Abstract
Linkage analysis combined with whole-exome sequencing in a large family with congenital and stable non-syndromic unilateral and asymmetric hearing loss (NS-UHL/AHL) revealed a heterozygous truncating mutation, c.286_303delinsT (p.Ser96Ter), in KITLG. This mutation co-segregated with NS-UHL/AHL as a dominant trait with reduced penetrance. By screening a panel of probands with NS-UHL/AHL, we found an additional mutation, c.200_202del (p.His67_Cys68delinsArg). In vitro studies revealed that the p.His67_Cys68delinsArg transmembrane isoform of KITLG is not detectable at the cell membrane, supporting pathogenicity. KITLG encodes a ligand for the KIT receptor. Also, KITLG-KIT signaling and MITF are suggested to mutually interact in melanocyte development. Because mutations in MITF are causative of Waardenburg syndrome type 2 (WS2), we screened KITLG in suspected WS2-affected probands. A heterozygous missense mutation, c.310C>G (p.Leu104Val), that segregated with WS2 was identified in a small family. In vitro studies revealed that the p.Leu104Val transmembrane isoform of KITLG is located at the cell membrane, as is wild-type KITLG. However, in culture media of transfected cells, the p.Leu104Val soluble isoform of KITLG was reduced, and no soluble p.His67_Cys68delinsArg and p.Ser96Ter KITLG could be detected. These data suggest that mutations in KITLG associated with NS-UHL/AHL have a loss-of-function effect. We speculate that the mechanism of the mutation underlying WS2 and leading to membrane incorporation and reduced secretion of KITLG occurs via a dominant-negative or gain-of-function effect. Our study unveils different phenotypes associated with KITLG, previously associated with pigmentation abnormalities, and will thereby improve the genetic counseling given to individuals with KITLG variants.
Introduction
Familial sensorineural non-syndromic unilateral and asymmetric hearing loss (NS-UHL/AHL) (UHL [MIM: 125000]) is rare; only a few families affected by it are described in literature, and no disease-associated genes or loci are known to date.1, 2, 3 Like symmetric hearing loss (HL), UHL/AHL can negatively affect affected individuals’ lives because of cognitive and social developmental delay. UHL/AHL can be inherited as part of a syndrome, e.g., Waardenburg syndrome (WS [MIM: PS193500]) or Pendred syndrome (MIM: 274600), or it can be induced by environmental factors such as prematurity, trauma, or meningitis.
WS is a dominantly inherited auditory-pigmentary syndrome with an estimated prevalence of 5–10:100,000. To date, four types of WS (1–4) have been distinguished, and seven genes have been found to be associated with the syndrome. The classification of WS is based on the presence of specific clinical features in addition to pigmentary abnormalities of hair, skin, and eyes and congenital HL.4 These specific, additional clinical features are craniofacial defects in the case of WS1, craniofacial and limb defects in in the case of WS3, and Hirschsprung disease in the case of WS4. WS2 is not associated with additional clinical features. All WS-associated genes function in the same pathways involved in the development of neural-crest-derived melanocytes by influencing Mitf expression and/or activity.5, 6, 7, 8, 9
Besides WS, there are other HL syndromes accompanied by hypopigmentation due to melanocyte defects.10 One of these is the piebald trait (MIM: 172800). Piebaldism is characterized by a congenital regional absence of melanocytes in the skin and hair that is caused by heterozygous loss-of-function mutations in KIT (MIM: 164920).10 Very occasionally, the piebald trait has been found to be associated with congenital HL, indicating that they might be part of the same disorder.11, 12
By using a combined strategy of linkage analysis and whole-exome sequencing (WES) in a large Dutch family affected by NS-UHL/AHL, we identified a truncating mutation, c.286_303delinsT (p.Ser96Ter [GenBank: NM_003994.5 and NM_000899.4]), of KITLG (MIM: 184745). Targeted gene testing of a panel of NS-UHL/AHL-affected individuals identified an in-frame deletion, c.200_202del (p.His67_Cys68delinsArg [GenBank: NM_003994.5 and NM_000899.4]), in a family of Spanish origin. Due to the dual regulation of melanocyte development by KIT-KITLG signaling and MITF,6, 8, 13 a cohort of WS2-affected individuals was tested for mutations in KITLG, which revealed a missense mutation, c.310C>G (p.Leu104Val [GenBank: NM_003994.5 and NM_000899.4]), that was segregating with WS2 in another family from the Netherlands. We analyzed the effect of these mutations on subcellular localization and secretion of the transmembrane and soluble KITLG isoforms, respectively.
Subjects and Methods
Subject Evaluation
The current study was approved by the medical ethics committees of the Radboud University Medical Center and the Hospital Universitario Ramón y Cajal and the institutional review board of the Baylor College of Medicine. Written informed consent was obtained from all participating subjects.
Medical histories were taken from all participants, and special attention was paid to hearing impairment, vestibular symptoms, and possible causes of acquired deafness. In family W09-1628 (Figure 1A), 26 people (II:4, III:1, III:3, III:4, III:5, III:6, III:7, III:9, IV:1, IV:3, IV:4, IV:5, IV:6, IV:7, IV:8, V:1, V:2, V:3, V:4, V:5, V:6, V:7, V:8, V:9, V:10, and V:11) participated in the ear, nose, and throat (ENT) evaluation. In family S1489, the index case, individual III:1, and his father, individual II:1, took part in an ENT examination (Figure 1B). From family 12-01744, two individuals, the index case, individual II:1, and his mother, individual I:2, both affected by WS2, participated in an ENT evaluation (Figure 1C). The affection status of the other family members was based on information provided by close relatives.
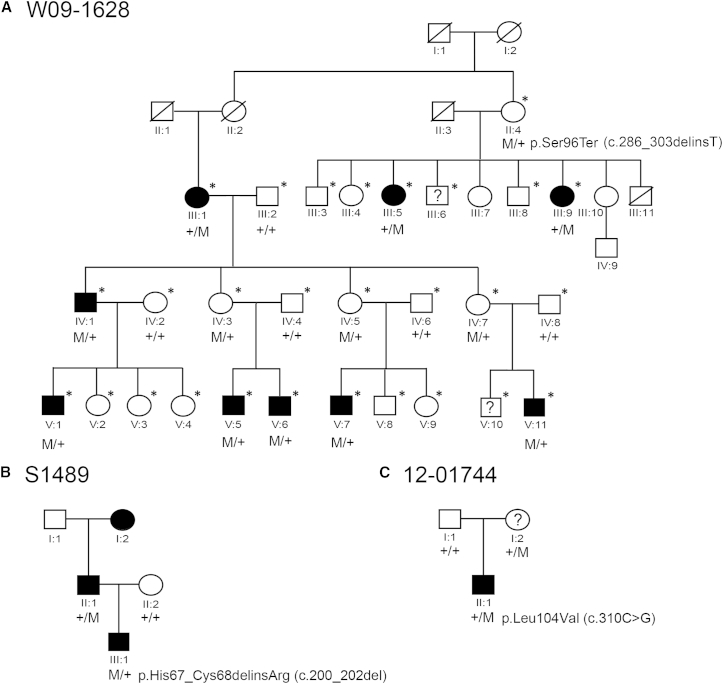
Figure 1.
Families Affected by KITLG Mutations
(A) Pedigree of family W09-1628, affected by NS-UHL/AHL. Asterisks indicate individuals who were genotyped with HumanOmniExpress BeadChip arrays. For privacy reasons, the genotype for the c.286_303delinsT variant has not been indicated for unaffected individuals, except for those with affected offspring. Abbreviations are as follows: M, mutant allele c.286_303delinsT; +, the wild-type allele; ?, unclear affection status.
(B) Pedigree of family S1489, affected by NS-AHL. Abbreviations are as follows: M, the c.200_202del variant; +, the wild-type allele.
(C) Pedigree of family 12-01744, affected by WS2. Abbreviations are as follows: M, the variant c.310C>G; +, the wild-type allele; ?, unclear affection status.
Individuals were considered to be affected if at least one ear showed three pure-tone thresholds below the 95th percentile (P95) at the last evaluation. HL was indicated as asymmetric when the difference between the hearing thresholds of both ears was more than 10 dB at three or more frequencies or more than 15 dB at two or more frequencies. HL was indicated as unilateral if three or more frequencies were beyond the P95 value in only one ear and there was normal hearing in the contralateral ear.
Audiometric examination comprised conventional pure-tone audiometry in a sound-treated room, in accordance with the International Organization for Standardization (ISO) standards 389:1985 and 8253-1:1989. Air-conduction thresholds were measured in dB of HL at 0.25, 0.5, 1, 2, 4, and 8 kHz. Bone-conduction thresholds were measured to exclude conductive or mixed HL. The P95 threshold values in relation to the person’s age and sex were derived for each frequency via the ISO 7029:1984 method.14 In family W09-1628, speech audiometry was performed under the above-mentioned conditions with standard monosyllabic (consonant-vowel-consonant) Dutch word lists. For individual II:1 of family 12-01744, play audiometry with blocks was performed at the age of 3 years. Thresholds were measured in dB of HL at 0.5, 1, and 4 kHz.
