontario Foundation. We wish to acknowledge the contribution of the high-throughput sequencing platform of the McGill University and Génome Québec Innovation Centre in Montreal. We thank Megan T. Cho (GeneDx) for coordinating the exchange of exome sequencing data. This work was funded by the Government of Canada through Genome Canada, the CIHR, Génome Québec, and the Ontario Genomics Institute (OGI-049). J.L.M. is a National Scholar from the Fonds de Recherche du Québec – Santé (FRQS). M.S. holds a CIHR clinician-scientist training award. D.L. receives financial support from the FRSQ Réseau de Médecine Génétique Appliquée.
Published: October 15, 2015
Footnotes
Supplemental Data include four figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.09.009.
Contributor Information
Stavit Shalev, Email: stavit_sh@clalit.org.il.
Jacques L. Michaud, Email: jacques.michaud@recherche-ste-justine.qc.ca.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org/index.html
Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu/
Ensembl Genome Browser, http://www.ensembl.org
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
GeneMatcher, https://genematcher.org/
Human Splicing Finder, http://www.umd.be/HSF3/
MutationTaster, http://www.mutationtaster.org/
NCBI HomoloGene, http://www.ncbi.nlm.nih.gov/homologene
NCBI Nucleotide, http://www.ncbi.nlm.nih.gov/nuccore
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org/
UCSC Genome Browser, https://genome.ucsc.edu/
UniProt, http://www.uniprot.org/
Supplemental Data
References
- 1.Maria B.L., Hoang K.B., Tusa R.J., Mancuso A.A., Hamed L.M., Quisling R.G., Hove M.T., Fennell E.B., Booth-Jones M., Ringdahl D.M. “Joubert syndrome” revisited: key ocular motor signs with magnetic resonance imaging correlation. J. Child Neurol. 1997;12:423–430. doi: 10.1177/088307389701200703. [DOI] [PubMed] [Google Scholar]
- 2.Romani M., Micalizzi A., Valente E.M. Joubert syndrome: congenital cerebellar ataxia with the molar tooth. Lancet Neurol. 2013;12:894–905. doi: 10.1016/S1474-4422(13)70136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valente E.M., Rosti R.O., Gibbs E., Gleeson J.G. Primary cilia in neurodevelopmental disorders. Nat. Rev. Neurol. 2014;10:27–36. doi: 10.1038/nrneurol.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chih B., Liu P., Chinn Y., Chalouni C., Komuves L.G., Hass P.E., Sandoval W., Peterson A.S. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 2012;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- 5.Czarnecki P.G., Shah J.V. The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol. 2012;22:201–210. doi: 10.1016/j.tcb.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szymanska K., Johnson C.A. The transition zone: an essential functional compartment of cilia. Cilia. 2012;1:10. doi: 10.1186/2046-2530-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurrieri F., Franco B., Toriello H., Neri G. Oral-facial-digital syndromes: review and diagnostic guidelines. Am. J. Med. Genet. A. 2007;143A:3314–3323. doi: 10.1002/ajmg.a.32032. [DOI] [PubMed] [Google Scholar]
- 8.Joubert M., Eisenring J.J., Robb J.P., Andermann F. Familial agenesis of the cerebellar vermis. A syndrome of episodic hyperpnea, abnormal eye movements, ataxia, and retardation. Neurology. 1969;19:813–825. doi: 10.1212/wnl.19.9.813. [DOI] [PubMed] [Google Scholar]
- 9.Srour M., Schwartzentruber J., Hamdan F.F., Ospina L.H., Patry L., Labuda D., Massicotte C., Dobrzeniecka S., Capo-Chichi J.M., Papillon-Cavanagh S., FORGE Canada Consortium Mutations in C5ORF42 cause Joubert syndrome in the French Canadian population. Am. J. Hum. Genet. 2012;90:693–700. doi: 10.1016/j.ajhg.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srour M., Hamdan F.F., Schwartzentruber J.A., Patry L., Ospina L.H., Shevell M.I., Désilets V., Dobrzeniecka S., Mathonnet G., Lemyre E., FORGE Canada Consortium Mutations in TMEM231 cause Joubert syndrome in French Canadians. J. Med. Genet. 2012;49:636–641. doi: 10.1136/jmedgenet-2012-101132. [DOI] [PubMed] [Google Scholar]