Six affected members (III:5, IV:1, V:1, V:5, V:6, and V:7) of family W09-1628 underwent vestibular and ocular motor tests. Ocular motor evaluation comprised saccades, smooth pursuit, optokinetic nystagmus, and gaze-evoked and spontaneous nystagmus. Vestibulo-ocular reflexes (VOR) were evaluated via electronystagmography (ENG) with computer analysis. Vestibular stimulation comprised rotary and caloric tests.15 The diagnosis of hyporeflexia in the rotary test was based on finding parameter values below the 5th percentile of normal. When the response parameters had directional preponderance values of >25%, VOR asymmetry was considered to be significant. The finding of caloric asymmetry relates to a difference in response levels larger than 20%. Caloric hyporeflexia indicates a response level of <7°/sec for cold irrigation and <10°/sec for warm. In accordance with the American National Standards Institute procedures (1999), we have established normative laboratory data by using the 95% confidence limit for the normal population. To avoid underestimation, we have calculated the lower limit of acceptance by using 2.34 times the SD from the mean to define a hyporeactivity of the caloric responses (i.e., 7°/sec and 10°/sec for cold and warm irrigation, respectively).16
High-resolution spiral computed tomography (CT) images of the temporal bones of both ears, with 0.6 mm axial sections and coronal reconstructions, were obtained for detection of possible inner ear anomalies in one affected individual (V:7) of family W09-1628. The images were analyzed by an experienced head and neck radiologist.
In the index case, individual III:1, of family S1489, we performed MRI to exclude inner-ear defects and retrocochlear pathology.
To evaluate the presence of pigmentation abnormalities, we took histories and performed physical examinations in all three families: W09-1628, 12-01744, and S1489. Fourteen individuals of family W09-1628 participated in a dermatological evaluation, ten of whom were heterozygous for the mutation underlying HL in the family (eight had HL, in one of whom HL was not associated with the mutation in the family, and two did not show HL) and four of whom were control individuals without the mutation. Dermatological evaluation included determination of skin type (I–VI), eye color, hair color, poliosis of the head, cilia, and eyebrows, skin depigmentation, hypopigmentation, and hyperpigmentation, number of naevi, allergic reactions, other observed skin abnormalities, and treatment of skin abnormalities in the past. The whole body was examined and a Wood’s lamp was used to analyze hypo-, hyper-, and depigmentations. If necessary, skin abnormalities were reviewed with a dermatoscope. Examination with a Wood’s lamp was also performed for the affected individuals of family 12-01744.
To evaluate the presence of abnormalities in hematopoiesis, we collected blood samples from the 14 individuals who underwent dermatological evaluation. Blood samples were evaluated for hemoglobin concentration, mean corpuscular hemoglobin concentration (MCHC), erythrocyte counts, mean corpuscular volume (MCV), red-blood-cell-count distribution width (RDW), reticulocyte counts, hematocrit, trombocyte counts, leukocyte counts, and differentiated leukocytes (neutrophilic granulocytes, lymphocytes, eosinophilic granulocytes, basophil granulocytes, and monocytes) by microscopic analysis and an automated protocol. As a reference, the Radboud University Medical Center standard was used.17
Description of Subject Cohorts
The cohort of 64 probands of Dutch origin, with a clinical suspicion of WS2, consisted of three groups of affected individuals: (1) 31 subjects with bilateral symmetric HL and hair, eye, and/or skin pigmentation abnormalities, (2) 9 subjects with AHL, 5 of whom had additional hair, eye and/or skin pigmentation abnormalities, and (3) 24 subjects with UHL, 15 of whom had pigmentation abnormalities of hair, eye, and/or skin. Because AHL is described in WS2, MITF testing was requested in routine diagnostics, and MITF was excluded as a disease-associated gene. In the index individual in family 12-01744 of this cohort, all genes known to be involved in WS2 (MITF, SOX10, and SNAI2) were analyzed by Sanger sequencing, and no potentially pathogenic variants were found. The NS-UHL/AHL cohort consisted of 23 index cases, including 10 individuals with NS-UHL and 13 with NS-AHL. In all subjects of this cohort, the c.35delG GJB2 (MIM: 121011) mutation (p.Gly12Valfs∗2 [GenBank: NM_004004.5]) and the m.1555A>G mutation (GenBank: NC_012920.1) in MT-RNR1 (MIM: 561000) were excluded. These subjects were not pretested for MITF mutations.
Linkage Analysis
Genomic DNA was isolated from peripheral blood lymphocytes by standard procedures. The HumanOmniExpress BeadChip arrays (Illumina), with a total of 733,202 SNPs’ covering the genome at a median of 2.2 kb intervals, were used for genotyping DNA from individuals’ blood (Figure 1A). Superlink online SNP 1.1 software was employed for multipoint LOD score calculations, and a window size of ten SNPs was used.18 In order to select independent SNPs, pruning (r2 = 0.025) was performed with PLINK.19 The disease was assumed to be an autosomal-dominant disorder with a disease-allele frequency of 0.001. The penetrance of the disease allele was set at 70%.
In order to confirm the linkage region defined by Superlink, we performed genotyping of variable number of tandem repeat (VNTR) genetic markers.20 Touchdown PCR was used to amplify the VNTR markers. Marker heterozygosity, order, and genetic location were derived from the Marshfield genetic map. VNTR markers were selected to encompass the linkage interval, on chromosome 12, region q21.32-q23.1. The alleles were assigned with GeneMapper v.4.0 software (Applied Biosystems).
WES
WES was performed at the Human Genome Sequencing Center (HGSC) at the Baylor College of Medicine through the Baylor-Hopkins Center for Mendelian Genomics initiative. Using 0.5 μg of DNA, we constructed an Illumina paired-end pre-capture library in accordance with the manufacturer’s protocol (Illumina Multiplexing_SamplePrep_Guide_1005361_D), with modifications as described in the BCM-HGSC Illumina Barcoded Paired-End Capture Library Preparation protocol. Four pre-captured libraries were pooled and then hybridized in solution to the HGSC CORE design (52 Mb, NimbleGen), in accordance with the manufacturer’s protocol (NimbleGen SeqCap EZ Exome Library SR User’s Guide version 2.2), with minor revisions.21 The sequencing run was performed in paired-end mode with the Illumina HiSeq 2000 platform, and sequencing-by-synthesis reactions were extended for 101 cycles from each end and for an additional seven cycles for the index read. With a sequencing yield of 10.4 Gb, the samples achieved 91% of the targeted exome bases covered to a depth of 20× or greater. The overall coverage in the linkage interval was optimal, with 97.7% and 94.4% of base pairs covered ≥10× for individuals IV:1 and III:9, respectively. Illumina sequence analysis was performed with the HGSC Mercury analysis pipeline,22, 23 which moves data through various analysis tools, from the initial sequence generation on the instrument to annotated variant calls (SNPs and intra-read insertions and deletions).
Sanger Sequence Analysis
Primers for amplification of exons and exon-intron boundaries of KITLG (GenBank: NM_003994.5 and NM_000899.4) and KITLG rs642742 and rs12821256 were designed with ExonPrimer (Table S1). Amplification by PCR was performed on 40 ng of genomic DNA with Taq DNA polymerase (Roche or Invitrogen). PCR fragments were purified with NucleoFast 96 PCR plates (Clontech) or ExoI/FastAP (Fermentas), in accordance with manufacturers’ protocols. Sequence analysis was performed with the ABI PRISM BigDye Terminator Cycle Sequencing v.2.0 Ready Reaction kit and analyzed with the ABI PRISM 3730 DNA analyzer (Applied Biosystems). GenBank: NM_003994.5 and NM_000899.4 were employed as reference sequences.
The presence of the KITLG c.286_303delinsT mutation was determined in 153 ethnicity-matched control individuals. To do so, we amplified exon 4 of KITLG and analyzed fragments by agarose gel electrophoresis. The presence of the KITLG c.200_202del variant was investigated in 200 control individuals of Spanish origin by a size-based screening with one primer labeled with a 6-FAM fluorophore. Allele size was determined by capillary electrophoresis in an ABI PRISM 3100 genetic analyzer (Applied Biosystems) and analysis with the GeneScan software (Applied Biosystems). For all three KITLG variants, absence was verified in the following databases: the Exome Variant Server (>200,000 exomes), the Nijmegen WES database (5,031 exomes), the Exome Aggregation Consortium database (ExAC; 65,000 exomes), the “Centro de Investigación Biomédica en Red de Enfermedades Raras” (CIBERER) Exome Sever (403 exomes), and the WES database of the Baylor-Hopkins Center for Mendelian Genomics (∼5,000 exomes).
A possible effect of the KITLG variants was predicted with the following software tools included in Alamut (Interactive Biosoftware): Sorting Intolerant from Tolerant (SIFT), Polymorphism Phenotyping version 2 (PolyPhen2), MutationTaster, NetGene, the Berkeley Drosophila Genome Project splice prediction site, and Human Splice Finder. The conservation of the substituted amino acids was scored with the ConSeq online tool.
pCR4-Topo Cloning
Exon 4 of KITLG from individual IV:1 in family W09-1628 and the index case, individual III:1, from family S1489 was amplified (primers as for sequencing) and cloned into a pCR4-Topo vector (Invitrogen), in accordance with the manufacturer’s protocol. Plasmid DNA was isolated from transformed E. coli DH5α with the NucleoSpin Plasmid (NoLid) kit (Machery Nagel) and analyzed by Sanger sequence analysis as described above.
Expression of KITLG in NIH 3T3 Cells
Total RNA was extracted from human control peripheral blood with the QIAamp RNA Blood Mini Kit (QIAGEN), according to the manufacturer’s instructions. Reverse transcription was carried out with 1 μg of RNA and random hexamers with the Transcriptor First Strand cDNA Synthesis Kit (Roche). Full-length transcripts encoding the human KITLG transmembrane (GenBank: NM_003994.5) and soluble (GenBank: NM_000899.4) isoforms were amplified with modified primers; the forward primer contains a BamHI site and the reverse primers (R2-XhoI or R1-XhoI) contain a XhoI restriction site (Table S1). We cloned amplimers in the eukaryotic expression vector pIRES-hrGFP-1a (Stratagene) to obtain two different sets of plasmids suitable to generate, respectively, (1) human soluble KITLG (with the region encoded by exon 6) and (2) KITLG (without the region encoded by exon 6) fused to three FLAG epitopes at the C-terminal end.
Plasmids encoding the aberrant KITLG, p.His67_Cys68delinsArg, p.Ser96Ter, and p.Leu104Val, were generated by direct mutagenesis with the QuickChange Site-Directed Mutagenesis Kit (Stratagene) and verified by Sanger sequencing. Cell culture, transfection, and detection of proteins by immunofluorescence were performed as previously described.24 Cells were transiently transfected with 500 ng of wild-type or mutated constructs via Lipofectamine 2000 (111668, Invitrogen), according to the manufacturer’s protocol. As primary antibodies, the monoclonal anti-FLAG M2 mouse antibody (1:500; Sigma, F1804) or anti-KITLG antibody (1:1,000; Abcam, ab52603, anti-SCF) were used, and as secondary antibodies, the Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (1:1,000; Molecular Probes, A11017; Invitrogen) or Alexa Fluor 594-conjugated goat anti-rabbit antibody (1:500; Molecular Probes, A11037; Invitrogen) were employed. Slides were mounted with Fluorsave Reagent (Calbiochem). Images were acquired and analyzed by confocal microscopy (Nikkon C1 plus ECLIPSE Ti-e microscope and an 60X Plan Apo VC OIL objective with NA 1.4).
ELISA
In order to assess the levels of the soluble KITLG isoform, we employed a commercial ELISA kit (ab100636 SCF Human ELISA kit) according to manufacturer’s instructions (Abcam). NIH 3T3 cells were independently transfected with plasmids encoding soluble wild-type KITLG or soluble KITLG containing the mutations identified in this study (p.Ser96Ter, p.Leu104Val, and p.His67_Cys68delinsArg). After cells were cultured for 24 hr, we collected cell-free supernatants by centrifuging them at 15,000 rpm for 10 min at 4°C. Samples were stored at −70°C until the ELISAs were performed. Samples were diluted with one volume of 1× assay diluent B. The detection limit was 2 pg/ml and the mean intra- and inter-assay coefficients of variability were <10% and <12%, respectively. Samples were assayed in quadruplicate and at least four independent transfections were performed. Transfection levels of cells with the wild-type and mutant constructs were determined by detection of GFP and KITLG by fluorescence and immunofluorescence, respectively, of permeabilized cells. For detection of the latter, we used the anti-KITLG antibody (Abcam, ab52603, anti-SCF) (Figure S1). The dosage of the soluble form detected by ELISA in the medium was normalized to the quantity of transfected cells per well in each case. Cells were examined with a fluorescence inverted microscope (Olympus IX81) equipped with UPLFLN40X, PLAPON60XO, and UPLSAPO100XO objectives. Images were recorded with a cooled CCD-F View II camera (Soft Imaging System) and processed with Cell R software (Olympus). We verified that the ELISA kit allowed the identification of the aberrant KITLG proteins, including the truncated form (p.Ser96Ter), by using cellular extracts of permeabilized cells transfected with constructs encoding FLAG-tagged transmembrane KITLG aberrant isoforms (Figure S2).
Results
Clinical Characterization of Families with NS-UHL/AHL and WS2
Family W09-1628
A large Dutch family, W09-1628 (Figure 1A), with autosomal-dominant, congenital, non-syndromic sensorineural HL and a large inter- and intra-individual variation of severity and audiogram configuration was ascertained. Otoscopy was normal for all but two individuals, V:7 and V:8 (for details, see the Supplemental Note).
In individuals with UHL, either the left or the right ear was affected, and severity of the HL varied from mild to profound (Figure 2A and Figure S3). Overall, five individuals presented with NS-AHL (III:9, V:1, V:5, V:6, and V:11) and four individuals with NS-UHL (III:1, III:5, IV:1, and V:7) (Figure S3). In four ears, HL either only affected lower frequencies (<2 kHz) or was more pronounced in these frequencies. Interaural differences in audiogram configuration were observed in one individual (III:9, Figure S3). Additional information on individual cases is provided in the Supplemental Note.
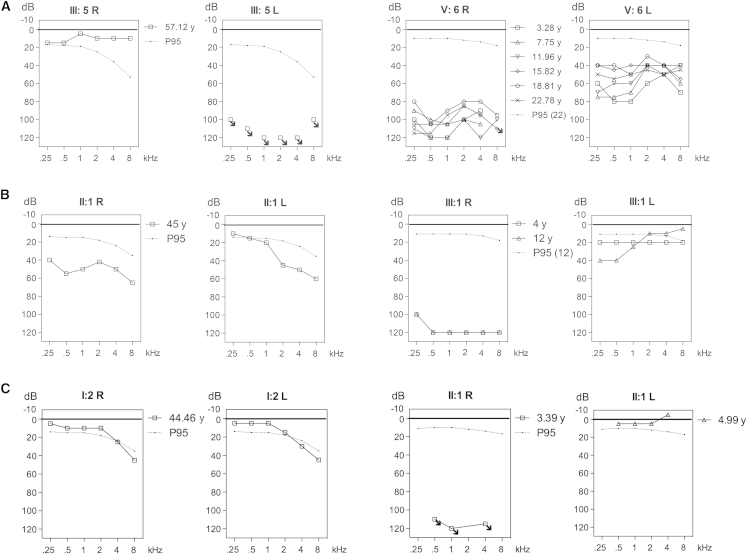
Figure 2.
Pure Tone Audiometry in Families Affected by KITLG Mutations
(A) Pure tone audiograms for family-W09-1628 individuals with UHL (III:5) and AHL (V:6).
(B) Pure tone audiograms for family S1489, affected by AHL.
(C) Pure tone audiograms for family 12-01744, affected by WS2. Abbreviations are as follows: R, right ear; L, left ear; P95, thresholds according to the P95 of presbyacusis (ISO 7029:1984).14
Analysis of longitudinal data revealed that HL was stable in all individuals. Only minor negative progression was seen in individual longitudinal data, which can probably be explained by the young age at which first thresholds were determined. The genetic defect underlying the NS-UHL/AHL in this family has a reduced penetrance, as evidenced by the fact that four individuals transmitting the disease to their offspring were unaffected (II:4, IV:3, IV:5, and IV:7) (Figure 1A).
Vestibular function was evaluated in six of the hearing-impaired family members (Table S2). Three individuals (V:1, IV:1, and III:5) displayed asymmetry on calorisation, corresponding with the side of the worst ear. One UHL-affected individual (V:7) displayed a marginal hyporeflexia on both sides; the other ear had a dip at 4 kHz only, although no history of excessive noise exposure was reported. In the remaining two individuals (V:5 and V:6), vestibular function was found to be normal (Table S2). Except for individual II:4, none of the individuals who carried the mutation reported vestibular symptoms. During history taking, individual II:4 indicated a possible Meniére disease.
Because KITLG functions in the proliferation, migration, and survival of hematopoietic stem cells and melanoblasts,25, 26, 27, 28 blood cell counts, as well as skin, hair, and eye color, were evaluated in family-W09-1628 members with and without the KITLG mutation. All determined hematological parameters were within the normal range, although individuals IV:1, IV:3, and V:4 were treated with ferrofumarate for anemia in the past (Table S3). These findings suggest that, in the heterozygous state, the KITLG mutation in the present family does not affect steady-state hematopoiesis.