- 11.Otto E.A., Ramaswami G., Janssen S., Chaki M., Allen S.J., Zhou W., Airik R., Hurd T.W., Ghosh A.K., Wolf M.T., GPN Study Group Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J. Med. Genet. 2011;48:105–116. doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baala L., Romano S., Khaddour R., Saunier S., Smith U.M., Audollent S., Ozilou C., Faivre L., Laurent N., Foliguet B. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am. J. Hum. Genet. 2007;80:186–194. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majewski J., Schwartzentruber J.A., Caqueret A., Patry L., Marcadier J., Fryns J.P., Boycott K.M., Ste-Marie L.G., McKiernan F.E., Marik I., FORGE Canada Consortium Mutations in NOTCH2 in families with Hajdu-Cheney syndrome. Hum. Mutat. 2011;32:1114–1117. doi: 10.1002/humu.21546. [DOI] [PubMed] [Google Scholar]
- 14.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bielas S.L., Silhavy J.L., Brancati F., Kisseleva M.V., Al-Gazali L., Sztriha L., Bayoumi R.A., Zaki M.S., Abdel-Aleem A., Rosti R.O. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat. Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edvardson S., Shaag A., Zenvirt S., Erlich Y., Hannon G.J., Shanske A.L., Gomori J.M., Ekstein J., Elpeleg O. Joubert syndrome 2 (JBTS2) in Ashkenazi Jews is associated with a TMEM216 mutation. Am. J. Hum. Genet. 2010;86:93–97. doi: 10.1016/j.ajhg.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valente E.M., Logan C.V., Mougou-Zerelli S., Lee J.H., Silhavy J.L., Brancati F., Iannicelli M., Travaglini L., Romani S., Illi B. Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat. Genet. 2010;42:619–625. doi: 10.1038/ng.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon-Salazar T., Silhavy J.L., Marsh S.E., Louie C.M., Scott L.C., Gururaj A., Al-Gazali L., Al-Tawari A.A., Kayserili H., Sztriha L., Gleeson J.G. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am. J. Hum. Genet. 2004;75:979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parisi M.A., Bennett C.L., Eckert M.L., Dobyns W.B., Gleeson J.G., Shaw D.W., McDonald R., Eddy A., Chance P.F., Glass I.A. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am. J. Hum. Genet. 2004;75:82–91. doi: 10.1086/421846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., International Joubert Syndrome Related Disorders Study Group Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 22.Sayer J.A., Otto E.A., O’Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 23.Arts H.H., Doherty D., van Beersum S.E., Parisi M.A., Letteboer S.J., Gorden N.T., Peters T.A., Märker T., Voesenek K., Kartono A. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- 24.Delous M., Baala L., Salomon R., Laclef C., Vierkotten J., Tory K., Golzio C., Lacoste T., Besse L., Ozilou C. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 25.Cantagrel V., Silhavy J.L., Bielas S.L., Swistun D., Marsh S.E., Bertrand J.Y., Audollent S., Attié-Bitach T., Holden K.R., Dobyns W.B., International Joubert Syndrome Related Disorders Study Group Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am. J. Hum. Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noor A., Windpassinger C., Patel M., Stachowiak B., Mikhailov A., Azam M., Irfan M., Siddiqui Z.K., Naeem F., Paterson A.D. CC2D2A, encoding a coiled-coil and C2 domain protein, causes autosomal-recessive mental retardation with retinitis pigmentosa. Am. J. Hum. Genet. 2008;82:1011–1018. doi: 10.1016/j.ajhg.