All evaluated individuals had skin type I or II, blond hair, and blue eyes (Table S4). Heterochromia iridis and dystopia canthorum were not observed. Hypo- and depigmentations of the skin were not found in the younger generation (generation V). In the older generations, hypo- and depigmentations were present in individuals with and without the KITLG mutation (Table S4). These pigmentation characteristics are age related and a common feature in the population.29 Our findings suggest that the heterozygous KITLG mutation in this family does not lead to pigmentation abnormalities of skin, eyes, or hair.
Family S1489
The index individual of family S1489 (Figure 1B) and his father presented with AHL and variability in severity and audiogram configuration (Figure 2B), as was observed in family W09-1628. The AHL in the propositus (III:1) was reported to be of prelingual onset without progression. The earliest clinical evidence of HL was obtained at the age of four years. HL was profound at all frequencies in the right ear and mild in the left ear, affecting the low frequencies. The father of the propositus (II:1) presented with moderate bilateral AHL. Audiogram configuration was downsloping for the left ear and flat for the right ear. No vestibular symptoms were reported by the affected father and son. Examination of the skin revealed no hypo- or hyperpigmentation features. Additional information is provided in the Supplemental Note.
Family 12-01744
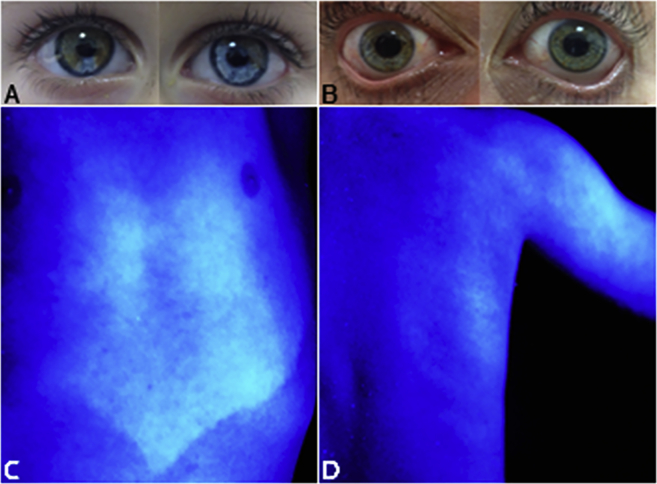
In family 12-01744 (Figure 1C), otoscopy demonstrated otitis media with effusion in the right ear of the index individual (II:1) at the age of almost five years. Therefore, reliable thresholds could only be measured for the left ear, and these were normal. At the age of three years, play audiometry had been performed elsewhere, which revealed profound HL for the right ear (Figure 2C). The mother of the index individual (I:2) did not show any abnormalities in otoscopy, and her hearing was normal with only two frequencies, 4 kHz and 8 kHz, below the P95 values (Figure 2C). The index individual had heterochromia iridis, which is suggested to be present in his mother as well by small blue spots in the iris (Figures 3A and 3B). Both individuals had skin type III. The index individual had brown-blond hair. On the thorax and upper arms, he had a partially sharply demarcated hypopigmented macule, which was already present at birth. Furthermore, a nummular sharply demarcated hyperpigmented macule was seen on his back (Figures 3C and 3D).
Figure 3.
Eye and Skin Hypopigmentation Phenotypes in Family 12-01744, Affected by WS2
(A) Heterochromia iridis in the index case, individual II:1.
(B) Signs of heterochromia iridis in his mother (I:2).
(C) A hypopigmented, sharply demarcated macule on the thorax of individual II:1 and
(D) on the upper arm. A nummular, sharply demarcated hyperpigmented macula at the back, detected by a Wood’s lamp.
A Locus for NS-UHL/AHL on Chromosome 12q21.32–q23.1
In family W09-1628, genotyping was performed with high-density SNP arrays (Figure 1A). Genome-wide multipoint LOD scores were calculated with 14,488 independent SNPs, which revealed a single linkage interval of 13.15 Mb on chromosome 12q21.32–q23.1, delimited by rs10459171 and rs35723 (chr12: 87,808,426–100,960,087; UCSC Genome Browser GRCh37/hg19). A maximum LOD score of 4.27 was calculated for rs7132875 and rs7309222 (Figure S4). There were no other regions with a LOD score suggestive of linkage (≥2.0) (Figure S4).
As a next step, we confirmed linkage of the disease to the 12q21.32–q23.1 region by genotyping VNTR genetic markers, and recombination events delimited the linkage interval to a region flanked by markers D12S88 and D12S346 (12q21.31–q23.1; chr12: 86,371,384–99,528,530; GRCh37/hg19) (Figure S5). According to the UCSC Genome Browser (GRCh37/hg19), 82 RefSeq genes were present in the critical region, but none were previously reported to be related to deafness in humans, and in regard to mice, only the ortholog of KITLG (Kitl) was associated with sensorineural HL (Mouse Genome Informatics database).
WES Revealed a Truncating Mutation in KITLG
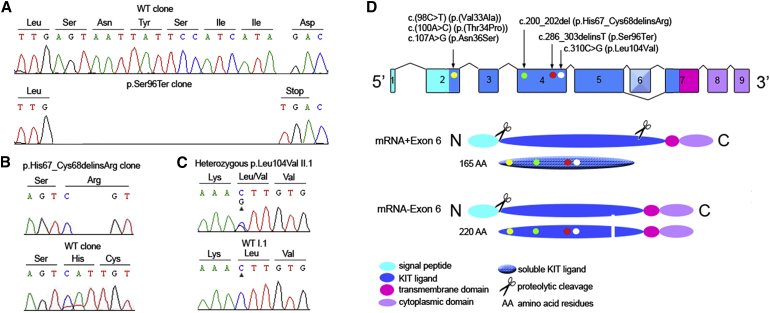
Because of the size of the critical region, we performed WES for the affected individuals III:9 and IV:1 to identify the genetic defect underlying the NS-UHL/AHL in family W09-1628. Multiple filter steps conducted on the initial 87,751 variants that were shared by individuals III:9 and IV:1 (Table 1) revealed two heterozygous candidate variants in exon 4 of KITLG, c.286_303del and c.303insT. Sanger sequencing after the cloning of KITLG exon 4 amplimers demonstrated that both variants are present in cis and thus are likely to represent one mutational event (c.286_303delinsT) (Figure 4A). The predicted effect of the c.286_303delinsT variant is a shift of the reading frame and a premature termination of protein synthesis, p.Ser96Ter. The variant was detected heterozygously in all affected individuals of the family, in all unaffected individuals who transmitted the mutation to their offspring, in one individual with an unknown affection status, and in an additional unaffected individual (Figure 1A). The KITLG variant c.286_303delinsT was neither present in 153 ethnicity-matched control individuals nor in exome databases (see Subjects and Methods). Copy-number variation (CNV) analysis of WES data did not reveal any CNV in the linkage interval or any CNVs that were shared by the two individuals (Table S5).
Table 1.
Filter Steps Applied on Sequence Variants Identified in WES of Individuals III:9 and IV:1 of Family W09-1628
| Filter Steps | No. of Variants |
|---|---|
| Shared exome | 87,751 |
| In linkage region 12q21.31–q23.1 | 241 |
| ≤0.5% Nijmegen in-house frequency | 50 |
| Exonic missense, nonsense, indels, non- and canonical splice sites | 9 |
| Confirmed by Sanger sequencing | 2 in KITLG |
Indicated are the number of variants as compared to reference genome GRCh37 (hg19) after each filter step.
Figure 4.
Genetic Defects Underlying UHL and AHL and Schematic Representation of the Genomic Structure of KITLG and of the Encoded KITLG Isoforms
(A) Electropherograms of pCR4-topo clones representing the wild-type and c.286_303delinsT alleles of KITLG exon 4 of individual IV:1 (family W09-1628).
(B) Electropherograms of pCR4-topo clones representing the wild-type and c.200_202del alleles of KITLG exon 4 of individual III:1 (family S1489).
(C) Partial nucleotide sequences of KITLG exon 4 of case II:1 of family 12-01744. The mutated nucleotide is marked by an arrowhead. GenBank: NM_003994.5 and NM_000899.4 were used as reference sequences.
(D) Exon 6 is alternatively spliced. The yellow dots represent the gain-of-function mutations c.98C>T, c.100A>C, and c.107A>G, causative of familial hyper- and hypopigmentation syndromes. The green and red dots represent the mutations identified to be causative of NS-UHL/AHL in the present study. The white dot represents the mutation identified in a family with WS2. All depicted variants affect the KIT ligand domain of both the soluble and transmembrane isoforms of KITLG. Abbreviation is as follows: WT, wild-type.
On the basis of the segregation pattern of the c.286_303delinsT variant in KITLG, we calculated a penetrance of 60%–67%. The SNPs rs642742 and rs12821256 are located in KITLG enhancers ∼326 kb and ∼350 kb upstream of the KITLG transcription start site, respectively, and are described to alter enhancer activity.30, 31 To evaluate a possible contribution of these SNPs to the penetrance of HL in the family, they were genotyped in all members with the mutation (Figure S5). No correlation was observed between the SNP genotypes and the penetrance of the mutation in this family.
KITLG Variants in NS-UHL/AHL and WS2
We further addressed involvement of KITLG mutations in both NS-UHL/AHL and in WS2. HL in individuals with WS2 is variable, given that it can be asymmetric and/or unilateral in some of the cases.32, 33 Furthermore, function of MITF (MIM: 156845), which is associated with WS2, is influenced by KITLG-KIT signaling.8 We selected two panels of affected individuals. The first panel consisted of 64 probands with a clinical suspicion of WS2. The second panel consisted of 23 unrelated probands with autosomal-dominant NS-UHL/AHL, mainly of Spanish origin. Further details on the panels are provided in Subjects and Methods. Two additional heterozygous KITLG variants were identified, namely an in-frame deletion, c.200_202del (p.His67_Cys68delinsArg; Figure 4B), in an index individual of Spanish origin with NS-AHL and a missense variant, c.310C>G (p.Leu104Val; Figure 4C), in a WS2-affected individual of Dutch origin with UHL.
The KITLG variant c.200_202del was heterozygously present in the father of the proband who also presented with NS-AHL (Figure 1B, Figure 2B). The c.200_202del variant was neither detected in 200 ethnicity-matched control alleles nor in exome databases (see Subjects and Methods). The predicted effect of the deletion of three nucleotides on the encoded protein is an amino acid deletion and a substitution (p.His67_Cys68delinsArg). Cys68 is highly conserved with a score of 9 in the ConSeq online bioinformatic tool (range of 1–9, where 9 is conserved and 1 is variable). The Cys68 residue forms an intramolecular Cys-Cys bond with Cys163, which is essential for the full biological activity of KITLG.25, 34, 35, 36 The loop in which Cys68 is located connects a β sheet to an α helix. Homology modeling of KITLG p.His67_Cys68delinsArg predicts that when residue Cys68 is not present and the Cys68-Cys163 bond thus cannot be formed, the local structure of KITLG around the loop is affected. Also, a reactive Cys (Cys163) is left without a binding partner, which could lead to even larger structural changes than predicted (Figures S6A and S6B). The interaction with the KIT receptor is predicted to be preserved (Figure S6B). In aggregate, these data suggest that the cysteine at position 68 has a critical role in maintaining the structural and functional integrity of the protein.
In family 12-01744, KITLG variant c.310C>G was inherited from the mother (Figure 1C). The variant is predicted to substitute valine for leucine at position 104 (p.Leu104Val) and is not present in any of the exome databases (see Subjects and Methods), which include at least 4,000 exomes of individuals of Dutch origin. The amino acid substitution is predicted to be deleterious for protein function according to PolyPhen2, with a score of 0.998 (range of 0–1, where 0 is benign and 1 is probably damaging), MutationTaster, with a score of 1 (range of 0–1, where 0 is benign and 1 is probably damaging), and SIFT, with a score of 0.04 (where ≤0.05 is probably damaging and >0.05 is predicted to be tolerated). Leu104 is highly conserved with a score of 9 in ConSeq. Leu104 is buried inside an α helix and therefore does not interact with the KIT receptor. The substitution Leu104Val is predicted to have a small, local effect on the structure given that valine is predicted not to be exactly at the same position as leucine (Figures S6C and S6D). Although Leu104 is buried in the protein, neighboring residues can contact the KIT receptor. Therefore, a small effect on the position of these contacting residues is expected, but because the contact surfaces of KITLG and the KIT receptor are much larger, the interaction is not predicted to be severely impaired by the Leu104Val substitution (Figures S6C and S6D).
KITLG encodes two KITLG isoforms, a transmembrane isoform and a soluble isoform, through alternative splicing and proteolytic processing, respectively (Figure 4D). In mice, it appears that soluble KITLG plays a role in melanocyte migration and/or early survival and that transmembrane KITLG plays a role in melanocyte survival at or close to the destination.26, 37 All three KITLG mutations identified in this study affect both KITLG isoforms (Figure 4D).
KITLG Variants Affect Protein Localization and/or Secretion
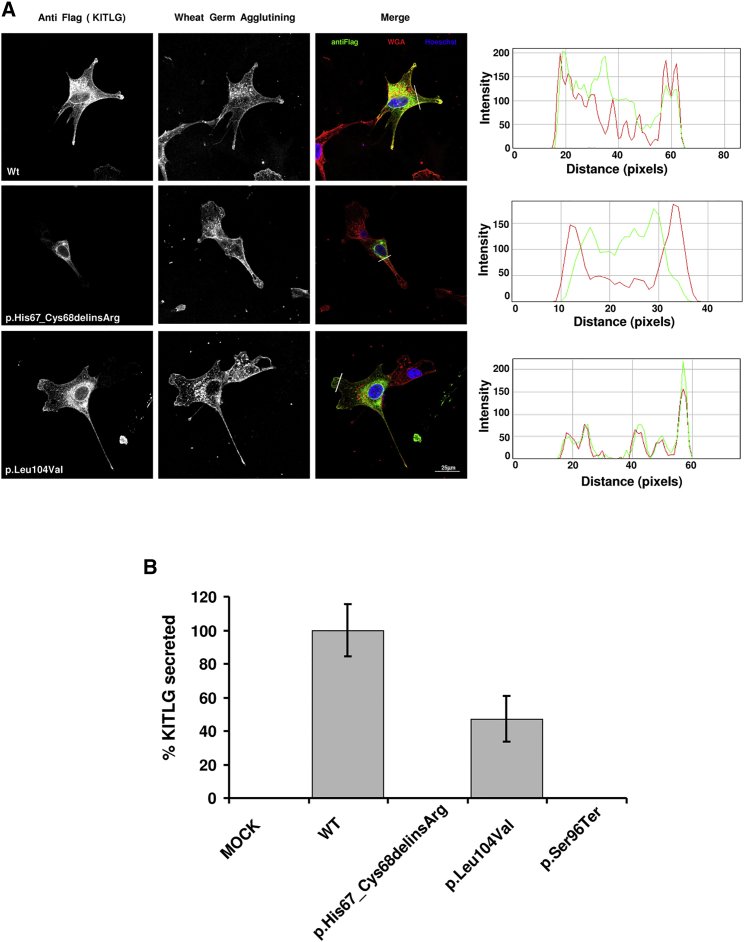
We studied the effect of the p.His67_Cys68delinsArg and p.Leu104Val variants on subcellular localization of transmembrane KITLG. NIH 3T3 cells were transiently transfected with a set of plasmids to express the FLAG-tagged wild-type or variant human transmembrane KITLG (Figure 5A). The levels of wild-type and aberrant variants of KITLG were verified by western blot analysis (Figure S7). We addressed localization of KITLG at the cell membrane by labeling the membrane with wheat germ agglutinin (WGA), a member of the lectin family, which binds to N-acetyl-D-glucosamine and sialic acid residues on the surface of cells. Both FLAG-tagged wild-type and p.Leu104Val KITLG were detected in the cytoplasm and at the cell membrane, as well as in lamellipodia and filipodia (Figure 5A). In contrast, p.His67_Cys68delinsArg KITLG staining did not overlap with the WGA labeling (Figure 5A). This indicates that the transmembrane p.His67_Cys68delinsArg KITLG fails to reach the cell membrane and therefore will not, or only to a very limited extent in comparison to controls, be integrated in the cell membrane, which supports functional consequences of the mutation and the pathogenic effect of this variant.
Figure 5.
Effect of KITLG Mutations on Subcellular Localization and Excretion of the Transmembrane and Soluble KITLG Isoforms, Respectively
(A) WGA staining of NIH 3T3 cells transfected with constructs encoding the FLAG-tagged wild-type, p.His67_Cys68delinsArg, and p.Leu104Val transmembrane isoform of KITLG. Cells were incubated with anti-FLAG antibodies and Alexa-488-conjugated goat anti-mouse (left column), and WGA Texas Red-X is visualized in the middle column. In the right column, the signals of the left and middle columns are merged. Co-localization of WGA with wild-type and p.Leu104Val KITLG was observed at the cell membrane but not with p.His67_Cys68delinsArg KITLG. WGA (red) and KITLG (green) fluorescence intensity profiles were plotted as a function of distance with ImageJ software. Scale bar represents 25 μm.
(B) The amount of soluble p.Leu104Val KITLG is significantly reduced (p < 10−3, Student’s t test) in the medium of transfected NIH 3T3 cells as measured by ELISA. No soluble KITLG was detected for the p.His67_CysdelinsArg and p.Ser96Ter mutations. Data are means ± SD of four independent experiments. Abbreviation is as follows: WT, wild-type.
As a next step, we investigated whether the p.His67_Cys68delinsArg, p.Leu104Val, and p.Ser96Ter variants affect proteolytic processing and subsequent secretion of soluble KITLG. After expression of wild-type and variant soluble KITLG in NIH 3T3 cells, secreted KITLG was measured by ELISA of the growth medium. The amount of soluble p.Leu104Val KITLG was significantly reduced as compared to that of wild-type soluble KITLG, whereas no p.His67_Cys68delinsArg and p.Ser96Ter soluble KITLG could be detected (Figure 5B). These findings support a pathogenic effect of all three KITLG variants.