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorden N.T., Arts H.H., Parisi M.A., Coene K.L., Letteboer S.J., van Beersum S.E., Mans D.A., Hikida A., Eckert M., Knutzen D. CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am. J. Hum. Genet. 2008;83:559–571. doi: 10.1016/j.ajhg.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dafinger C., Liebau M.C., Elsayed S.M., Hellenbroich Y., Boltshauser E., Korenke G.C., Fabretti F., Janecke A.R., Ebermann I., Nürnberg G. Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J. Clin. Invest. 2011;121:2662–2667. doi: 10.1172/JCI43639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sang L., Miller J.J., Corbit K.C., Giles R.H., Brauer M.J., Otto E.A., Baye L.M., Wen X., Scales S.J., Kwong M. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L., Szymanska K., Jensen V.L., Janecke A.R., Innes A.M., Davis E.E., Frosk P., Li C., Willer J.R., Chodirker B.N. TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am. J. Hum. Genet. 2011;89:713–730. doi: 10.1016/j.ajhg.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.E., Silhavy J.L., Zaki M.S., Schroth J., Bielas S.L., Marsh S.E., Olvera J., Brancati F., Iannicelli M., Ikegami K. CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat. Genet. 2012;44:193–199. doi: 10.1038/ng.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J.H., Silhavy J.L., Lee J.E., Al-Gazali L., Thomas S., Davis E.E., Bielas S.L., Hill K.J., Iannicelli M., Brancati F. Evolutionarily assembled cis-regulatory module at a human ciliopathy locus. Science. 2012;335:966–969. doi: 10.1126/science.1213506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuz K., Bachmann-Gagescu R., O’Day D.R., Hua K., Isabella C.R., Phelps I.G., Stolarski A.E., O’Roak B.J., Dempsey J.C., Lourenco C. Mutations in CSPP1 cause primary cilia abnormalities and Joubert syndrome with or without Jeune asphyxiating thoracic dystrophy. Am. J. Hum. Genet. 2014;94:62–72. doi: 10.1016/j.ajhg.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaheen R., Shamseldin H.E., Loucks C.M., Seidahmed M.Z., Ansari S., Ibrahim Khalil M., Al-Yacoub N., Davis E.E., Mola N.A., Szymanska K. Mutations in CSPP1, encoding a core centrosomal protein, cause a range of ciliopathy phenotypes in humans. Am. J. Hum. Genet. 2014;94:73–79. doi: 10.1016/j.ajhg.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akizu N., Silhavy J.L., Rosti R.O., Scott E., Fenstermaker A.G., Schroth J., Zaki M.S., Sanchez H., Gupta N., Kabra M. Mutations in CSPP1 lead to classical Joubert syndrome. Am. J. Hum. Genet. 2014;94:80–86. doi: 10.1016/j.ajhg.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thauvin-Robinet C., Lee J.S., Lopez E., Herranz-Pérez V., Shida T., Franco B., Jego L., Ye F., Pasquier L., Loget P. The oral-facial-digital syndrome gene C2CD3 encodes a positive regulator of centriole elongation. Nat. Genet. 2014;46:905–911. doi: 10.1038/ng.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romani M., Micalizzi A., Kraoua I., Dotti M.T., Cavallin M., Sztriha L., Ruta R., Mancini F., Mazza T., Castellana S. Mutations in B9D1 and MKS1 cause mild Joubert syndrome: expanding the genetic overlap with the lethal ciliopathy Meckel syndrome. Orphanet J. Rare Dis. 2014;9:72. doi: 10.1186/1750-1172-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roosing S., Hofree M., Kim S., Scott E., Copeland B., Romani M., Silhavy J.L., Rosti R.O., Schroth J., Mazza T. Functional genome-wide siRNA screen identifies KIAA0586 as mutated in Joubert syndrome. eLife. 2015;4:e06602. doi: 10.7554/eLife.06602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachmann-Gagescu R., Phelps I.G., Dempsey J.C., Sharma V.A., Ishak G.E., Boyle E.A., Wilson M., Marques Lourenço C., Arslan M., Shendure J., Doherty D., University of Washington Center for Mendelian Genomics KIAA0586 is Mutated in Joubert Syndrome. Hum. Mutat. 2015;36:831–835. doi: 10.1002/humu.22821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis E.E., Zhang Q., Liu Q., Diplas B.H., Davey L.M., Hartley J., Stoetzel C., Szymanska K., Ramaswami G., Logan C.V., NISC Comparative Sequencing Program TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat. Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bachmann-Gagescu R., Dempsey J.C., Phelps I.G., O’Roak B.J., Knutzen D.M., Rue T.C., Ishak G.E., Isabella C.R., Gorden N., Adkins J., University of Washington Center for Mendelian Genomics Joubert syndrome: a model for untangling recessive disorders with extreme genetic heterogeneity. J. Med. Genet. 2015;52:514–522. doi: 10.1136/jmedgenet-2015-103087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Gonzalo F.R., Corbit K.C., Sirerol-Piquer M.S., Ramaswami G., Otto E.A., Noriega T.R., Seol A.D., Robinson J.F., Bennett C.L., Josifova D.J. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coppieters F., Lefever S., Leroy B.P., De Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum. Mutat. 2010;31:1097–1108. doi: 10.1002/humu.21337. [DOI] [PubMed] [Google Scholar]
- 44.Jakobsen L., Vanselow K., Skogs M., Toyoda Y., Lundberg E., Poser I., Falkenby L.G., Bennetzen M., Westendorf J., Nigg E.A. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang K., Toedt G., Montenegro Gouveia S., Davey N.E., Hua S., van der Vaart B., Grigoriev I., Larsen J., Pedersen L.B., Bezstarosti K. A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 2012;22:1800–1807. doi: 10.1016/j.cub.2012.07.047. [DOI] [PubMed] [Google Scholar]
- 46.Satish Tammana T.V., Tammana D., Diener D.R., Rosenbaum J. Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during ciliary assembly. J. Cell Sci. 2013;126:5018–5029. doi: 10.1242/jcs.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Damseh N., Danson C.M., Al-Ashhab M., Abu-Libdeh B., Gallon M., Sharma K., Yaacov B., Coulthard E., Caldwell M.A., Edvardson S. A defect in the retromer accessory protein, SNX27, manifests by infantile myoclonic epilepsy and neurodegeneration. Neurogenetics. 2015;16:215–221. doi: 10.1007/s10048-015-0446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Damseh N., Simonin A., Jalas C., Picoraro J.A., Shaag A., Cho M.T., Yaacov B., Neidich J., Al-Ashhab M., Juusola J. Mutations in SLC1A4, encoding the brain serine transporter, are associated with developmental delay, microcephaly and hypomyelination. J. Med. Genet. 2015;52:541–547. doi: 10.1136/jmedgenet-2015-103104. [DOI] [PubMed] [Google Scholar]
- 50.Genome of the Netherlands Consortium Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 51.Veltman J.A., Brunner H.G. De novo mutations in human genetic disease. Nat. Rev. Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 52.Spektor A., Tsang W.Y., Khoo D., Dynlacht B.D. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 53.Tsang W.Y., Dynlacht B.D. CP110 and its network of partners coordinately regulate cilia assembly. Cilia. 2013;2:9. doi: 10.1186/2046-2530-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang W.Y., Bossard C., Khanna H., Peränen J., Swaroop A., Malhotra V., Dynlacht B.D. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye X., Zeng H., Ning G., Reiter J.F., Liu A. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc. Natl. Acad. Sci. USA. 2014;111:2164–2169. doi: 10.1073/pnas.1318737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kroes H.Y., Monroe G.R., van der Zwaag B., Duran K.J., de Kovel C.G., van Roosmalen M.J., Harakalova M., Nijman I.J., Kloosterman W.P., Giles R.H. Joubert syndrome: genotyping a Northern European patient cohort. Eur. J. Hum. Genet. 2015 doi: 10.1038/ejhg.2015.84. Published online April 29, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.