Because KITLG can be detected in human blood,34 we assessed by western blot analysis whether the levels of KITLG were significantly reduced in peripheral blood samples of affected individuals with the c.286_303delinsT, c.200_202del, and c.310C>G mutations as compared to levels in control individuals. No clear differences were observed (Figure S8). Moreover, the truncated protein encoded by the c.286_303delinsT KITLG allele was not detected. This suggests that either the truncated protein is not stable and/or that the transcript is degraded by nonsense-mediated decay (NMD). Because the truncating variant is in exon 4, not in the last or penultimate exon, the transcript is predicted to be subject to NMD.38
Kitl Expression in the Mouse Cochlea
Because expression of KITLG or Kitl in cochlear tissue was not demonstrated previously, we assessed transcription of Kitl in mouse cochlea by RT-PCR, employing cDNA derived from RNA of P2 and P28 mice as templates. Two amplification products were obtained that corresponded to Kitl transcripts with and without exon 6. Moreover, qPCR demonstrated that the relative levels of both Kitl alternative transcripts were higher at postnatal day 28 (P28) than at P2 (Figure S9). Although blood in cochlear vessels might have contributed to the detected Kitl transcripts, our data suggest that Kitl functions postnatally in the mouse inner ear.
Discussion
Here, we present variant alleles of KITLG, alternatively called SCF (stem cell factor) or MGF (mast cell growth factor), and provide evidence that these disease-associated alleles underlie dominantly inherited NS-UHL/AHL and WS2. A combined strategy of linkage analysis and WES revealed a heterozygous truncating mutation, c.286_303delinsT, in KITLG in a large Dutch family (W09-1628) affected by NS-UHL/AHL. The HL is inherited as a dominant trait with incomplete penetrance. Through further testing of KITLG, two additional mutations were identified in a small family affected by NS-UHL/AHL and in a family affected by WS2. The HL in all three families with mutations in KITLG was congenital, stable, and variable with regard to the affected ear as well as audiogram configuration and severity. Based on the variable expression of KITLG mutations, bilateral symmetric HL might be seen as a phenotypic outcome. However, no potentially pathogenic variants were identified in WES data of 231 individuals with bilateral and symmetric HL (87 autosomal-dominant NS-HL and 144 isolated cases). This indicates that KITLG mutations are not a frequent cause of bilateral symmetric sensorineural HI.
KITLG-KIT signaling plays a role in proliferation, migration from the neural crest, and survival and differentiation of hematopoietic precursor cells, primordial germ cells, and melanoblasts.25, 26, 27, 28, 39 In mice, severe mutations in Kitl (Sl locus) and Kit (W locus) are embryonic lethal when homozygous, but viable when compound heterozygous with less severe defects, as are semi-dominant mutations.40, 41, 42 Mice homozygous for the semi-dominant alleles Wv of Kit and Sld of Kitl are viable and display similar phenotypes, i.e. macrocytic anemia, sterility, absence of coat pigmentation, and abnormal pigmentation of the iris and of the stria vascularis in the inner ear.28, 31, 43, 44, 45, 46, 47 Absence or reduction of highly specialized melanocytes, the intermediate cells, in the stria vascularis leads to no or reduced endocochlear potential (EP) and consequently to HL. This EP is the driving force for the transduction of sound into hair cells via influx of K+ and other cations.48, 49 Steel et al. (1992)47 demonstrated that in Sld/Sld mice, both the survival and targeting of melanoblasts to the developing inner ear is affected, but early melanoblast differentiation and migration is not. At 11 days of gestation, lower numbers of melanoblasts could be detected near the otic vesicle in mutant animals than in control animals, and these numbers further decreased during development.26, 47
In most of the studied inner ears of 6-day-old Wv/Wv mice, no melanocytes could be detected in the stria vascularis, which explains why no EP was measured. However, the penetrance of this phenotype was incomplete, as evidenced by the fact that, in some ears, the EP was nearly normal and intermediate cells were present, as in control mice.28 This variability in severity of the hearing impairment and affected ear is reminiscent of the phenotype in the presented families with KITLG mutations. In Wv/Wv mice, this variability in inner-ear phenotype was found to reflect the number of melanocytes present and how far they migrated along each cochlea.28 Also, a stria vascularis with structural anomalies was found to be capable of maintaining a normal EP,50 which could contribute to the variable expressivity of the KITLG mutations in families W09-1628 and 12-01744. Findings on secondary hair-cell degeneration have been reported in Kit and Kitl mutant mice compound heterozygous for a null allele and an allele with the less severe mutations, Wv and Sld, respectively.27, 51 We have no indications of secondary hair-cell loss in the presented families because the HL was found or reported to be stable. However, we cannot exclude progressive degeneration of hair cells in the prenatal stage, especially in those individuals with severe to profound HL in one ear.
In family W09-1628, vestibular testing in six individuals with the mutation and with HL revealed abnormal, vestibular responses in four of the subjects. However, these subjects did not report vestibular symptoms during history taking. The only individual who complained about balance problems was not tested on vestibular function because of her high age. In conclusion, it seems that vestibular dysfunction could be part of the inner-ear phenotype associated with KITLG mutations. However, this seems to lead to subclinical abnormal function, given that the majority of individuals with the mutation did not report vestibular symptoms, which suggests sufficient compensatory mechanisms.
In families W09-1628 and S1489, there are no indications for phenotypes of the tissues that are affected in the Kit and Kitl mutant mice, i.e. hematopoietic tissues, gonads, eye, hair, and skin, other than the inner ear. This suggests dominance of the wild-type KITLG allele over the mutant allele in those tissues. For the KITLG mutation in family 12-01744, this does not hold true for the iris and skin.
Interestingly, activation of KITLG mutations and variants in regulatory sequences of KITLG affects pigmentation of skin and hair. First, missense mutations in KITLG are causative for familial progressive 2 hyperpigmentation and/or hypopigmentation syndromes (Figure 4D) (MIM: 145250). Mutations underlying these syndromes are demonstrated (c.107A>G, p.Asn36Ser [GenBank: NM_003994.5 and NM_000899.4]) or predicted (c.107A>G, p.Val33Ala; and c.100A>C, p.Thr34Pro) to have an activating effect because all three are in the conserved ValThrAsnAsn motif.52, 53 Protein modeling suggests that mutations in this domain might affect the affinity of KITLG for KIT.52 Second, SNPs in KITLG regulatory regions are associated with skin and hair pigmentation (rs642742 and rs12821256 [MIM: 611664]).31 These data, together with the facts that KIT signaling regulates MITF function13, 54, 55 and that genes associated with hypopigmentation-deafness disorders function in the KIT-MITF pathway,8, 56 pinpointed KITLG as a candidate gene for WS2. Indeed, we have identified a missense mutation in KITLG to be associated with WS2 in family 12-01744. The HL phenotype in this family is similar to that in families W09-1628 and S1489. The pigmentation abnormalities of skin and iris in this WS2-affected family suggest that the p.Leu104Val substitution has a more severe effect in these tissues than the p.Ser96Ter and the p.His67_Cys68delinsArg mutations do; the mechanism behind this greater severity might be a gain-of-function or dominant-negative effect of either the transmembrane isoform of p.Leu104Val KITLG, the soluble isoform, or both. To activate KIT, dimerization occurs for both isoforms.57, 58 The p.Ser96Ter and the p.His67_Cys68delinsArg defects are likely to have a loss-of-function effect because the transmembrane isoform of p.His67_Cys68delinsArg KITLG is not detectably integrated into the membrane; this will also be the case for p.Ser96Ter KITLG because it lacks the transmembrane domain. Also, for both defects, the soluble isoform could not be detected in ELISA of the medium of cultured cells.
Our findings add KITLG to the family of genes associated with pigmentation-deafness disorders, i.e., KIT, PAX3 (MIM: 606597), SOX10 (MIM: 602229), EDN3 (MIM: 131242), EDNRB (MIM: 131244), and MITF. As presented here for KITLG, the phenotypic variability of mutations in several of these hypopigmentation-deafness genes depends on whether the effect of the mutation is loss-of-function, presumably dominant-negative, dominant- negative, or activating. Loss-of-function mutations in MITF, for example, are dominant and cause WS2. Mutations in MITF that underlie Tietz syndrome are assumed to have a dominant-negative effect. Tietz syndrome (MIM: 103500) is more severe than WS2 and is characterized by fully penetrant bilateral, congenital, and profound HL, fair skin, and blond hair.59
In conclusion, we demonstrate that allelic mutations in KITLG can underlie different clinical conditions, and these findings extend the set of genes known to be associated with hypopigmentation-deafness disorders given that KITLG is now shown to be associated with WS2 and with NS-UHL/AHL. Analogous to the phenotype in mice, the severity of the human phenotype is likely to reflect the number of melanocytes that have reached the inner ear and skin and/or survived in these tissues.
Acknowledgments
The authors thank Mrs. Saskia van der Velde-Visser for technical assistance and Dr. Christian Gilissen for assistance in bioinformatics. We are grateful to Rafael Gonzalo-Gobernado (Microscopy Unit, Instituto Ramón y Cajal de Investigación Sanitaria) for the technical assistance and to Prof. Guy Richardson for kindly providing mRNA of mouse cochlea for RT-PCR experiments. We would like to thank Dr. Andy Beynon for the fruitful discussions about audiometric phenotypes. This work was supported by the Oticon Foundation (09-3742 to H.K.), by ZonMw (40-00812-98-09047 to H.K. and 90700388 to R.J.E.P.), by the Heinsius Houbolt Foundation (to H.K. and H.P.K.), by the Instituto de Salud Carlos III (PI11/1215 and PI14/0948 to M.A.M.-P.) and Fundación Ramón Areces (to M.A.M.-P.), and by the National Human Genome Research Institute and National Heart, Lung, and Blood Institute Baylor Hopkins Center for Mendelian Genomics (HG006542). L.S.d.C. was supported by a post-doctoral training grant (201399/2012-1) from the Programa Ciência sem Fronteiras and Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Published: October 29, 2015
Footnotes
Supplemental Data include a Supplemental Note, nine figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.09.011.
Accession Numbers
The accession numbers for the c.310C>G, c.200_202del, and c.286_303delinsT variants reported in this paper are LOVD: 0000065309, 0000065310, and 0000065311, respectively.
Web Resources
The URLs for data presented herein are as follows:
Alamut Visual, http://www.interactive-biosoftware.com/alamut-visual/
CIBERER Exome Server, http://bioinfo.cipf.es/apps-beta/exome-server/beta/
ConSeq Server, http://conseq.tau.ac.il/
ExAC Browser, http://exac.broadinstitute.org/
ExonPrimer, http://ihg.gsf.de/ihg/ExonPrimer.html
HGSC Mercury Analysis Pipeline, https://www.hgsc.bcm.edu/software/mercury
Mammalian Genotyping Service (Marshfield Map), http://research.marshfieldclinic.org/genetics/GeneticResearch/compMaps.asp
Mouse Genome Informatics, https://www.jax.org/research-and-faculty/tools/hereditary-hearing-impairment/hearing-mice-table-two
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
OMIM Phenotypic Series, http://www.omim.org/phenotypicSeriesTitle/all
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org/
Superlink Online SNP 1.1, http://cbl-hap.cs.technion.ac.il/superlink-snp/
UCSC Genome Browser, http://genome.ucsc.edu
WHAT IF Twinset, http://swift.cmbi.ru.nl/whatif/
Yasara, http://www.yasara.org/
Supplemental Data
References
- 1.Everberg G. Hereditary unilateral deafness. Acta Otolaryngol. 1957;47:303–311. doi: 10.3109/00016485709130346. [DOI] [PubMed] [Google Scholar]
- 2.Everberg G. Further studies on hereditary unilateral deafness. Acta Otolaryngol. 1960;51:615–635. doi: 10.3109/00016486009124539. [DOI] [PubMed] [Google Scholar]
- 3.Dikkers F.G., Verheij J.B., van Mechelen M. Hereditary congenital unilateral deafness: a new disorder? Ann. Otol. Rhinol. Laryngol. 2005;114:332–337. doi: 10.1177/000348940511400414. [DOI] [PubMed] [Google Scholar]
- 4.Pingault V., Ente D., Dastot-Le Moal F., Goossens M., Marlin S., Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Losada J., Sánchez-Martín M., Rodríguez-García A., Sánchez M.L., Orfao A., Flores T., Sánchez-García I. Zinc-finger transcription factor Slug contributes to the function of the stem cell factor c-kit signaling pathway. Blood. 2002;100:1274–1286. [PubMed] [Google Scholar]
- 6.Sato-Jin K., Nishimura E.K., Akasaka E., Huber W., Nakano H., Miller A., Du J., Wu M., Hanada K., Sawamura D. Epistatic connections between microphthalmia-associated transcription factor and endothelin signaling in Waardenburg syndrome and other pigmentary disorders. FASEB J. 2008;22:1155–1168. doi: 10.1096/fj.07-9080com. [DOI] [PubMed] [Google Scholar]
- 7.Read A.P., Newton V.E. Waardenburg syndrome. J. Med. Genet. 1997;34:656–665. doi: 10.1136/jmg.34.8.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou L., Pavan W.J. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res. 2008;18:1163–1176. doi: 10.1038/cr.2008.303. [DOI] [PubMed] [Google Scholar]
- 9.Lang D., Lu M.M., Huang L., Engleka K.A., Zhang M., Chu E.Y., Lipner S., Skoultchi A., Millar S.E., Epstein J.A. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 10.Fleischman R.A., Saltman D.L., Stastny V., Zneimer S. Deletion of the c-kit protooncogene in the human developmental defect piebald trait. Proc. Natl. Acad. Sci. USA. 1991;88:10885–10889. doi: 10.1073/pnas.88.23.10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comings D.E., Odland G.F. Partial albinism. JAMA. 1966;195:519–523. [PubMed] [Google Scholar]
- 12.Spritz R.A., Beighton P. Piebaldism with deafness: molecular evidence for an expanded syndrome. Am. J. Med. Genet. 1998;75:101–103. doi: 10.1002/(sici)1096-8628(19980106)75:1<101::aid-ajmg20>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 13.Hemesath T.J., Price E.R., Takemoto C., Badalian T., Fisher D.E. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 14.De Leenheer E.M., Huygen P.L., Coucke P.J., Admiraal R.J., van Camp G., Cremers C.W. Longitudinal and cross-sectional phenotype analysis in a new, large Dutch DFNA2/KCNQ4 family. Ann. Otol. Rhinol. Laryngol. 2002;111:267–274. doi: 10.1177/000348940211100312. [DOI] [PubMed] [Google Scholar]
- 15.Theunissen E.J., Huygen P.L., Folgering H.T. Vestibular hyperreactivity and hyperventilation. Clin. Otolaryngol. Allied Sci. 1986;11:161–169. doi: 10.1111/j.1365-2273.1986.tb00123.x. [DOI] [PubMed] [Google Scholar]
- 16.Wuyts F.L., Furman J., Vanspauwen R., Van de Heyning P. Vestibular function testing. Curr. Opin. Neurol. 2007;20:19–24. doi: 10.1097/WCO.0b013e3280140808. [DOI] [PubMed] [Google Scholar]
- 17.Pekelharing J.M., Hauss O., de Jonge R., Lokhoff J., Sodikromo J., Spaans M., Brouwer R., de Lathouder S., Hinzmann R. Haematology reference intervals for established and novel parameters in healthy adults. Diagnostic Perspectives. 2010;1:1–11. [Google Scholar]
- 18.Silberstein M., Weissbrod O., Otten L., Tzemach A., Anisenia A., Shtark O., Tuberg D., Galfrin E., Gannon I., Shalata A. A system for exact and approximate genetic linkage analysis of SNP data in large pedigrees. Bioinformatics. 2013;29:197–205. doi: 10.1093/bioinformatics/bts658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schraders M., Haas S.A., Weegerink N.J., Oostrik J., Hu H., Hoefsloot L.H., Kannan S., Huygen P.L., Pennings R.J., Admiraal R.J. Next-generation sequencing identifies mutations of SMPX, which encodes the small muscle protein, X-linked, as a cause of progressive hearing impairment. Am. J. Hum. Genet. 2011;88:628–634. doi: 10.1016/j.ajhg.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bainbridge M.N., Wang M., Wu Y., Newsham I., Muzny D.M., Jefferies J.L., Albert T.J., Burgess D.L., Gibbs R.A. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morín M., Bryan K.E., Mayo-Merino F., Goodyear R., Mencía A., Modamio-Høybjør S., del Castillo I., Cabalka J.M., Richardson G., Moreno F. In vivo and in vitro effects of two novel gamma-actin (ACTG1) mutations that cause DFNA20/26 hearing impairment. Hum. Mol. Genet. 2009;18:3075–3089. doi: 10.1093/hmg/ddp249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broudy V.C. Stem cell factor and hematopoiesis. Blood. 1997;90:1345–1364. [PubMed] [Google Scholar]
- 26.Wehrle-Haller B., Weston J.A. Soluble and cell-bound forms of steel factor activity play distinct roles in melanocyte precursor dispersal and survival on the lateral neural crest migration pathway. Development. 1995;121:731–742. doi: 10.1242/dev.121.3.731. [DOI] [PubMed] [Google Scholar]
- 27.Schrott A., Melichar I., Popelár J., Syka J. Deterioration of hearing function in mice with neural crest defect. Hear. Res. 1990;46:1–7. doi: 10.1016/0378-5955(90)90134-b. [DOI] [PubMed] [Google Scholar]
- 28.Cable J., Barkway C., Steel K.P. Characteristics of stria vascularis melanocytes of viable dominant spotting (Wv/Wv) mouse mutants. Hear. Res. 1992;64:6–20. doi: 10.1016/0378-5955(92)90164-i. [DOI] [PubMed] [Google Scholar]
- 29.Rerknimitr P., Disphanurat W., Achariyakul M. Topical tacrolimus significantly promotes repigmentation in idiopathic guttate hypomelanosis: a double-blind, randomized, placebo-controlled study. J. Eur. Acad. Dermatol. Venereol. 2013;27:460–464. doi: 10.1111/j.1468-3083.2012.04462.x. [DOI] [PubMed] [Google Scholar]
- 30.Miller C.T., Beleza S., Pollen A.A., Schluter D., Kittles R.A., Shriver M.D., Kingsley D.M. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guenther C.A., Tasic B., Luo L., Bedell M.A., Kingsley D.M. A molecular basis for classic blond hair color in Europeans. Nat. Genet. 2014;46:748–752. doi: 10.1038/ng.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandya A., Xia X.J., Landa B.L., Arnos K.S., Israel J., Lloyd J., James A.L., Diehl S.R., Blanton S.H., Nance W.E. Phenotypic variation in Waardenburg syndrome: mutational heterogeneity, modifier genes or polygenic background? Hum. Mol. Genet. 1996;5:497–502. doi: 10.1093/hmg/5.4.497. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds J.E., Meyer J.M., Landa B., Stevens C.A., Arnos K.S., Israel J., Marazita M.L., Bodurtha J., Nance W.E., Diehl S.R. Analysis of variability of clinical manifestations in Waardenburg syndrome. Am. J. Med. Genet. 1995;57:540–547. doi: 10.1002/ajmg.1320570405. [DOI] [PubMed] [Google Scholar]
- 34.Langley K.E., Wypych J., Mendiaz E.A., Clogston C.L., Parker V.P., Farrar D.H., Brothers M.O., Satygal V.N., Leslie I., Birkett N.C. Purification and characterization of soluble forms of human and rat stem cell factor recombinantly expressed by Escherichia coli and by Chinese hamster ovary cells. Arch. Biochem. Biophys. 1992;295:21–28. doi: 10.1016/0003-9861(92)90482-c. [DOI] [PubMed] [Google Scholar]
- 35.Jones M.D., Narhi L.O., Chang W.C., Lu H.S. Refolding and oxidation of recombinant human stem cell factor produced in Escherichia coli. J. Biol. Chem. 1996;271:11301–11308. doi: 10.1074/jbc.271.19.11301. [DOI] [PubMed] [Google Scholar]
- 36.Nishikawa M., Tojo A., Ikebuchi K., Katayama K., Fujii N., Ozawa K., Asano S. Deletion mutagenesis of stem cell factor defines the C-terminal sequences essential for its biological activity. Biochem. Biophys. Res. Commun. 1992;188:292–297. doi: 10.1016/0006-291x(92)92383-9. [DOI] [PubMed] [Google Scholar]
- 37.Tabone-Eglinger S., Calderin-Sollet Z., Pinon P., Aebischer N., Wehrle-Haller M., Jacquier M.C., Boettiger D., Wehrle-Haller B. Niche anchorage and signaling through membrane-bound Kit-ligand/c-kit receptor are kinase independent and imatinib insensitive. FASEB J. 2014;28:4441–4456. doi: 10.1096/fj.14-249425. [DOI] [PubMed] [Google Scholar]
- 38.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 39.Høyer P.E., Byskov A.G., Møllgård K. Stem cell factor and c-Kit in human primordial germ cells and fetal ovaries. Mol. Cell. Endocrinol. 2005;234:1–10. doi: 10.1016/j.mce.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Huang E., Nocka K., Beier D.R., Chu T.Y., Buck J., Lahm H.W., Wellner D., Leder P., Besmer P. The hematopoietic growth factor KL is encoded by the Sl locus and is the ligand of the c-kit receptor, the gene product of the W locus. Cell. 1990;63:225–233. doi: 10.1016/0092-8674(90)90303-v. [DOI] [PubMed] [Google Scholar]
- 41.Copeland N.G., Gilbert D.J., Cho B.C., Donovan P.J., Jenkins N.A., Cosman D., Anderson D., Lyman S.D., Williams D.E. Mast cell growth factor maps near the steel locus on mouse chromosome 10 and is deleted in a number of steel alleles. Cell. 1990;63:175–183. doi: 10.1016/0092-8674(90)90298-s. [DOI] [PubMed] [Google Scholar]
- 42.Rajaraman S., Davis W.S., Mahakali-Zama A., Evans H.K., Russell L.B., Bedell M.A. An allelic series of mutations in the Kit ligand gene of mice. II. Effects of ethylnitrosourea-induced Kitl point mutations on survival and peripheral blood cells of Kitl(Steel) mice. Genetics. 2002;162:341–353. doi: 10.1093/genetics/162.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zsebo K.M., Williams D.A., Geissler E.N., Broudy V.C., Martin F.H., Atkins H.L., Hsu R.Y., Birkett N.C., Okino K.H., Murdock D.C. Stem cell factor is encoded at the Sl locus of the mouse and is the ligand for the c-kit tyrosine kinase receptor. Cell. 1990;63:213–224. doi: 10.1016/0092-8674(90)90302-u. [DOI] [PubMed] [Google Scholar]
- 44.Geissler E.N., Cheng S.V., Gusella J.F., Housman D.E. Genetic analysis of the dominant white-spotting (W) region on mouse chromosome 5: identification of cloned DNA markers near W. Proc. Natl. Acad. Sci. USA. 1988;85:9635–9639. doi: 10.1073/pnas.85.24.9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geissler E.N., Ryan M.A., Housman D.E. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell. 1988;55:185–192. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 46.Chabot B., Stephenson D.A., Chapman V.M., Besmer P., Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;335:88–89. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- 47.Steel K.P., Davidson D.R., Jackson I.J. TRP-2/DT, a new early melanoblast marker, shows that steel growth factor (c-kit ligand) is a survival factor. Development. 1992;115:1111–1119. doi: 10.1242/dev.115.4.1111. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi S., Ando M., Kakigi A. Mechanism generating endocochlear potential: role played by intermediate cells in stria vascularis. Biophys. J. 2000;79:2572–2582. doi: 10.1016/S0006-3495(00)76497-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hudspeth A.J. Integrating the active process of hair cells with cochlear function. Nat. Rev. Neurosci. 2014;15:600–614. doi: 10.1038/nrn3786. [DOI] [PubMed] [Google Scholar]
- 50.Brown P.G., Ruben R.J. The endocochlear potential in the Shaker-1 (sh-1-sh-1) mouse. Acta Otolaryngol. 1969;68:14–20. doi: 10.3109/00016486909121538. [DOI] [PubMed] [Google Scholar]
- 51.Deol M.S. The relationship between abnormalities of pigmentation and of the inner ear. Proc. R. Soc. Lond. B Biol. Sci. 1970;175:201–217. doi: 10.1098/rspb.1970.0019. [DOI] [PubMed] [Google Scholar]
- 52.Amyere M., Vogt T., Hoo J., Brandrup F., Bygum A., Boon L., Vikkula M. KITLG mutations cause familial progressive hyper- and hypopigmentation. J. Invest. Dermatol. 2011;131:1234–1239. doi: 10.1038/jid.2011.29. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z.Q., Si L., Tang Q., Lin D., Fu Z., Zhang J., Cui B., Zhu Y., Kong X., Deng M. Gain-of-function mutation of KIT ligand on melanin synthesis causes familial progressive hyperpigmentation. Am. J. Hum. Genet. 2009;84:672–677. doi: 10.1016/j.ajhg.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Opdecamp K., Nakayama A., Nguyen M.T., Hodgkinson C.A., Pavan W.J., Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development. 1997;124:2377–2386. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- 55.Wu M., Hemesath T.J., Takemoto C.M., Horstmann M.A., Wells A.G., Price E.R., Fisher D.Z., Fisher D.E. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- 56.Sommer L. Generation of melanocytes from neural crest cells. Pigment Cell Melanoma Res. 2011;24:411–421. doi: 10.1111/j.1755-148X.2011.00834.x. [DOI] [PubMed] [Google Scholar]
- 57.Paulhe F., Wehrle-Haller M., Jacquier M.C., Imhof B.A., Tabone-Eglinger S., Wehrle-Haller B. Dimerization of Kit-ligand and efficient cell-surface presentation requires a conserved Ser-Gly-Gly-Tyr motif in its transmembrane domain. FASEB J. 2009;23:3037–3048. doi: 10.1096/fj.09-129577. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z., Zhang R., Joachimiak A., Schlessinger J., Kong X.P. Crystal structure of human stem cell factor: implication for stem cell factor receptor dimerization and activation. Proc. Natl. Acad. Sci. USA. 2000;97:7732–7737. doi: 10.1073/pnas.97.14.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith S.D., Kelley P.M., Kenyon J.B., Hoover D. Tietz syndrome (hypopigmentation/deafness) caused by mutation of MITF. J. Med. Genet. 2000;37:446–448. doi: 10.1136/jmg.37.6.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.